Abstract
An Escherichia coli oligonucleotide microarray based on three sequenced genomes was validated for comparative genomic microarray hybridization and used to study the diversity of E. coli O157 isolates from human infections and food and animal sources. Among 26 test strains, 24 (including both Shiga toxin [Stx]-positive and -negative strains) were found to be related to the two sequenced E. coli O157:H7 strains, EDL933 and Sakai. However, these strains showed much greater genetic diversity than those reported previously, and most of them could not be categorized as either lineage I or II. Some genes were found more often in isolates from human than from nonhuman sources; e.g., ECs1202 and ECs2976, associated with stx2AB and stx1AB, were in all isolates from human sources but in only 40% of those from nonhuman sources. Some (but not all) lineage I-specific or -dominant genes were also more frequently associated with isolates from human. The results suggested that it might be more effective to concentrate our efforts on finding markers that are directly related to infection rather than those specific to certain lineages. In addition, two Stx-negative O157 cattle isolates (one confirmed to be H7) were significantly different from other Stx-positive and -negative E. coli O157:H7 strains and were more similar to MG1655 in their gene content. This work demonstrates that not all E. coli O157:H7 strains belong to the same clonal group, and those that were similar to E. coli K-12 might be less virulent.
Escherichia coli O157:H7 is an important food-borne pathogen and has been a significant public health concern since it was identified in 1983 (30). It causes diarrhea and bloody diarrhea in humans, which can sometimes lead to hemolytic uremic syndrome and even death. Although many other serotypes of E. coli are capable of producing Shiga toxin, E. coli O157:H7 is the most frequently isolated serotype during outbreak situations. Human infections with E. coli O157:H7 have been associated with a variety of contaminated food items and water, as well as person-to-person transmission. However, ruminants and especially cattle are considered the primary source of this organism (12, 18, 45).
A number of virulence factors have been identified for this bacterium, of which the Shiga toxins, Stx1 and in particular Stx2 and its variants, are responsible for bloody diarrhea and hemolytic uremic syndrome. Other virulence determinants include the locus of enterocyte effacement (LEE), which encodes a type III secretion system, EspA filaments, intimin and its receptor, Tir, and other factors essential for adherence to epithelial cells (22). In addition, E. coli O157:H7 carries other potential virulence factors, such as a second type III secretion system named ETT2 (E. coli type III secretion 2) (25), and an extensive repertoire of type III secretion effectors (34). Most strains also carry a virulence-associated plasmid, pO157, that encodes a number of potential virulence determinants, such as a secreted serine protease (EspP) (7), StcE (9), ToxB (33), an enterohemolysin, and catalase peroxidase (22). However, sorbitol-fermenting O157:H− strains lack some of these plasmid-borne genes (6). Current knowledge of E. coli O157 virulence factors still cannot fully explain the high level of virulence associated with this organism.
The published genomic sequences of two E. coli O157:H7 strains have provided clues to many other potential virulence determinants (13, 28). These two sequences are highly similar to each other. Both strains have the same fundamental backbone of E. coli K-12 but with additional sequences inserted at various parts of the backbone, often accompanied by small deletions on the backbone. These insertion sequences, termed O islands (28) or S loops (13), often originate from bacteriophages. Based on the sequence data, comparative genomic hybridization (CGH) microarray analysis of E. coli O157:H7 isolates has been carried out with a small number of strains from human infections (27, 41) and recently with isolates from both human and cattle (44). At the Veterinary Laboratories Agency (VLA), we have an extensive collection of E. coli O157 isolates from farms, abattoirs, food sources, and human infections. In order to understand the genetic diversity of these isolates and identify any genetic elements that may be associated with human infections, we set up and validated a three-genome panarray platform and used it to study a selection of those E. coli O157 isolates with a view to increasing our understanding of the basis of the pathogenicity and identifying potential markers of virulence.
MATERIALS AND METHODS
Composition of the oligonucleotide microarray.
The array-ready oligonucleotide set for the E. coli genome, version 1.0, was purchased from Operon Biotechnologies GmbH (Cologne, Germany). It contains 5,978 probes (70-mer) representing open reading frames of three E. coli genomes comprising strain K-12 (MG1655) and O157:H7 (EDL933 and RIMD 0509952 Sakai). In addition, 110 oligonucleotides representing genes from two pO157 plasmids (from EDL933 [AF074613] and Sakai [NC_002128]) and pOSAK1 (from Sakai [NC_002127]) were also included on the array.
Array printing.
Oligonucleotide probes were dissolved in Pronto! universal spotting solution (Corning Life Sciences, Koolhovenlaan, The Netherlands) at a concentration of 40 μM. The oligonucleotides were spotted on Corning UltraGAPS slides with a BioRobotics MicroGrid II arrayer (Genomic Solutions). TAS application software was used to design the printing pattern and to control the printing process. Each subgrid was 21 by 21 (columns by rows) with 0.21-mm spacing. Each oligonucleotide was printed at least in duplicate on a glass slide, with the average diameter of spots being 120 μm. The printed slides were cross-linked with 600 mJ of UV energy and stored in a desiccator at room temperature until use.
Isolates used in the study. The E. coli O157 test strains used in this work were from the VLA collection and were used in a previous study (3). All strains were confirmed as E. coli with an API 20E kit (bioMerieux) and as serotype O157 with a latex agglutination kit (Oxoid) The H7 type was established by PCR following the methods of Wang et al. (37). Two O157 strains (1583/00 and 1176/00) fermented sorbitol within 24 h, whereas all other O157 strains did not ferment sorbitol. As the purpose of this work was to detect strain diversity and to identify genetic markers for virulence, we selected a panel of strains that were not known to be linked epidemiologically or geographically, possessed different pulse-field gel electrophoresis macrorestriction profiles (2), and contained similar numbers of isolates from humans, animals, and food. The sources of the isolates are shown in Fig. 2. Strains were grown routinely on 5% sheep blood agar.
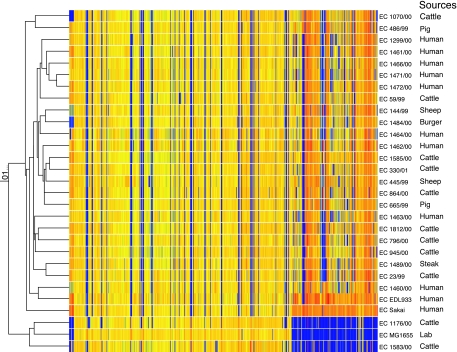
FIG. 2.
Pearson correlation coefficient comparison of the CGH microarray results. The strain number and source of each strain are also given. Orange or yellow indicates that a gene is present. The orange and yellow color difference reflects the different amount of control DNA in each spot, as control DNA was the mixture of DNAs from three control strains, where some genes are common to all three strains and others are present in only one or two strains. Blue indicates that a gene is absent. Gray and different shades of green indicate divergent or partial identity, as many phage-related genes fall into this category. Red may represent genes in multiple copies, which needs further confirmation. Genes are arranged from left to right in the order of genes from plasmids, MG1655 and O islands and S loops. Two major groups of O157 isolates were identified during this study.
Genomic DNA preparation, labeling, and hybridization. Genomic DNA was isolated from overnight cultures of bacteria grown aerobically in LB broth (31). DNA was extracted using a DNeasy tissue kit (no. 69504; Qiagen). DNA samples were kept at −20°C until used. For the control sample, the DNA from three sequenced strains (MG1655, EDL933, and Sakai) was mixed, and each strain contributed to a third of the total DNA (i.e., 0.66 μg each). Two micrograms of the control or test DNA (from a single strain) was used for labeling. The DNA was labeled using the protocol of the Institute of Food Research (http://www.ifr.bbsrc.ac.uk/Safety/Microarrays/protocols.html), with some minor adjustments. Briefly, 2 μg of genomic DNA in 23.5 μl of H2O was mixed with 20 μl of 2.5× random primer-reaction buffer mix from the BioPrime DNA labeling system (18094-011; Invitrogen). The mixture was boiled for 5 min and cooled on ice for 5 min. Then, 5 μl of 10× deoxynucleoside triphosphates (1.2 mM each dATP, dGTP, and dTTP and 1.1 mM dCTP; Amersham Biosciences) and 0.5 μl of Cy5 or Cy3 dCTP (1 mM stock; Amersham Biosciences) were added. Finally, 1 μl of Klenow enzyme from the kit was added, and the reaction was carried out at 37°C for 1.5 h. Excess Cy3 and Cy5 dCTP were removed from the labeled DNA with a MinElute PCR purification kit (28006; Qiagen). Hybridization was performed with a protocol similar to the one developed by the BμG@S group (http://bugs.sgul.ac.uk/bugsbase/index.php). The slide was submerged in prehybridization solution (3.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS], and 10 mg/ml bovine serum albumin) at 65°C for 20 min. Then, it was rinsed in 400 ml of water for 1 min and followed by 400 ml of propan-2-ol for 1 min before being dried by centrifugation at 1,500 rpm for 5 min. The labeled DNA was prepared in hybridization solution (4× SSC, 3% SDS), heated at 95°C for 2 min, and allowed to cool at room temperature before being added to a slide covered with a LifterSlip. Hybridization was carried out in a Genetix hybridization chamber and incubated at 65°C in a hybridization oven for 16 to 20 h. After the hybridization, the slide was washed twice gently in wash buffer A (1× SSC, 0.05% SDS) at 65°C for at least 5 min each time. The slide was then washed in 400 ml wash buffer B (0.06× SSC) for 2 min at room temperature.
Data acquisition and analysis.
Processed slides were scanned using a Genetix 4000B scanner (Axon Instruments, Inc.) with GenePix Pro 4.1 software. A good spot was defined as spot with a minimum of 65% of its pixels having intensities that were greater than the background plus 2 standard deviations.
After the raw data were cleaned, the data were normalized using a print-tip control gene median correction. Control genes were defined as those known to be present in all three sequenced strains in a single copy. During normalization, ratio intensity diagnostic plots were performed to ensure that the effect of normalization was consistent between hybridization experiments. After normalization, each gene was associated with a latent variable of presence or divergence/absence. The latent variable was a summary of each of the replicates for each gene. Finally, the data were validated by comparing the BLASTN information with the latent variable associated with presence or divergence/absence (B. Cater, G. Wu, M. Woodward, and M. F. Anjum, submitted for publication) and with Genespring (version 7.1; Agilent Technologies).
Plasmid profiling and PCRs.
Plasmid profiling was carried out according to Kado and Liu (15). PCR primers for espP (5′ TTTGCGAAAAATGGCGGAACTC 3′ and 5′ GCTGACGGGGCATTGACTG 3′), etpD (5′ CGACTGCACCTGTTCCTGATT3′ and 5′ CGTCAGGAGGATGTTCAG3′), hlyAC (5′ TGTCTTGCGTCATATCCATTCTCA 3′ and 5′ CGCTATGGGCCTGTTCTCCTCTG3′), and katP (5′ CTTCCTGTTCTGATTCTTCTGG 3′ and 5′ AACTTA TTTCTCGCATCATCC 3′) were used. PCRs were carried out with Taq DNA polymerase from Promega according to the manufacturer's instructions. Annealing temperatures were 52°C for etp and katP and 58°C for espP and hlyAC. Supernatants (2 μl) from the boiled overnight cultures were used as templates. The reaction mixtures were heated to 94°C for 5 min, and then 25 cycles of the following reactions were carried out: 94°C for 30 s, 52 or 58°C for 30 s, and 72°C for 2 min. Finally, a further extension was carried out at 72°C for 7 min.
Microarray data accession numbers.
All data were deposited in ArrayExpress and were assigned accession numbers E-MEXP-622 and E-MEXP-1059.
RESULTS AND DISCUSSION
H7 typing.
A PCR method based on the sequence conservation among H7 antigens (29, 37) was used. PCR fragments generated with the H7-specific primers (p1806-p1809) that are located within the fliC gene of H7 were at the expected size with genomic DNA from all E. coli O157 strains used in this work except for 1583/00 (results not shown). For further confirmation, PCRs were carried out with DNAs from pure colonies of Sakai, 1176/00, and 1583/00 as templates. No PCR product was generated with DNA from 1583/00. PCR products of the expected sizes were obtained with E. coli O157- and O55-specific H7 primer pairs (p1696-p1809 and p1697-p1806, all located within fliC) and with DNA from 1176/00 and Sakai. PCR products of expected sizes were also obtained with the primer pair p2648 (within the fliA gene next to fliC of H7)-p1806 (within fliC) for both Sakai and 1176/00. The primer pair p2650 (within the fliD gene)-p1809 (within fliC) produced a PCR product for Sakai but failed to produce a product for 1176/00, possibly due to a single nucleotide mutation within fliD (37). Importantly, both ends of the PCR products generated with H7-specific primers (p1806-p1809) from four strains (Sakai, NCTC12900, 1585/00, and 1176/00) were sequenced and found to be identical to the H7 sequence of EDL933 (results not shown). These results demonstrated that all E. coli O157 strains used in this work have the H7 type of fliC, apart from 1583/00.
Validation of the genomic microarray.
The microarray method was validated against three sequenced E. coli strains: a K-12 strain (MG1655) and two O157 strains (EDL933 and Sakai). All probe sequences (i.e., oligonucleotides printed on the slide) were compared against the complete nucleotide sequence of the above three strains by BLASTN to predict for cross-hybridization and determine probe specificity (data not shown). Probes showing >80% identity with any region of the three genomes were counted as theoretically present (data not shown.), as this cutoff value matched well with the experimental results (see below).
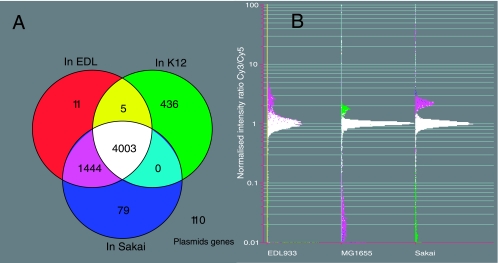
Using this criterion, 4,003 probe sequences were found in all three strains. Five probe sequences were found in EDL933 and MG1655 but not in Sakai, while the sequences of 1,444 probes were found only in EDL933 and Sakai (Fig. 1A). One hundred ten probes were derived from pOSAK1- and pO157-encoded sequences (Fig. 1A; see Table S1 in the supplemental material).
FIG. 1.
(A) Venn diagram to show the distribution of oligonucleotide probes among the three sequenced E. coli strains according to BLASTN results. (B) CGH microarray results, expressed as the normalized log signal intensity ratios between the test (Cy3) and control (Cy5) channels. The data were normalized using positive control genes present in single copy and shared by all three strains.
Due to the presence of multiple copies of certain genes or sequence similarities between some regions of the chromosome, 872 probes had more than 80% identity with more than one region on the chromosome (see Table S2 in the supplemental material). A large number of these regions were phage genes or IS element-related or intergenic regions of unknown functions. The majority of probes with multiple hits (586 probes) were found in EDL933 and Sakai, and less than one-fourth (240 probes) were found in all three strains. The BLASTN results of the coding regions represented by these probes are shown in Table S3 in the supplemental material, where the matches to the noncoding regions are ignored.
These theoretical calculations were compared with the experimental results from the CGH microarray data using the three control strains. The distributions of experimental hybridization signal intensities are shown in Fig. 1B, in which the hybridization signal intensities produced by probes common to all three strains are around 1, after normalization with control genes that are single-copy genes present in all three strains. For those probes that are common to EDL933 and Sakai, hybridizations with the genomic DNA from either EDL933 or Sakai produced signal intensities greater than 1, while the same probe, when hybridized with the genomic DNA from MG1655, generated signal intensities that were less than 0.1. Similarly, when probes that were unique to MG1655 were hybridized with MG1655 genomic DNA, the hybridization signals were larger than 1 (3/1 in theory), while the same probes, when hybridized with the genomic DNA from either EDL 933 or Sakai, generated hybridization signal intensities that were less than 0.1. Relatively few hybridization signals were between 0.1 and 0.6. A specificity of 94.18 to 99.18 and sensitivity of 98.93 to 99.45 were achieved. A detailed description of the mathematical process established in this study to analyze CGH microarray data which also determined the cutoff point for the presence/conserved and absence/divergent of genes has been submitted elsewhere (B. Carter et al., submitted for publication).
A proper validation is vital for the correct interpretation of the microarray results and interpretation of their biological significance. The information provided in Tables S1 to S3 in the supplemental material will be invaluable for both CGH and gene expression microarray result analysis using O157 genomes in the future.
It should be mentioned that the limitation of using a single oligonucleotide probe for each gene is that we cannot be certain whether we are identifying a gene fragment or a complete coding region within the test genome. Nevertheless, it gives a good idea of genes likely to be present.
Comparative genomic microarray results.
The validated CGH microarray process was thus used to analyze the genomic composition of field and clinical isolates of human and nonhuman origin and generated normalized log2 (Cy3/Cy5) intensities for each slide (Table S4 in the supplemental material). From the hybridization data, each probe sequence was computed as either present or absent/divergent in a given strain. These were translated into binary presence and absence/divergence (data not shown).
Clustering analysis of E. coli O157 isolates.
The hybridization results were summarized as condition trees using the clustering programs in Genespring (version 7.1). We noted that irrespective of the clustering programs used, two major clusters were always identified. The majority of E. coli O157:H7 (including both Stx-positive and Stx-negative) strains clustered with EDL933 and Sakai in one cluster, whereas two Stx-negative E. coli O157 strains (1583/00 and 1176/00) clustered with MG1655 in another cluster (Fig. 2). Both the parametric Pearson's correlation coefficient and the nonparametric Spearman's rank correlation coefficient were applied; however, the subclusters could not be reported with a degree of confidence.
This work has uncovered greater diversity among E. coli O157:H7 isolates than previously shown (27, 44). Among the 26 E. coli O157 test strains, 24 were similar to the two sequenced E. coli O157:H7 strains; two were similar to MG1655, with no known virulence factors detected in them (see below). It was surprising to find that an MG1655-like strain, 1176/00, carried both O157 and H7 antigens, since it is generally thought that E. coli O157:H7 consists of a group of closely related bacteria (38-40).
Identification of variable genes among the EDL933 and Sakai-like E. coli O157 isolates and genes associated with human infections.
Among strains studied here, 562 genes on the O islands and S loops were variably absent or present (VAP) in EDL933 and Sakai-like strains (see Table S4 in the supplemental material). This number of VAP genes was much larger than numbers found in other studies (27, 44). To make the results comparable, we recalculated the number of VAP genes reported by Zhang et al. (44) to be 407, based on the data in additional file 1 accompanying that publication. In this calculation, genes found in MG1655 were excluded from the VAP genes. This represents variable genes from O islands and S loops. This comparison showed that strains used in this study were more diverse than those reported so far.
However, we do not know whether isolates that appear to be distantly related to EDL933 and Sakai, as determined by the presence of fewer VAP genes, have smaller genomes or have other sets of VAP genes about which we know nothing. The data generated by this analysis may give clues as to which isolates (i.e., distant as determined by VAP relatedness) need to be sequenced or have subtractive hybridizations performed to assess the presence of unknown variable regions.
The majority of VAP genes (∼80%) were functionally unknown and phage or insertion element related. However, some of the most important virulence genes were among VAP genes, e.g., Shiga-toxin genes (stx1 and stx2) and genes for a putative cytotoxin (Z4332, Z4333, ECs3860, and ECs3861). There were also a number of genes for transcriptional regulators, e.g., a BfpM-like protein (ECs1391) and C4-type zinc finger proteins (TraR family, ECs1250), present in this set. The regions encoding the Shiga toxin genes appeared to be particularly dynamic, as some strains carried only stx1, some carried only stx2, and some carried both, while others carried none. Strains 1070/00 (cattle isolate), 1484/00 (meat isolate), 665/99 (pig isolate), 486/99 (pig isolate), 864/00 (cattle isolate), 144/99 (sheep isolate), 1464/00 (human), and 445/99 (sheep isolate) may carry a variant of stx2, as we detected only weak hybridization signals with the stx2B probe when the genomic DNA from these strains was used for hybridization. It was interesting that only one human isolate belonged to this group. All isolates from human infections that harbored stx1AB also contained 10 additional genes associated with prophage CP933V, which included genes for a putative tail fiber protein (Z3311), a putative transcription antitermination protein N (Z3361), a putative superinfection exclusion protein B (Z3362 and ECs2995), a putative repressor protein C1 (Z3358 and ECs2990) and some unknown proteins (see below for ECs2976). However, these 10 genes were missing from a cattle isolate (23/99) and a steak isolate (1489/00) that also harbored stx1AB.
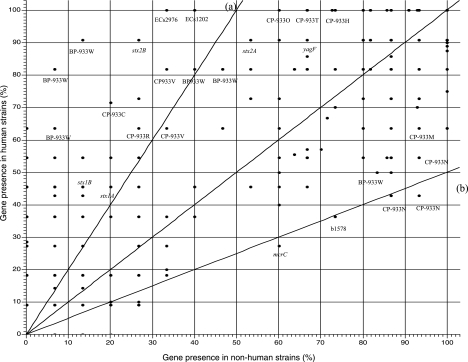
The distribution of genes in strains from human sources versus those from nonhuman sources was plotted. The results (Fig. 3) showed that some genes were found more frequently in isolates from human sources than in those from nonhuman sources. Notably, the presence of stx2B was highly correlated with human infection. Genes on bacteriophage BP-933W appeared to be frequently associated with human infections, whereas genes associated with prophage CP-933N were less frequently associated with human infections. We were unable to identify genes associated with isolates from nonhuman sources due to the limitation of the array, as it contained only genes from EDL933, Sakai, and MG1655, which are all from human sources.
FIG. 3.
Association of genes with strains from human and nonhuman sources. Genes close to the 45-degree line were distributed equally among isolates from human and nonhuman sources. Lines a and b are twofold lines drawn to help the visualization. Genes at the left side of line a were found more than twice as often in isolates from human as in those from nonhuman sources. Genes below line b are found more than twice as often in isolates from nonhuman sources as in those from human. Many dots represent more than one gene; e.g., BP933W indicates the location of a few genes related to BP933W.
Two genes, ECs2976 and ECs1202, were found in 100% of human isolates and in less than 40% of those from nonhuman sources (Fig. 3). These two genes were also found in EDL933, and they were 100% identical to those in Sakai, although they were not annotated as open reading frames. Both genes were also present in isolates from humans in an earlier study (27). Further study revealed that these two genes have 97% identities with each other within the 195-nucleotide sequence of the whole gene. The translated 65-amino-acid sequences of the two genes were identical. Therefore, they would cross-hybridize with each other, and we would be unable to distinguish ECs2976 and ECs1202 with the array method.
The locations of these two genes on the chromosomes are particularly interesting: ECs2976 is found upstream of the antiterminator Q gene (ECs2975, Z3345) of prophage CP 933V, which carries stx1AB (see Fig. S1A in the supplemental material), and ECs1202 is located upstream of the antiterminator Q gene of bacteriophage BP933W (ECs1203, Z1459), which carries stx2AB (see Fig. S1B in the supplemental material). The proteins encoded by these two genes have 62% identities with S0725 of Shigella flexneri 2a 2457T, whose gene is also located upstream of a putative Q gene. What is intriguing is that these two genes were missing from some Stx-positive isolates from nonhuman sources.
The importance of the antiterminator Q gene in Shiga toxin expression and human infection has been reported (23). Unfortunately, we did not manage to identify the association of the antiterminator Q gene with human infection in this work; this may be due to the probes cross-hybridizing with more than one region of the chromosome (see Table S2 in the supplemental material). PCR screenings are being carried out in order to study the distribution of ECs1202 and ECS2976 in a large number of human and nonhuman isolates, and the role(s) of these two genes in stx gene expression is under investigation in our laboratory.
Distribution of the non-LEE-encoded type III effectors.
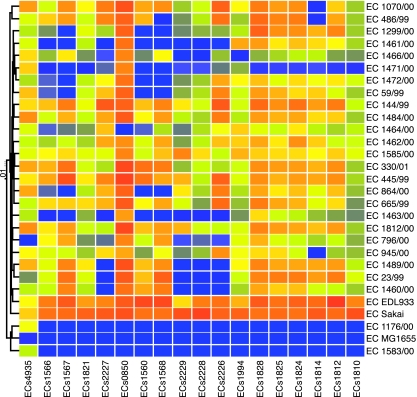
Although the majority of genes that encode the type III secretion effectors (34) are present in all EDL933- and Sakai-like E. coli O157:H7 strains, some were variably present in these strains (Fig. 4). ECs1560 (EspX7), ECs1567 (EspO1-1), and ECs1568 (EspK) are found in >85% of isolates from nonhuman sources and about 50% of isolates from human sources. The significance of this distribution is unknown.
FIG. 4.
Conservation of non-LEE-encoded type III secretion effectors. Only genes showing variable presence in EDL933 and Sakai-like strains are shown. Color coding is the same as in Fig. 2.
E. coli O157 lineage analysis.
Using octamer-based genome scanning, Kim et al. found that Stx-producing, β-glucuronidase- and sorbitol-negative E. coli O157 strains have diverged into two distinct lineages, lineages I and II (17). Lineage I was thought to be more commonly associated with human disease than lineage II (42). Recently, Zhang et al. (44) performed CGH analysis of two lineages of E. coli O157 and identified genes that they considered specific or dominant in each lineage. Their results are compared with those of our study (Fig. 5). Genes that are identified as specific or dominant in lineage I indeed dominated in some human isolates, i.e., EDL933, Sakai, and 1460/00. These genes were mostly absent from cattle isolate 1070/00 and pig isolate 486/00 (Fig. 5A). The first three strains appeared to fit in lineage I, and the latter two strains may belong to lineage II. For the genes that are specific to lineage II according to Zhang et al. (44), a backbone gene, b1202, which was reported to be absent in lineage I, appeared to be present in all strains studied here, including EDL933 and Sakai (Fig. 5B). Most other genes (seven of nine) within lineage II, including ymfL, ymfM, b1152, b2361, b2362, b2363, and b1201, were absent from the two known lineage I strains (i.e., EDL933 and Sakai) and present in our putative lineage II strains 1070/00 and 486/99. These two clusters of strains had the most differences from each other within our selection of strains according to the clustering analysis. However, the assignment of lineage based on this gene list for the rest of strains was much harder. This again showed that the strains used in the present study were more diverse than those used by Zhang et al. (44). This greater diversity probably resulted from the selection of strains mentioned earlier.
FIG. 5.
Distribution of lineage-specific genes within our strains. Genes were identified as specific or dominant in lineage I (A) or lineage II (B) according to Zhang et al. (44). The clustering analysis was done as for Fig. 2, based on all genes on the array, but only results from the selected genes are displayed. As two different sets of oligonucleotide probes were used in these two studies, not all genes identified by Zhang et al. (44) were represented on our array and vice versa. Color coding is the same as in Fig. 2.
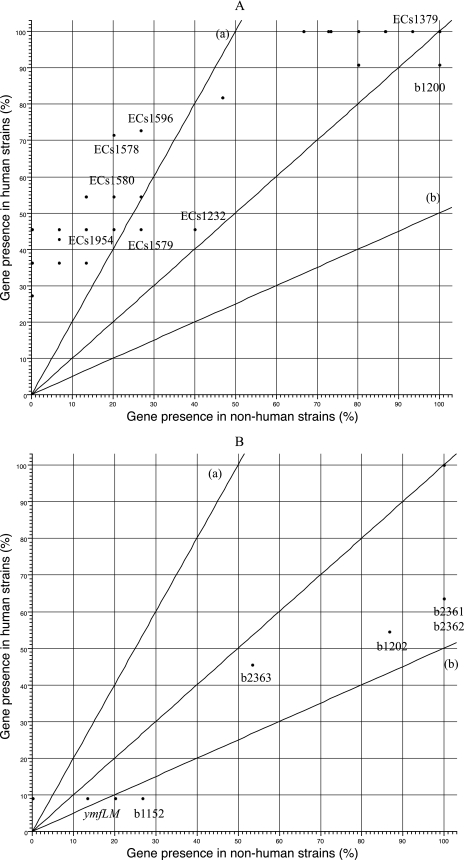
Interestingly, a subset of genes within the lineage I list from Zhang et al. (44) were found more frequently in our human isolates, while other genes from lineage I were present equally in isolates from human and nonhuman sources (Fig. 6A). Similarly, a subset of genes specific to lineage II were found less frequently in our isolates from human than nonhuman sources (Fig. 6B). It may be more informative to identify genes that are specific to strains from human infections than to identify those specific to a particular lineage. Due to the inherent limitation of both studies, as the arrays used in both studies consisted of genes from only virulent E. coli O157 isolates, we were unable to identify genes that are specific to E. coli O157:H7 isolates that are less virulent or avirulent.
FIG. 6.
The presence of lineage-specific or -dominant genes in isolates from human and nonhuman sources. Genes identified as specific or dominant in lineage I (A) and lineage II (B) according to Zhang et al. (44) were plotted as described for Fig. 3.
Identification of core components among EDL933- and Sakai-like O157 isolates.
Figure 2 shows that all EDL933- and Sakai-like E. coli O157 strains used in this study shared common genetic elements, with genes on many O islands or S loops being conserved. The genetic elements present throughout all EDL933- and Sakai-like E. coli O157:H7 strains but absent from MG1655 and other putative commensal strains of E. coli can be defined as the core genes.
As more independent strains are included in a study, the number of core genes will decrease and the number of VAP genes will increase. Although the core can be calculated with a single strain, the most informative core will be obtained after all of the strains are considered. Here, we wished not only to predict the actual core for each data set but also to compare the final asymptote between curves generated based on data from the previous paper by Zhang et al. (44) and those produced in this work. If strains are genetically close, having fewer variable genes, the core will stabilize faster.
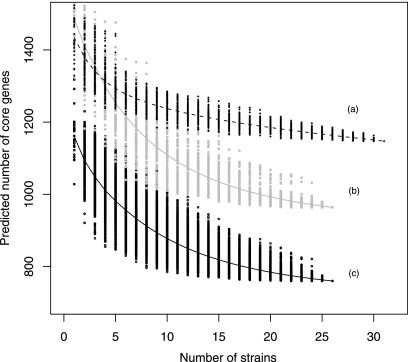
To ascertain whether any differences in core are due to atypical strains within one of the data sets, we used a resampling technique called bootstrapping (5, 10, 11) to provide variability for the order in which the strains are included in their respective studies. After each data set had been bootstrapped 100 times, the core was generated after inclusion of each additional strain. The data were plotted (Fig. 7), and a general linear model was fitted to each data set to provide an indication of the line of best fit.
FIG. 7.
Calculation of theoretical core genes by bootstrapping sampling. The data were generated by bootstrapping (5, 10, 11) to determine how the core decreased in size as more strains were included. The points surrounding the predicted lines are the individual bootstrap core sizes. The line of best fit was calculated using a linear model. Line a was calculated based on data from Zhang et al. (44); genes found in MG1655 were excluded. Line b was calculated in the same way as line a but with the data generated in this work. Line c was also generated with data from this work, but genes found in MG1655 and two MG1655-like strains were all excluded.
Figure 7 shows bootstrapping simulation calculating the core component based on data from Zhang et al. (44) and those from this work. The data sets were comparable when fewer than five strains were considered (Fig. 7, lines a and b). A quicker flattening of the line indicates a lack of independence within the data, or greater similarity between strains within the data set. In short, this proved that our strains were more diverse than those used by Zhang et al. (44). Genes found in two MG1655-like strains were excluded from line c, which showed that using a larger pool of commensal strains gave a better understanding of the E. coli O157:H7 core.
There were about 760 core genes among EDL933- and Sakai-like strains (data not shown). Among these, half were unknown, phage/prophage, or Rhs element related. Among protein coding sequences with known functions, there were virulence-related genes, such as genes for LEE, type III secretion system, putative RTX family exoprotein (Z0615, ECs0542), invasin (Z5932, ECs5290), and long polar fimbrial operon (lpf) (35) and genes for a urease (ureD, -A, -C, -E, and -G). Although, the RTX family exoprotein, invasin, and urease genes have been classed as virulence-related genes, there is still no experimental evidence to suggest that they play a role in the pathogenesis of E. coli O157.
All genes for the secondary type III secretion system (ETT2) were present in the EDL933- and Sakai-like E. coli O157 strains (25, 43). Another fimbrial operon (E100005285-6) (24) in O island 61 was present in 23 strains but not in cattle isolate 796/00. The invariably present core genes also included a number of genes for putative transcriptional regulators, such as the putative LysR-like transcriptional regulators (Z0346, Z0371, Z0885, ECs0309, ECs0333, and ECs0755), a putative transcriptional regulator of l-sorbose uptake (ECs4941 and Z5613) and utilization (Z5613, Z5614, Z5615, Z5616, Z5617, ECS4941, ECS4999, ECs5000, ECs5001, and ECs5002) genes, as well as genes for several transporters, including a putative iron and heme/hemoglobin transporter (chuS), a putative ribose-specific transporter (Z5690 and ECs5072), and a putative antibiotic efflux pump (Z3494 and ECs3132). Among the E. coli O157 core genes, there were genes that confer the ability to utilize sorbose (sor), although in Sakai the sor genes showed insertion of a phage disrupting the operon. Genes putatively involved in the modification of the structure or function of DNA (ECs4281, ECs5263, ECs5264, Z5901, and Z5902), outer membrane proteins (ybgQ, Z2322, Z3058, ECs2007, and ECs2703), and O-antigen biosynthesis (wbdR, manC, fcl, wbdP, per, wzx, wbdO, wzy, and wbdN) were also in the core set. It was hypothesized that the remaining genes in the core set may play a role in other cellular processes, such as the genes for different subunits of a putative dimethyl sulfoxide reductase (Z3783, Z3784, Z3785, ECs3382, ECs3383, and ESc3384), a sulfatase (Z1089 and ECs0942), and two subunits of a glutamate mutase (Z0893, Z0895, ECs0762, and ECs0764).
We have yet to determine whether these genes are functional and whether they play any role in bacterial physiology. As CGH microarray studies only detect the presence or absence of genes, the functional roles of these genes may be inferred by expression studies.
Identification of genes not present in Sakai- and EDL933-like E. coli O157 isolates.
All EDL933- and Sakai-like E. coli O157:H7 used in this study had lost genes at many identical positions corresponding to the MG1655 chromosome (Fig. 2), which suggested that they all originated from a common ancestor. It is known that different serotypes of E. coli have different MG1655 genes (K islands) in their genomes (1). The clonal groups of E. coli may be linked to their loss or possession of K-island genes as well as the possession of the additional genes.
The reason for the gene loss is unknown, but it may be that these genes are not essential in the environmental niche that EDL933- and Sakai-like E. coli O157 strains occupy. However, in some cases, gene loss augments virulence (26). It is known that pathogens not only acquire virulence determinants but also lose genes to increase their virulence, and this phenomenon is called a black hole (8). However, the role played by gene loss in the pathogenicity of E. coli O157 is still unknown, but the genes identified in this study are candidates for future studies and are likely to play an important role in understanding E. coli O157 virulence.
In some instances, the missing genes were replaced by others with similar functions. For example, fimbrial genes (yadC, yadK, yadL, yadM, htrE, ecpD, and yadN) in MG1655 were replaced by different fimbrial genes in EDL933-like (Z0147, Z0148, Z0149, Z0150, Z0151, and Z0152) and Sakai-like (ECs0139, ECs0140, ECs0141, ECs0142, ECs0143, ECs0144, and ECs0145) strains, and the bacterioferrin gene (bfr) in MG1655 was replaced by a different bacterioferrin gene (Z4695 and ECs4189) in EDL933- and Sakai-like strains (see Table S4 in the supplemental material).
Analysis of the K-12-like E. coli O157 isolates.
The two E. coli O157 strains (1583/00 and 1176/00) that were similar to MG1655 lacked most genes specific to EDL933 and Sakai (Fig. 2), and both were Stx negative. Both had genes (wbdR, manC, fcl, wbdP, per, wzx, wbdO, wzy, and wbdN) specific for the O157 antigen (36), which replaced the nonfunctional O-antigen cluster of MG1655. fliC was present in all strains apart from 1583/00. Strains 1176/00 and 1583/00 contained about 4.8% and 14.4% of O-island and S-loop genes, respectively.
Like other EDL- and Sakai-like strains, both 1176/00 and 1583/00 had lost genes in the idnR-to-idnK region, which is involved in l-idonate catabolism. It is known that MG1655 can use idonate as the sole carbon and energy source (4), so we can predict that all E. coli O157 isolates have lost their ability to use idonate as a sole carbon and energy source. Like other E. coli O157 isolates used in this study, these strains were also missing a 12-gene region from intB to yjhE, the b4286-to-fecI regions that are involved in ferric citrate transport, a 14-gene region from yjhU to yjhR, and genes around mcrDCB. Strain 1583/00 was also missing hsdSMR, yjiW, and mrr, the restriction and modification system. Many missing regions appeared to be associated with insertion elements or integrase genes.
The presence of other virulence genes was also tested in these two strains with a miniaturized microarray chip developed in our laboratory containing representative virulence genes for each pathotype of E. coli (2). None of the virulence genes present on our array was detected in these strains (results not shown).
Presence of plasmid genes.
Most genes on both pO157 plasmids and pOSAK1 were not present in either of the two MG1655-like E. coli O157 strains. Genes on pO157 were present in most EDL933- and Sakai-like E. coli O157:H7 strains. The exceptions were strains 1070/00 and 1484/00: DNA from both strains had failed to hybridize with most genes on pO157 present in our microarray. A human isolate, 1464/00, harbored only some pO157 genes; genes missing from this strain included those encoding the type II secretion system (etp), stcE (L7031) (9, 19-21), and the gene for a metalloprotease which plays a role in adhesion (see Table S4 in the supplemental material).
Plasmid profiling (see Fig. S2 in the supplemental material) showed the heterogeneities of plasmids. Most strains contained a 90-kb plasmid, as found in Sakai. Some also had the additional 3-kb plasmid, while others had a plasmid of about 7.8 kb. Some contained one or more plasmids from 60 to 90 kb. Plasmids were found in both 1070/00 (90 kb) and 1484/00 (60 kb and 7 kb). Strain 1464/00 appeared to have a truncated pO157 plasmid according to the sizes of its plasmids, microarray results, and the PCR results (see below).
PCRs were carried out to detect the presence of selected plasmid genes (hlyAC, katP, espP, and etpD). Sakai and MG1655 were used as positive and negative controls, respectively. Primers for hlyAC, katP, and espP but not etpD generated products of the expected sizes for 1464/00; the results agreed with the CGH microarray and plasmid profiling results (see Fig. S2 in the supplemental material). Products of the correct sizes were obtained with PCR primers for hlyAC, katP, and etpD but not espP for 1484/00 (results not shown), but the CGH microarray had failed to identify those plasmid genes in 1484/00. All four plasmid genes were found in 1070/00 by PCR (results not shown); however, the CGH microarray did not identify any of them. Although we had a certain degree of success in identifying genes on plasmids by microarray analysis, it completely failed to identify genes carried on plasmids when an alternative method was used for the genomic DNA isolation (R. Tozzoli, personal communication). This could be due to the loss of plasmids during genomic DNA preparation, whereas the PCR method is more sensitive. Therefore, when genes from plasmids are not detected by microarrays, we should exercise caution by verifying their absence by PCR.
Implications of CGH work for strain identification.
Currently, pathogenic E. coli O157 is identified based on a few established markers such as O157 antigen, H7 antigen, the inability to ferment sorbitol, and β-glucuronidase activity. However, a number of unrelated E. coli strains can have the O157 antigen (39), and strains from the related genera Citrobacter and Salmonella can also cross-react with the O157 antigen (32). Also, many E. coli O157 strains ferment sorbitol and are β-glucuronidase positive, such as those found in continental Europe and recently in Scotland (14, 16). Therefore, testing for these established markers can be misleading, and this increases the importance of identifying new ones. We believe some of the markers identified in this study will be useful in this respect, and we have already started to look at their potential as markers. Such markers, in conjunction with subtyping of the H7 antigen (37), are likely to provide more comprehensive and useful information in the future.
Supplementary Material
Acknowledgments
The work was supported by VLA seedcorn funding SC0145.
We thank S. Avery and Clare Cassar for providing us with strains and Roberto La Ragione for helping with the category III work.
Editor: A. Camilli
Footnotes
Published ahead of print on 10 December 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Anjum, M. F., S. Lucchini, A. Thompson, J. C. Hinton, and M. J. Woodward. 2003. Comparative genomic indexing reveals the phylogenomics of Escherichia coli pathogens. Infect. Immun. 714674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anjum, M. F., M. Mafura, P. Slickers, K. Ballmer, P. Kuhnert, M. J. Woodward, and R. Ehricht. 2007. Pathotyping Escherichia coli by using miniaturized DNA microarrays. Appl. Environ Microbiol. 735692-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avery, S. M., E. Liebana, C. A. Reid, M. J. Woodward, and S. Buncic. 2002. Combined use of two genetic fingerprinting methods, pulsed-field gel electrophoresis and ribotyping, for characterization of Escherichia coli O157 isolates from food animals, retail meats, and cases of human disease. J. Clin. Microbiol. 402806-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bausch, C., N. Peekhaus, C. Utz, T. Blais, E. Murray, T. Lowary, and T. Conway. 1998. Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for L-idonic acid catabolism in Escherichia coli. J. Bacteriol. 1803704-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley, E. 1979. Computers and theory of statistics: thinking the unthinkable. SIAM Rev. 21460-480. [Google Scholar]
- 6.Brunder, W., H. Karch, and H. Schmidt. 2006. Complete sequence of the large virulence plasmid pSFO157 of the sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H- strain 3072/96. Int. J. Med. Microbiol. 296467-474. [DOI] [PubMed] [Google Scholar]
- 7.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24767-778. [DOI] [PubMed] [Google Scholar]
- 8.Day, W. A., and A. T. Maurelli. 2006. Black holes and antivirulence genes: selection for gene loss as part of the evolution of bacterial pathogens, p. 109-122. In H. S. Seifert and V. J. DiRita (ed.), Evolution of microbial pathogens. ASM Press, Washington, DC.
- 9.Grys, T. E., M. B. Siegel, W. W. Lathem, and R. A. Welch. 2005. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect. Immun. 731295-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, P. 1986. On the number of bootstrap simulations required to construct a confidence interval. Ann. Stat. 141453-1462. [Google Scholar]
- 11.Hall, P., L. Peng, and N. Tajvidi. 1999. On prediction intervals based on predictive likelihood or bootstrap methods. Biometrika 86871-880. [Google Scholar]
- 12.Hancock, D. D., T. E. Besser, M. L. Kinsel, P. I. Tarr, D. H. Rice, and M. G. Paros. 1994. The prevalence of Escherichia coli O157.H7 in dairy and beef cattle in Washington State. Epidemiol. Infect. 113199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 811-22. [DOI] [PubMed] [Google Scholar]
- 14.Janka, A., M. Bielaszewska, U. Dobrindt, L. Greune, M. A. Schmidt, and H. Karch. 2003. Cytolethal distending toxin gene cluster in enterohemorrhagic Escherichia coli O157:H− and O157:H7: characterization and evolutionary considerations. Infect. Immun. 713634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 1451365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karch, H., and M. Bielaszewska. 2001. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J. Clin. Microbiol. 392043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J., J. Nietfeldt, and A. K. Benson. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. USA 9613288-13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1997. Characterization of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli serotypes isolated from sheep. J. Clin. Microbiol. 35892-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lathem, W. W., T. Bergsbaken, and R. A. Welch. 2004. Potentiation of C1 esterase inhibitor by StcE, a metalloprotease secreted by Escherichia coli O157:H7. J. Exp. Med. 1991077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lathem, W. W., T. Bergsbaken, S. E. Witowski, N. T. Perna, and R. A. Welch. 2003. Acquisition of stcE, a C1 esterase inhibitor-specific metalloprotease, during the evolution of Escherichia coli O157:H7. J. Infect. Dis. 1871907-1914. [DOI] [PubMed] [Google Scholar]
- 21.Lathem, W. W., T. E. Grys, S. E. Witowski, A. G. Torres, J. B. Kaper, P. I. Tarr, and R. A. Welch. 2002. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45277-288. [DOI] [PubMed] [Google Scholar]
- 22.Law, D. 2000. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J. Appl. Microbiol. 88729-745. [DOI] [PubMed] [Google Scholar]
- 23.Lejeune, J. T., S. T. Abedon, K. Takemura, N. P. Christie, and S. Sreevatsan. 2004. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg. Infect. Dis. 101482-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Low, A. S., F. Dziva, A. G. Torres, J. L. Martinez, T. Rosser, S. Naylor, K. Spears, N. Holden, A. Mahajan, J. Findlay, J. Sales, D. G. Smith, J. C. Low, M. P. Stevens, and D. L. Gally. 2006. Cloning, expression, and characterization of fimbrial operon F9 from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 742233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino, S., T. Tobe, H. Asakura, M. Watarai, T. Ikeda, K. Takeshi, and C. Sasakawa. 2003. Distribution of the secondary type III secretion system locus found in enterohemorrhagic Escherichia coli O157:H7 isolates among Shiga toxin-producing E. coli strains. J. Clin. Microbiol. 412341-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, R. A., S. Reckseidler-Zenteno, H. Kim, W. Nierman, Y. Yu, A. Tuanyok, J. Warawa, D. DeShazer, and D. E. Woods. 2004. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 724172-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogura, Y., K. Kurokawa, T. Ooka, K. Tashiro, T. Tobe, M. Ohnishi, K. Nakayama, T. Morimoto, J. Terajima, H. Watanabe, S. Kuhara, and T. Hayashi. 2006. Complexity of the genomic diversity in enterohemorrhagic Escherichia coli O157 revealed by the combinational use of the O157 Sakai OligoDNA microarray and the Whole Genome PCR scanning. DNA Res. 133-14. [DOI] [PubMed] [Google Scholar]
- 28.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409529-533. [DOI] [PubMed] [Google Scholar]
- 29.Reid, S. D., R. K. Selander, and T. S. Whittam. 1999. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J. Bacteriol. 181153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308681-685. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 32.Samuel, G., J. P. Hogbin, L. Wang, and P. R. Reeves. 2004. Relationships of the Escherichia coli O157, O111, and O55 O-antigen gene clusters with those of Salmonella enterica and Citrobacter freundii, which express identical O antigens. J. Bacteriol. 1866536-6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 696660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tobe, T., S. A. Beatson, H. Taniguchi, H. Abe, C. M. Bailey, A. Fivian, R. Younis, S. Matthews, O. Marches, G. Frankel, T. Hayashi, and M. J. Pallen. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA 10314941-14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 705416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 663545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, L., D. Rothemund, H. Curd, and P. R. Reeves. 2000. Sequence diversity of the Escherichia coli H7 fliC genes: implication for a DNA-based typing scheme for E. coli O157:H7. J. Clin. Microbiol. 381786-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittam, T. S., I. K. Wachsmuth, and R. A. Wilson. 1988. Genetic evidence of clonal descent of Escherichia coli O157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome. J. Infect. Dis. 1571124-1133. [DOI] [PubMed] [Google Scholar]
- 39.Whittam, T. S., and R. A. Wilson. 1988. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect. Immun. 562467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 611619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wick, L. M., W. Qi, D. W. Lacher, and T. S. Whittam. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 1871783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, Z., J. Kovar, J. Kim, J. Nietfeldt, D. R. Smith, R. A. Moxley, M. E. Olson, P. D. Fey, and A. K. Benson. 2004. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 706846-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, L., R. R. Chaudhuri, C. Constantinidou, J. L. Hobman, M. D. Patel, A. C. Jones, D. Sarti, A. J. Roe, I. Vlisidou, R. K. Shaw, F. Falciani, M. P. Stevens, D. L. Gally, S. Knutton, G. Frankel, C. W. Penn, and M. J. Pallen. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 727282-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y., C. Laing, M. Steele, K. Ziebell, R. Johnson, A. K. Benson, E. Taboada, and V. P. Gannon. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, T., M. P. Doyle, J. Shere, and L. Garber. 1995. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ Microbiol. 611290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.