Abstract
In all Yersinia pestis strains examined, the adhesin/invasin yadA gene is a pseudogene, yet Y. pestis is invasive for epithelial cells. To identify potential surface proteins that are structurally and functionally similar to YadA, we searched the Y. pestis genome for open reading frames with homology to yadA and found three: the bicistronic operon yadBC (YPO1387 and YPO1388 of Y. pestis CO92; y2786 and y2785 of Y. pestis KIM5), which encodes two putative surface proteins, and YPO0902, which lacks a signal sequence and likely is nonfunctional. In this study we characterized yadBC regulation and tested the importance of this operon for Y. pestis adherence, invasion, and virulence. We found that loss of yadBC caused a modest loss of invasiveness for epithelioid cells and a large decrease in virulence for bubonic plague but not for pneumonic plague in mice.
Adherence of bacterial pathogens to host cells is an early step in the infectious process and allows exploitation of host cell signaling pathways or cell entry (4). Yersinia pseudotuberculosis and Yersinia enterocolitica, both of which are food-borne pathogens, express two adhesins, YadA and InvA, that support bacterial docking at the mucosal surface and provide the intimate contact needed for functioning of the Ysc type III secretion system (12, 14, 21). Yersinia pestis lacks functional YadA and InvA (37), yet this pathogen adheres tightly to epithelial cells and macrophages and invades these cells as avidly as the enteropathogenic yersiniae (8, 44). One adhesin/invasin unique to Y. pestis is the surface aspartyl protease Pla; however, absence of Pla did not eliminate all invasiveness for epithelial cells and resulted in little reduction in adherence, indicating that additional adhesins/invasins must be present (8).
Inspection of the genome sequences available for Y. pestis has revealed the presence of open reading frames that encode putative structural analogs of YadA (19). YadA and the structurally analogous Moraxella UspAs belong to a class of nonfimbrial adhesins called oligomeric coiled-coil adhesins (Oca) (19). The YadA molecule consists of five major domains: head, neck, stalk, coil-coil segment, and membrane anchor. Crystallography of the collagen-binding head domain of YadA resolved at 1.55 Å showed a stable trimeric locknut structure that is required for collagen binding (33, 34). More recently, the structure of the C-terminal membrane anchor, which forms a β-barrel, was resolved at 3.8 Å, and this anchor was shown to contain a helical part at its N terminus (51). Model studies have proposed a pore assembly scheme in which a 12-strand β-barrel is assembled by trimerization (41) of the four transmembrane β-strands, forming an opening through which the N-terminal head, neck, stalk, and coiled helical domains of the three monomer chains exit to the cell surface (23). The Oca family of proteins is now viewed as a subset of autotransporters, the type Vc or trimeric autotransporters (7, 18). The N-terminal domains are not cleaved, which is commonly true for “conventional” (“type Va”) autotransporters, but they function together at the bacterial surface to provide trivalent (high-avidity) ligands that can cluster receptors on eukaryotic cells (7, 18).
The present study characterized the Y. pestis yadBC operon, which encodes two proteins with limited sequence similarity to YadA. We evaluated the transcriptional and translational expression of these proteins, their possible function as adhesins and invasins for epithelioid cells, and their importance for lethality in bubonic plague and pneumonic plague, and we showed that they are new Oca family members that are essential for lethality of bubonic plague.
MATERIALS AND METHODS
Bacterial strains and cultivation.
All bacterial strains used in this study are described in Table 1. Y. pestis CO99-3015 (a derivative of strain CO92) lacking an Lcr plasmid or containing the Lcr plasmid pCD2 but lacking the pgm locus (Δpgm) was obtained from Scott Bearden, Centers for Disease Control and Prevention Division of Vector-Borne Infectious Diseases, Fort Collins, CO. Lcr− Y. pestis strain CO92 was obtained from Luther Lindler, Walter Reed Army Institute of Research, Silver Spring, MD. Escherichia coli strain DH5α (Life Technologies, Gaithersburg, MD) was used for routine cloning of DNA. E. coli strains were routinely grown in Luria-Bertani broth or on Luria-Bertani agar (28) at 37°C. Y. pestis strains were routinely grown at 26 or 37°C in heart infusion broth (HIB) (Difco Laboratories, Detroit, MI) and on HIB agar or tryptose blood agar base (Difco) as indicated below. HIB was supplemented with 2.5 mM CaCl2 and 0.2% xylose when the Lcr plasmid was present. Yersinia selective agar (Difco) was used during plasmid conjugation. Where appropriate (unless otherwise indicated), ampicillin (100 μg/ml) or carbenicillin, kanamycin (50 μg/ml), streptomycin (50 μg/ml), spectinomycin (50 μg/ml), tetracycline (12 μg/ml), and chloramphenicol (20 μg/ml) were added to cultures. Carbenicillin was used at a concentration of 50 μg/ml for growth of Y. pestis strains containing pCD2Ap (Table 1). The presence of the pigmentation locus (37) was confirmed by the formation of red colonies on Congo red agar (46) at 28°C. The presence of a functional Lcr virulence plasmid was confirmed by the absence of growth at 37°C on HIB agar containing 0.2 M MgCl2 and 0.2 M sodium oxalate and by the presence of the Lcr phenotype (37) in defined medium TMH (45) (see below). Cell growth was monitored with a Spectronic Genesys 5 spectrophotometer at 620 nm.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Key properties | Reference or source |
|---|---|---|
| Y. pestis strains | ||
| KIM6+ | Pgm+ Lcr− | 15 |
| KIM6-2109+ | Pgm+ Lcr−; ΔluxS::kan ΔypsIR ΔytbIR | A. Bobrov, S. W. Bearden, J. D. Fetherston, and R. D. Perry |
| CO99-3015.S1 | CO92 Pgm+ Lcr−; pFra pPCP1; Lcr− derivative of CDC strain CO99-3015 of virulent Y. pestis (a subclone of CO92) | Scott Bearden, CDC, Ft. Collins, CO |
| CO99-3015.S4 | invA::promoterless lacZ Lcr−; made from CO99-3015.S1 by single-crossover integration of pSucinv::lacZ | Spencer Leigh |
| CO99-3015.S5 | CO92 Δpgm; pCD2 pFra pPCP1; Δpgm derivative of CDC strain CO99-3015 of virulent Y. pestis (a subclone of CO92); used to make pCD2Ap | Scott Bearden, CDC, Ft. Collins, CO |
| CO99-3015.S6 | CO92 Apr Δpgm; pCD2 pFra pPCP1; contains pCD2Ap | Spencer Leigh |
| CO99-3015.S10 | CO92 Apr Pgm+ Lcr−; pFra pPCP1; invA::PyadBC-lacZ; single-crossover integration of pSucinv::yadBC-lacZ into CO99-3015.S1 | This study |
| CO92.S1 | Pgm+ Lcr−; pFra pPCP1; Lcr− derivative of virulent Y. pestis CO92; molecular group 1.ORI | Luther Lindler |
| CO92.S2 | CO92 Pgm+ Lcr−; pFra Δcaf1; F1− derivative made by Red/Flp-mediated allelic exchange into CO92.S1; entire caf1 coding sequence and upstream 12 bp deleted | This study |
| CO92.S6 | CO92 Apr Pgm+ Lcr+; pCD2Ap pFra pPCP1; reconstituted virulent strain made by introduction of pCD2Ap into CO92.S1 | This study |
| CO92.S7 | CO92 Apr Pgm+ Lcr+; pCD2Ap pFra Δcaf1 pPCP1; reconstituted conditionally virulent F1− strain made by introduction of pCD2Ap into CO92.S2a | This study |
| CO92.S8 | CO92 Pgm+ Lcr−; pFra pPCP1 ΔyadBC; derived from CO92.S1 by allelic exchange | This study |
| CO92.S9 | CO92 Apr Pgm+ Lcr+; pCD2Ap pFra pPCP1 ΔyadBC; potentially virulent derivative of CO92.S8 containing pCD2Ap | This study |
| CO92.S10 | CO92 Pgm+ Lcr−; pFra Δcaf1 pPCP1 ΔyadBC; F1− derivative of CO92.S8 made by Red/Flp-mediated allelic exchange; entire caf1 coding sequence and upstream 12 bp deleted | This study |
| CO92.S11 | CO92 Apr Pgm+ Lcr+; pCD2Ap pFra Δcaf1 pPCP1 ΔyadBC; F1− potentially conditionally virulent derivative of CO92.S10 containing pCD2Apa | This study |
| CO92.S12 | CO92 Kmr Pgm+ Lcr−; pFra Δcaf1 pPCP1::Kan-fsPla ΔyadBC ΔdegP; Pla− DegP− strain derived from CO92.S11 by allelic exchange and plasmid exchange | This study |
| CO92.S15 | CO92 Pgm+ Lcr−; pFra pPCP1; yadBC CO92.S8 strain reconstituted to yadBC+ by allelic exchange | This study |
| CO92.S16 | Reconstituted virulent CO92 Apr Pgm+ Lcr+; pCD2Ap pFra pPCP1; reconstituted virulent strain made by introducing pCD2Ap into CO92.S15 | This study |
| CO92.S17 | CO92 Pgm+ Lcr−; pFra pPCP1 ΔyadC; derived from CO92.S1 by allelic exchange | This study |
| CO92.S18 | CO92 Apr Pgm+ Lcr+; pCD2Ap pFra pPCP1 ΔyadC strain made Lcr+ by introducing pCD2Ap into CO92.S17 | This study |
| E. coli strains | ||
| DH5α | F− Δφ80dlacZΔM15 endA1 recA1 hsdR17(rM− mK+) supE44 thi-1 gyrA96 Δ(lacZYA-argF)U169 | Life Technologies |
| DH5α λpir | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR Δ(lacZYA-argF)U169 λpir+ | Lab stock |
| S17-1 | Smr Spr Tra+; pro thi hsdR recA; chromosomal integration of RP4-2-Tc::Mu-Kan::Tn7 | 11 |
| BW20767 | RP4-2-Tc::Mu-Kan::Tn7 integrant, leu-63::IS10 recA1 zbf-5 creB510 hsdR17 endA1 thi uidA(ΔMluI)::pir+ | 28 |
| HB101 | recA23 hsdS20 supE44 (rB− mB−) ara-14 lacY1 galK2 xyl-5 rpsL20 mtl-1 proA2 leu | 1 |
| MG4(pKDT17) | E. coli MG4 with reporter plasmid pKDT17 | 39 |
| Plasmids | ||
| pKD3 | Template plasmid; cat flanked by FRT sites; Cmr Apr | 9 |
| pKD4 | Template plasmid; kan flanked by FRT sites; Kmr Apr | 9 |
| pKD46 | Red recombinase expression plasmid; Apr | 9 |
| pCP20 | Suicide plasmid with temperature-sensitive replication and thermally induced expression of FLP recombinase; Apr Cmr | 9 |
| pLD55 | Suicide vector with oriR6Kγ and fusaric acid selection; Apr Tcr | 28 |
| pLD55ΔyadBC | Fragment Y1-4 (∼1.5 kb) inserted into SpeI site of pLD55; deletes bp 1 to 2904 of yadBC; Apr Tcr | This study |
| pLD55yadBC-L | Entire yadBC operon flanked by 560 bp upstream and 551 bp downstream cloned into pLD55 (4,146-bp insertion); restores yadBC to a ΔyadBC mutant or the individual genes to single yadB or yadC mutants | This study |
| pWSK29 | 5.4 kb, Apr Kmr, low-copy-number cloning vector | 48 |
| pYadBC | 5.8-kb BglII fragment (bp −2352 to 3494) cloned into BglII site of pWSK29; 11.2 kb; Apr Kmr | This study |
| pBAD24 | 4.5 kb, Apr, expression vector using ParaBAD to drive gene expression | 17 |
| pBAD24YadBC | 3.0-kb yadBC cloned into EcoRI/HindIII-cut pBAD24; 7.5 kb; Apr | This study |
| pLD55ΔDegP | Fragment ΔdegPUp1-ΔdegPdown2 (ca. 2 kb) inserted into XhoI/SacI site of pLD55; deletes 1,523 bp, from −50 bp upstream of degP through degP and 27 bp downstream; Apr | This study |
| pPCP1::Kan | 10.6 kb; Kmr; kan gene from pKD4 inserted into pPCP1 at bp 2135 to 2221 in the intergenic region downstream from the IS100 ATP-binding protein (YPPCP1.02); construction deleted sequence between 2135 and 2221 | This study |
| pPCP1::Kan-fsPla | Derivative of pPCP1::Kan in which a frameshift in pla was created by filling in the BamHI site at codon 79; Kmr Pla− | This study |
| pBSlacZMCS | Cloning vector with rrnBT1 transcriptional terminator and lacZ gene from pEU730; Apr | 36 |
| pBS-PyadBC-lacZ | 207-bp Y. pestis yadBC promoter region (bp −205 to 3 of yadBC) inserted into KpnI/Acc65I-digested pBSlacZMCS | This study |
| pSucinv | Suicide vector with R6K ori and sacB selection containing the 2-kb StuI/XhoI fragment (contains bp 709 to 2706 [amino acids 237 to 902] of Y. pseudotuberculosis invA; Apr | 38 |
| pSucinv::yadBC-lacZ | 4.4-kb PyadBC-lacZ EagI fragment of pBS-PyadBC-lacZ inserted into EagI site of pSucinv | This study |
| pSucinv::lacZ | Promoterless lacZ cloned into the EagI site within invA in pSucinv; Apr | Spencer Leigh |
| pCD2Ap | Lcr virulence plasmid of Y. pestis CO99-3015.S5 into which the bla gene from pRL494e was inserted within the yadA pseudogene (pCD2 yadA::bla); Aprb | Spencer Leigh |
| pGEX-3X | lacIqlacP-gst; Apr | Pharmacia |
| pGEX-YadB25-150 | 378-bp EcoRI/SmaI yadB fragment (amplified by BA11 and BA22) inserted into EcoRI/SmaI-digested pGEX-3X | This study |
| pGEX-YadC137-409 | 819-bp EcoRI/SmaI yadB fragment (amplified by CA11 and CA22) inserted into EcoRI/SmaI-digested pGEX-3X | This study |
Strain and plasmid construction.
The plasmids used in this study are listed in Table 1, and the primers used are shown in Table S1 in the supplemental material. Plasmids were purified from overnight cultures by alkaline lysis (3) or with a Qiaprep spin miniprep kit (Qiagen Inc., Valencia, CA), and they were purified further when necessary by polyethylene glycol precipitation (20) or were column purified and concentrated (Zymo Research). A standard CaCl2 protocol was used to introduce plasmids into E. coli (43). Y. pestis cells were transformed by electroporation as previously described (38). All plasmid DNA and PCR products used for construction of mutants were sequenced by IDT or Elim Biopharmaceuticals to ensure that the correct mutation had been introduced.
The allelic exchange plasmid pLD55ΔyadBC was constructed from the suicide vector pLD55 (27) and used to delete both cistrons of the yadBC operon. The upstream region of yadB and the downstream region of yadC (∼700 bp each) were PCR amplified with primers Y1 and Y2 and primers Y3 and Y4, respectively (see Table S1 in the supplemental material). The Y1-2 and Y3-4 PCR fragments were phosphorylated and ligated prior to a second amplification with primers Y1 and Y4 to produce fragment Y1-4 spanning the region of interest, eliminating the yadBC operon (genome bases 1565090 to 1568070 corresponding to 78 bp of the promoter region and bases 1 to 2904 of the 2,963-bp operon). SpeI-digested PCR fragment Y1-4 was introduced into the SpeI site of pLD55. Upon electroporation into Y. pestis strain CO92.S1 (Lcr−), the first crossover event was selected on tryptose blood agar containing ampicillin, and final plasmid-free chromosome mutant recombinants were identified on HIB or Congo red medium containing fusaric acid, chlortetracycline, and ZnCl2 as previously described (27, 31). After initial checks by PCR, the identity of the expected Y. pestis ΔyadBC deletion mutant strain (CO92.S8) was confirmed by Southern blot analysis carried out with NdeI- or BglII-digested genomic DNA and with digoxigenin-labeled (Roche Applied Science, Indianapolis, IN) probe Y1-2 (data not shown).
The ΔyadBC strain CO92.S8 was restored to yadBC+ by allelic exchange. The yadBC operon flanked upstream by 560 bp and downstream by 551 bp was copied from genomic DNA by PCR using Herculase DNA polymerase (Stratagene, La Jolla, CA) and primers Long-BC-A and Long-BC-B (see Table S1 in the supplemental material), which encoded SalI and SacI sites, respectively. The resulting 4,146-bp fragment was cloned into SalI- and SacI-digested pLD55, creating pLD55yadBC-L. Then 950 ng of pLD55yadBC-L was electroporated into Y. pestis CO92.S8, and the yersiniae were allowed to recover at 37°C for 5 h. The entire 1-ml suspension was plated onto HIB plates containing 50 μg/ml carbenicillin and 10 μg/ml tetracycline. The remainder of the allelic exchange procedure was carried out, resulting ultimately in a single tetracycline-sensitive isolate that contained both yadB and yadC, as indicated by positive PCRs with primer sets for the two genes (primers YadBEcoRISD-5′ and YadBPstI-3′ and primers YadCEcoRI-5′ and YadCHindIII-3′).
Because of the difficulty of detecting YadB and YadC expression at the protein level without overexpression (see below), the functionality of the reconstituted yadBC operon was confirmed by reverse transcription (RT)-PCR with primers 1387U and 1387L for yadB and with primers 1388U and 1388L for yadC (see Table S1 in the supplemental material). Total bacterial RNA was obtained using RNeasy minicolumns (Qiagen). cDNA synthesis and quantification were performed with a LightCycler RNA Master SYBR green II kit (Roch) and a LightCycler 2.0 instrument equipped with version 4.0 software (Roche). The RT and PCR conditions were as follows: 61°C for 20 min, 95°C for 30 s, and then 30 cycles of 5 s at 95°C, 5 s at 60°C, and 15 s at 72°C. The potential DNA contamination in each RNA preparation was assessed by RT-PCR using a sample that had been treated with RNAse A, followed by SuperaseIN (Ambion, Inc., Austin, TX) to inhibit the RNase A.
Production of the F1 protein capsule was eliminated by deleting the caf1 gene in the ΔyadBC Y. pestis mutant strain CO92.S8 and in the parent strain CO92.S1. Primers 449 and 450 were used to generate a PCR fragment containing the FRP site-flanked cat gene from pKD3 flanked by the caf1 sequence, and caf1 was deleted by allelic exchange by using the Red recombinase-expressing plasmid pKD46 and the flipase-expressing plasmid pCP20 as described previously (9). The resulting deletion started at bp −12 upstream of the caf1 translation initiation codon and extended through the last translated codon. The absence of F1 expression was verified by immunoblotting yersiniae grown at 37°C using monoclonal antibody YPF1 (Research Diagnostics, Inc., Flanders, NJ), and the absence of cat in the final strain was confirmed by PCR and by the inability of the strain to grow on chloramphenicol-containing plates (data not shown).
Red- and Flp-mediated recombination were similarly employed to construct a nonpolar deletion of yadC in Y. pestis CO92.S1, using yadC gene-specific primers CF01 and CF02 (see Table S1 in the supplemental material). The presence of the cam cassette in selected isolates was confirmed by PCR, and the flipase-expressing plasmid pCP20 was used to create the final deletion. Colonies sensitive to chloramphenicol and ampicillin (excised chloramphenicol cassette and pCP20 cured) were isolated, and the mutation was confirmed by PCR. The deletion excised genomic bp 1566262 to 1568130, exactly removing yadC. Quantitative RT-PCR was used to determine whether the deletion had a polar effect on yadB expression. The level of mRNA for yadB was compared to the mRNA level of the reference gene proS (C. R. Wulff, A. M. Uittenbogaard, and S. C. Straley, unpublished data), and the results showed that the yadC deletion caused a slight decrease in the net message abundance for yadB; the normalized ratios of the crossing points for the target gene to the crossing points for the reference gene were 0.17 ± 0.09 for parent strain Y. pestis CO92.S1 and 0.13 ± 0.09 for the yadC mutant (P = 0.0377, as determined by a two-tailed paired Student t test).
A Y. pestis strain lacking functional surface protease Pla and periplasmic protease DegP (GsrA; YPO3382) in addition to YadB, YadC, and F1 was derived from Y. pestis CO92.S11. degP was deleted (bp −50 with respect to the translational start to 27 bp downstream of the translational termination codon; 1,523 bp) by allelic exchange using pLD55 and primers ΔdegP Up1, ΔdegP Up2, ΔdegP Down1, and ΔdegP Down2 to create flanking DNA, as described previously (52). ΔdegPscreen1 and ΔdegPscreen2 were used to verify the absence of degP in the final strain. Plasmid pPCP1::Kan-fsPla was initially created in Y. pestis CO99-3015.S1. The FRP site-flanked kan gene from pKD4 was inserted into pPCP1 by the Red recombinase between bp 2135 and 2221 in the intergenic region downstream from the IS100 ATP-binding protein (YPPCP1.02) to create pPCP1::Kan; the construction procedure deleted the sequence between bp 2135 and 2221. A frameshift at codon 79 of pla was then created by digesting pPCP1::Kan with BamHI and filling in with T4 DNA polymerase, resulting in pPCP1::Kan-fsPla. Both steps in the construction procedure were confirmed by DNA sequencing. pPCP1::Kan-fsPla was electroporated into ΔdegP ΔyadBC Δcaf1 Y. pestis (pPCP1+), and pPCP1::Kan-fsPla was allowed to homogenotize by segregation during three sequential growth periods on media containing kanamycin. The absence of functional Pla in the final Y. pestis CO92.S13 strain was confirmed by the inability of the strain to digest a fibrin film (2) (data not shown).
Two plasmids containing yadBC were used in this study. The Y. pestis CO92 wild-type yadBC operon was isolated as a 5,846-bp BglII-digested genomic DNA fragment and cloned into the 5,438-bp BglII-opened vector pWSK29 (48). The PCR-verified final plasmid contained the full-length 2,963-bp yadBC operon under the control of its native promoter and was designated pYadBC. To create pBAD24YadBC, yadBC was amplified from genomic DNA by PCR with primer YadBEcoRISD-5′, which included a 5′ EcoRI site and Shine-Dalgarno sequence, and primer YadCHindIII-3′, which had an additional 3′ TAA codon followed by a HindIII site downstream of the natural YadC termination codon. The product was digested with these enzymes and cloned into similarly digested pBAD24 (17). YadB and YadC were expressed from the plasmid-encoded ParaBAD promoter by induction for 4 h at 37°C with 0.25% (wt/vol) arabinose. The cells were pelleted, suspended at an optical density at 620 nm (OD620) of 10 in lysis loading buffer (0.4 M Tris-HCl [pH 6.8], 8% [wt/vol] sodium dodecyl sulfate [SDS], 5% [wt/vol] dithiothreitol, 25% [vol/vol] glycerol, 0.4% [wt/vol] bromphenol blue), boiled for 4 min, and analyzed by immunoblotting.
To create a yadBC promoter-lacZ fusion, the putative promoter region of the Y. pestis yadBC operon was amplified with primers P1 and P2 from SpeI-digested genomic DNA and cloned into KpnI/Acc65I-digested pBSlacZMCS (36). The resulting fusion in pBS-PyadBC-lacZ was sequenced, excised as a 4.4-kb EagI fragment, and subcloned into the EagI site of the suicide vector pSucinv (36) for subsequent chromosomal integration as pSucinv::yadBC-lacZ. lacZ was similarly cloned to obtain pSucinv::lacZ. pSucinv::yadBC-lacZ was first transformed into the E. coli host S17-1 (11) or BW20767 (27) and then conjugated into Y. pestis CO92.S1 to obtain strain CO92.S10 or into Y. pestis CO99-3015.S1 to obtain strain CO99-3015.S4.
An ampicillin resistance marker was inserted at bp 312 of the yadA pseudogene on virulence plasmid pCD2, which encodes the Yops and their cognate type III secretion system (6; S. A. Leigh and S. C. Straley, unpublished data). Briefly, a 1,700-bp fragment of the yadA gene was copied by PCR and cloned into pLD55. Then the β-lactamase (bla) gene from pRL494e (13), starting 40 bp upstream of its promoter, was copied by PCR. The resultant plasmid was transformed into E. coli BW20767 (27), and the plasmid was conjugated into Y. pestis CO99-3015.S5. The allelic exchange was carried out as previously described (31, 52), creating strain CO99-3015.S6.
Potential virulence was reconstituted in a biosafety level 3 containment facility with select-agent security for some Lcr− Y. pestis strains by introducing pCD2Ap by electroporation. Briefly, each attenuated strain was electroporated with total plasmid DNA from Y. pestis CO99-3015.S6. Transformed strains were selected for carbenicillin resistance and screened for the Pgm+ and Lcr+ phenotypes. Pgm+ Lcr+ isolates were subcultured at 26°C in TMH containing carbenicillin for ca. 10 generations and then for 2 h in TMH with or without 2.5 mM CaCl2 (no carbenicillin); then they were incubated at 37°C for 4 h. A 500-μl sample of each culture was removed and briefly placed on ice to encourage cellular aggregation, and the cells were pelleted in a microcentrifuge. The top 250 μl of supernatant was removed, and the secreted proteins were precipitated overnight at 4°C with 5% (wt/vol) trichloroacetic acid and collected by centrifugation. Samples of whole cells and secreted proteins were made equivalent to a culture having an OD620 of 2.0 in lysis loading buffer and heated at 95°C for 15 min. One-half of each heat-treated sample was plated and incubated for 5 days at 28°C to confirm the absence of viable cells before the samples were removed from the containment facility.
The resultant cellular extracts and supernatant proteins were then analyzed by immunoblotting to determine the presence of secreted LcrV and YopM as described previously (52) in order to determine whether the type III secretion system was fully functional (data not shown). These tests proved to be crucial, as we did obtain some isolates with defects that were unmasked by this assay after they tested positive by PCR analysis for the presence of a unique Lcr gene (lcrQ) and by crude Lcr determination on plates containing HIB agar with 0.2 M MgCl2 and 0.2 M sodium oxalate (data not shown). Several isolated colonies of each strain that was confirmed by the Yop secretion test were pooled and used for 50% lethal dose (LD50) studies.
To produce YadB and YadC antigens, portions of the corresponding genes were amplified as 378- and 819-bp DNA fragments with primers BA11 and BA22 and primers CA11 and CA22, respectively, and with the wild-type copy of the yadBC operon on pYadBC as a template. Both antigenic fragments were digested with EcoRI/SmaI and cloned into appropriate sites in expression vector pGEX-3X to form glutathione S-transferase (GST) fusion proteins that contained amino acids 25 to 150 of YadB and amino acids 137 to 409 of YadC fused at their N termini to GST for further purification.
Promoter-lacZ expression assays.
yadBC promoter activity was monitored by determining the β-galactosidase activity of PyadBC-lacZ. The control strain was Y. pestis CO92.S1 carrying promoterless invA::lacZ integrated into the chromosome (CO99-3015.S4). The cultures to be tested were routinely freshly inoculated, grown overnight in HIB at 26 or 37°C to an OD620 of ca. 3.0, and then back-diluted to obtain an OD620 of 0.1 to start the experiment. Samples used to measure the OD620 were taken as needed (usually at 1-h intervals), and samples were pelleted and frozen at −80°C for further analysis. Thawed samples were resuspended in Z-buffer, and β-galactosidase activity was measured as previously described and expressed in Miller units (28).
β-Galactosidase activity was measured similarly when PyadBC-lacZ was tested for regulation by quorum sensing. The bacteria were grown in HIB at 37°C to an OD620 of 0.5 and then back-diluted to obtain an OD620 of 0.1 and supplemented with 0.25 volume of culture supernatant obtained fresh from cultures producing or lacking an autoinducer grown overnight in HIB at 37°C to an OD620 of ca. 3.0. The numbers of Miller units were then determined using samples taken from the supplemented cultures at hourly intervals for 6 h.
Immunoblot analysis.
Equal protein concentrations were resolved on polyacrylamide gels containing SDS prior to transfer onto polyvinylidene difluoride membranes (Immobilon P; Millipore). A modification of the procedure of Towbin et al. (47) was used for immunodetection. Briefly, polyvinylidene difluoride membranes were blocked with 5% nonfat dry milk or 1.5% bovine serum albumin in 10 mM Tris-HCl (pH 7.6)-137 mM NaCl with 0.1% Tween 20 and then incubated with the appropriate antibody diluted in 10 mM Tris-HCl (pH 7.6)-137 mM NaCl with 0.1% Tween 20. Antibodies against the GST-YadB and GST-YadC fusion proteins described above were generated in rabbits by Animal Pharm Services, Inc. Following incubation with horseradish peroxidase-conjugated protein A (Amersham Pharmacia Biotech), the immunoreactive proteins were detected by enhanced chemiluminescence (Amersham Pharmacia Biotech). Alternatively, proteins interacting with alkaline phosphatase-labeled anti-rabbit immunoglobulin G molecules were detected with disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate (CSPD) (Roche Inc.).
The GST-YadB and GST-YadC antigens were overexpressed from the pGEX-3X constructs in E. coli strain HB101. Following purification on GST-Sepharose (Sigma Chemical Co., St. Louis, MO) the antigens were dialyzed against phosphate-buffered saline (PBS) and used for immunization. The GST part of the fusion proteins was successfully cleaved by factor Xa (New England BioLabs) when necessary for testing purposes.
Adherence and invasion.
WI-26 (human type 1 pneumocyte) cells or HeLa epithelioid cells (ATCC, Manassas, VA) were grown to ∼95% confluence in six-well culture dishes. Thirty minutes prior to infection, each well was washed twice with serum-free medium (minimal essential medium or RPMI 1640 [Life Technologies, Grand Island, NY]) and incubated with serum-free medium until infection. Overnight cultures of Y. pestis grown at 37°C were used to inoculate minimal essential medium or RPMI 1640 culture medium. WI-26 or HeLa cell cultures in six-well plates were infected with Y. pestis in appropriate media at a multiplicity of infection of 10. After 5 min of centrifugation at 500 × g to facilitate the primary contact phase, the cells were incubated in a CO2 incubator for 15 min and for 1 h at 37°C for adherence and invasion assays, respectively. To determine the number of gentamicin-protected CFU (invasion), gentamicin (15 μg/ml) was added to wells and the plate was incubated for an additional 60 min. The cells were then washed twice with PBS, subjected to water lysis, and plated. The results were expressed as a percentage based on the number of output CFU relative to the number of input CFU from triplicate wells for each experiment.
Infection of mice: virulence tests.
F1+ parent strain CO92.S1, F1− parent strain CO92.S2, F1+ yadBC mutant strain CO92.S8, F1− yadBC strain CO92.S11, F1+ yadBC+ reconstituted strain CO92.S15, and F1+ yadC strain CO92.S17 were reconstituted to Lcr+ by introduction of pCD2Ap from Y. pestis CO99-3015.S6. The F1+ strains were tested to determine their lethality in a mouse model of bubonic plague. The Lcr+ parent and ΔyadBC mutants (CO92.S6 and CO92.S9) were grown at 37°C to mid-exponential phase, and the same bacterial cultures were used for both the intranasal and subcutaneous routes of infection. However, in a separate experiment, the Lcr+ parent and the ΔyadC mutant (CO92.S18) were grown at 28°C. The lower temperature more closely resembles the temperature likely to be found in the flea vector and might have had a small effect on the LD50 due to different initial states of the bacterial dose, but it did not impact the conclusions of this study. Groups of four female C57BL/6 mice were anesthetized with ketamine and xylazine and given one of a set of four 10-fold increasing doses of Y. pestis subcutaneously in 100 μl of PBS by injection under the skin on the back of the neck. A single-dose test with seven mice was performed for the Lcr+ yadBC+ strain (CO92.S16) grown at 28°C to mid-exponential phase. After infection, the anesthesia was reversed by intraperitoneal injection of yohimbine, and an optical lubricant was applied to the eyes. The mice were monitored for signs of illness and survival for 14 days. Mice that lost their righting reflex were deemed morbid and unlikely to survive until the next observation time and were humanely killed. Actual doses given to the mice were determined by plating to determine the number of CFU. LD50s were calculated by the method of Reed and Muench (40).
The lethalities of the Lcr+ F1− (caf1) parent and yadBC mutant strains were compared using a pneumonic plague mouse model. Groups of mice were anesthetized as described above and given 10-fold increasing doses of Y. pestis grown at 37°C intranasally in 20 μl of PBS delivered to the nares using a micropipette. Otherwise, mice were handled as described above. Single-dose tests were performed with the Lcr+ F1+ ΔyadBC, ΔyadC, and reconstituted yadBC+ strains, also grown at 37°C. Protocols approved by the Institutional Animal Care and Use Committee were used for all mouse handling procedures, and the procedures were monitored by a licensed veterinarian. Work with select-agent nonexempt strains was done in a biosafety level 3 containment facility with select-agent security.
Statistics.
Experiments were done two or more times unless indicated otherwise. The mean time to death was calculated for each group of mice, and the values for groups were compared to determine significant differences by using the Student unpaired t test. Survival distributions were compared to determine relative risk and significant differences by using the web-based interactive statistical calculation program on the Dartmouth College Biostat server (http://statpages.org/). Significant differences in gene expression were determined by using the Student t test (paired versus unpaired, as indicated below). The sign test was used to determine the significance of invasion assay results.
RESULTS
yadBC operon and predicted proteins.
We screened the genome of Y. pestis CO92 and identified a set of chromosomal genes that encode putative proteins likely to be expressed on the bacterial surface and to have virulence-related functions, based on similarities to genes that encode surface proteins in other pathogens. Among these genes were YPO1387 and YPO1388, both of which are structurally related to the YadA gene (pfam-3895), as noted by Hoiczyk et al. (19). We designated these genes yadB and yadC, respectively. yadBC appears to be a bicistronic operon with overlapping termination and initiation codons in its two genes (Fig. 1), and it is separated by 300 bp from the upstream gene ansB (encoding a putative l-asparaginase II precursor) and by 366 bp from the downstream putative operon serC-aroA. yadC is AT rich (38.7% G+C) compared to the overall composition of the Y. pestis CO92 chromosome (46.7% G+C [35]); yadB is less AT rich (40.5% G+C).
FIG. 1.
Schematic diagram of the yadBC operon, with coordinates for the open reading frames, signal sequences, and antigenic regions used in this study.
yadB encodes a putative 364-residue preprotein and a 35-kDa mature protein with a pI of 4.5. yadC's predicted product is a 622-amino-acid preprotein and a 61.6-kDa mature protein with a pI of 4.0. The amino acid sequences of YadA (Y. enterocolitica O:3), YadB, and YadC were aligned by T-coffee (32) and manually corrected to align the “head” of YadB with the analogous regions of YadA and YadC. The similarity among the three proteins was similarity to the C-terminal half of YadA (14% identical residues and 20% very similar residues shared by all three proteins over 264 amino acids), and there was a locus with greater similarity within the short neck domain (10 of 21 residues were identical in all three proteins). In YadA, the neck domain functions as a platform for the head region and helps stabilize the trimeric structure. The periodicities found in the stalk sequence of YadC resembled those determined in a REPPER analysis of YadA (16; data not shown), supporting the hypothesis that the two proteins may be structurally similar in this region. However, the “head” region of YadC had no sequence similarity to YadA or to any protein in organisms other than Y. pestis and Y. pseudotuberculosis, and although REPPER analysis did indicate the presence of a repeat structure within this region, the repeats had a different periodicity than that of the repeats in YadA. The corresponding region of YadB was only 62 amino acids long and 29% identical to the aligned stretch of YadC.
At the nucleotide level, yadB and yadC sequences specifying the putative neck to the C terminus are only 55% identical (as determined by Align version 2.0 [30]); global alignment score, 587) and have low G+C contents (39.8 and 35.3%, respectively). (The G+C content of the sequence specifying this region of YadA in either Y. enterocolitica O:3 or Y. pestis CO92 is 40.5%.) The DNA encoding the putative head region of YadC has a G+C content of 46.8%, and the short “head” of YadB is encoded by a sequence with a G+C content of 41.8%. The differences between yadB and yadC indicate that these two genes are not likely to represent a recent simple duplication event, with yadB being a truncated version of yadC. Nor are they simple variants of the YadA gene.
The YadB- and YadC-encoding sequences of Y. pestis biovar Medievalis strain KIM, biovar Microtus strain 91001, biovar Antiqua strains Antiqua and Nepal516, and biovar Orientalis strains IP275 and CO92 are identical, although the annotations indicate that different methionines are translation initiation codons. Y. pestis Angola has a mutation (F239L) in the YadC sequence. Both available Y. pseudotuberculosis genome sequences (IP32953 and IP31758) contain counterpart yadBC operons with several amino acid substitutions in both predicted proteins (Table 2). Interestingly, there are no yadBC orthologs in the genome of Y. enterocolitica 8081 (http://www.sanger.ac.uk/Projects/Y_enterocolitica/).
TABLE 2.
Comparison of YadB and YadC orthologs in strains of Y. pestis and Y. pseudotuberculosis
| Strain | Comparison with Y. pestis CO92a
|
|||
|---|---|---|---|---|
| YadB (364 amino acids)
|
YadC (622 amino acids)
|
|||
| Relatedness | Locus tag | Relatedness | Locus tag | |
| Y. pestis KIM | Identical | y2786 | Identical | y2785 |
| Y. pestis Antiqua | Identical | YPA_0678 | Identical | YPA_679 |
| Y. pestis Nepal516 | Identical | YPN_2590 | Identical | YPN_2589 |
| Y. pestis 91001 | Identical | YP_1206 | Identical | YP_1205 |
| Y. pestis IP275 | Identical | YpesB_01000518 | Identical | YpesB_01000519 |
| Y. pestis Angola | Identical | YpesA_01000661 | Similar | YpesA_01000660b |
| Y. pseudotuberculosis IP32953 | Similar | YPTB1412c | Similar | YPTB1413d |
| Y. pseudotuberculosis IP31758 | Similar | YpseI_02002348e | Similar | YpseI_02002347f |
The locus tags for Y. pestis CO92 YadB and YadC are YPO1387 and YPO1388, respectively.
Of the 622 residues, 621 were positive and identical; the single change was F239L.
Of the 364 residues, 359 were positive and 354 were identical. The similar residues were V57I, V85I, R105Q, M109I, and I162L, and the changes were S5R, F31L, I67T, T220N, and G226E.
Of the 622 residues, 617 were positive and 616 were identical. The similar residue was D86N, and the changes were T25I, V29A, Q69L, M100T, and T313I.
Of the 364 residues, 359 were positive and 355 were identical. The similar residues were V85I, R105Q, M109I, and I162L, and the changes were S5R, F31L, I67T, T220N, and G226E.
Of the 622 residues, 618 were positive and 615 were identical. The similar residues were D86N, N405S, and V489M, and the changes were T25I, V29A, Q69L, and M100T.
Regulation of yadBC expression.
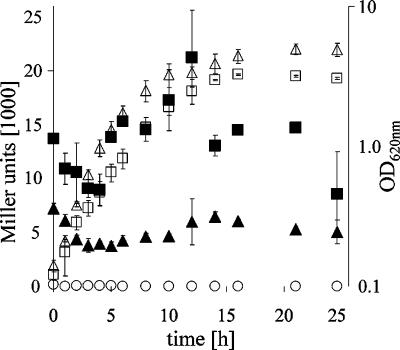
To obtain clues about natural environments in which YadB and YadC might function, we determined in vitro conditions that optimize yadBC gene expression. We constructed a yadBC::lacZ transcriptional fusion and introduced it into the Y. pestis invA pseudogene in the chromosome. Expression of β-galactosidase by the resulting Y. pestis CO99-3015.S10 strain was measured during growth at 26 and 37°C in complex medium (Fig. 2). In all phases of growth, the expression of the yadBC promoter was up to three times higher at 37°C than at 26°C (Fig. 2) (P = 0.004, as determined by a paired Student's t test comparing values for the exponential phase [3 and 4 h] and values for the stationary phase [15 and 20 h] at the two temperatures). There was a reproducibly significant increase in expression as the bacteria approached stationary phase at both temperatures (P = 0.0101 at 26°C and P = 0.0227 at 37°C, as determined by an unpaired t test comparing values for 3 and 4 h with values for 15 and 20 h at each temperature).
FIG. 2.
Growth and yadBC promoter activity from yadBC-lacZ integrated within invA in the chromosome of Y. pestis CO99-3015.S10. Y. pestis CO99-3015.S10 and the negative control strain Y. pestis CO99-3015.S4 with promoterless lacZ integrated into invA were grown at 26 and 37°C, and the OD620 and β-galactosidase activity were determined at the indicated times. β-Galactosidase levels (expressed in Miller units) were determined for yersiniae grown at 26°C (▴) and at 37°C (▪); corresponding OD620 values are also shown (▵ and □, respectively). The open circles show β-galactosidase activity at 37°C for Y. pestis CO99-3015.S4. Essentially identical data were obtained for this strain grown at 26°C, and the growth data for this strain overlapped those for Y. pestis CO99-3015.S10 (data not shown). The experiments were done three times on different days with similar results. The data shown are data from one experiment, and the error bars indicate the ranges for duplicate samples.
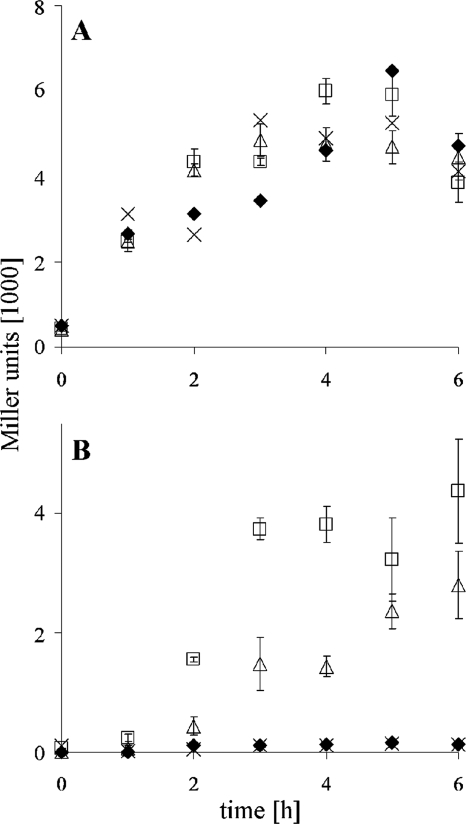
This finding led us to investigate whether quorum sensing might be involved in yadBC promoter regulation. Y. pestis CO92.S1 and Y. pestis KIM6+ were grown at 37°C until entry into stationary phase, and the cell-free culture medium was used as a source of homoserine lactone (HSL) autoinducers. Y. pestis KIM6-2109+, which lacks all three Y. pestis quorum-sensing systems, was similarly grown to provide a negative control culture supernatant. The test strains were the PyadBC reporter strain Y. pestis CO99-3015.S10 and a reference strain, E. coli MG4(pKDT17) (39) containing lasR-lacZ and lasB-lacZ fusions that respond to 3-oxo-C12-HSL. Both strains were adapted to exponential-phase growth at 37°C in order to obtain basal expression of their reporter constructs. Replicate cultures of each test strain were supplemented with 0.25 volume of a freshly prepared culture supernatant. None of the culture supernatants had an effect on yadBC promoter activity (Fig. 3A). In contrast, the reference E. coli strain MG4(pKDT17) was responsive to both Y. pestis HSL-containing culture supernatants but was not stimulated by the supernatant from the quorum-sensing mutant KIM6-2109+, as expected (Fig. 3B). Therefore, the early-stationary-phase increase in expression activity of the yadBC promoter was not due to regulation by any of the three Y. pestis quorum-sensing systems.
FIG. 3.
Test for possible quorum-sensing effect of HSL molecules present in the culture supernatant on yadBC promoter activity. (A) Promoter activity resulting from single-crossover integration of pSucinv::yadBC-lacZ in Y. pestis strain CO99-3015.S10 in response to fresh medium (negative control) (×) and to cell-free culture supernatants of Y. pestis strains KIM6+ (positive control supernatant) (□), CO99-3015.S1 (▵), and KIM6-2109+ (“quorum-sensing triple mutant,” negative control supernatant) (⧫). The data are averages ± standard deviations of triplicate assays. (B) Promoter responsiveness of the E. coli MG4(pKDT17) reporter strain (positive control). The medium and symbols are the same as those described above for panel A.
Expression of YadB and YadC in Y. pestis.
Rabbit antisera were raised against GST-YadB and GST-YadC fusion proteins containing peptide portions of mature YadB and YadC that spanned the predicted head and neck regions (126 and 273 amino acids, respectively) (Fig. 1). Both antisera reacted with the antigenic portion of the corresponding GST fusion protein upon cleavage with factor Xa (data not shown); however, we were unable to detect YadB and YadC in Y. pestis whole-cell extracts collected at either the exponential or early stationary phase of growth. Upon purification of crude membrane fractions and prolonged mild denaturation (incubation with SDS buffer at 37°C), potential trimeric forms of YadB and YadC were only faintly detected (data not shown).
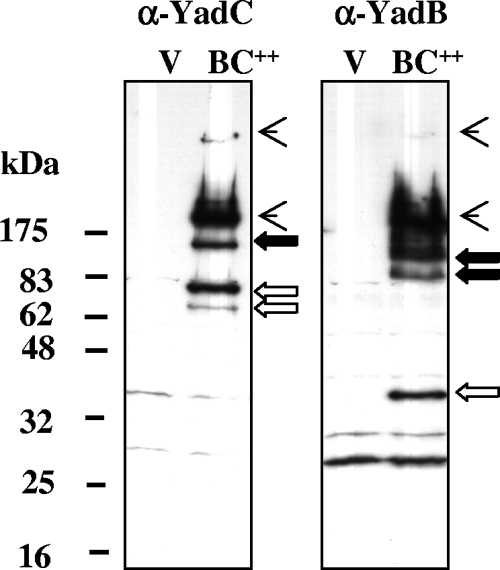
To improve detection, we expressed yadBC in trans from an inducible promoter in ΔyadBC strain Y. pestis CO92.S13, which lacks the periplasmic protease DegP (GsrA) and the very abundant surface protease Pla. Figure 4 shows immunoblots prepared from whole cells of the yersiniae containing pBAD24YadBC or the empty vector. YadB was detected as three species unique to the pBAD24YadBC-containing strain, which migrated at masses consistent with the hypothesis that a monomer (35 kDa) and a trimer (105 kDa) were present. A more slowly migrating second putative oligomer of YadB also was seen. In addition, the anti-YadB antibody recognized two larger species (Fig. 4). Anti-YadC antibody detected a pair of bands in the vicinity of the monomer size (62 kDa) but at a higher apparent molecular mass. There also was a species that could represent a trimer (186 kDa) of YadC, and there were two larger species that migrated like species detected by the anti-YadB antibody. These findings indicate that both YadB and YadC have the potential to form oligomers, including trimers, as suggested from their similarity to YadA, and raise the possibility that they may interact in a multimeric complex.
FIG. 4.
Expression of pBAD24YadBC in a ΔyadBC Y. pestis strain. Y. pestis CO92.S13(pBAD24YadBC) and Y. pestis CO92.S13(pBAD24) (vector control) were grown at 37°C, 0.25% arabinose was added, and incubation was continued for 4 h to induce expression of YadB and YadC. Whole cells were solubilized and analyzed by using immunoblots probed with anti-YadB (right panel) or anti-YadC (left panel) antibodies. Lane BC++, Y. pestis CO92.S13(pBAD24YadBC); lane V, vector control [Y. pestis CO92.S13(pBAD24)]. The positions of molecular mass markers are indicated on the left. Open arrows indicate bands thought to represent monomeric forms of YadB and YadC; putative trimeric forms are indicated by solid arrows; and the arrowheads indicate oligomeric forms that react with both anti-YadB and anti-YadC antibodies.
Tests for roles of YadB and YadC in adherence to and invasion of epithelial cells.
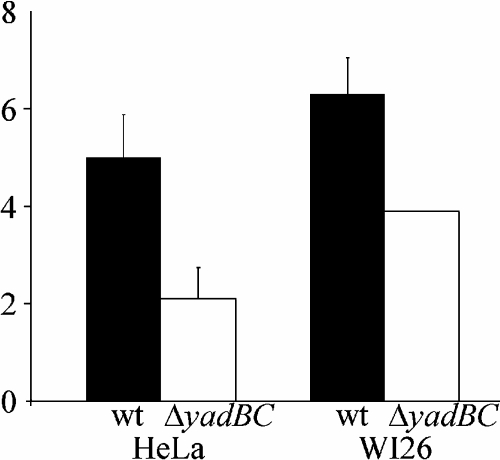
Because all Oca family proteins studied so far are adhesins, we wondered if YadB and YadC also make an important contribution to the adherence and invasive capabilities of Y. pestis for epithelial cells. We tested the yadBC double mutant Y. pestis CO92.S8 along with the parent Y. pestis strain for adherence to and invasion of WI-26 type 1 pneumocytes and HeLa epithelioid cells. There was no statistically significant difference between the strains in terms of adherence measured 15 min after centrifugation of the bacteria onto the cells (data not shown). In the invasion assay, with gentamicin protection treatment, we repeatedly observed a small but statistically significant defect in invasion by the double-mutant strain (Fig. 5). In three independent experiments with WI-26 cells, carried out in triplicate, and in two experiments with the HeLa cell line there was statistical significance as determined by the sign test. Thus, on average, the ΔyadBC Y. pestis mutant exhibited slightly reduced invasion (60% of the invasion shown by the parental strain).
FIG. 5.
Invasiveness of the parent strain of Y. pestis CO92.S1 and the yadBC deletion mutant strain CO92.S8 with WI-26 and HeLa epithelioid cell lines. Invasion was measured by performing gentamicin protection assays in triplicate, and the data are averages and standard deviations. The three independent WI-26 experiments and the two HeLa experiments were positive for significance as determined by the sign test. wt, wild type.
Tests for importance of yadBC in bubonic plague and pneumonic plague in mice.
We infected groups of C57BL/6 mice intranasally with F1− strain Y. pestis CO92.S7 or F1− ΔyadBC strain Y. pestis CO92.S10. The F1− derivatives were used to maximally expose YadB and YadC on the bacterial surface and to exaggerate the effects of loss of these proteins in the ΔyadBC mutant. However, loss of these proteins had no significant effect on virulence in pneumonic plague. The two infections followed similar time courses, with the earliest deaths occurring at day 4 postinfection. The LD50 of the parent strain was 2.0 × 102 ± 0.1 × 102 bacteria, consistent with findings reported by others (24, 50), and the LD50 of the ΔyadBC mutant was 3.7 × 102 ± 2.9 × 102 bacteria. Two separate tests were performed with single doses of the F1+ ΔyadBC strain Y. pestis CO92.S9 given to mice intranasally. A dose of 5,100 bacteria was 100% lethal (four mice tested), and in a separate experiment, the LD50 was 500 bacteria (eight mice in the group), indicating that the LD50 of the F1+ ΔyadBC strain is similar to that of the F1− ΔyadBC strain. Likewise, an intranasal dose of 5,400 F1+ ΔyadC strain Y. pestis CO92.S18 cells was 100% lethal (four mice were tested). The reconstituted yadBC+ strain CO92.S16 killed six of seven mice given an intranasal dose of 1,800 bacteria, which again was consistent with full virulence. These findings indicated that YadB and YadC are not required for virulence in pneumonic plague, whether F1 is present or not.
The virulence of F1+ Y. pestis CO92.S6 administered by the subcutaneous route was tested in one experiment, and the results showed that the LD50 was between 4 and 5 bacteria, which again is consistent with the high lethality documented previously for bubonic plague in Swiss Webster mice (LD50, 1.7 bacteria) (49). A separate test with a single dose of 20 bacteria killed three of four mice (on days 7, 8, and 9 postinfection). The reconstituted yadBC+ strain CO92.S16 was tested using a subcutaneous dose of 270 bacteria, which killed all seven mice (on days 5 through 9 postinfection), which was also consistent with full virulence. However, when the subcutaneous route was used, the F1+ Y. pestis ΔyadBC mutant CO92.S9 did not kill mice at doses as high as 10,000 bacteria per animal. Likewise, the ΔyadC single mutant CO92.S18 was attenuated and did not kill mice at the highest dose tested (930 bacteria). These findings showed that yadBC is a new major virulence property for bubonic plague and indicate that the YadC component may be essential for full lethality.
DISCUSSION
This study identified Y. pestis YadB and YadC as two new members of the Oca family of proteins that have the ability to form trimers and increase invasion of epithelioid cells. Accordingly, they likely are trimeric autotransporters, analogous to YadA. However, the functional head domains of YadA and YadC are distinct, and YadB appears to have only a small head domain (7.3 kDa). We do not know the basis for the forms of YadC that migrated more slowly in SDS-polyacrylamide gel electrophoresis than the predicted monomer and were too small to represent dimers (Fig. 4). Sometimes we observed the true 62-kDa monomer size, but its prominence depended on the expression construct (unpublished data). We hypothesize that the forms that migrated more slowly in SDS-polyacrylamide gel electrophoresis than the predicted monomer and were too small to represent dimers represent different folding states of YadC. The arrangement of two putative trimeric autotransporters in a bicistronic operon is uncommon and likely indicates functional relatedness. The data in Fig. 4 and our unpublished data from experiments with other expression constructs raise the possibility that YadB and YadC may form a complex at the bacterial surface. Studies are under way to test this idea and determine whether YadB and YadC may interact during biogenesis or function.
The expression of yadBC is upregulated in vitro upon entry into stationary phase and is two- to threefold greater at 37°C than at 26°C, suggesting that this operon has a function in a mammalian host with plague. Although in vitro expression patterns do not necessarily predict expression during plague (25), yadBC expression has been detected during the late stages of pneumonic plague in mice (C. R. Wulff, A. M. Uittenbogaard, and S. C. Straley, unpublished data). Moreover, the proteins likely are displayed on the bacterial surface during pneumonic plague, because mice immunized subcutaneously with the GST-YadC136-409 fragment that was created in this study for production of antibody in rabbits were partially protected against pneumonic plague due to virulent Y. pestis (29). Interestingly, our LD50 test showed that deletion of yadBC had no significant effect on virulence when the intranasal route of infection was used, whereas the virulence of the yadBC mutant was severely compromised when the skin route was used for challenge. These findings add YadB and YadC to the growing list of properties, such as the capsule F1, that play a role in bubonic plague but are not required for lethality in pneumonic plague (5, 10), and they suggest that the initial niches inhabited by the bacteria in the two diseases are very different and determine the subsequent course of infection.
The relatively minor effect of the ΔyadBC mutation on the invasive phenotype of Y. pestis CO92 does not rule out the possibility that YadB and YadC have a role in invasion, but it is consistent with the failure of previous signature-tagged mutagenesis attempts to identify a major adhesin (22, 26). We suggest that YadB and YadC are not dominant mediators of tight adherence or invasion. In a skin infection, where dissemination to internal organs and eventual bacteremia are paramount for survival in nature, factors that would make the bacteria strongly adherent to an extracellular matrix or phagocytes would be detrimental. Consistent with this idea, expression of the strong adhesin YadA in Y. pestis actually decreases virulence (42).
Nonetheless, although we predict that YadB and YadC have an invasin function, the yadBC deletion has a greater effect on virulence than we would predict if it were simply one of multiple adhesins/invasins that function redundantly. For example, deletion of the surface-localizing (i.e., antiphagocytic) adhesin PsaA causes an increase of only ca. 100-fold in the subcutaneous LD50 (5) (see the supplemental material) and a delay in colonization of organs (5). In the case of the yadBC strain, the subcutaneous LD50 increased more than 2,000-fold compared to the parent Y. pestis strain, suggesting that YadB and YadC must serve a unique, critical role. Threading analysis and database searches have failed to provide a possible clue to an enzymatic activity for YadC; however, this protein could inhibit a host enzyme. This and other possible activities are currently under investigation.
In summary, this study identified yadBC as a new virulence property for bubonic plague. The fact that yadBC is unique to Y. pestis and Y. pseudotuberculosis could implicate this operon in the more highly disseminatory character of these yersiniae compared to Y. enterocolitica, and acquisition of this operon could represent one important step in the evolution of Y. pestis as a flea-borne pathogen. YadB and YadC are new members of the Oca family of adhesins, but their main roles likely are novel. Their genetic arrangement suggests that they act in concert; accordingly, additional study of these proteins could provide new insights into the biogenesis and structure of trimeric autotransporters.
Supplementary Material
Acknowledgments
This study was initiated with funding from Public Health Service (NIAID) grant AI48491 and was completed with support from project 4.1 in 1 UF4 AI057175 to SERCEB (Region IV Center of Excellence for Biodefense and Emerging Infectious Diseases) and from the University of Kentucky.
We thank Michael Gray for his excellent technical help with creating pPCP1::Kan and pPCP1::Kan-fsPla. Spencer Leigh (USDA ARS Poultry Science Research Unit, Mississippi State, MS) created pCD2Ap (in Y. pestis CO99-3015.S6) and control strain CO99-3015.S4 used in this study. Annette Uittenbogaard carried out the RT-PCR tests, and she and Amanda Gorman helped carry out the animal experiments.
Editor: D. L. Burns
Footnotes
Published ahead of print on 19 November 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 2.Beesley, E. D., R. R. Brubaker, W. A. Janssen, and M. J. Surgalla. 1967. Pesticins. III. Expression of coagulase and mechanism of fibrinolysis. J. Bacteriol. 9419-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 71513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, E. C., and B. B. Finlay. 2003. Bacterial pathogenesis: exploiting cellular adherence. Curr. Opin. Cell Biol. 15633-639. [DOI] [PubMed] [Google Scholar]
- 5.Cathelyn, J. S., S. D. Crosby, W. W. Lathem, W. E. Goldman, and V. L. Miller. 2006. RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. USA 10313514-13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelis, G. R. 2002. Yersinia type III secretion: send in the effectors. J. Cell Biol. 158401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13199-205. [DOI] [PubMed] [Google Scholar]
- 8.Cowan, C., H. A. Jones, Y. H. Kaya, R. D. Perry, and S. C. Straley. 2000. Invasion of epithelial cells by Yersinia pestis: evidence for a Y. pestis-specific invasin. Infect. Immun. 684523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, K. J., D. L. Fritz, M. L. Pitt, S. L. Welkos, P. L. Worsham, and A. M. Friedlander. 1996. Pathology of experimental pneumonic plague produced by F1-positive and F1-negative Yersinia pestis in African green monkeys. Arch. Pathol. 120156-163. [PubMed] [Google Scholar]
- 11.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 135386-405. [DOI] [PubMed] [Google Scholar]
- 12.Eitel, J., and P. Dersch. 2002. The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions under which invasin is repressed. Infect. Immun. 704880-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elhai, J., and C. P. Wolk. 1988. A versatile set of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68119-138. [DOI] [PubMed] [Google Scholar]
- 14.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaced Yersinia adhesin. Int. J. Med. Microbiol. 291209-218. [DOI] [PubMed] [Google Scholar]
- 15.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 62693-2704. [DOI] [PubMed] [Google Scholar]
- 16.Gruber, M., J. Soding, and A. N. Lupas. 2005. REPPER-repeats and their periodicities in fibrous proteins. Nucleic Acids Res. 33W239-W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. N. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 195989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphreys, G. O., G. A. Willshaw, and E. S. Anderson. 1975. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim. Biophys. Acta 383457-463. [DOI] [PubMed] [Google Scholar]
- 21.Isberg, R. R., and P. Barnes. 2001. Subversion of integrins by enteropathogenic Yersinia. J. Cell Sci. 11421-28. [DOI] [PubMed] [Google Scholar]
- 22.Karlyshev, A. V., P. C. Oyston, K. Williams, G. C. Clark, R. W. Titball, E. A. Winzeler, and B. W. Wren. 2001. Application of high-density array-based signature-tagged mutagenesis to discover novel Yersinia virulence-associated genes. Infect. Immun. 697810-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koretke, K. K., P. Szczesny, M. Gruber, and A. N. Lupas. 2006. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J. Struc. Biol. 155154-161. [DOI] [PubMed] [Google Scholar]
- 24.Lathem, W. W., S. D. Crosby, V. L. Miller, and W. E. Goldman. 2005. Progression of pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. USA 10217786-17791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawson, J. N., C. R. Lyons, and S. A. Johnston. 2006. Expression profiling of Yersinia pestis during mouse pulmonary infection. DNA Cell Biol. 25608-616. [DOI] [PubMed] [Google Scholar]
- 26.Leigh, S. A., S. Forman, R. D. Perry, and S. C. Straley. 2005. Unexpected results from the application of signature-tagged mutagenesis to identify Yersinia pestis genes required for adherence and invasion. Microb. Pathog. 38259-266. [DOI] [PubMed] [Google Scholar]
- 27.Metcalf, W. W., W. Jiang, L. L. Daniels, S.-K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 351-13. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 29.Murphy, B. S., C. R. Wulff, B. A. Garvy, and S. C. Straley. 2007. Yersinia pestis YadC: a novel vaccine candidate against plague. Adv. Exp. Med. Biol. 603400-414. [DOI] [PubMed] [Google Scholar]
- 30.Myers, E. W., and W. Miller. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 411-17. [DOI] [PubMed] [Google Scholar]
- 31.Nilles, M. L., K. A. Fields, and S. C. Straley. 1998. The V antigen of Yersinia pestis regulates Yop vectorial targeting as well as Yop secretion through effects on YopB and LcrG. J. Bacteriol. 1803410-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Notredame, C., D. G. Higgins, and J. Heringa. 2000. A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302205-217. [DOI] [PubMed] [Google Scholar]
- 33.Nummelin, H., M. C. Merckel, J. C. Leo, H. Lankinen, M. Skurnik, and A. Goldman. 2004. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel β-roll. EMBO J. 23701-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nummelin, H., Y. El Tahir, P. Ollikka, M. Skurnik, and A. Goldman. 2002. Expression, purification and crystallization of a collagen-binding fragment of Yersinia adhesin YadA. Acta Crystallogr. Sect. D 581042-1044. [DOI] [PubMed] [Google Scholar]
- 35.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413523-527. [DOI] [PubMed] [Google Scholar]
- 36.Perry, R. D., A. Bobrov, O. Kirillina, H. A. Jones, L. Pedersen, J. Abney, and J. D. Fetherston. 2004. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 1861638-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 1035-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry, R. D., M. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 1725929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ralling, G., S. Bodrug, and T. Linn. 1985. Growth rate-dependent regulation of RNA polymerase synthesis in Escherichia coli. Mol. Gen. Genet. 201379-386. [DOI] [PubMed] [Google Scholar]
- 40.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 41.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 1853735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosqvist, R., M. Skurnik, and H. Wolf-Watz. 1988. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature 364522-524. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Straley, S. C., and P. A. Harmon. 1984. Growth in mouse peritoneal macrophages of Yersinia pestis lacking established virulence determinants. Infect. Immun. 45649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Straley, S. C., and W. S. Bowmer. 1986. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect. Immun. 51445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surgalla, M. J., and E. D. Beesley. 1969. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl. Microbiol. 18834-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Towbin, H., T. Staehelin, and G. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci., USA 764350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100195-199. [PubMed] [Google Scholar]
- 49.Welkos, S. L., A. M. Friedlander, and K. J. Davis. 1997. Studies on the role of plasminogen activator in systemic infection by virulent Yersinia pestis strain CO92. Microb. Pathog. 23211-223. [DOI] [PubMed] [Google Scholar]
- 50.Welkos, S. L., K. M. Davis, L. M. Pitt, P. M. Worsham, and A. M. Friedlander. 1995. Studies on the contribution of the F1 capsule-associated plasmid pFra to the virulence of Yersinia pestis. Contrib. Microbiol. Immunol. 13299-305. [PubMed] [Google Scholar]
- 51.Wollmann, P., K. Zeth, A. N. Lupas, and D. Linka. 2006. Purification of the YadA membrane anchor for secondary structure analysis and crystallization. Int. J. Biol. Macromol. 393-9. [DOI] [PubMed] [Google Scholar]
- 52.Wulff-Strobel, C. R., A. W. Williams, and S. C. Straley. 2002. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol. Microbiol. 43411-423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.