Abstract
Mycoplasma hyopneumoniae causes swine pneumonia and contributes significantly to the porcine respiratory disease complex. The mechanisms of pathogenesis are difficult to address, since there is a lack of genetic tools, but microarrays are available and can be used to study transcriptional changes that occur during disease as a way to identify important virulence-related genes. Mycoplasmas were collected from bronchial alveolar lavage samples and compared to broth-grown cells using microarrays. Bronchial alveolar lavage was performed on pigs 28 days postinfection, and mycoplasmas were isolated by differential centrifugation. Mycoplasma RNA-enriched preparations were then obtained from total RNA by subtracting eucaryotic ribosomal and messenger RNAs. Labeled cDNAs were generated with mycoplasma open reading frame-specific primers. Nine biological replicates were analyzed. During lung infection, our analysis indicated that 79 M. hyopneumoniae genes were differentially expressed (P < 0.01), at a false-discovery rate of <2.7%. Of the down-regulated genes, 28 of 46 (61%) lacked an assigned function, in comparison to 21 of 33 (63%) of up-regulated genes. Four down-regulated genes and two up-regulated genes encoded putative lipoproteins. secA (mhp295) (P = 0.003) and two glycerol transport permease genes (potA [mhp380; P = 0.006] and ugpA [mhp381; P = 0.003]) were up-regulated in vivo. Elongation factor EF-G (fusA [mhp083]) (P = 0.002), RNA polymerase beta chain (rpoC [mhp635]) (P = 0.003), adenylate kinase (adk [mhp208]) (P = 0.001), prolyl aminoacyl tRNA synthetase (proS [mhp397]) (P = 0.009), and cysteinyl-tRNA synthetase (cysS [mhp661]) (P < 0.001) were down-regulated in vivo.
The reactions that invading pathogens have toward host responses determine their ability to colonize and cause disease. Most bacteria have inducible regulons whose products enhance colonization and assist in the disease process. With mycoplasmas, the simplest of bacterial pathogens, the story of virulence factors and their regulation has been poorly studied and is less well known. Some mycoplasma species can generate high rates of diversity in selected genes or gene families through small sequence changes and recombination events resulting in changes at the genetic level (32). This variation is usually expressed by the generation of new chimeric surface molecules with high rates of antigenic diversity that are thought to help evade the host immune system. Phase switching may result when variation occurs within poly(A) tracts in promoter regions or in structural gene sequences or by DNA inversion. The generation of chimeric genes by intragenic recombination also occurs. This variation is carried through subsequent generations. There is no evidence that any of these mechanisms are operative in Mycoplasma hyopneumoniae, however. Some surface molecules in M. hyopneumoniae undergo posttranslational modification through proteolytic processing as a form of variation identifiable by immunoblotting (4, 7).
Previous studies have shown that gene regulation does occur in M. hyopneumoniae in regard to heat shock (13), iron deprivation (14), and exposure to hydrogen peroxide (23). Since these growth conditions only partially mimic those found in vivo, we wanted to determine how exposure to the host during disease alters the transcriptional profiles of M. hyopneumoniae. Thus, we obtained organisms from infected pig lungs and compared their transcriptome with that from organisms grown in the laboratory using microarray technology. This provided a “global” snapshot of the steady-state concentrations of mRNAs, a good indicator of gene expression. Surprisingly, our results indicate that more genes are down-regulated during lung infection than during broth growth.
MATERIALS AND METHODS
Mycoplasma strains and culture conditions.
Pathogenic M. hyopneumoniae strain 232, a derivative of strain 11, was used in this study. In vitro cultures consisted of 125 ml of Friis medium (10) in 250-ml Erlenmeyer flasks incubated at 37°C with slow agitation until the culture reached mid-log phase as indicated by color change and turbidity. Mycoplasmas were pelleted by centrifugation at 24,000 × g and resuspended in 0.5 ml RNALater (Ambion, Inc., Austin, TX), and the cells were stored at −70°C until the total RNA was isolated. For in vivo cultures, pigs were infected with M. hyopneumoniae with 10 ml of mid-log-phase broth culture (105 color changing units/ml) intratracheally at 4 weeks of age (27). Pigs were evaluated daily and scored for degree of severity of respiratory disease (0 to 6), sneezing (0 to 3), coughing (0 to 3), lethargy (0 to 3), wasting (0 to 3), and icterus (0 to 3), and a summary clinical score was recorded. At 28 days postinfection, pigs were necropsied and the lungs were lavaged as described previously (16). Bronchial alveolar lavage fluids (BALF) were quickly cooled to 4°C and centrifuged at 250 × g for 5 min (4°C) to remove the cellular debris and large host cells from the fluid. The mycoplasmas were isolated by high-speed centrifugation (25,000 × g, 15 min, 4°C). Mycoplasma pellets were then resuspended in 0.5 ml RNALater and stored at −70°C until RNA was extracted. The BALF from each pig represented a single experimental unit.
RNA isolation.
RNA was isolated using the Versagene RNA purification system (Gentra Systems, Minneapolis, MN) according to the manufacturer's protocol. The optional step of DNase treatment was performed on a column according to the manufacturer's recommendation. Contaminating pig RNA was removed from each sample using the MICROBEnrich kit (Ambion), and the samples were quantified using the Nanodrop ND-1000 spectrophotometer (Nanodrop, Wilmington, DE). The broth-grown control samples were treated in an identical fashion. The efficiency of removal of contaminating pig nucleic acids was verified by PCR using primers specific for pig cyclophilin (cyclo-f, TAACCCCACCGTCTTCTT; cyclo-r, TGCCATCCAACCACTCAG). In addition, to evaluate the possibility of having false signals on the arrays from potential residual swine lung RNAs in the BALF preparations, a hybridization was performed with total RNA isolated from whole-lung tissues harvested from mycoplasma-free control pigs using the Versagene RNA purification tissue kit (Gentra Systems). As a control for these studies, total pig lung RNA was labeled with M. hyopneumoniae open reading frame (ORF)-specific hexamer oligonucleotide primers selected to bind only to M. hyopneumoniae mRNA as previously described (13).
Microarray.
The M. hyopneumoniae microarray consists of PCR products (probes) spotted to Corning UltraGAPS glass substrates as described previously (13). The array represents 89% (620/698) of the total ORFs of M. hyopneumoniae strain 232 as PCR products of approximately 125 to 350 bp in length. Each probe is spotted in triplicate using a nonadjacent, well-spaced format. Each slide contains two complete arrays, one at each end of the substrate.
Microarray experimental design.
Nine independent BALF samples were processed for total RNA along with nine in vitro cultures. Each in vivo BALF sample was paired with an independent in vitro culture sample for hybridization on nine two-color arrays. For five of the arrays, the control in vitro sample was labeled with Alexa 555 and compared to an Alexa 647-labeled BALF sample. The dye assignment to control samples was reversed for the other four arrays. The slides were hybridized under identical conditions as described below.
Hybridization, image acquisition, and normalization.
Fluorescently labeled cDNA targets were generated, purified, hybridized to arrays, scanned, and analyzed as follows. Briefly, 10 μg of M. hyopneumoniae enriched RNA was incubated at 70°C for 10 min with 5 μl of M. hyopneumoniae ORF-specific hexamer oligonucleotide primers (750 ng/μl) in a total reaction volume of 15.5 μl, chilled on ice for 10 min, and then combined with 14.5 μl of reverse transcription reaction mix (3 μl Superscript III 200U/μl [Invitrogen Corp., Carlsbad, CA], 6 μl 5× First Strand buffer, 0.6 μl 50× amino allyl-dUTP/deoxynucleoside triphosphate mix, 3 μl RNase-free water). Reactions were incubated at 42°C for at least 2 h. RNA hydrolysis was performed using 3 μl 0.1 M ETDA and 3.5 μl 0.1 M NaOH at 65°C for 10 min. To neutralize the pH, 36.5 μl of 500 mM HEPES (pH 7.0) was added. The hydrolyzed reaction mixtures were purified using the Mo Bio UltraClean PCR cleanup kit (MO BIO Laboratories, Inc., Carlsbad, CA). Reaction mixtures were quantified using an ND-1000 spectrophotometer (Nanodrop, Wilmington, DE), lyophilized, and resuspended in 6.5 μl RNase-free water. After 10 min, 1.5 μl sodium bicarbonate (pH 8.7) and 2 μl of Alexa 555 or Alexa 647 (Invitrogen, Carlsbad, CA) were added. Dyes were prepared by resuspending a single tube of dye in 2 μl of dimethyl sulfoxide. Dye-coupled reactions were purified and quantified as described above. The frequency of incorporation was determined as described elsewhere (http://Promega.com/applications/arrays/calculator/#ResultsView). Hybridization, scanning, and analysis were preformed as described previously (13).
Our analysis employed three scans using various laser powers and photomultiplier tube gain settings to increase the dynamic range of expression measurement (8). Spot signal intensities were quantified from images, and the values were log transformed, background subtracted, and normalized as described previously (13). The normalized values for triplicate spots were averaged within each array to produce 1 normalized measure of expression for each of the 620 probe sequences and each of the 9 RNA samples.
A separate mixed-linear-model analysis was conducted for each probe sequence using the normalized data (29). Each mixed model included fixed effects for treatment (broth versus in vivo), slide region (upper versus lower), and dye (Alexa 555 versus Alexa 647) and random effects for slide and slide-by-region interaction. A t test for differential expression across treatments was conducted for each probe as part of our mixed-linear-model analysis. The 620 P values from these t tests were converted to q values using the method of Storey and Tibshirani (25). These q values can be used to obtain approximate control of the false discovery rate at a specified value.
qRT-PCR.
Quantitative reverse transcriptase PCRs (qRT-PCRs) were performed as described by the manufacturer. Six genes were analyzed using the Brilliant SYBR Green QRT-PCR kit (Stratagene, La Jolla, CA). The gene mhp345 was chosen as the housekeeping gene for these studies based on its consistent expression levels in several microarray studies (13, 14, 23). Cycling conditions were based upon the manufacturer's recommendations (Stratagene) using a Bio-Rad (Hercules, CA) MyiQ real-time PCR cycler. In all cases, the same RNA samples were used for both the microarray and qRT-PCR studies. The primers used for amplification are described in Table 1. The qRT-PCR data were analyzed according to the method of Gallup and Ackermann (11).
TABLE 1.
RT-PCR primer sequences
| Gene ID | Primer name | Primer sequence (5′-3′) |
|---|---|---|
| mhp083 | 083-F | CGACTTGCACTTGCAGAAGCAGTT |
| 083-R | TAAGTTCGCAGCCCGAATTGCAG | |
| mhp092 | 092-F | TTTGCGGGCTTATGCTGACTTCTG |
| 092-R | AAATTTGCAACAAGGGCGGCGT | |
| mhp345 | 345-F | TGAAGCGCTTATGCTACTGAGG |
| 345-R | ATTGCGGTTGTACGAGCGACCTTA | |
| mhp366 | 366-F | AGGTGTTAATTTCCGCAGGAACCC |
| 366-R | AGGCTCCTGGTCCATTAGAAGCAA | |
| mhp381 | 381-F | GATCACAATTGGAATCATCGGCG |
| 381-R | AGATTGCTAATGTTCCTGCCTGGG | |
| mhp670 | 670-F | ACAGATACGGTTCCGGGAATTGCT |
| 670-R | AAAGTGGTAGCTCCAGGCCAGATT | |
| mhp674 | 674-F | AGGGATTCATCCAACTTTGGTCGC |
| 674-R | AATCAAGACCGACTTCGCCGAT |
Microarray data accession number.
The microarray data can be accessed through the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE9057.
RESULTS AND DISCUSSION
In vivo model and statistical analyses.
These studies were performed to obtain a snapshot of transcriptional activity in M. hyopneumoniae during infection. It is the first analysis of its kind for mycoplasmas and has rarely been reported for other bacterial pathogens for several reasons. First, for most bacteria, it is difficult to obtain sufficient numbers of organisms from infected tissues to provide adequate levels of RNA for microarray analysis. Second, even if RNA amplification is performed, the approach used to produce cDNA with random primers results in poor or false signal levels due to contaminating host RNA. Finally, often the natural host is not available for studies of this type or is difficult to obtain, preventing in vivo studies of this type from being performed. One of the few examples of in vivo expression analysis of a bacterial pathogen by microarrays has been performed with Vibrio cholerae isolated from human stools (1, 24). A hyperinfectious phenotype for mice was observed with stool-isolated Vibrio and correlated with gene expression changes (17), but significant differences in gene expression patterns resulted when human stool samples and rabbit infection samples were compared (31). This suggests that use of the normal host is critical to global transcriptome studies. Of course, working with a host interjects additional confounding problems into the analysis, such as genetic differences of the host and the variability of immune responses to a specific pathogen (2).
Our M. hyopneumoniae-pig infection model circumvents many of these problems. First, with swine we have ready access to large numbers of in vivo-grown organisms during the height of infection simply by washing the lung with sterile buffer. Second, the labeling protocol uses M. hyopneumoniae mRNA-specific primers. This prevents the generation of false signals if host RNA is present in the samples. We are also using a natural host-pathogen model with normal infection protocols. Finally, the removal of contaminating pig RNAs results in highly enriched mycoplasma RNA preparations that label well under the conditions used.
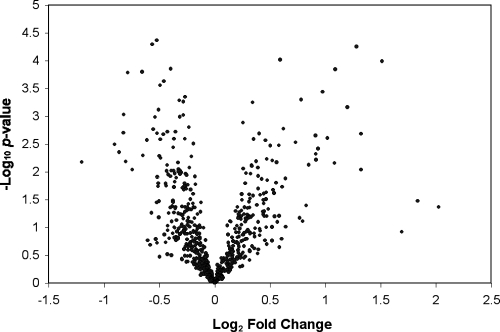
We were unable to deal with the host genetic differences effect, however. By adding additional biological replicates to the analysis, we were able to reduce the false discovery rate to <2.7%, a strong indicator of the quality and reproducibility of our data. We confirmed our data by qRT-PCR using the genes and primer pairs indicated in Tables 1 and 2. Although several of the genes showed increased transcript levels over our microarray data, all of the genes showed concordance in the direction of variation with the microarray data (Table 2). The values for significant transcript level differences in our microarray data (Fig. 1) are relatively low (Table 2). Recent evidence, however, indicates that the direction of change is more important than actual value correlations (20, 30). Also, since mycoplasmas are considered to be among the smallest organisms, with a volume of less than 0.1 μm3, smaller changes in transcript levels may have more impact on physiological processes in them than in larger organisms with significantly larger volumes (Escherichia coli has an approximate volume of 3 μm3).
TABLE 2.
Transcriptional differences between M. hyopneumoniae grown in vivo and in vitro
| Category or gene IDa | Gene or label | Description of product | Comparison of in vivo and in vitro transcription
|
|||
|---|---|---|---|---|---|---|
| P value | q value | Fold change | qRT-PCR | |||
| Genes up-regulated in BAL cells relative to expression in broth-grown cells | ||||||
| mhp019 | UH | Unique hypothetical | 0.0001 | 0.004 | 2.13 | |
| mhp027 | UH | Unique hypothetical | 0.0060 | 0.024 | 1.89 | |
| mhp029 | gatA | Glutamyl-tRNA amidotransferase subunit A | 0.0005 | 0.007 | 1.27 | |
| mhp036 | gap | Glyceraldehyde 3-phosphate dehydrogenase | 0.0074 | 0.025 | 1.26 | |
| mhp084 | rpsG | 30S ribosomal protein S7 | 0.0047 | 0.021 | 1.88 | |
| mhp093 | UH | Unique hypothetical | 0.0003 | 0.007 | 1.97 | |
| mhp142 | UH | Unique hypothetical | 0.0020 | 0.014 | 2.50 | |
| mhp197 | rpS17 | Ribosomal protein S17 | 0.0091 | 0.027 | 2.50 | |
| mhp219 | UH | Unique hypothetical | 0.0083 | 0.027 | 1.31 | |
| mhp222 | abc | ATP-dependent transport protein | 0.0001 | 0.004 | 1.50 | |
| mhp292 | CH | Conserved hypothetical, putative serine protease | 0.0074 | 0.025 | 1.80 | |
| mhp295 | secA | Preprotein translocase subunit | 0.0033 | 0.016 | 1.50 | |
| mhp324 | CH | Conserved hypothetical | 0.0063 | 0.024 | 1.30 | |
| mhp354 | UH | Unique hypothetical | 0.0067 | 0.024 | 1.47 | |
| mhp357 | CH | Conserved hypothetical | 0.0012 | 0.012 | 1.19 | |
| mhp360 | UH | Unique hypothetical | 0.0016 | 0.014 | 1.54 | |
| mhp366 | CH | Conserved hypothetical (lipoprotein) | 0.0001 | 0.004 | 2.43 | 3.44 |
| mhp371 | CH | Conserved hypothetical, P37 precursor (lipoprotein) | 0.0058 | 0.024 | 1.38 | |
| mhp380 | potA | Hypothetical ABC transporter ATP-binding protein | 0.0062 | 0.024 | 1.43 | |
| mhp381 | ugpA | sn-glycerol-3-phosphate transport system permease | 0.0026 | 0.014 | 1.37 | 10.40 |
| mhp395 | CH | Conserved hypothetical | 0.0021 | 0.014 | 1.88 | |
| mhp460 | CH | Conserved hypothetical | 0.0006 | 0.008 | 2.30 | |
| mhp468 | CH | Conserved hypothetical | 0.0024 | 0.014 | 2.02 | |
| mhp499 | oppD | Oligopeptide transport ATP-binding protein | 0.0005 | 0.007 | 1.72 | |
| mhp522 | UH | Unique hypothetical | 0.0078 | 0.026 | 1.26 | |
| mhp548 | pgiB | Phosphoglucose isomerase B | 0.0069 | 0.024 | 2.12 | |
| mhp552 | pyrH | Uridylate kinase | 0.0029 | 0.015 | 1.66 | |
| mhp575 | CH | Conserved hypothetical | 0.0020 | 0.014 | 1.32 | |
| mhp576 | UH | Unique hypothetical | 0.0025 | 0.014 | 1.27 | |
| mhp629 | CH | Conserved hypothetical | 0.0001 | 0.004 | 2.85 | |
| mhp654 | CH | Conserved hypothetical | 0.0033 | 0.016 | 1.42 | |
| mhp674 | CH | Conserved hypothetical | 0.0038 | 0.017 | 1.91 | 22.88 |
| mhp678 | p115 | P115 protein | 0.0086 | 0.027 | 1.20 | |
| Genes down-regulated in BAL cells relative to expression in broth-grown cells | ||||||
| mhp083 | fusA | Chain elongation factor EF-G | 0.0020 | 0.014 | 1.38 | 13.63 |
| mhp092 | CH | Conserved hypothetical | 0.0025 | 0.014 | 1.29 | 3.52 |
| mhp104 | truB | tRNA pseudouridine synthase B | 0.0090 | 0.027 | 1.68 | |
| mhp118 | CH | Conserved hypothetical | 0.0094 | 0.027 | 1.24 | |
| mhp130 | CH | Conserved hypothetical, DNA processing protein | 0.0015 | 0.014 | 1.18 | |
| mhp131 | UH | Unique hypothetical | 0.0083 | 0.027 | 1.17 | |
| mhp133 | trsE | Transfer complex protein | 0.0050 | 0.022 | 1.57 | |
| mhp136 | CH | Conserved hypothetical | 0.0068 | 0.024 | 1.26 | |
| mhp151 | CH | Conserved hypothetical | 0.0001 | 0.004 | 1.44 | |
| mhp158 | nrdE | Ribonucleoside-diphosphate reductase alpha chain | 0.0005 | 0.007 | 1.22 | |
| mhp166 | oppF | Oligopeptide transport system permease protein | 0.0036 | 0.017 | 1.30 | |
| mhp170 | UH | Unique hypothetical (lipoprotein) | 0.0018 | 0.014 | 1.35 | |
| mhp181 | CH | Conserved hypothetical (ORF 5 in cilium adhesin operon) | 0.0001 | 0.004 | 1.32 | |
| mhp200 | rpL5 | Ribosomal protein L5 | 0.0097 | 0.028 | 1.25 | |
| mhp202 | rpS8 | Ribosomal protein S8 | 0.0095 | 0.027 | 1.35 | |
| mhp208 | adk | Adenylate kinase | 0.0009 | 0.010 | 1.22 | |
| mhp211 | rpS13 | Ribosomal protein S13 | 0.0019 | 0.014 | 1.77 | |
| mhp228 | CH | Conserved hypothetical | 0.0031 | 0.016 | 1.14 | |
| mhp231 | rpe | Ribulose-5-phosphate-3-epimerase | 0.0025 | 0.014 | 1.21 | |
| mhp235 | CH | Conserved hypothetical | 0.0007 | 0.009 | 1.42 | |
| mhp247 | CH | Conserved hypothetical | 0.0089 | 0.027 | 1.25 | |
| mhp273 | UH | Unique hypothetical | 0.0005 | 0.007 | 1.25 | |
| mhp303 | UH | Unique hypothetical | 0.0009 | 0.010 | 1.77 | |
| mhp304 | gtp1 | GTP-binding protein | 0.0065 | 0.024 | 2.30 | |
| mhp305 | rps18 | 30S ribosomal protein S18 | 0.0055 | 0.023 | 1.40 | |
| mhp309 | UH | Unique hypothetical | 0.0031 | 0.016 | 1.87 | |
| mhp310 | UH | Unique hypothetical | 0.0001 | 0.004 | 1.72 | |
| mhp312 | UH | Unique hypothetical (lipoprotein) | 0.0044 | 0.020 | 1.82 | |
| mhp341 | CH | Conserved hypothetical | 0.0094 | 0.027 | 1.31 | |
| mhp356 | UH | Unique hypothetical | 0.0018 | 0.014 | 1.28 | |
| mhp362 | CH | Conserved hypothetical | 0.0026 | 0.014 | 1.53 | |
| mhp397 | proS | Prolyl aminoacyl-tRNA synthetase | 0.0094 | 0.027 | 1.29 | |
| mhp457 | rluD | Ribosomal large subunit pseudouridine synthase D | 0.0051 | 0.022 | 1.41 | |
| mhp466 | CH | Conserved hypothetical (lipoprotein) | 0.0020 | 0.014 | 1.44 | |
| mhp472 | CH | Conserved hypothetical | 0.0001 | 0.004 | 1.58 | |
| mhp474 | CH | Conserved hypothetical | 0.0010 | 0.010 | 1.45 | |
| mhp480 | CH | Conserved hypothetical | 0.0002 | 0.006 | 1.41 | |
| mhp495 | yx1 | YX1 | 0.0064 | 0.024 | 1.75 | |
| mhp517 | UH | Unique hypothetical (lipoprotein) | 0.0004 | 0.007 | 1.20 | |
| mhp526 | CH | Conserved hypothetical | 0.0010 | 0.010 | 1.24 | |
| mhp583 | UH | Unique hypothetical | 0.0074 | 0.025 | 1.35 | |
| mhp634 | CH | Conserved hypothetical | 0.0002 | 0.005 | 1.38 | |
| mhp635 | rpoC | DNA-directed RNA polymerase beta chain | 0.0025 | 0.014 | 1.41 | |
| mhp637 | rplL | 50S ribosomal protein L7/L12 | 0.0016 | 0.014 | 1.47 | |
| mhp661 | cysS | Cysteinyl-tRNA synthetase | 0.0001 | 0.004 | 1.48 | |
| mhp670 | CH | Conserved hypothetical | 0.0052 | 0.022 | 1.16 | 9.61 |
ID, identifier.
FIG. 1.
Volcano plot of transcriptional differences in M. hyopneumoniae during growth in vivo. Data represent individual gene responses plotted as log2 n-fold changes versus negative log10 P values. Points above the negative log10 P value of 2 are significant at P values of <0.01, with a negative change representing the down-regulated genes and a positive change representing the up-regulated genes.
The variability in the host disease state at necropsy may contribute to these low values. The extent of lung lesions in these pigs varied, as is usually seen with this pathogen and infection model (27). Another factor that may contribute to low levels of change is that the organisms isolated by bronchial lavage may represent organisms from numerous environments within the pig lung, i.e., organisms attached to ciliated epithelium, nonadherent organisms growing in tissue secretions, organisms in different phases of growth, organisms from lesions that have altered pHs, etc. The spot intensity values would then represent an average of mRNA levels within the population. Our microarray studies involved isolating mycoplasmas from lavage fluids after centrifugation to remove host cellular debris. In effect, this could have removed attached mycoplasmas as well. Further study will be needed to differentiate between these possibilities.
Membrane transport genes.
Our comparative analysis of transcripts from in vivo-grown versus in vitro-grown M. hyopneumoniae indicated that 33 genes were up-regulated in vivo (P < 0.01) relative to expression in vitro (Table 2). A similar proportion (63%) of these were hypothetical genes, as was the case for those that were down-regulated (61%). Also, a larger proportion of genes associated with membrane transport processes were up-regulated in vivo, including secA (mhp295), abc (mhp222), potA (mhp380), and ugpA (mhp381). The role of SecA in mycoplasmal physiology is assumed to be in membrane transport processes as part of the translocase machinery, although this has not been determined experimentally. Increasing protein translocation during disease might provide for more-rapid responses to host immune defenses. Both abc and potA have an ATP/GTP binding site motif A (also called a P loop), and potA also has a strong ABC transporter family signature. The role of PotA seems to be in transport of spermidine, a molecule involved in ribosome binding, translation stimulation, and DNA stability (12). Spermidine has also been reported to affect nitric oxide-mediated immune functions (5, 26), a major host innate immune component during neutrophil and macrophage stimulation. Perhaps increased spermidine transport enhances the survival of M. hyopneumoniae in vivo. Enhanced uptake of glycerol in the infected lung as a result of increased transcript levels of ugpA (sn-glycerol-3-phosphate transport permease) might indicate a need for glycerol as an energy source, or alternatively, glycerol might provide osmotic protection within the various environments of the swine lung as the organism shifts from cell association on mucosal surfaces through its release into mucosal secretions. Interestingly, the presence of glycerol in the growth medium has been reported to enhance the virulence of Mycoplasma mycoides subsp. mycoides through enhanced hydrogen peroxide production (21). Whether M. hyopneumoniae also responds in this way is not known, since it has not been examined for hydrogen peroxide production, which usually involves growth on blood agar; M. hyopneumoniae grows poorly on agar surfaces, if at all. Inclusion of glycerol in broth medium had no effect on the growth of M. hyopneumoniae (data not shown). Further study will be needed to better understand the role of these gene products in pathogenesis.
The down-regulation of trsE (mhp133) in vivo is an interesting observation. The product of this gene has a conserved motif (VirD4/VirB4) that is present in members of the type IV secretory pathway. From the genome annotation (19) (available at http://mycoplasma.genome.uab.edu/), it appears that mhp133 is part of a five-gene operon (mhp130 to mhp134). Only one other member of that putative operon has a product with a conserved domain or similarity with other proteins, mhp130. The Mhp130 SMF motif is found in proteins involved in DNA uptake (15). Like mhp133, mhp130 is down-regulated in vivo (P = 0.001). Since DNA nucleotides are essential nucleic acid precursors for mycoplasmas (9, 18), this operon may function as a DNA transport complex stimulated by the presence of high concentrations of DNA in the surrounding environment, such as in DNA-rich broth.
Lipoprotein genes.
Three genes identified in this study coded for lipoproteins. mhp366 and mhp371 were up-regulated and mhp170 was down-regulated in vivo relative to results for broth-grown organisms. The relatively low levels of transcriptional variation in lipoprotein genes is not surprising, since mycoplasmas in general do not regulate their lipoprotein genes by transcriptional control mechanisms (32). Also, M. hyopneumoniae lacks the types of lipoprotein gene families that undergo phase switching and size variation, although the latter would not necessarily alter transcription. This was evident from the analysis of the M. hyopneumoniae genome sequence (19, 28) and not from biological studies. The classical approach to identifying surface variable antigens could not be used with M. hyopneumoniae due to its poor growth on agar surfaces.
Transcription-translation genes.
More genes involved in transcription and translation were down-regulated in vivo than was the case with broth-grown organisms. These included fusA (mhp083), nrdE (mhp158), rps18 (mhp305), proS (mhp397), rpoC (mhp635), rplL (mhp637), and cysS (mhp661). It is not unusual to find groups of these genes regulated in microarray studies, including tRNA synthases, ribosomal proteins, and rRNA genes (3, 6, 13, 14, 23). The reason for this regulation is not known, but it is commonly reported. For M. hyopneumoniae, the answer might be simple. Perhaps it is a nutritional issue; there are more nutrients in the rich broth media, while the in vivo environment is highly competitive for nutrients such as iron, amino acids, purines, etc. The up-regulation of fusA, which codes for chain elongation factor EF-G, might simply reflect faster growth in vitro. A similar argument could be made for the other transcription-translation related genes.
General metabolism genes.
Interestingly, two genes with greater changes and P values of <0.05, mhp396 (P = 0.042; 4.07-fold change) and mhp070 (P = 0.033; 3.56-fold change) (data not shown) (see Table S1 in the supplemental material), were up-regulated in vivo. The gene mhp396 codes for thioredoxin, a protein involved in reduction of thiodisulfide bonds. Serving as an antioxidant, thioredoxin may play important roles in protecting the organism in vivo during active host immune responses. The product of gene mhp070 is excinuclease ABC subunit C. It has an active role in excising DNA lesions during DNA repair and is sometimes referred to as UvrABC nuclease. The up-regulation of subunit C in vivo may indicate that DNA damage may be occurring more frequently in vivo relative to that in in vitro conditions. The two other components of the excinuclease ABC enzyme, subunit A (encoded by mhp288) and subunit B (encoded by mhp669), are constitutively expressed under these conditions (data not shown). The C subunit, however, seems to be the excision-controlling component (22). Alternatively, this exonuclease may be involved in the uptake of DNA precursors. Mycoplasma broth has relatively high concentrations of purines and pyrimidines from the yeast extract component, whereas organisms in the swine lung may have limited access to free bases. Thus, more digestion of chromosomal DNA may be required for in vivo survival, and thus, higher transcript levels of the mhp070 gene result.
In summary, our studies have given insight into the transcript levels of M. hyopneumoniae genes in vivo during disease. We were unable, however, to identify specific gene targets that might be related to virulence. Almost 60% of the differentially expressed genes were classified as hypotheticals, and virulence-related genes might eventually be found among this group. Further refinement of the sampling of mycoplasmas from lung tissues might provide additional evidence of the regulation of key genes in response to swine tissues and the host immune response. This could be accomplished by laser capture microdissection of mycoplasmas in association with tracheal epithelial cells. These studies would necessarily involve additional complex technical barriers, mainly amplification of mycoplasma RNAs. To date this has not been accomplished, but ongoing studies are moving in this direction. Further, our experimental design involved comparing mid-log-phase broth-grown organisms with those isolated from infected pigs. This time point was chosen because we have no information regarding what stage of growth M. hyopneumoniae is in during infection. The mid-log phase of growth yields sufficient quantities of intact RNA for microarray analysis. Future studies comparing growth phases in vitro are planned.
Supplementary Material
Acknowledgments
We thank members of Eileen Thacker's laboratory (Nancy Upchurch and Barb Erickson) for the mycoplasma growth medium. Michael J. Oneal assisted with the qRT-PCR.
This work was partially funded by a grant from The Center for Integrated Animal Genomics at Iowa State University to F.C.M.
Editor: A. Camilli
Footnotes
Published ahead of print on 10 December 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Bina, J., J. Zhu, M. Dziejman, S. Faruque, S. Calderwood, and J. Mekalanos. 2003. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc. Natl. Acad. Sci. USA 1002801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 706871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2004. Genomic-scale analysis of Pasteurella multocida gene expression during growth within liver tissue of chickens with fowl cholera. Microbes Infect. 6290-298. [DOI] [PubMed] [Google Scholar]
- 4.Burnett, T. A., K. Dinkla, M. Rohde, G. S. Chhatwal, M. Srivastava, S. J. Cordwell, S. Geary, F. C. Minion, M. J. Walker, and S. P. Djordjevic. 2006. P159 is a proteolytically processed, surface adhesin of Mycoplasma hyopneumoniae: defined domains of P159 bind heparin and promote adherence to eukaryotic cells. Mol. Microbiol. 60669-686. [DOI] [PubMed] [Google Scholar]
- 5.Bussiere, F. I., R. Chaturvedi, Y. Cheng, A. P. Gobert, M. Asim, D. R. Blumberg, H. Xu, P. Y. Kim, A. Hacker, R. A. Casero, Jr., and K. T. Wilson. 2005. Spermine causes loss of innate immune response to Helicobacter pylori by inhibition of inducible nitric-oxide synthase translation. J. Biol. Chem. 2802409-2412. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich, G., S. Kurz, C. Hèubner, C. Aepinus, S. Theiss, M. Guckenberger, U. Panzner, J. Weber, and M. Frosch. 2003. Transcriptome analysis of Neisseria meningitidis during infection. J. Bacteriol. 185155-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djordjevic, S. P., S. J. Cordwell, M. A. Djordjevic, J. Wilton, and F. C. Minion. 2004. Proteolytic processing of the Mycoplasma hyopneumoniae cilium adhesin. Infect. Immun. 722791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudley, A. M., J. Aach, M. A. Steffen, and G. M. Church. 2002. Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc. Natl. Acad. Sci. USA 997554-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finch, L. R., and A. Mitchell. 1992. Sources of nucleotides, p. 211-230. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, DC.
- 10.Friis, N. F. 1975. Some recommendations concerning primary isolation of Mycoplasma hyopneumoniae and Mycoplasma flocculare, a survey. Nord. Veterinaermed. 27337-339. [PubMed] [Google Scholar]
- 11.Gallup, J. M., and M. R. Ackermann. 2006. Addressing fluorogenic real-time qPCR inhibition using the novel custom Excel file system ′Focusfield2-6GallupqPCRSet-upTool-001′ to attain consistently high fidelity qPCR reactions. Biol. Proced. Online 887-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igarashi, K., and K. Kashiwagi. 1999. Polyamine transport in bacteria and yeast. Biochem. J. 344633-642. [PMC free article] [PubMed] [Google Scholar]
- 13.Madsen, M. L., D. Nettleton, E. L. Thacker, R. Edwards, and F. C. Minion. 2006. Transcriptional profiling of Mycoplasma hyopneumoniae during heat shock using microarrays. Infect. Immun. 74160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madsen, M. L., D. Nettleton, E. L. Thacker, and F. C. Minion. 2006. Transcriptional profiling of Mycoplasma hyopneumoniae during iron depletion using microarrays. Microbiology 152937-944. [DOI] [PubMed] [Google Scholar]
- 15.Marchler-Bauer, A., J. B. Anderson, M. K. Derbyshire, C. DeWeese-Scott, N. R. Gonzales, M. Gwadz, L. Hao, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, D. Krylov, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, N. Thanki, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35D237-D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mengeling, W. L., K. M. Lager, and A. C. Vorwald. 1995. Diagnosis of porcine reproductive and respiratory syndrome. J. Vet. Diagn. Investig. 73-16. [DOI] [PubMed] [Google Scholar]
- 17.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miles, R. J. 1992. Cell nutrition and growth, p. 23-40. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, DC.
- 19.Minion, F. C., E. L. Lefkowitz, M. L. Madsen, B. J. Cleary, S. M. Swartzell, and G. G. Mahairas. 2004. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J. Bacteriol. 1867123-7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morey, J. S., J. C. Ryan, and F. M. Van Dolah. 2006. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online 8175-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pilo, P., E. M. Vilei, E. Peterhans, L. Bonvin-Klotz, M. H. Stoffel, D. Dobbelaere, and J. Frey. 2005. A metabolic enzyme as a primary virulence factor of Mycoplasma mycoides subsp. mycoides small colony. J. Bacteriol. 1876824-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rupp, W. D. 1996. DNA repair mechanisms, p. 2277-2294. In F. C. Neidhardt, R. I. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Lo, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and E. H. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, DC.
- 23.Schafer, E. R., M. J. Oneal, M. L. Madsen, and F. C. Minion. 2007. Global transcriptional analysis of Mycoplasma hyopneumoniae following exposure to hydrogen peroxide. Microbiology 1533785-3790. [DOI] [PubMed] [Google Scholar]
- 24.Shelburne, S. A., and J. M. Musser. 2004. Virulence gene expression in vivo. Curr. Opin. Microbiol. 7283-289. [DOI] [PubMed] [Google Scholar]
- 25.Storey, J. D., and R. Tibshirani. 2003. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol. Biol. 224149-157. [DOI] [PubMed] [Google Scholar]
- 26.Szabo, C., G. J. Southan, C. Thiemermann, and J. R. Vane. 1994. The mechanism of the inhibitory effect of polyamines on the induction of nitric oxide synthase: role of aldehyde metabolites. Br. J. Pharmacol. 113757-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thacker, E. L., P. G. Halbur, R. F. Ross, R. Thanawongnuwech, and B. J. Thacker. 1999. Mycoplasma hyopneumoniae potentiation of porcine reproductive and respiratory syndrome virus-induced pneumonia. J. Clin. Microbiol. 37620-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasconcelos, A. T., H. B. Ferreira, C. V. Bizarro, S. L. Bonatto, M. O. Carvalho, P. M. Pinto, D. F. Almeida, L. G. Almeida, R. Almeida, L. Alves-Filho, E. N. Assunðcäao, V. A. Azevedo, M. R. Bogo, M. M. Brigido, M. Brocchi, H. A. Burity, A. A. Camargo, S. S. Camargo, M. S. Carepo, D. M. Carraro, J. C. de Mattos Cascardo, L. A. Castro, G. Cavalcanti, G. Chemale, R. G. Collevatti, C. W. Cunha, B. Dallagiovanna, B. P. Dambrâos, O. A. Dellagostin, C. Falcäao, F. Fantinatti-Garboggini, M. S. Felipe, L. Fiorentin, G. R. Franco, N. S. Freitas, D. Frâias, T. B. Grangeiro, E. C. Grisard, C. T. Guimaräaes, M. Hungria, S. N. Jardim, M. A. Krieger, J. P. Laurino, L. F. Lima, M. I. Lopes, E. L. Loreto, H. M. Madeira, G. P. Manfio, A. Q. Maranhäao, C. T. Martinkovics, S. R. Medeiros, M. A. Moreira, M. Neiva, C. E. Ramalho-Neto, M. F. Nicolâas, S. C. Oliveira, R. F. Paixäao, F. O. Pedrosa, S. D. Pena, M. Pereira, L. Pereira-Ferrari, I. Piffer, L. S. Pinto, D. P. Potrich, A. C. Salim, F. R. Santos, R. Schmitt, M. P. Schneider, A. Schrank, I. S. Schrank, A. F. Schuck, H. N. Seuanez, D. W. Silva, R. Silva, S. C. Silva, C. M. Soares, K. R. Souza, R. C. Souza, C. C. Staats, M. B. Steffens, S. M. Teixeira, T. P. Urmenyi, M. H. Vainstein, L. W. Zuccherato, A. J. Simpson, and A. Zaha. 2005. Swine and poultry pathogens: the complete genome sequence of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 1875568-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comp. Biol. 8625-637. [DOI] [PubMed] [Google Scholar]
- 30.Wurmbach, E., T. Yuen, and S. C. Sealfon. 2003. Focused microarray analysis. Methods 31306-316. [DOI] [PubMed] [Google Scholar]
- 31.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 1001286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yogev, D., G. F. Browning, and K. S. Wise. 2002. Genetic mechanisms of surface variation, p. 417-443. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.