Abstract
The virulence mechanisms leading Streptococcus pneumoniae to convert from nasopharyngeal colonization to a tissue-invasive phenotype are still largely unknown. Proliferation of infection requires penetration of the extracellular matrix, which occurs by recruitment of host proteases to the bacterial cell surface. We present evidence supporting the role of choline-binding protein E (CBPE) (a member of the surface-exposed choline-binding protein family) as an important receptor for human plasminogen, the precursor of plasmin. The results of ligand overlay blot analyses, solid-phase binding assays, and surface plasmon resonance experiments support the idea of an interaction between CBPE and plasminogen. We have shown that the phosphorylcholine esterase (Pce) domain of CBPE interacts with the plasminogen kringle domains. Analysis of the crystal structure of the Pce domain, followed by site-directed mutagenesis, allowed the identification of the plasminogen-binding region composed in part by lysine residues, some of which map in a linear fashion on the surface of the Pce domain. The biological relevance of the CBPE-plasminogen interaction is supported by the fact that, compared to the wild-type strain, a mutant of pneumococcus with the cbpE gene deleted (i) displays a reduced level of plasminogen binding and plasmin activation and (ii) shows reduced ability to cross the extracellular matrix in an in vitro model. These results support the idea of a physiological role for the CBPE-plasminogen interaction in pneumococcal dissemination into human tissue.
Streptococcus pneumoniae is a gram-positive commensal inhabitant of the human respiratory tract and the most common cause of noninvasive (otitis, sinusitis, pneumonia) and invasive (sepsis and meningitis) diseases. Pneumococcal infections are prevalent throughout the world, accounting yearly for the death of 1.6 million people, according to the World Health Organization. Most pneumococcal infection patients are children under the age of 5, the elderly, and immunocompromised individuals (37). The current prevention strategies based on immunogenic properties of capsular polysaccharides rely on a 23-valent and a 7-valent conjugate vaccine, even though the latter does not offer complete serotype coverage (3, 10, 26). Pneumococcal infection treatments are impaired by the increased number of clinical strains resistant to single or multiple antibiotherapies (11, 39). There is a pressing need for the development of a vaccine with a wider serotype coverage spectrum as well as for the development of new drugs. Surface-exposed pneumococcal proteins are considered to be good new therapeutic target candidates for two main reasons: (i) they play roles in colonization and/or dissemination of S. pneumoniae within the host organism, and (ii) their high amino-acid-sequence conservation among serotypes may lead to new vaccine strategies protecting all age groups against a large spectrum of serotypes (15, 23, 24).
Hundreds of proteins are exposed on the S. pneumoniae surface through different attachment modes (25, 50). The choline-binding proteins (CBPs) bind to the phosphorylcholine of cell wall (lipo)teichoic acids through the choline-binding domain, which consists of 2 to 10 repeats of a 20-amino-acid sequence (19). CBPs can fulfill a variety of functions, including cleavage of peptidoglycan bonds (LytA, LytB, LytC, and CBPD) and proteolysis (CBPG) (2, 14, 21, 42, 44). A role in pneumococcal virulence has been attributed for most CBPs: PspA inhibits complement activation; CBPA is a major adhesin and interferes with the host immune response; and CBPE and CBPG are involved in nasopharynx colonization and adherence to human epithelial cells (22, 27, 31, 33, 36, 40, 45, 46, 52, 54).
Proteolytic cleavage of the extracellular matrix (ECM) is a prerequisite for bacterial invasion into host tissues. For invasion to occur, pathogenic bacteria must recruit host proteases, among them the plasminogen (Plg)-plasmin system (28, 49). On gram-negative bacteria, fimbrial and flagellar surface appendages form a major group of Plg receptors (29). Among group A and C streptococci, the M-like protein (PAM) and the α-enolase have been identified as Plg receptors (4, 34, 53). The interaction of S. pneumoniae with Plg has been shown to be mediated by the glycolytic enzymes GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and α-enolase (5-8, 16).
Plasmin is a broad-spectrum serine protease playing central roles in fibrinolysis, inflammation, wound healing, tissue remodeling, and cell migration through ECM degradation (1, 13, 28, 38). The proenzyme Plg is a single-chain glycoprotein of 92 kDa composed of five kringle modules at the N terminus of the plasmin catalytic domain. Plg binding to fibrin, to laminin (Lm) (a major glycoprotein of basement membranes), and to mammalian cells receptors is mediated by the Plg kringle domains, the first three of which are called Lysine-Binding Site 1 (LBS-1) (28, 38).
This report focuses on the involvement of the CBPE in the pneumococcal virulence process (22, 52). Interactions with ECM components and blood circulating factors were investigated, and recognition of recombinant purified CBPE with respect to Lm and Plg was observed. The CBPE-Plg interaction was further characterized as being restricted to the Pce and LBS-1 domains, and some residues of Pce involved in this interaction have been identified. It was shown that Plg bound to CBPE at the bacterial surface and was competent for plasmin activation. Finally, in an in vitro model of the ECM, CBPE-dependent bound plasmin allowed the pneumococcus to cross the ECM. Taken together, our results suggest that CBPE plays a role in pneumococcal dissemination into human tissue through interaction with the Plg-plasmin system.
MATERIALS AND METHODS
Pneumococcal strains.
Streptococcus pneumoniae R6 is a nonencapsulated derivative of strain R36A (Rockefeller University). The cbpE-inactivated R6 strain was constructed using the approach described by Fadda et al. (18) by inserting the cat cassette in the cbpE locus, leading to the deleted mutant cbpE::cat strain. The cbpE gene inactivation does not generate a polar effect on downstream gene expression, according to a previous report (52). Wild-type and mutant pneumococcal strains were cultured in Todd-Hewitt broth (BD Sciences). Whenever required, pneumococcal bacteria were labeled with fluorescein isothiocyanate (FITC). Bacteria from mid-exponentially grown cultures were washed with phosphate-buffered saline (PBS) before incubation with FITC at 1 mg/ml in PBS for 20 min at room temperature. Bacteria were extensively washed five times with PBS before use.
Production and purification of recombinant proteins.
Cloning of the pce gene encoding the catalytic domain of CBPE in pHIS8 has been previously described (20). Cloning of the cbpE gene, which codes for the Pce and the choline-binding domain, was performed using the same protocol. Genomic DNA from the R6 strain of S. pneumoniae was used as a template to amplify the gene by conventional PCR methodology. Subsequently, the resulting PCR product was cloned into the pLIM01 vector (Protein'eXpert SA, Grenoble, Switzerland) that had been modified from pQE80 by insertion of patented sequences, allowing a ligase-independent cloning procedure. The resulting construct, pLIM01/cbpE, encodes the CBPE protein from Glu27 to Gln627 fused to a His6 tag at the N terminus. A tobacco etch virus protease cleavage site was inserted between the His6 tag and the N-terminal sequence of CBPE. DNA sequencing (Genome Express, Grenoble, Switzerland) confirmed that no mutations had been introduced during PCR. The pHIS8/pce construct was employed as the template DNA for site-directed mutagenesis (51). All the mutant constructs were subsequently sequenced; the results showed that only expected mutations were introduced during PCR.
An overnight culture of the Escherichia coli BL21(DE3)-CodonPlus-RIL (Stratagene) strain transformed with the expression vectors was used for inoculation with 250 ml of Terrific Broth medium (Euromedex) supplemented with 100 μg/ml of ampicillin and 34 μg/ml of chloramphenicol and was cultured at 37°C. Protein expression was induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside for 16 h at 15°C. After cell lysis, recombinant proteins were recovered from the soluble fraction and loaded onto a 1-ml prepacked HisTrap HP column (catalog no. 17-5247-01; GE Healthcare). Lysis and column equilibration were performed using 50 mM Tris-HCl (pH 8.0)-200 mM NaCl-20 mM imidazole. After extensive washing, recombinant proteins were eluted with 20 to 500 mM imidazole gradient in 50 mM Tris-HCl (pH 8.0)-200 mM NaCl buffer and subsequently dialyzed against PBS (pH 7.4) before use for biological tests. The degree of purity of each Pce variant was checked by silver-staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-polyacrylamide gel electrophoresis). Protein concentrations were determined by absorbance measurement at 280 nm and by using the Bradford assay (9).
Phosphorylcholine esterase activity.
Enzymatic activities of wild-type and mutant Pce variants were measured using p-nitrophenyl-phosphorylcholine (pNP-PC) (catalog no. N5879; Sigma) as the substrate at 37°C in PBS (pH 7.4) in a total volume of 200 μl. The activity was measured by use of a Fluostar Optima luminescence reader (BMG) to monitor the increase in absorbance at 405 nm due to production of pNP, whose quantification as a reaction product of Pce activity had been previously calibrated with a standard curve. The concentration of pNP-PC ranged from 0.1 to 2 mM, and each purified protein was used at 150 nM. Values for the kinetic parameters kcat and Km were determined by fitting the data to a Lineweaver-Burk plot.
Fluorescence-based protein thermal stability assay.
Assays were performed using an IQ5 96-well format real-time PCR instrument (Bio-Rad) over a temperature range starting at 293 K (20°C) and ranging up to 362.6 K (89.6°C) with a heating rate of 1 K min−1. Assay samples of 25 μl consisted of 3.5 μg of each Pce variant, 5 mM TCEP [Tris(2-carboxyethyl) phosphine], and 2 μl of 5,000× Sypro Orange solution (Molecular Probes, Eugene, OR) diluted 1/100 in water. Excitation and emission wavelengths of SYPRO Orange are 470 nm and 570 nm, respectively. Protein thermal unfolding curves were monitored through closed-circuit device camera detection of changes in fluorescence for the SYPRO Orange environmentally sensitive dye (35). To determine the inflection point of the melting transition and thus more accurately determine the melting temperature (Tm), the first derivative was calculated.
Recombinant assay of binding of Pce to mammalian proteins.
Solid-phase binding assays were performed to measure binding of Pce to different proteins. White 96-well microtiter plates (catalog no. 655074; Greiner Bio One) were coated with 1 μg each of collagen IV, fibronectin, fibrinogen, Lm, and Plg (catalog no. C5533, F3879, L2020, and P5661 [Sigma] and catalog no. 1051-407 [Boehringer]) and bovine serum albumin (BSA) as a control (catalog no. R369D; Promega) in 100 μl PBS at 4°C overnight. Saturation was performed by adding 200 μl of PBS-2% BSA per well for 1 h. His-tagged Pce (200 pmol) was added to each well, and the mixture was incubated for 2 h. Three washes were performed for a total period of 30 min using 200 μl of PBS-0.03% Tween. His-tagged bound Pce was detected by adding 100 μl of horseradish peroxidase-conjugated anti-His antibody (catalog no. A7058; Sigma) (1:1,000 dilution) in PBS-0.03% Tween 20-0.2% BSA per well for 1 h. Four washes with 200 μl PBS-0.03% Tween 20 were performed. ECL solution (catalog no. RPN2209; GE Healthcare) (100 μl) was added to each well, and chemiluminescence was measured using a multiwell luminescence reader (Fluostar Optima; BMG).
Ligand blotting overlay assay.
A mixture of Plg (33 pmol), LBS-1 (catalog no. P1667; Sigma) (20 pmol), and BSA (33 pmol) was electrophoresed on 10% or 12.5% SDS-polyacrylamide gels and transferred onto nitrocellulose membranes, which were subsequently saturated with PBS containing 0.3% Tween 20-5% milk (blocking buffer). Membranes were then incubated with 1 μM of purified Pce or CBPE in blocking buffer for 1 h. Successive 1-h incubations were conducted with anti-Pce antibody and horseradish peroxidase-conjugated anti-rabbit antibody (catalog no. A0545; Sigma), both diluted 1:10,000 in the blocking buffer, before chemiluminescence detection with enhanced chemiluminescence.
SPR measurements.
Surface plasmon resonance (SPR) measurements were performed using a Biacore X instrument (Biacore AB, Uppsala, Sweden). Plg (1.5 μg) or LBS-1 (2.5 μg) was covalently immobilized to the dextran matrix of a CM5 sensor chip via the primary amine groups (amine coupling kit; Biacore AB). The carboxymethylated dextran surface was activated by the injection of a mixture of 0.2 M N-ethyl-N′-(diethylamino-propyl)carbodiimide and 0.05 M N-hydroxysuccinimide. Ligands were injected in 10 mM sodium acetate buffer (pH 4.0). Activation time, ligand concentrations, and contact time were adjusted according to the desired extent of immobilization. The remaining N-hydroxysuccinimide esters were blocked by the injection of 1 M ethanolamine hydrochloride (pH 8.5). All immobilization steps were performed at a flow rate of 10 μl/min. Immobilization levels for Plg and LBS-1 were 1,879 and 2,101 resonance units (RU), respectively, for binding assays and 8,440 RU and 9,205 RU, respectively, when conditions of limitation of mass transport were required. No protein was immobilized on the control flow cell that underwent the activation and blocking steps.
The active concentrations of bound CBPE and Pce were determined using high-density Plg and LBS-1 sensor chips. These experiments were done under conditions of mass transport limitation, because the rate of mass transport is directly proportional to the ligand concentration such that when the rate of mass transport is limiting, the rate of ligand binding to the surface provides a direct measure of the ligand concentration and the signal is independent of the analyte-ligand affinity at the surface (12). Various concentrations of CBPE and Pce were injected during 1 min at flow rates ranging from 10 to 30 μl/min. Theoretical calibration curves relating the initial slope to the concentration of CBPE or Pce were established with BIAsimulation software (Biacore AB) and were used to calculate the concentration of protein in the samples.
Binding assays were performed at 25°C in PBS buffer (pH 7.4) containing 0.005% (vol/vol) P20 surfactant. CBPE and Pce were injected at different concentrations ranging from 10 nM to 8 μM at a flow rate of 30 μl/min. In all experiments, association and dissociation phases ran for 120 s. The surface was then regenerated with pulses of guanidinium chloride ranging from 0.1 M up to 1 M. Control sensorgrams were automatically subtracted from those obtained with immobilized Plg and LBS-1.
Binding inhibition experiments were carried out using increasing concentrations of a lysine analog, 6-amino hexanoic acid (6-AHA) (catalog no. A7824; Sigma).
Bacterium-Plg binding assay.
Solid-phase binding assays were performed to measure binding of the R6 strain and the cbpE::cat mutant strains to Plg. Black 96-well microtiter plates (catalog no. 655077; Greiner Bio One) were coated with 1 μg of Plg or BSA in PBS at 4°C overnight. Following two washes with PBS, the wells were blocked with 100 μl of PBS-1% BSA for 1 h. Identical washes were performed, and 5·107 or 5·108 CFU of R6 and cbpE::cat FITC-labeled bacteria in PBS-0.2% BSA were added to the wells for 2 h at 37°C. After five washes with PBS, FITC fluorescence (excitation wavelength, 490 nm; emission wavelength, 520 nm) was measured on a multiwell fluorescence reader (Fluostar Optima; BMG).
Plasmin activity assay.
Different amounts of R6 and cbpE::cat strains (5·107 or 5·108 CFU) or 20 μg of BSA (as a negative control) was added to transparent 96-well microtiter plates (catalog no. 675801; Greiner Bio One), and efficient deposition was ensured by low-speed centrifugation (5 min at 300 × g) before a 1-h incubation. After three washes with PBS, wells were blocked with 100 μl PBS-1% BSA for 1 h. In each well, 2 μg of Plg was added in a 50-μl reaction volume and incubated for 2 h at 37°C. After four washes with PBS, 50 μl of 50 nM streptokinase (catalog no. S8026; Sigma) was added and the mixture was incubated for 30 min. After four washes with PBS, the plasmin chromogenic substrate N-(p-Tosyl)-Gly-Pro-Lys 4-nitroanilide acetate salt (catalog no. T6140; Sigma) (500 μM) was added in a final volume of 50 μl and incubated for 30 min at 37°C. Absorbance at 406 nm was measured using a multiwell Fluostar Optima spectrophotometer. In order to estimate the amount of plasmin activity generated on the surface of the bacteria, the maximal plasmin activity (used as positive control) was measured in a final volume of 50 μl in a test tube as follows: 2 μg of Plg was incubated with 25 μl of 100 nM streptokinase for 30 min before addition of 25 μl of 1 mM plasmin substrate and subsequent incubation for 30 min at 37°C before absorbance was read.
In vitro ECM transmigration assay.
A solution of reconstituted basement membranes from mouse Engelbreth Holm Swarm sarcoma cells (catalog no. 326234; Matrigel BD Sciences) was diluted 1/10 in ice-cold Dulbecco's modified Eagle's medium (DMEM); 150 μl was layered on a 3-μm transparent membrane insert (catalog no. 662630; Greiner Bio One) and allowed to gel for 1 h at 37°C. FITC-labeled R6 or cbpE::cat mutant strains (1·108 CFU in 100 μl of DMEM) were incubated with or without 10 μg of Plg for 1 h at 37°C. After one wash, bacteria were resuspended in DMEM and added to the upper compartment of the transwell whereas the lower compartment contained 700 μl of DMEM supplemented with 10% bovine calf serum. Addition of Plg activator was not necessary, since Matrigel contains approximately 4.7 IU/mg of activators (32), leading to a final concentration of 0.47 IU/mg in our experiments. The culture chambers were incubated at 37°C with 5% CO2. FITC fluorescence present in the lower compartment was read at different time intervals using a Fluostar Optima fluorimeter.
Statistical analyses.
Error bars correspond to standard errors of the means. The statistical significance of the differences obtained in group comparisons was determined by a Student's t test. A P value < 0.01 was considered statistically significant.
RESULTS
CBPE binds to Plg.
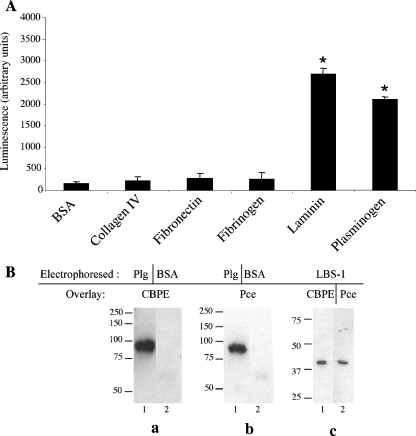
The interactions between CBPE and various mammalian proteins commonly recognized by pathogenic bacteria have been tested to investigate the role of this pneumococcal surface-exposed protein in the virulence process. The recombinant purified catalytic domain Pce was tested for interactions in a solid-phase binding assay with various ECM components and blood circulating factors (Fig. 1A). The results show that Pce binds to Lm and Plg whereas responses to collagen IV, fibronectin, and fibrinogen are not significantly different from those seen with BSA, the negative control (Fig. 1A).
FIG. 1.
Binding of soluble CBPE variants to immobilized Plg by solid-phase binding assay and ligand blot overlay assay. (A) His-tagged Pce (200 pmol) was incubated for 2 h on 1 μg each of proteins (collagen IV, fibronectin, fibrinogen, Lm, Plg, and BSA) coated on 96-well plate. Pce binding was quantified by chemiluminescence reading after extensive washes and incubation with horseradish peroxidase-conjugated anti-His antibody. This assay was performed twice in triplicate wells; data representing the results of a representative experiment are shown. The Lm and Plg values were significantly different from the BSA values as indicated (*, P < 0.01). (B) Plg (33 pmol), BSA (33 pmol), and LBS-1 (20 pmol) were subjected to electrophoresis on 10% or 12.5% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane. Panel a, lane 1, immobilized Plg; lane 2, immobilized BSA (1 μM CBPE overlay). Panel b, lane 1, immobilized Plg; lane 2, immobilized BSA (1 μM Pce overlay). Panel c (immobilized LBS-1), lane 1, 1 μM CBPE overlay; lane 2, 1 μM Pce overlay. The molecular mass values (in kilodaltons) are represented at the left of each Western blot.
Due to the important role played by the eukaryotic host's Plg system in bacterial pathogenicity (28), we have selected the CBPE-Plg interaction for a detailed analysis. Ligand blot overlay assays were used to confirm the interaction (Fig. 1B). Plg, which migrates on an SDS gel as a single polypeptide chain of 92 kDa, was blotted onto a nitrocellulose membrane which was incubated with purified recombinant proteins CBPE and Pce. Both proteins recognize immobilized Plg, while no signal was observed with BSA (67 kDa), a nonrelated protein (Fig. 1B, panels a and b). Using the same approach, we observed that the LBS-1 region, i.e., the three first kringle domains of Plg, was also recognized by both CBPE and Pce (Fig. 1B, panel c, lanes 1 and 2, respectively). These results indicate that CBPE and Plg interact through their Pce and LBS-1 domains, respectively.
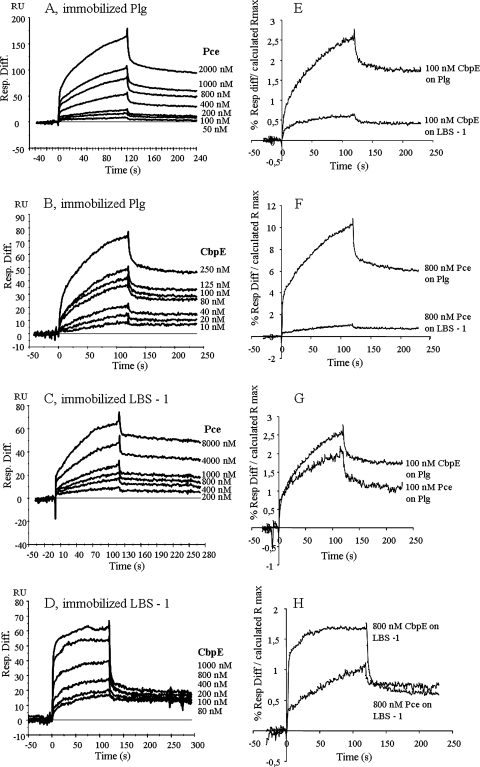
SPR experiments also confirmed the interactions between CBPE and Pce and between Plg and LBS-1 (Fig. 2). Binding of Pce and CBPE to Plg (Fig. 2A and B) and LBS-1 (Fig. 2C and D) was concentration dependent, and the sensorgrams are simple exponentials showing curvature in the association phase and signal decay in the dissociation phase. Since these data do not fit to the simple 1:1 interaction model proposed by the BIAevaluation software, calculation of kinetic parameters was not pursued (41). However, in order to evaluate the relative affinities of the interactions, we compared the individual binding profiles. The data were normalized, taking into account the differences of the molecular weights of the analytes and the different levels of ligand immobilization. Rmax was calculated by using the equation:
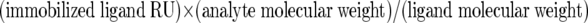 |
and the binding response was expressed as the percentage of the differential response/calculated Rmax (Fig. 2E, F, G, and H). The end point of the association phase was chosen to compare the binding responses. The binding of CBPE or Pce to Plg is 5 or 10 times higher than to LBS-1, respectively (Fig. 2E and F). When comparing the binding level of CBPE and Pce to Plg and to LBS-1, in both cases only a very small binding difference is observed (Fig. 2G and H). Taken together, the SPR experiments indicate that the LBS-1 region is recognized by both CBPE and Pce proteins and that the strongest interactions are established between CBPE and Plg through the Pce domain.
FIG. 2.
SPR measurements of CBPE/Pce interactions with Plg/LBS-1. Plg and LBS-1 were coated on a Biacore CM5 sensor chip; CBPE and Pce were the analytes (flow rate, 30 μl/min; injected volumes of 60 μl). Data represent overlays of sensorgrams resulting from the injection of different concentrations of recombinant CBPE and Pce over immobilized Plg and LBS-1 as follows: Pce (50 nM to 2 μM) over Plg (A); CBPE (10 nM to 250 nM) over Plg (B); Pce (200 nM to 8 μM) over LBS-1 (C); CBPE (80 nM to 1 μM) over LBS-1 (D). The blank run was subtracted from each sensorgram. (E, F, G, and H) SPR results showing binding of CBPE or Pce to immobilized Plg or LBS-1 were compared. Data are expressed as percentages of the Rmax calculated as follows: (differential response [Resp. Diff.]/calculated Rmax) × 100. The Rmax was calculated as follows: (immobilized ligand RU) × (analyte molecular weight)/(ligand molecular weight).
SPR experiments are usually designed to calculate kinetic parameters. The sensorgrams presented in this article fulfill biosensor criterion expectations (41). However, despite various trials to optimize experimental conditions and controls (i.e., surface binding capacity after regeneration procedure and determination of active concentrations), no mathematical data fit could be obtained with the 1:1 simple interaction model proposed by the BIAevaluation software. Even though satisfying statistics for Chi2 values were obtained with complex interaction data fit models, these were not employed in the absence of independent experimental evidence for a complex model of interaction.
Since ligand binding to the kringle domains of Plg usually involves lysine residues, this possibility was investigated in the CBPE system. We employed SPR to measure the binding of CBPE and Pce to immobilized Plg and LBS-1 in the presence of different concentrations of 6-AHA, a lysine analog (29) (Table 1). The RU responses reached at the end of the association phase were used to calculate the relative levels of inhibition. A 6-AHA dose-dependent inhibition of CBPE and Pce binding to Plg was observed, with the level of inhibition reaching 74% and 73%, respectively, when a 200:1 ratio of 6-AHA:protein was employed (Table 1). Higher inhibition values were measured when LBS-1 was challenged, since binding of Pce and CBPE was inhibited by up to 98% and 95%, respectively (Table 1). These observations suggest that the recognition between CBPE and Pce and between Plg and LBS-1 is mediated by lysine residues.
TABLE 1.
SPR inhibition experimentsa
| 6-AHA:protein molar ratio | Inhibition of Pce binding to immobilized Plg (%) | Inhibition of CBPE binding to immobilized Plg (%) | Inhibition of Pce binding to immobilized LBS-1 (%) | Inhibition of CBPE binding to immobilized LBS-1 (%) |
|---|---|---|---|---|
| 10:1 | 20/31 | 20/39 | 65/73 | 67/73 |
| 30:1 | 43/46 | 44/44 | 86/82 | 82/85 |
| 50:1 | 56/65 | 49/51 | 90/85 | 84/87 |
| 100:1 | 67/68 | 58/64 | 93/94 | 91/92 |
| 200:1 | 74/72 | 74/74 | 100/95 | 97/92 |
Pce (800 nM and 8 μM) and CBPE (250 nM and 2 μM) were injected with various concentrations of 6-AHA corresponding to molar inhibitor:protein ratios of 10:1, 30:1, 50:1, 100:1, and 200:1 over immobilized Plg or LBS-1. The results are expressed as percentages of inhibition of the binding measured in the absence of competing 6-AHA. The percentage values obtained from the results of two independent experiments are reported as paired data.
Identification of the Plg binding site harbored by the Pce domain.
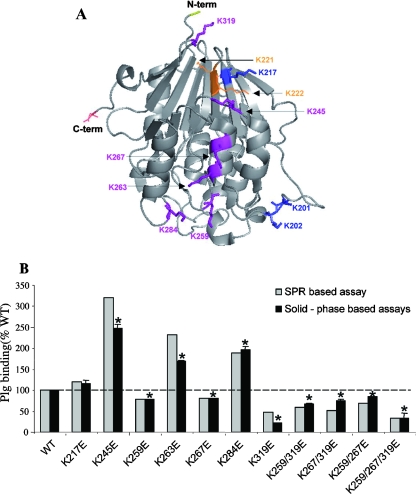
We then set out to identify with respect to Pce the lysine residues involved in binding to Plg/LBS-1 by mutating a series of lysine residues. Initial attempts to do alanine scanning resulted in mutated proteins that were insoluble; therefore, we decided to mutate selected lysine residues into glutamate residues. This replacement was selected on the basis of a procedure involving conserving of the lateral chain length while altering the side chain charge. Analysis of the three-dimensional structure of Pce (20) allowed us to exclude solvent-inaccessible lysine residues (i.e., lysines 134 and 250) as well as those whose accessibility is impaired by the vicinity of the choline-binding domain (lysines 84, 96, 156, 308, and 330). In addition, isolated exposed residues such as lysines 32, 102, 135, 238, and 296 were not considered due to the observation that the Plg binding sites are notably formed by clusters of lysine residues (43) (Fig. 3A).
FIG. 3.
Lysine residues of Pce domain involved in Plg binding. (A) Key lysine residues on the Pce domain involved in Plg binding are represented on a Pce structure. Lysine residues not analyzed due to protein insolubility are colored orange and shown as lines. Mutated lysines which did not influence the Plg binding are shown in blue and as sticks. Lysine residues defining the linear cluster of the Plg binding site are figured in magenta and shown as sticks (Protein Data Bank entry 1WRA). (B) Pce mutants binding to immobilized Plg was measured by SPR and solid-phase binding assays. Pce proteins (500 nM) were injected over immobilized Plg (flow rate, 30 μl/min; injected volumes of 60 μl) in the SRP experiments. Binding of wild-type (WT) Pce to Plg at the end of the association phase was defined as 100%. In the solid-phase binding assays, 200 pmol of His-tagged wild-type and mutant Pce was incubated for 2 h on Plg coated onto a 96-well plate. Binding of wild-type Pce to Plg was defined as 100%. The solid-phase binding assay was performed three times in triplicate wells. The standard error was derived from the results of three independent experiments. Plg binding values of Pce mutants were significantly different from that of wild-type Pce (100%) as indicated (*, P < 0.01). N-term, N terminus; C-term, C terminus.
Initially, single- and double-mutated proteins of the Lys 221-Lys 222 and Lys 201-Lys 202 pairs were constructed. Mutations at sites 221 and 222 yielded insoluble proteins, while those at sites 201 and 202 generated molecules whose properties of binding to Plg were unchanged in comparison with wild-type Pce properties (data not shown). Subsequently, lysine residues 217, 245, 259, 263, 267, 284, and 319, which are clustered and form a solvent-exposed linear patch (Fig. 3A), were mutated either alone or in combination.
All the recombinant mutant proteins were expressed in E. coli in soluble form and purified by nickel affinity chromatography. Thermal stability measurements and phosphorylcholine esterase activity tests were performed for all variants in order to assess proper folding and stability (Table 2). The fluorescence-based thermal stability assay distinguishes folded and unfolded proteins through recognition of exposed hydrophobic areas (17, 35). Consequently, the unfolding process is monitored as a function of temperature, with the Tm value being defined as the midpoint of the folded protein-unfolded protein transition. Small differences were observed for all Pce mutants compared to the wild-type protein, which undergoes a minor transition at 46°C and a major unfolding stage at 64°C (Table 2). The observed Tm values are still within the range of stability measured for protein of mesophyl organisms and are unlikely to affect substantially the behavior of the mutated proteins. We have also measured the kinetic parameters of all the Pce variants by use of the pNP-PC substrate (Table 2). Some significant differences were observed, likely originating from a charge effect resulting from interaction between the mutated residues (Lys to Glu) and the positively charged choline substrate (20). In conclusion, all individual lysine replacements for glutamate in Pce that were investigated maintained the overall protein structure.
TABLE 2.
Kinetic parameters and thermal stability of the Pce variant proteinsa
| Protein | Tm (°C) | Km (mM) | kcat (s−1) |
|---|---|---|---|
| Wild type | 46/64 | 0.631 | 5.11 |
| K217E | 44/64 | 0.608 | 6.38 |
| K245E | 44/67 | 0.106 | 1.28 |
| K259E | 44/65 | 0.795 | 6.38 |
| K263E | 42/66 | 0.470 | 6.38 |
| K267E | 43/65 | 0.608 | 6.38 |
| K284E | 44/63 | 0.278 | 1.91 |
| K319E | 43/64 | 0.743 | 5.11 |
| K259E/K319E | 43/62 | 0.222 | 3.83 |
| K267E/K319E | 40/64 | 0.743 | 6.38 |
| K259E/K267E | 38/62 | 0.8 | 11.49 |
| K259E/K267E/K319E | 46/64 | 0.608 | 8.94 |
Tm values were determined from a protein thermal stability assay using 3.5 μg of protein and 5 mM TCEP, with SYPRO orange as the environmentally reporter dye. PCho esterase activity measurements were performed using PBS (pH 7.4) at 37°C, and data were fitted using the Michaelis-Menten equation and the Lineweaver Burk plot from which Km and kcat values were determined.
The binding response of the Pce variants was tested with immobilized Plg by SPR and by solid-phase binding assays; the data were expressed as percentages of the wild-type Pce value, and the results show that these independent, distinct techniques gave similar responses (Fig. 3B). Single-point mutations of lysines 245, 263, and 284 gave rise to an increase in the level of Plg binding to 280% for Lys245Glu and 200% for Lys263Glu and Lys284Glu (average values as obtained using both techniques). These observations suggest that glutamate residues introduced at position 245, 263, or 284 of Pce stabilize its interaction with Plg (Fig. 3B). Three mutations induced a decrease in Plg binding: Lys259Glu and Lys267Glu Pce mutants bound Plg with 80% of the wild-type efficiency, while Lys319Glu displayed much larger reductions of 50% and 21% as detected by SPR and solid-phase binding assays, respectively. Double Pce mutants bound Plg at 70% compared with wild-type protein results, while the triple mutant Lys259Glu/Lys267Glu/Lys319Glu displayed 30% of Plg binding capability. These results support the idea of a role for lysine residues 259, 267, and 319 in the Plg recognition by Pce (Fig. 3A).
Native CBPE exposed at the surface of the pneumococcus is a Plg receptor.
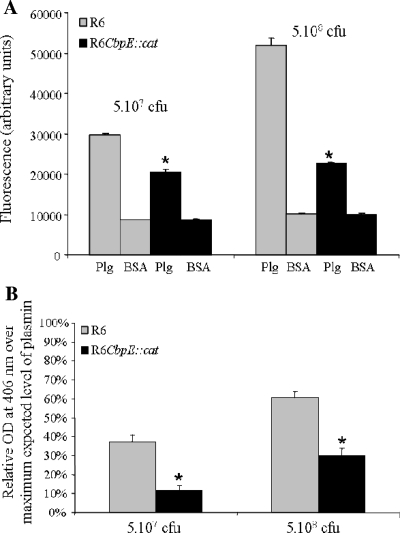
The specific binding of FITC-labeled strain R6 and the specific binding of cbpE-deleted pneumococcus to Plg were compared using a solid-phase binding assay. Increasing bacterial concentrations led to a dose-dependent binding to Plg for R6 but not when Plg was replaced by BSA as a negative control (Fig. 4A). Incubation with 5·108 CFU led to a level of mutant binding reduced by half compared to the wild-type binding (Fig. 4A). As expected, binding to Plg was not totally abolished in the cbpE mutant strain, as other pneumococcal surface proteins are known to interact with Plg (5).
FIG. 4.
Binding of Plg to pneumococcus and activation into plasmin. (A) FITC-labeled R6 wild-type cells (gray bars) and cbpE::cat mutant cells (black bars) (5·107 and 5·108 CFU) were incubated for 2 h at 37°C on 1 μg of Plg (black bars) or BSA (gray bars) coated onto 96-well plates. After five washes, the fluorescence of FITC was measured. All experimental point determinations were repeated eight times in each single assay, and four independent binding assays were performed. The standard error was derived from the results of the four independent experiments. The Plg binding values of the cbpE mutant strain were significantly reduced compared to the values of the parental strain R6 as indicated (*, P < 0.01). (B) R6 wild-type cells (gray bars) and cbpE mutant cells (black bars) (5·107 and 5·108 CFU) were deposited on 96-well plates and incubated with 2 μg of Plg for 2 h at 37°C. Streptokinase was added, and the proteolytic activity was measured at 406 nm after addition of the plasmin substrate. The positive control value obtained when 2 μg of Plg was activated into plasmin in a test tube by use of streptokinase in the absence of bacteria was defined as the 100% activity. The absorbance value of the negative control (20 μg of BSA instead of bacteria) was subtracted from the reading absorbance values obtained under the different tested conditions. The plasmin activity assay was performed three times, and each point determination was repeated eight times in each assay. The standard error was derived from the results of three independent experiments. The absorbance values reflecting plasmin activity on the cbpE mutant strain were significantly reduced compared to the values of the plasmin activity on the parental R6 strain as indicated (*, P < 0.01). OD, optical density.
Plg bound through CBPE at the pneumococcal surface can be activated into plasmin.
An assay dedicated to the measurement of plasmin activity was set up in order to check whether bacterium-associated Plg could be activated into plasmin. Different amounts of FITC-labeled R6 or cbpE mutant strains were coated on 96-well plates. Plg was added, and streptokinase was used to activate Plg into plasmin, whose enzymatic activity was measured using a synthetic substrate (Fig. 4B). The results indicate that Plg bound at the surface of the R6 parental strain can be activated into plasmin, reaching up to 60% of the maximum expected plasmin activity (Fig. 4B). When the cbpE mutant was assayed, the plasmin activity generated was reduced by at least 50% compared with the wild-type pneumococcal strain results (Fig. 4B). These experiments show that about half of the plasmin activity associated with the S. pneumoniae surface is mediated by the interaction of CBPE with Plg. In the course of these experiments, no conversion of Plg into plasmin was detected in the absence of streptokinase, indicating that S. pneumoniae does not express a functional endogenous Plg activator (data not shown).
Plg/plasmin bound to CBPE stimulates pneumococcus transmigration through the ECM.
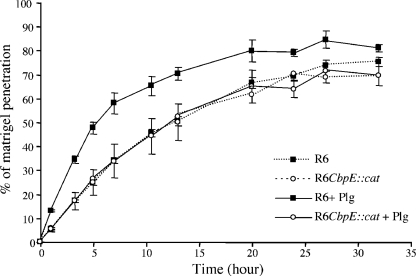
Degradation of ECM by the plasmin bound to the surface of pneumococcus was monitored using Matrigel, a commercially reconstituted basement membrane composed of Lm, collagen, heparan sulfate, and nidogen. Semipermeable transwell membranes were coated with Matrigel, and FITC-labeled pneumococcal cells were added in the upper compartment in the presence or absence of Plg. Activation of Plg in plasmin was unnecessary, since Matrigel harbors Plg activators (32). The migration kinetic of the bacteria across Matrigel was measured by monitoring, over time, bacterial fluorescence in the lower compartment. We verified in a control experiment that FITC is not released from the bacterial surface upon activation of Plg in plasmin. Therefore, the level of fluorescence in the lower compartment reflects the number of bacteria. The fraction of bacteria migrating across Matrigel was calculated as the ratio of the fluorescence level measured in the lower compartment to the level of the fluorescence present in the upper compartment at the beginning of the experiment (Fig. 5). In the presence of Plg, the wild-type R6 strain crossed the reconstituted basement membrane more rapidly than the cbpE mutant: 50% of wild-type bacteria reached the lower compartment in about 5 h compared to about 13 h for the mutant strain (Fig. 5). In addition, the rate of transmigration by the mutant strain bound to Plg was similar to that of the R6 strain in the absence of Plg (Fig. 5). These results demonstrate that plasmin linked to the pneumococcal surface stimulates migration across the reconstituted basement membrane, likely by cleaving ECM components.
FIG. 5.
Pneumococcus transmigration across Matrigel. FITC-labeled R6 and cbpE mutant strains (1·108 CFU in 100 μl of DMEM) were incubated with or without 10 μg Plg. After washes, bacteria were added to the upper well of the transwells whose membranes were coated with Matrigel. The culture chambers were incubated at 37°C with 5% CO2, and FITC fluorescence levels in the lower well were read at time intervals to monitor bacteria Matrigel penetration. The percentage of bacteria capable of degrading Matrigel was calculated as the ratio between the fluorescence measured in the lower compartment and total bacterial fluorescence. Filled symbols, wild-type R6 strain; empty symbols, cbpE-deleted mutant; solid lines, with Plg; dashed lines, without Plg. The assay was performed three times using triplicate wells.
DISCUSSION
S. pneumoniae is the main causative agent of respiratory infections leading yearly to the death of about 1.6 million people. Due to the significant levels of drug resistance developed by this human pathogen, the need of new antibiotics and/or vaccines is pressing. Surface-exposed proteins, which play multiple roles in the virulence process, are considered potential target candidates, and their therapeutical validation requires structural and functional characterization.
In this report, we show that CBPE is the first member of the choline-binding protein family characterized as a receptor for human Plg/plasmin. Interaction between the Pce domain of CBPE and Plg involves lysine residues, as shown in vitro using a lysine analogue molecule. Mutagenesis experiments support the idea of a role in the interaction with Plg for three neighboring lysines (319, 259, and 267) pointing in the same direction and aligned on the surface of Pce (20). The geometry of this lysine alignment appears to be significant, as lysines 201 and 202 and lysine 217, located away from and misaligned with respect to the linear cluster, respectively, do not contribute to the interaction. Surprisingly, we observed that the presence of glutamate in place of Lys245, Lys263, and Lys284 increased Plg recognition, suggesting that negative charges may be involved in the interaction, as described for the pneumococcal enolase. Indeed, mutations of the charged residues in the binding motif of the internal nonapeptide, alone or in combination with the lysine residues, abolished the enolase-based Plg binding property of the bacteria (8). Another example concerns the Plg-binding group A streptococcal M protein (PAM) Plg binding site: the three-dimensional structure of the complex formed with the recombinant kringle-2 domain of human Plg was solved previously (43). Structural study combined with a site-directed mutagenesis approach identified arginine, histidine, and glutamate, in addition to the expected lysine, as key residues mediating high-affinity binding of PAM to Plg (43, 48). Similar results were observed with the group A streptococcal M protein-related protein Prp, which binds Plg via arginine and histidine residues additionally to lysine residues (47).
The identification of the internal nonapeptide Plg binding site in the pneumococcal enolase was performed by use of spot-synthesized peptides, and the minimal Plg-binding motif, acting as a linear peptide, was determined previously (8). The crystal structure of pneumococcal enolase showed, a posteriori, that the C-terminal lysines are buried within the oligomer and are thus unlikely to be involved in Plg recognition whereas the internal nonapeptide is exposed at the surface and could be accessible to Plg (5, 8, 16). In this work, a different approach was used to identify residues involved in the Pce-Plg binding site as follows: (i) lysine residues were investigated, as the binding on LBS-1 was abrogated by the lysine analog 6-AHA; and (ii) analysis of the three-dimensional structure of Pce identified solvent-accessible lysine residues which, in addition, were aligned in a cluster (20, 43). This screening procedure identified some residues involved in the interaction of CBPE with Plg and, more importantly, mapped the Pce structural region whose charged residues, likely to participate in the interaction with Plg, need to be further investigated.
Two pneumococcal proteins, enolase and GAPDH, have been previously described as Plg receptors. The affinity values for enolase and GAPDH, as measured by SPR using the heterogenous ligand model, were KD1 (equilibrium dissociation constant 1) = 0.55 nM to KD2 = 86.2 nM and KD1 = 0.43 μM to KD2 = 0.16 nM, respectively, indicating a lower affinity of GAPDH for Plg (5-8).
Genetic deletions of enolase and GAPDH genes are not possible because of their essential roles in pneumococcus metabolism. Therefore, we took advantage of the dispensability of CBPE to analyze the relevance of Plg interaction with the surface of the bacteria. Indeed, Plg binds to the native surface-exposed CBPE in a manner suitable for subsequent activation into plasmin. In addition, we have observed that both Plg binding and plasmin activation were reduced by about half in the absence of the cbpE gene, confirming the effect of other surface proteins in these processes. Indeed, when the enolase Plg internal binding site was mutated in S. pneumoniae D39, Plg binding was reduced to about 50% compared to the results seen with the wild type (8). Because independent inactivation of Plg binding properties of enolase or CBPE led to a 50% reduction of Plg binding, it is likely that these two proteins are the dominant Plg receptors at the pneumococcus surface. However, in addition to GAPDH, enolase, and now CBPE, other pneumococcal proteins could be involved in Plg binding, as overlay experiments detected eight proteins (5). Plg recognition by S. pneumoniae most probably depends on several receptors whose expression may be regulated by factors such as tissue localization, infection stage, and host immune response.
CBPE harbors phosphorylcholine esterase activity which removes the phosphorylcholine residues from cell wall teichoic acid and lipoteichoic acid. Consequently, in the cbpE-deleted strain, a higher level of phosphorylcholine residues might increase the level of CBPs exposed on the surface of the bacteria. It is difficult to predict the consequences of these alterations of the phosphorylcholine and CBP molecular landscape at the pneumococcal surface for the capture of Plg and for the resulting interactions with the extracellular matrix and cell surfaces.
Pneumococcus transmigration across the basement membrane in an in vitro model was specifically stimulated when the bacterial surface was loaded with Plg/plasmin, and this effect required the cbpE gene. Transmigration likely results from hydrolysis of the Matrigel ECM components by plasmin. A similar observation has been reported by Bergmann et al. (7), by whom plasmin, associated to pneumococcus, was shown to degrade the ECM in an enolase-dependent manner. Our observation of a slower penetration through the ECM in the absence of CBPE-mediated surface-bound Plg/plasmin may suggest that endogenous pneumococcal surface or secreted proteases such as IgA, PrtA, the heat shock serine protease HtrA, and the putative zinc metalloproteinase ZmpB may participate cooperatively in the process.
Our results support the idea of a role for the CBPE-Plg/plasmin complex on the pneumococcus surface in the progression and the dissemination of the bacteria into the human host. The capture by the pneumococcus surface of Plg from the plasma and subsequent maturation in the active protease plasmin may facilitate the migration of bacteria through tissue barriers, such as the epithelium and endothelium layers, via degradation of the ECM. Furthermore, CBPE may interact with the ECM through Lm, as we have shown that it binds to this component of the ECM. This observation is reminiscent of the laminin-binding property of enterobacterial fimbriae, which also function as a Plg receptor (30).
The multiplicity of virulence factors associated with pneumococcus that interact with the Plg/plasmin system (enolase, GAPDH, and CBPE) necessitates a deeper understanding of the pneumococcus-Plg relationship. Future work will address the question of the selective advantage during host colonization and infection of the Plg/plasmin system recruitment and of the relevance of the various receptors for host tissue localization and invasion.
Acknowledgments
We thank Marjolaine Noirclerc-Savoye (Fondation Rhône-Alpes Futur) and Benoît Gallet (IBS/LIM) of the RoBioMol platform for their assistance with cloning procedures. Nicole Thielens (IBS/LEM), Sylvie Ricard-Blum (IBCP, Lyon, France), and Danièle Altschuh (ESBS, Strasbourg, France) are acknowledged for their support with SPR experiments. We thank Andrea Dessen for critical reading of the manuscript.
This work was funded in part by an ANR Jeunes Chercheurs 2005 grant (ANR-05-JCJC-0049-01) to A.M.D.G.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 10 December 2007.
REFERENCES
- 1.Andreasen, P. A., R. Egelund, and H. H. Petersen. 2000. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell. Mol. Life Sci. 5725-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28234-237. [DOI] [PubMed] [Google Scholar]
- 3.Beall, B. 2007. Vaccination with the pneumococcal 7-valent conjugate: a successful experiment but the species is adapting. Expert Rev. Vaccines 6297-300. [DOI] [PubMed] [Google Scholar]
- 4.Berge, A., and U. Sjobring. 1993. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J. Biol. Chem. 26825417-25424. [PubMed] [Google Scholar]
- 5.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 401273-1287. [DOI] [PubMed] [Google Scholar]
- 6.Bergmann, S., M. Rohde, and S. Hammerschmidt. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 722416-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmann, S., M. Rohde, K. T. Preissner, and S. Hammerschmidt. 2005. The nine residue plasminogen-binding motif of the pneumococcal enolase is the major cofactor of plasmin-mediated degradation of extracellular matrix, dissolution of fibrin and transmigration. Thromb. Haemost. 94304-311. [DOI] [PubMed] [Google Scholar]
- 8.Bergmann, S., D. Wild, O. Diekmann, R. Frank, D. Bracht, G. S. Chhatwal, and S. Hammerschmidt. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49411-423. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 10.Briles, D. E., R. C. Tart, E. Swiatlo, J. P. Dillard, P. Smith, K. A. Benton, B. A. Ralph, A. Brooks-Walter, M. J. Crain, S. K. Hollingshead, and L. S. McDaniel. 1998. Pneumococcal diversity: considerations for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA). Clin. Microbiol. Rev. 11645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, S. D., D. J. Farrell, and I. Morrissey. 2004. Prevalence and molecular analysis of macrolide and fluoroquinolone resistance among isolates of Streptococcus pneumoniae collected during the 2000-2001 PROTEKT US Study. J. Clin. Microbiol. 424980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choulier, L., N. Rauffer-Bruyere, M. Ben Khalifa, F. Martin, T. Vernet, and D. Altschuh. 1999. Kinetic analysis of the effect on Fab binding of identical substitutions in a peptide and its parent protein. Biochemistry 383530-3537. [DOI] [PubMed] [Google Scholar]
- 13.Coleman, J. L., and J. L. Benach. 1999. Use of the plasminogen activation system by microorganisms. J. Lab. Clin. Med. 134567-576. [DOI] [PubMed] [Google Scholar]
- 14.De Las Rivas, B., J. L. Garcia, R. Lopez, and P. Garcia. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-β-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 1844988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Guilmi, A. M., and A. Dessen. 2002. New approaches towards the identification of antibiotic and vaccine targets in Streptococcus pneumoniae. EMBO Rep. 3728-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehinger, S., W. D. Schubert, S. Bergmann, S. Hammerschmidt, and D. W. Heinz. 2004. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 343997-1005. [DOI] [PubMed] [Google Scholar]
- 17.Ericsson, U. B., B. M. Hallberg, G. T. Detitta, N. Dekker, and P. Nordlund. 2006. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357289-298. [DOI] [PubMed] [Google Scholar]
- 18.Fadda, D., C. Pischedda, F. Caldara, M. B. Whalen, D. Anderluzzi, E. Domenici, and O. Massidda. 2003. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J. Bacteriol. 1856209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernández-Tornero, C., R. Lopez, E. Garcia, G. Gimenez-Gallego, and A. Romero. 2001. A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol. 81020-1024. [DOI] [PubMed] [Google Scholar]
- 20.Garau, G., D. Lemaire, T. Vernet, O. Dideberg, and A. M. Di Guilmi. 2005. Crystal structure of phosphorylcholine esterase domain of the virulence factor choline-binding protein E from Streptococcus pneumoniae: new structural features among the metallo-beta-lactamase superfamily. J. Biol. Chem. 28028591-28600. [DOI] [PubMed] [Google Scholar]
- 21.Garcia, P., M. Paz Gonzalez, E. Garcia, J. L. Garcia, and R. Lopez. 1999. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 33128-138. [DOI] [PubMed] [Google Scholar]
- 22.Gosink, K. K., E. R. Mann, C. Guglielmo, E. I. Tuomanen, and H. R. Masure. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 685690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammerschmidt, S. 2006. Adherence molecules of pathogenic pneumococci. Curr. Opin. Microbiol. 912-20. [DOI] [PubMed] [Google Scholar]
- 24.Hanniffy, S. B., A. T. Carter, E. Hitchin, and J. M. Wells. 2007. Mucosal delivery of a pneumococcal vaccine using Lactococcus lactis affords protection against respiratory infection. J. Infect. Dis. 195185-193. [DOI] [PubMed] [Google Scholar]
- 25.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu, K. K., and S. I. Pelton. 2003. Heptavalent pneumococcal conjugate vaccine: current and future impact. Exp. Rev. Vaccines 2619-631. [DOI] [PubMed] [Google Scholar]
- 27.Jedrzejas, M. J. 2001. Pneumococcal virulence factors: structure and function. Microbiol. Mol. Biol. Rev. 65187-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lähteenmäki, K., S. Edelman, and T. K. Korhonen. 2005. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 1379-85. [DOI] [PubMed] [Google Scholar]
- 29.Lahteenmaki, K., P. Kuusela, and T. K. Korhonen. 2001. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25531-552. [DOI] [PubMed] [Google Scholar]
- 30.Lähteenmäki, K., P. Kuusela, and T. K. Korhonen. 2000. Plasminogen activation in degradation and penetration of extracellular matrices and basement membranes by invasive bacteria. Methods 21125-132. [DOI] [PubMed] [Google Scholar]
- 31.Luo, R., B. Mann, W. S. Lewis, A. Rowe, R. Heath, M. L. Stewart, A. E. Hamburger, S. Sivakolundu, E. R. Lacy, P. J. Bjorkman, E. Tuomanen, and R. W. Kriwacki. 2005. Solution structure of choline binding protein A, the major adhesin of Streptococcus pneumoniae. EMBO J. 2434-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuire, P. G., and N. W. Seeds. 1989. The interaction of plasminogen activator with a reconstituted basement membrane matrix and extracellular macromolecules produced by cultured epithelial cells. J. Cell. Biochem. 40215-227. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, T. J. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. 1219-230. [DOI] [PubMed] [Google Scholar]
- 34.Pancholi, V., and V. A. Fischetti. 1998. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 27314503-14515. [DOI] [PubMed] [Google Scholar]
- 35.Pantoliano, M. W., E. C. Petrella, J. D. Kwasnoski, V. S. Lobanov, J. Myslik, E. Graf, T. Carver, E. Asel, B. A. Springer, P. Lane, and F. R. Salemme. 2001. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 6429-440. [DOI] [PubMed] [Google Scholar]
- 36.Paton, J. C., P. W. Andrew, G. J. Boulnois, and T. J. Mitchell. 1993. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins. Annu. Rev. Microbiol. 4789-115. [DOI] [PubMed] [Google Scholar]
- 37.Pichichero, M. E., M. A. Shelly, and J. J. Treanor. 1997. Evaluation of a pentavalent conjugated pneumococcal vaccine in toddlers. Pediatr. Infect. Dis. J. 1672-74. [DOI] [PubMed] [Google Scholar]
- 38.Plow, E. F., T. Herren, A. Redlitz, L. A. Miles, and J. L. Hoover-Plow. 1995. The cell biology of the plasminogen system. FASEB J. 9939-945. [DOI] [PubMed] [Google Scholar]
- 39.Reinert, R. R., S. Reinert, M. van der Linden, M. Y. Cil, A. Al-Lahham, and P. Appelbaum. 2005. Antimicrobial susceptibility of Streptococcus pneumoniae in eight European countries from 2001 to 2003. J. Antimicrob. Chemother. 492903-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren, B., M. A. McCrory, C. Pass, D. C. Bullard, C. M. Ballantyne, Y. Xu, D. E. Briles, and A. J. Szalai. 2004. The virulence function of Streptococcus pneumoniae surface protein A involves inhibition of complement activation and impairment of complement receptor-mediated protection. J. Immunol. 1737506-7512. [DOI] [PubMed] [Google Scholar]
- 41.Rich, R. L., and D. G. Myszka. 2006. Survey of the year 2005 commercial optical biosensor literature. J. Mol. Recognit. 19478-534. [DOI] [PubMed] [Google Scholar]
- 42.Rigden, D. J., M. J. Jedrzejas, and M. Y. Galperin. 2003. Amidase domains from bacterial and phage autolysins define a family of γ-d,l-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28230-234. [DOI] [PubMed] [Google Scholar]
- 43.Rios-Steiner, J. L., M. Schenone, I. Mochalkin, A. Tulinsky, and F. J. Castellino. 2001. Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A streptococcal surface protein. J. Mol. Biol. 308705-719. [DOI] [PubMed] [Google Scholar]
- 44.Ronda, C., J. L. Garcia, E. Garcia, J. M. Sanchez-Puelles, and R. Lopez. 1987. Biological role of the pneumococcal amidase. Cloning of the lytA gene in Streptococcus pneumoniae. Eur. J. Biochem. 164621-624. [DOI] [PubMed] [Google Scholar]
- 45.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25819-829. [DOI] [PubMed] [Google Scholar]
- 46.Sánchez-Beato, A. R., R. Lopez, and J. L. Garcia. 1998. Molecular characterization of PcpA: a novel choline-binding protein of Streptococcus pneumoniae. FEMS Microbiol. Lett. 164207-214. [DOI] [PubMed] [Google Scholar]
- 47.Sanderson-Smith, M. L., M. Dowton, M. Ranson, and M. J. Walker. 2007. The plasminogen-binding group A streptococcal M protein-related protein Prp binds plasminogen via arginine and histidine residues. J. Bacteriol. 1891435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanderson-Smith, M. L., M. J. Walker, and M. Ranson. 2006. The maintenance of high affinity plasminogen binding by group A streptococcal plasminogen-binding M-like protein is mediated by arginine and histidine residues within the a1 and a2 repeat domains. J. Biol. Chem. 28125965-25971. [DOI] [PubMed] [Google Scholar]
- 49.Sun, H. 2006. The interaction between pathogens and the host coagulation system. Physiology 21281-288. [DOI] [PubMed] [Google Scholar]
- 50.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293498-506. [DOI] [PubMed] [Google Scholar]
- 51.van den Ent, F., and J. Lowe. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 6767-74. [DOI] [PubMed] [Google Scholar]
- 52.Vollmer, W., and A. Tomasz. 2001. Identification of the teichoic acid phosphorylcholine esterase in Streptococcus pneumoniae. Mol. Microbiol. 391610-1622. [DOI] [PubMed] [Google Scholar]
- 53.Winram, S. B., and R. Lottenberg. 1996. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology 1422311-2320. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, J. R., K. E. Mostov, M. E. Lamm, M. Nanno, S. Shimida, M. Ohwaki, and E. Tuomanen. 2000. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell 102827-837. [DOI] [PubMed] [Google Scholar]