Abstract
Yersinia pestis, the causative agent of plague, must survive in blood in order to cause disease and to be transmitted from host to host by fleas. Members of the Ail/Lom family of outer membrane proteins provide protection from complement-dependent killing for a number of pathogenic bacteria. The Y. pestis KIM genome is predicted to encode four Ail/Lom family proteins. Y. pestis mutants specifically deficient in expression of each of these proteins were constructed using lambda Red-mediated recombination. The Ail outer membrane protein was essential for Y. pestis to resist complement-mediated killing at 26 and 37°C. Ail was expressed at high levels at both 26 and 37°C, but not at 6°C. Expression of Ail in Escherichia coli provided protection from the bactericidal activity of complement. High-level expression of the three other Y. pestis Ail/Lom family proteins (the y1682, y2034, and y2446 proteins) provided no protection against complement-mediated bacterial killing. A Y. pestis ail deletion mutant was rapidly killed by sera obtained from all mammals tested except mouse serum. The role of Ail in infection of mice, Caenorhabditis elegans, and fleas was investigated.
Yersinia pestis is the etiologic agent of plague, an often fatal disease of mammals (30). The pathogenicity of Y. pestis results largely from its ability to thwart the defenses of its host and to overwhelm it with massive growth (7). Transmission of plague normally occurs via the bite of an infected flea (16). Regurgitated bacteria initially journey to the draining lymph nodes, which enlarge to form the characteristic buboes. Bacteria subsequently spread to the blood and colonize the spleen, liver, and other organs. Eventually, a terminal, high-density septicemia occurs, leading to shock, disseminated intravascular coagulation, peripheral gangrene, and death.
The ability to survive in blood is critical for Y. pestis to cause disease, to infect its flea vector, and to be transmitted from host to host. To survive in diverse mammalian tissues and blood, Y. pestis actively blocks bacterial phagocytosis and resists complement-mediated bacteriolysis (30). A plasmid pCD1-encoded type III secretion system plays a key role in preventing bacterial phagocytosis (41); however, the mechanism by which Y. pestis evades complement-mediated lysis is not fully understood.
The enteropathogenic yersiniae (Y. enterocolitica and Y. pseudotuberculosis) are fully resistant to complement when they are grown at 37°C but not when they are grown at 26°C (5, 30). In contrast, Y. pestis is constitutively resistant to complement at both 26 and 37°C, but it is sensitive when it is grown at 6°C (1). Resistance to complement in Y. enterocolitica has been studied in detail and has been shown to involve YadA, Ail, and lipopolysaccharide (LPS) O antigen, with YadA playing a dominant role (4). Both YadA and Ail are outer membrane proteins that function as adhesins/invasins, as well as complement resistance proteins. Interestingly, Y. pestis does not express YadA and expresses only rough LPS (30). Previous studies have suggested that Y. pestis LPS may mediate serum resistance (34); however, there is no evidence of a direct role for LPS in serum resistance.
Members of the Ail/Lom family, which include Ail of Y. enterocolitica and Y. pseudotuberculosis, Rck and PagC of Salmonella enterica, and OmpX of Escherichia coli, are outer membrane proteins that share significant amino acid sequence similarity and identity and are predicted to have similar membrane topologies (15, 24, 26, 36, 42, 44). These proteins are all predicted to possess eight transmembrane amphipathic β-strands and four extracellular loops, with the regions of greatest amino acid sequence similarity and identity concentrated in the membrane-spanning segments. Amino acid residues located in extracellular loops 2 and 3 have been shown to play a critical role in Ail-mediated attachment, invasion, and serum resistance of Y. enterocolitica (25). Although members of this family of proteins exhibit diverse functions, several of them, including Ail of Y. enterocolitica and Y. pseudotuberculosis, as well as Rck of S. enterica, function, at least in part, to protect bacteria from complement-mediated lysis. The Rck protein has been shown to protect S. enterica from complement by inhibiting C9 polymerization and subsequent assembly of a functional membrane attack complex (14). Other Ail/Lom family members are required for survival in macrophages (PagC) or have no confirmed function (OmpX and Lom).
In this study, we investigated the role of Ail/Lom family members in the pathogenesis of Y. pestis. The genomes of Y. pestis KIM and CO92 both contain four genes predicted to encode Ail/Lom family proteins (11, 29). We demonstrated that one of these proteins, designated Ail, is required for resistance to complement-mediated killing at both 26 and 37°C. The Ail protein was expressed at high levels at both 26 and 37°C; however, expression of Ail was minimal at 6°C. Serum-sensitive E. coli DH5α transformed with an Ail expression vector became serum resistant, confirming that the Ail protein can provide serum resistance independent of other Y. pestis virulence factors. A Y. pestis ail deletion mutant was rapidly killed by exposure to all animal sera tested except mouse serum. In accordance with these findings, a Y. pestis KIM ail deletion mutant was fully virulent in an intravenous mouse model of infection. Interestingly, the ail mutant was attenuated in a Caenorhabditis elegans infection model, but it exhibited no defect in colonization or blockage of the flea digestive tract.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains used in this study are described in Table 1. All Y. pestis KIM strains except Y. pestis KIM6+ and derivatives of this strain are Pgm− and avirulent when peripheral routes of infection are used (40). Y. pestis KIM6+ lacks the pCD1 virulence plasmid that is required for virulence in mammals (37). These strains and their derivatives were routinely grown in heart infusion broth (HIB) or on tryptose blood agar (TBA) base plates (Difco, Detroit, MI). For serum survival experiments Y. pestis and E. coli strains were grown at 6, 26, or 37°C in HIB in the presence of 2.5 mM calcium chloride. E. coli DH5α (6) was used for routine cloning experiments.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristicsb | Reference |
|---|---|---|
| Y. pestis strainsa | ||
| KIM5 (Pla+ parent) | pCD1, pPCP1, pMT1 | 40 |
| KIM5 (Δy1324) (Δail) | pCD1, pPCP1, pMT1 Δail | This study |
| KIM5 (Δy1682) | pCD1, pPCP1, pMT1 Δy1682 | This study |
| KIM5 (Δy2034) | pCD1, pPCP1, pMT1 Δy2034 | This study |
| KIM5 (Δy2446) | pCD1, pPCP1, pMT1 Δy2446 | This study |
| KIM8-E (ΔyopE-sycE::dhfr) (Pla− parent)c | pCD1 (sycE yopE::Km), pPCP1−, pMT1 | 2 |
| KIM8-E (Δy1324) (Δail) | pCD1 (sycE yopE::Km), pPCP1−, pMT1 Δail | This study |
| KIM8-E (Δy1682) | pCD1 (sycE yopE::Km), pPCP1−, pMT1 Δy1682 | This study |
| KIM8-E (Δy2034) | pCD1 (sycE yopE::Km), pPCP1−, pMT1 Δy2034 | This study |
| KIM8-E (Δy2446) | pCD1 (sycE yopE::Km), pPCP1−, pMT1 Δy2446 | This study |
| KIM8-E (Δ4) (Δy1324 Δy1682 Δy2034 Δy2446) | pCD1 (sycE yopE::Km), pPCP1−, pMT1 Δy1324 Δy1682 Δy2034 Δy2446 | This study |
| KIM6+ | pCD1−, pPCP1, pMT1 | 37 |
| KIM6+ Δail | pCD1−, pPCP1, pMT1 Δail | This study |
| E. coli DH5α | F−recA endA gyrA thi hsdR supE relA Δ(lacZYA-argF) deoR Φ80lac(ΔlacZ)M15 | 6 |
All Y. pestis strains except KIM6+ and KIM6+ Δail are avirulent due to deletion of the pgm locus (40). Y. pestis KIM6+ and derivatives of this strain have been cured of plasmid pCD1.
The native plasmids of Y. pestis include pCD1 (31), pPCP1 (35) encoding the outer membrane plasminogen activator (Pla) protease that has been shown to degrade exported Yops, and pMT1 encoding the capsular protein (21).
Y. pestis KIM8-E was previously designated KIM8-Δ4 (2).
Deletion of Y. pestis KIM5, KIM6+, or KIM8-E (2) chromosomal DNA sequences encoding amino acid residues 20 to 122 of the y1324 (ail) protein, amino acid residues 17 to 121 of the y1682 protein, amino acid residues 37 to 144 of the y2034 protein, and amino acid residues 48 to 154 of the y2446 protein and insertion of a kanamycin resistance (Kmr) or chloramphenicol resistance (Cmr) cassette flanked by FLP recognition sequences were accomplished using lambda Red-mediated recombination essentially as described by Datsenko and Wanner (10). PCR products used to construct gene replacements were generated using template plasmids pKD3 (Cmr) and pKD4 (Kmr). Oligonucleotide primers used for amplification of the PCR products are listed in Table 2. PCR products were gel purified, ethanol precipitated, and resuspended in 10 μl of distilled H2O. Y. pestis strains carrying plasmid pKD46, encoding the Red recombinase, were induced with 0.2% l-arabinose for 2 h prior to harvest. Electrocompetent cells were electroporated with the purified PCR products and plated on TBA plates containing chloramphenicol (20 μg/ml) or kanamycin (50 μg/ml). Plasmid pCP20, which encodes the FLP recombinase, was electroporated into the single-deletion strains to facilitate removal of the FLP recognition target-flanked cassettes. In addition to the single-deletion mutants, a strain with all four identified genes deleted was constructed using lambda Red-mediated recombination to sequentially delete the y1682, y2034, and y2446 genes from KIM8-E (Δy1324). Plasmids pKD46 and pCP20 were cured from the Y. pestis strains by overnight growth at 39°C. All gene replacements were confirmed by PCR analysis using CTL primers listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| y1324-P1 | TCTCTTCTTTAATTGCATGTTTATCAATTGCGTCTGTTATGTGTAGGCTGGAGCTGCTTC |
| y1324-P2 | TTTACTTCTACTCTCTGATTGACCAAATATTGAGGAAAACATATGAATATCCTCCTTAGT |
| y1682-P1 | ATTGCATGTCTTTCAGCGGTAGCAGCGTGTGTGTTAGCCTGTGTAGGCTGGAGCTGCTTC |
| y1682-P2 | GGTGAAGCCGTAGTCACTGGTGCTGTGTTTATCACCAACCATATGAATATCCTCCTTAGT |
| y2034-P1 | CAGTGATGCTAGCAACACGGTATCTTTCGGTTACGCTCATGTGTAGGCTGGAGCTGCTTC |
| y2034-P2 | AGCTGTATCAATAGCAACGTGTTTTACCGGATTGAACTGCATATGAATATCCTCCTTAGT |
| y2446-P1 | CTCAGGGAGATGTAAGACTCGGTGATGGAAATCGAAAAGTGTGTAGGCTGGAGCTGCTTC |
| y2446-P2 | AGTTTTGTATACTCGTATGAAGCATCAATAGCAATATTGCATATGAATATCCTCCTTAGT |
| y1324-KpnI | TTTGGTACCTACTGTATTAGGTATTGTTATAAC |
| y1324-XbaI | TTTTCTAGAAGGACGTTAGAACGGTAACCC |
| y1682-KpnI | TTTGGTACCTTGTGTCACTCAATAATTTGCGTC |
| y1682-XbaI | TTTTCTAGATCAGGCTATCACTTAGAAAGTGTA |
| y2034-KpnI | TTTGGTACCCCTTATCAAAAATTAACTTTTAAT |
| y2034-XbaI | TTTTCTAGAATCAACAGAATATTAGAAGCGGTA |
| y2446-KpnI | TTTGGTACCTCTAAGATAATAATTTAAGGATAA |
| y2446-XbaI | TTTTCTAGAATAGAAAGTATTTTATTAAAAACG |
| y1324-CTL1 | TACTGTATTAGGTATTGTTATA |
| y1324-CTL2 | AGGAGGACGTTAGAACCGGTA |
| y1682-CTL1 | TTTGGTACCTTGTGTCACTCAATAATTTGCGTC |
| y1682-CTL2 | TTTTCTAGATCAGGCTATCACTTAGAAAGTGTA |
| y2034-CTL1 | GAAAATTGGAAAAGATAATAAAGG |
| y2034-CTL2 | ACAGTGTTGCTTACCATACGCGGC |
| y2446-CTL1 | CCATAAGTTGAGTGAAATGTTCGG |
| y2446-CTL2 | AATTGCATCCCAACACCATATGCC |
Construction of Ail/Lom family expression plasmids.
Plasmids pAil and pTRC-Ail were constructed by inserting a ca. 0.6-kb PCR fragment generated with oligonucleotide primers y1324-KpnI and y1324-XbaI (Table 2) into KpnI- and XbaI-digested pBluescript KS(−) and pTRC99a, respectively. PCR fragments containing the entire y1682, y2034, and y2446 genes, including at least 50 bp upstream of the start codon, were amplified with oligonucleotide primer pairs y1682-KpnI/y1682-XbaI, y2034-KpnI/y2034-XbaI, and y2446-KpnI/y2446-XbaI (Table 2). The resultant ca. 0.6-kb PCR fragments were digested with KpnI and XbaI and inserted into KpnI- and XbaI-digested pTRC99a, generating plasmids pTRC-y1682, pTRC-y2034, and pTRC-y2446.
Serum survival assay.
Normal human serum (NHS) was pooled from at least five healthy volunteers. Animal sera were purchased from Valley Biomedical Products and Services, Inc. (Winchester, VA) or Sigma (St. Louis, MO). Heat-inactivated serum was used as a control in all experiments. Harvested and washed Y. pestis or E. coli was diluted in phosphate-buffered saline (PBS) (pH 7.4) to obtain an optical density at 620 nm (OD620) of 0.2 (∼1 × 108 bacteria/ml). Fifty microliters of the diluted bacteria (∼5 × 106 bacteria) was added to 200 μl of NHS, normal goat serum (NGS), normal sheep serum (NSS), normal rabbit serum (NRS), normal mouse serum (NMS), normal guinea pig serum (GPS), heat-inactivated serum (HIS), or PBS (final concentration of serum, 80%). The samples were incubated at 37°C with shaking for 1 h. The number of surviving bacteria (CFU) in each sample was determined by dilution and plating on TBA plates at 30°C for 1 or 2 days. The percent survival was calculated as follows: average number of bacteria that survived exposure to NHS at 1 h/number of bacteria that survived exposure to HIS at 1 h × 100.
Outer membrane preparations.
Outer membranes preparations were obtained essentially as described by Biedzka-Sarek et al. (4). Bacteria were grown overnight in HIB at 26 or 37°C. Bacterial cultures (5 ml) were harvested by centrifugation at 8,000 × g for 10 min at 4°C. Pellets were resuspended in 2 ml of 10 mM Tris-HCl-5 mM MgCl2 (pH 8.0) (buffer A), and the bacteria were lysed by two passages through an ice-cold French pressure cell. Unlysed cells and large debris were removed by centrifugation as described above. Bacterial membranes were collected by ultracentrifugation at 100,000 × g for 1 h at 4°C. Membrane pellets were resuspended in buffer A containing 2% Triton X-100. The Triton X-100-resistant outer membrane protein fraction was pelleted by ultracentrifugation and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
SDS-PAGE analysis of Y. pestis and E. coli proteins.
Volumes of cellular fractions corresponding to equal numbers of bacteria or equal amounts of protein were mixed 1:1 (vol/vol) with 2× electrophoresis sample buffer and analyzed by SDS-PAGE essentially as previously described (33). SDS-PAGE-separated proteins were stained with Coomassie brilliant blue R-250 or silver stained.
Mouse infection assays.
Female 6- to 8-week-old Swiss-Webster mice were used for infections. Y. pestis KIM5 and KIM5 Δail were grown overnight in HIB at 26°C. Fresh cultures were inoculated to obtain an OD620 of 0.2 and grown with shaking to an OD620 of 1.0. Bacterial cultures were harvested by centrifugation and resuspended in PBS. Mice were inoculated with Y. pestis via the retroorbital sinus using blunt feeding needles. Plate counting was performed to verify the number of CFU in the infectious doses.
C. elegans infection assays.
Groups of 40 C. elegans N2 (wild-type) young adults were exposed to Y. pestis strains grown on modified NG agar plates at 25°C. Worm mortality was scored over time. C. elegans survival was plotted using the PRISM computer program. A survival curve was considered significantly different from the control when the P value was <0.05.
Flea infection assays.
Xenopsylla cheopis fleas were infected with Y. pestis KIM6+ or KIM6+ Δail using an artificial feeding system essentially as described previously (17). One group of 50 male and 50 female fleas that took an infected blood meal were saved and examined for blockage immediately after they were allowed to feed on an uninfected mouse on days 6, 9, 13, 16, 20, 23, and 27 after infection. Additional samples of 20 female fleas were taken at the beginning of the experiment (time zero), 9 days after infection, and at the end of the experiment (day 28) to determine the infection rate and the average bacterial load per positive flea by CFU counting (17).
RESULTS
Generation of Y. pestis strains deficient in expression of Ail/Lom family proteins.
Previous analyses of available Y. pestis genome sequences identified four genes (the Y. pestis KIM y1324, y1682, y2034, and y2446 genes) predicted to encode Ail/Lom family proteins (11, 29). All four of the predicted gene products share significant amino acid sequence similarity and identity with each other and with Ail of Y. enterocolitica (Fig. 1). The Y. pestis y1682 gene encodes a protein that is 71% identical to the E. coli OmpX protein. In contrast, the y1324, y2034, and y2446 gene products share only 41, 41, and 31% amino acid sequence identity with OmpX, respectively. The y1682 gene and the E. coli ompX gene are also both located adjacent to the glnH, dps, and rhtA genes in the Y. pestis KIM and E. coli K-12 chromosomes, respectively. Thus, the y1682 gene appears to encode a Y. pestis OmpX homolog.
FIG. 1.
Amino acid sequence alignment of Y. pestis KIM Ail with other Yersinia Ail/Lom family proteins. The amino acid sequences of Y. pestis KIM Ail (Yp Ail) and Y. enterocolitica Ail (Ye Ail) and the predicted amino acid sequences of the Y. pestis KIM Ail/Lom family y1682, y2034, and y2446 proteins are aligned. Amino acid residues in the four predicted surface-exposed loops are enclosed in boxes (loops 1 to 4). Identical residues are indicated by asterisks, strongly similar residues are indicated by colons, and weakly similar residues are indicated by periods. The alignment was generated using the CLUSTALW multiple-sequence alignment program (39).
To investigate the role of the y1324, y1682, y2034, and y2446 gene products in the serum resistance and virulence of Y. pestis KIM, we used lambda Red-mediated recombination (10) to create Y. pestis strains KIM5 (Pla+) and KIM8-E (Pla− ΔyopE-sycE::dhfr) specifically deficient in expression of the four Ail/Lom family proteins (Table 1). The DNA fragments encoding amino acid residues 20 to 122 (y1324), 17 to 121 (y1682), 37 to 144 (y2034), and 48 to 154 (y2446) were removed, and a Kmr cassette flanked by FLP recognition sequences was inserted. Plasmid pCP20, which encodes the FLP recombinase (10), was electroporated into the single-deletion strains to facilitate removal of the FLP recognition target-flanked Kmr cassette. In addition to the individual deletion mutants, a strain with all four identified genes deleted was constructed using lambda Red-mediated recombination to sequentially delete the y1682, y2034, and y2446 genes from KIM8-E (Δy1324).
Y. pestis Ail (y1324) is required for resistance to complement-mediated killing.
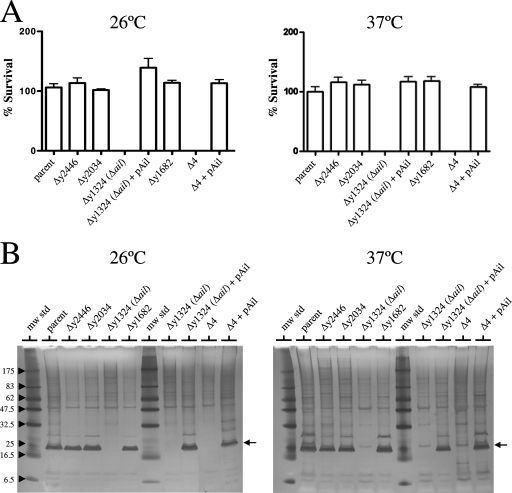
The abilities of the parent Y. pestis KIM8-E strain and the isogenic mutants lacking one or all four of the genes encoding Ail/Lom family proteins to survive in NHS or HIS were determined. Bacteria were grown at 26 or 37°C and diluted in PBS to obtain an OD620 of 0.2 (approximately 1 × 108 bacteria/ml), and 50 μl of bacteria (approximately 5 × 106 bacteria) was added to 200 μl of NHS, HIS, or PBS. The samples were incubated at 37°C for 60 min, and the numbers of surviving bacteria (CFU) were determined by dilution and plating on TBA base plates at 30°C.
The parent Y. pestis strain and the isogenic strains with deletions in the y1682, y2034, or y2446 gene were completely resistant to the killing action of NHS at both 26 and 37°C (Fig. 2A). In contrast, the Y. pestis strain with a deletion in the y1324 gene and the strain with all four genes deleted (Δ4) were susceptible to complement-mediated killing at both temperatures, suggesting that the y1324 gene product was required for survival in NHS. The survival of each of the strains was unaffected by incubation with HIS or PBS. To confirm that the y1324 gene product was required for serum resistance, the y1324 gene was PCR amplified and inserted into pBluescript KS(−) (Stratagene) downstream of the vector Plac promoter, and the resulting plasmid was moved into the y1324 deletion strain and into the multiple-deletion strain (Δ4). Providing the y1324 gene in trans completely restored the ability to survive complement-mediated killing at both 26 and 37°C in these strains, confirming that the y1324 gene is required for serum resistance of Y. pestis. Use of NHS treated with 10 mM EGTA and 5 mM MgCl2 (which inactivates the Ca2+-dependent classical and lectin pathways) instead of NHS provided essentially identical results (data not shown), suggesting that the alternative pathway was sufficient to mediate killing of the serum-sensitive Y. pestis mutants.
FIG. 2.
Y. pestis Ail is highly expressed and required for survival in NHS. (A) Survival of wild-type and mutant strains of Y. pestis in 80% NHS. Bacteria grown overnight at 26 or 37°C (∼5 × 106 bacteria) were incubated with 80% NHS or HIS for 1 h at 37°C. Aliquots were diluted and plated on TBA plates at 30°C. The percent survival was calculated as follows: average number of bacteria that survived exposure to NHS at 1 h/number of bacteria that survived exposure to HIS at 1 h × 100. (B) Silver-stained SDS-PAGE gel of outer membrane proteins isolated from the parent and mutant strains of Y. pestis. The location of the Ail (y1324) protein is indicated by an arrow. The positions of molecular mass standards (in kilodaltons) are indicated on the left. The strains used were Y. pestis strains KIM8-E (parent), KIM8-E Δy2446, KIM8-E Δy2034, KIM8-E Δy1324 (Δail), KIM8-E Δy1324 (Δail)/pAil, KIM8-E Δy1682, KIM8-E Δ4, and KIM8-E Δ4/pAil. mw std, molecular mass standards.
Y. pestis Ail is highly expressed at 26 and 37°C but not at 6°C.
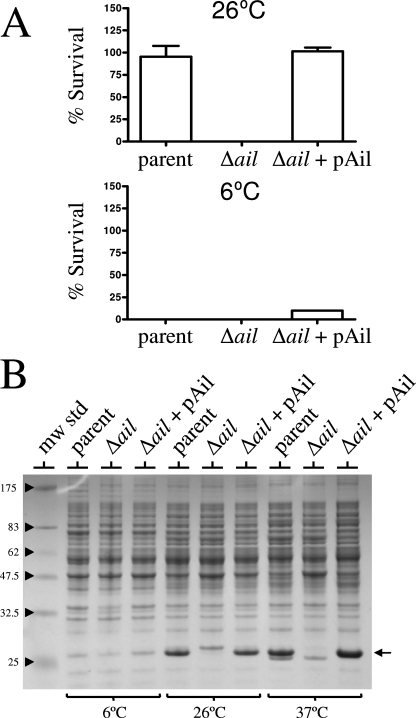
The outer membrane protein profile of each of the strains was examined to determine the expression of the Ail/Lom family proteins essentially as described by Biedzka-Sarek et al. (4). Isolated total membrane fractions from 5-ml overnight cultures of each strain grown at either 26 or 37°C were treated with 2% Triton X-100, and the Triton X-100-insoluble fractions were analyzed by SDS-PAGE and silver staining (Fig. 2B). A highly expressed putative outer membrane protein with an apparent molecular mass of ca. 20 kDa was found in the parent strain and in the strains having deletions in the y1682, y2034, and y2446 genes but not in the y1324 gene deletion strain or in the strain with all four genes deleted (Δ4). Complementation with the y1324 gene expression plasmid (pAil) restored expression of the ca. 20-kDa protein, confirming that this protein was the y1324 gene product. No consistent protein band corresponding to the y1682, y2034, or y2446 gene product was observed, suggesting that these proteins were not highly expressed under the growth conditions used. The y1324 gene product was required for serum resistance of Y. pestis at both 26 and 37°C and exhibited 77 and 98% amino acid sequence identity to the Y. enterocolitica and Y. pseudotuberculosis Ail proteins, respectively. Furthermore, the chromosomal locations of the Y. pestis y1324 gene and the Y. pseudotuberculosis ail gene were strictly conserved; therefore, the y1324 protein was designated the Y. pestis Ail protein.
The high-level expression of Y. pestis Ail at both 26 and 37°C provides an explanation for the ability of Y. pestis to resist the killing action of complement at both temperatures. Interestingly, Anisimov et al. (1) demonstrated that Y. pestis strains that are serum resistant when they are grown at 25 or 37°C become serum sensitive when they are grown at 6°C. We confirmed that Y. pestis KIM8-E and KIM8-E Δail were both serum sensitive when they were grown at 6°C (Fig. 3A). Analysis of whole-cell proteins from Y. pestis strains grown at 6, 26, and 37°C (Fig. 3B) confirmed that Ail was expressed well at both 26 and 37°C; however, Ail was not as highly expressed in strains grown at 6°C. Thus, serum resistance of Y. pestis explicitly correlated with high-level expression of Ail.
FIG. 3.
Ail is not expressed by Y. pestis strains grown at 6°C. (A) Survival of wild-type and mutant strains of Y. pestis in 80% NHS. Bacteria grown overnight at 6 or 26°C (∼5 × 106 bacteria) were incubated with 80% NHS or HIS for 1 h at 37°C. Aliquots were diluted and plated on TBA plates at 30°C. The percent survival was calculated as follows: average number of bacteria that survived exposure to NHS at 1 h/number of bacteria that survived exposure to HIS at 1 h × 100. (B) Coomassie blue R-250-stained SDS-PAGE gel of whole bacterial cell proteins isolated from wild-type and mutant strains of Y. pestis. The location of the Ail protein is indicated by an arrow. The positions of molecular mass standards (in kilodaltons) are indicated on the left. The strains used were Y. pestis strains KIM8-E (parent), KIM8-E Δy1324 (Δail), and KIM8-E Δy1324 (Δail)/pAil. mw std, molecular mass standards.
E. coli DH5α expressing Ail is serum resistant.
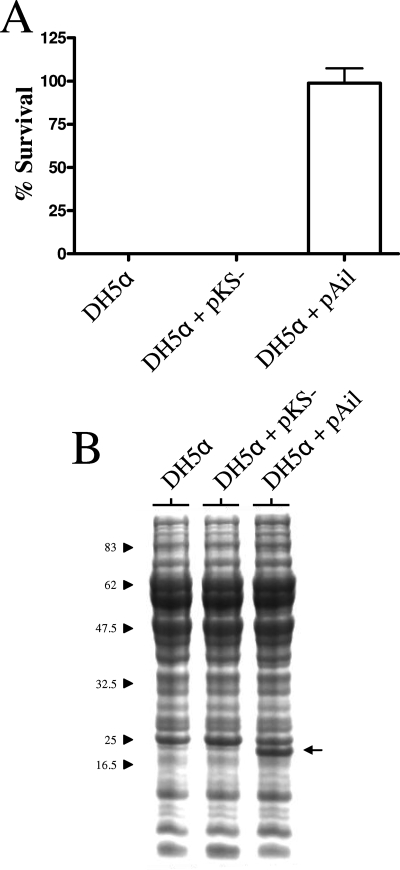
To confirm that expression of Ail alone is sufficient to mediate serum resistance, we moved the Ail expression plasmid (pAil) into a serum-sensitive strain of E. coli. E. coli DH5α strains carrying no plasmid, pBluescript KS(−) (vector control), or pAil were grown overnight at 37°C, and the serum resistance of the strains was determined as described above for the Y. pestis strains (Fig. 4A). DH5α alone and DH5α carrying pBluescript KS(−) were both rapidly killed by NHS but not by HIS. In contrast, DH5α carrying the pAil plasmid was resistant to the bactericidal action of NHS. Analysis of whole-cell proteins by SDS-PAGE and Coomassie blue R-250 staining confirmed that DH5α carrying pAil expressed the ∼20-kDa Ail protein (Fig. 4B). These results confirm that the Y. pestis Ail protein can confer resistance to complement-dependent killing independent of other Y. pestis-specific virulence factors.
FIG. 4.
Expression of Ail in E. coli provides protection from complement-dependent killing. (A) Survival of E. coli with or without plasmid pAil in 80% NHS. Bacteria grown overnight at 37°C (∼5 × 106 bacteria) were incubated with 80% NHS or HIS for 1 h at 37°C. Aliquots were diluted and plated on LB plates at 37°C. The percent survival was calculated as follows: average number of bacteria that survived exposure to NHS at 1 h/number of bacteria that survived exposure to HIS at 1 h × 100. (B) Coomassie blue R-250-stained SDS-PAGE gel of whole bacterial cell proteins isolated from E. coli with or without plasmid pAil. The location of the Ail protein is indicated by an arrow. The positions of molecular mass standards (in kilodaltons) are indicated on the left. The strains used were E. coli strains DH5α, DH5α/pBluescript KS(−) (pKS−) (vector control), and DH5α/pAil.
High-level expression of the Ail/Lom family y1682, y2034, and y2446 proteins does not protect Y. pestis from complement-mediated killing.
Y. pestis strains defective in expression of the Ail protein were sensitive to complement-mediated killing at both 26 and 37°C, whereas strains having a deletion in one of the genes encoding other Ail/Lom family proteins were serum resistant. These results suggest that the y1682, y2034, and y2446 genes were not expressed under the conditions used or that the expressed proteins were unable to mediate resistance to complement-directed bacteriolysis. Analysis of the proteins expressed by these strains (Fig. 2 and 3) indicated that the y1682, y2034, and y2446 proteins were not expressed at a level that allowed detection under the experimental conditions utilized.
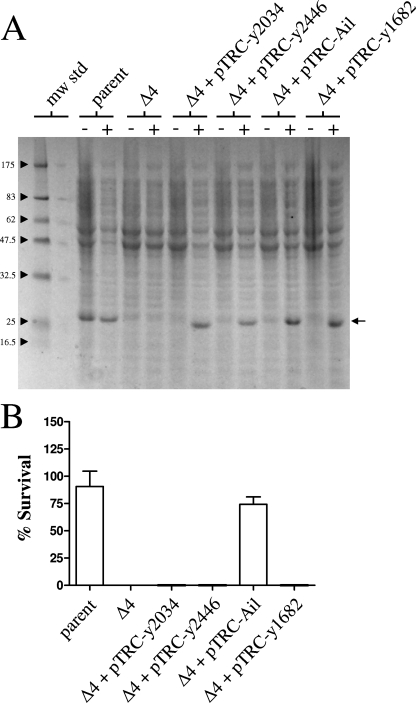
To determine if the y1682, y2034, and y2446 gene products were capable of mediating resistance to complement-mediated killing, expression vectors for each Y. pestis Ail/Lom family protein were constructed. The ail, y1682, y2034, and y2446 genes were inserted downstream of the strong trc promoter in plasmid pTRC99a (Amersham Biosciences). The resulting constructs, designated pTRC-Ail, pTRC-y1682, pTRC-y2034, and pTRC-y2446, were moved into the serum-sensitive multiple-Ail/Lom-deletion mutant (Y. pestis KIM8-E Δ4), and the resulting strains were grown at 26°C in the presence or absence of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce expression of the Ail/Lom family proteins. SDS-PAGE and Coomassie blue R-250 staining demonstrated that each of the Ail/Lom family proteins was highly expressed following induction with IPTG (Fig. 5A). Selective solubilization studies (data not shown) demonstrated that the majority of each Ail/Lom family protein was present in the Y. pestis outer membrane.
FIG. 5.
Expression of Ail, but not expression of other Y. pestis Ail/Lom family proteins, provides protection against complement-dependent killing. (A) Coomassie blue R-250-stained SDS-PAGE gel of whole bacterial cell proteins isolated from Y. pestis strains induced (lanes +) or not induced (lanes −) with 1 mM IPTG. The location of the various Ail/Lom family proteins is indicated by an arrow. The positions of molecular mass standards (in kilodaltons) are indicated on the left. The strains used were Y. pestis strains KIM8-E (parent), KIM8-E Δ4, and KIM8-E Δ4 containing pTRC-y2034, pTRC-y2446, pTRC-Ail, or pTRC-y1682. mw std, molecular mass standards. (B) Survival of the parent strain, the Δ4 strain, and the Δ4 strain carrying plasmid pTRC-y2034, pTRC-y2446, pTRC-Ail, or pTRC-y1682 in 80% NHS. Bacterial cultures were grown at 26°C and induced or not induced with 1 mM IPTG 3 h prior to harvest. Bacteria (∼5 × 106 cells) were incubated with 80% NHS or HIS for 1 h at 37°C. Aliquots were diluted and plated on TBA plates at 30°C. The percent survival was calculated as follows: average number of bacteria that survived exposure to NHS at 1 h/number of bacteria that survived exposure to HIS at 1 h × 100.
The ability of each of the Y. pestis strains expressing the different Ail/Lom family proteins (IPTG-induced cultures) to survive in NHS was determined (Fig. 5B). The parent strain and the Δ4 strain expressing the Ail protein were resistant to complement-mediated killing. In contrast, the Δ4 strain and the Δ4 strain carrying plasmid pTRC-y1682, pTRC-y2034, or pTRC-y2446 were sensitive to complement-mediated killing. These results suggest that the Y. pestis Ail protein is the sole Ail/Lom family protein responsible for the serum resistance of Y. pestis. However, these results do not rule out alternative roles for the other Ail/Lom family proteins in the pathogenesis of Y. pestis.
Y. pestis Ail mediates resistance to the bactericidal activity of a wide variety of animal sera.
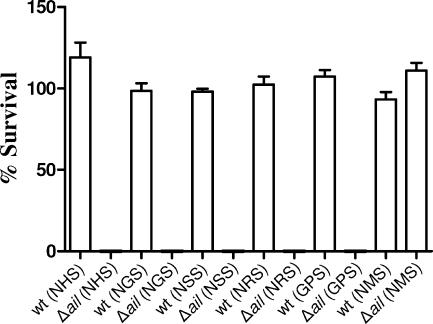
Sera obtained from different species of mammals possess different levels of bactericidal activity for gram-negative bacteria. For example, previous studies have shown that NMS is not bactericidal for E. coli or Y. enterocolitica (13, 23, 43). Furthermore, NMS had no effect on a Y. enterocolitica ail or yadA deletion mutant (43). The abilities of Y. pestis parent and Y. pestis ail deletion mutant strains to survive in sera obtained from various mammals were determined essentially as described above for NHS. Y. pestis KIM8-E and KIM8-E Δail that had been grown overnight at 26°C were prepared as described above and incubated with sera obtained from humans, goats (NGS), sheep (NSS), rabbits (NRS), guinea pigs (GPS), and mice (final concentration of serum, 80%) for 1 h at 37°C (Fig. 6). The viability of the parent strain Y. pestis KIM8-E was unaffected by any of the sera tested. The NHS, NGS, NSS, NRS, and GPS efficiently killed the ail deletion mutant. In contrast, the NMS had no affect on the parent or the ail deletion mutant. These results confirm that the ail deletion mutant is serum sensitive; however, as reported previously, NMS is not bactericidal, and exposure of the ail deletion mutant to NMS did not alter the viability of this strain in vitro.
FIG. 6.
Ail is required for survival in sera obtained from many diverse mammals but not for survival in mouse serum: survival of the parent (wt) and Δail strains of Y. pestis in 80% animal sera. Bacteria grown overnight at 26°C (∼5 × 106 bacteria) were incubated with 80% animal serum or HIS for 1 h at 37°C. Aliquots were diluted and plated on TBA plates at 30°C. The percent survival was calculated as follows: average number of bacteria that survived exposure to NHS at 1 h/number of bacteria that survived exposure to HIS at 1 h × 100.
Y. pestis Ail is not required for virulence in an intravenous mouse model of plague.
To begin to evaluate the role that Y. pestis Ail plays during an infection, preliminary virulence studies were conducted using a murine model of plague. Although NMS is not bactericidal, Ail could have alternate functions or additional roles that are important for Y. pestis virulence in mice. Intravenous injection of approximately 100 CFU (six mice) or 1,000 CFU (six mice) of the Y. pestis KIM5 ail deletion mutant (Pgm−) resulted in 100% fatality by day 6 postinfection. As expected, almost all mice inoculated with the Y. pestis KIM5 parent strain (Pgm−) also succumbed to the infection (five of the six mice inoculated with 100 CFU and all six mice inoculated with 1,000 CFU). These data indicate that that the Ail protein is not required for virulence in mice infected via an intravenous route; however, these results do not rule out a role for Ail in mice infected via alternate routes. In addition, Ail is likely required to cause infection in other higher mammals, such as humans, that possess a complement system capable of rapidly killing Ail-deficient strains of Y. pestis.
Y. pestis Ail is required for virulence in a C. elegans infection model.
Previous studies have demonstrated that Y. pestis can kill the nematode C. elegans by both biofilm-dependent and biofilm-independent mechanisms (9, 38). Y. pestis strains that carry the hmsHFRS genes produce a biofilm that is required for flea blockage and subsequent transmission of plague (17). The production of biofilm also blocks feeding of C. elegans. In contrast, Y. pestis strains that lack the hmsHFRS genes (for example, Pgm− strains) are capable of killing C. elegans by a biofilm-independent process that involves direct ingestion of the pathogen and accumulation of bacteria in the intestine (38).
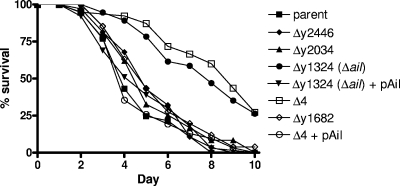
To determine if any of the Y. pestis Ail/Lom family proteins play a role in the biofilm-independent infection of C. elegans, we exposed groups of 40 C. elegans N2 young adults to the parent strain (KIM8-E) and to each of the Ail/Lom family deletion mutants (Fig. 7). As shown previously (38), ingestion of the Pgm− parent strain produced a lethal infection at 25°C that resulted in the death of all 40 worms by day 8 of infection. Similar results were obtained when the worms were fed Y. pestis strains specifically deficient in expression of the y1682, y2034, or y2446 gene. In contrast, worms exposed to the Δail strain or to the strain with all four ail-like genes deleted (Δ4) showed significantly increased survival (P < 0.0001). Providing the ail gene in trans (pAil) to the Δail or Δ4 strain restored the ability of these strains to efficiently kill C. elegans. These results indicate that the Y. pestis Ail protein plays an important role in the biofilm-independent killing of C. elegans. Exposure of C. elegans to biofilm-competent (Pgm+) Y. pestis KIM6+ or KIM6+ Δail resulted in rapid killing of the nematodes, indicating that Ail plays no significant role in the biofilm-dependent killing of C. elegans (data not shown).
FIG. 7.
Ail is required for efficient biofilm-independent killing of C. elegans. Groups of 40 C. elegans N2 young adults were exposed to Y. pestis KIM8-E (parent), KIM8-E Δy2446, KIM8-E Δy2034, KIM8-E Δy1324 (Δail), Δy1324 (Δail)/pAil, KIM8-E Δ4, KIM8-E Δ4/pAil, and Δy1682. Animal survival was plotted using the PRISM computer program.
Y. pestis Ail is not required to infect or block fleas.
Previously, the C. elegans infection model has been used to identify and/or characterize gene products that are important for Y. pestis to infect and/or block fleas (8, 9). C. elegans infection experiments are performed at 25°C; therefore, Y. pestis gene products expressed in the C. elegans infection model are likely to also be expressed in the flea (21°C). Therefore, we investigated if the Y. pestis Ail protein is required to infect or block fleas.
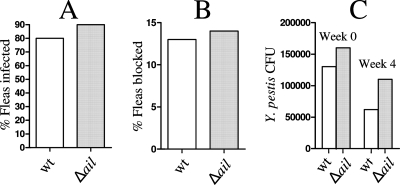
X. cheopis rat fleas were infected with mouse blood containing Y. pestis KIM6+ or an isogenic ail deletion mutant (KIM6+ Δail) (Fig. 8). Infected fleas were monitored for 4 weeks following the initial infection. During this time, 13% of the fleas infected with KIM6+ and 14% of the fleas infected with KIM6+ Δail became blocked. Similarly, no significant difference in the infection rate or bacterial load per flea was observed between the two strains, as determined by a t test. These data indicate that Ail does not play a significant role in the infection or blockage of fleas.
FIG. 8.
Ail is not required for infection or blockage of fleas. (A) Colonization of fleas after an infected-blood meal. (B) Flea blockage during a 4-week period following a single infected-blood meal. (C) Y. pestis CFU associated with infected fleas on day 0 (week 0) and day 28 (week 4). wt, parent strain KIM6+; Δail, strain KIM6+ Δail.
DISCUSSION
The success of Y. pestis as a vector-borne pathogen is dependent upon its ability to produce high-level septicemia in its host. The ability to reliably generate bacterial loads in the blood that can approach 109 bacteria/ml ensures that a feeding flea ingests a sufficient number of bacteria to establish an infection (12, 16, 22). To survive in the blood of its mammalian host and in the blood meal of its flea vector, Y. pestis must be resistant to complement-dependent killing at both 26 and 37°C. We determined that the Y. pestis Ail protein is required for resistance to complement-dependent killing at both temperatures. Thus, Ail is an important virulence factor that would be expected to be essential for infection of mammalian hosts that possess a fully functional bactericidal complement system. Indeed, Ail may represent one of the key factors that has enabled Y. pestis to infect a wide range of mammals, including humans.
Anisimov et al. (1) demonstrated that Y. pestis strains that are resistant to complement at 26 and 37°C become sensitive to complement when they are grown at 6°C. It has been hypothesized that this response represents part of a coordinated down regulation of virulence attributes that enables Y. pestis to coexist with its host during periods of winter hibernation. Our studies indicate that strong down regulation of expression of Ail at low temperatures is likely responsible for the dramatic increase in the susceptibility of the bacteria to serum when they are grown at 6°C; however, other factors, including temperature-dependent changes in LPS structure, may also be involved in regulating serum resistance in response to low temperature (1).
Previous studies have indicated that LPS may play a critical role in the resistance of Y. pestis to complement-mediated killing (1, 34); however, the fact that Y. pestis strains deficient in expression of Ail are extremely sensitive to complement supports the hypothesis that Y. pestis LPS provides little or no intrinsic protection against complement. Alternatively, alterations in the structure of LPS may have dramatic effects on the expression, surface exposure, or activity of Ail. Indeed, in Y. enterocolitica alterations in LPS structure have been demonstrated to alter both Ail expression and Ail function (3, 32). Thus, the structure of Y. pestis LPS is likely optimized to promote Ail function and may be modified, at least in part, to regulate Ail function in response to changes in environmental conditions.
Initial infection experiments indicated that Ail is not required to produce a lethal infection in mice when an intravenous route of infection is used. This was not surprising, considering that mouse serum is not bactericidal for Yersinia spp. (13) and thus had no effect on the ail deletion mutant. In contrast, sera from guinea pigs, rabbits, sheep, goats, and humans were all able to rapidly kill the ail deletion mutant. These results strongly indicate that the ail deletion mutant should be unable to survive in these animals and indicate that Ail is an important virulence factor; however, confirmation of the role of Ail during an infection will likely require an alternative animal model. Guinea pigs have previously been used to evaluate the virulence of Y. pestis (18) and may represent an alternative host for evaluation of the role of Ail during an infection.
Surprisingly, the ail deletion mutant exhibited a significant defect in virulence in the biofilm-independent C. elegans infection model (38). Interestingly, the C. elegans genome appears to encode only a limited number of putative complement components (28), such as factor H, a regulator of the alternative pathway (20). However, the unique combinations of mammalian complement domains required for a functional pathway are not found in nematodes, suggesting that Ail may provide protection against other antibacterium products or that Ail has alternative functions that are important in C. elegans virulence. Interestingly, the Y. enterocolitica Ail protein functions as an adhesin/invasin, in addition to its role in serum resistance (27). However, no role in attachment to, or invasion of, eukaryotic cells was associated with the Y. pseudotuberculosis Ail protein (44). Future studies will be aimed at determining the specific role of Ail in C. elegans.
Y. enterocolitica Ail was originally identified as an outer membrane protein that promoted bacterial invasion of eukaryotic cells (26). Subsequent studies revealed that Ail also plays a role in serum resistance (5). The mechanism by which Y. enterocolitica Ail protects the bacterium from complement-dependent lysis is not understood. In fact, studies with Y. enterocolitica have suggested that Ail alone is not sufficient to fully protect Y. enterocolitica from complement-dependent killing in vitro (4). Instead, evidence suggests that the Y. enterocolitica YadA protein is the dominant serum resistance factor expressed by this bacterium. In contrast to serum resistance in Y. enterocolitica, in Y. pestis Ail appears to be the sole complement resistance factor required to fully protect the bacterium from complement in vitro (44). This suggests that the Y. pestis Ail protein functions differently, is more efficient, or is expressed at a higher level than the Y. enterocolitica Ail protein.
The Y. enterocolitica Ail and Y. pestis Ail proteins share significant amino acid sequence identity throughout their lengths (75% overall identity); however, the level of amino acid sequence identity in the four surface-exposed loops is much less (53%) than the level in the regions located outside these loops (87%). Results of site-directed mutagenesis studies of ail of Y. enterocolitica suggest that the residues located in loop 2 are particularly important for Ail function; however, only 5 of the 19 residues found in loop 2 are identical to residues found in loop 2 of Y. pestis Ail (Fig. 1). Therefore, even though preliminary experiments suggest that the Y. pestis Ail protein is expressed at a higher level than the Y. enterocolitica Ail protein (data not shown), it is also possible that amino acid sequence differences in the surface-exposed loops account, in part, for the ability of the Y. pestis Ail protein to fully protect the bacterium from complement-dependent killing. It is also possible that the Y. pestis Ail protein, which closely resembles Ail of Y. pseudotuberculosis, represents a protein optimized and dedicated to serum resistance. Indeed, previous studies have suggested that the Y. pseudotuberculosis Ail protein has no role in bacterial attachment or invasion; however, recent studies of Kolodziejek et al. (19) indicated that the Y. pestis Ail protein functions as both an adhesin and an invasin. These studies suggested that Y. pestis Ail, like Y. enterocolitica Ail, is a multifunctional protein with roles in serum resistance, attachment, and invasion.
Acknowledgments
We thank Hans Wolf-Watz for helpful discussions and Lisa Plano for advice and technical assistance.
This work was funded by grants R01 AI039575 and R21 AI074820-02 from the National Institutes of Health to G.V.P.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 19 November 2007.
REFERENCES
- 1.Anisimov, A. P., S. V. Dentovskaya, G. M. Titareva, I. V. Bakhteeva, R. Z. Shaikhutdinova, S. V. Balakhonov, B. Lindner, N. A. Kocharova, S. N. Senchenkova, O. Holst, G. B. Pier, and Y. A. Knirel. 2005. Intraspecies and temperature-dependent variations in susceptibility of Yersinia pestis to the bactericidal action of serum and to polymyxin B. Infect. Immun. 737324-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartra, S. S., M. W. Jackson, J. A. Ross, and G. V. Plano. 2006. Calcium-regulated type III secretion of Yop proteins by an Escherichia coli hha mutant carrying a Yersinia pestis pCD1 virulence plasmid. Infect. Immun. 741381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bengoechea, J. A., H. Najdenski, and M. Skurnik. 2004. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol. Microbiol. 52451-469. [DOI] [PubMed] [Google Scholar]
- 4.Biedzka-Sarek, M., R. Venho, and M. Skurnik. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 732232-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bliska, J. B., and S. Falkow. 1992. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc. Natl. Acad. Sci. USA 893561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cambau, E., F. Bordon, E. Collatz, and L. Gutmann. 1993. Novel gyrA point mutation in a strain of Escherichia coli resistant to fluoroquinolones but not to nalidixic acid. Antimicrob. Agents Chemother. 371247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis, G. R. 2000. Molecular and cell biology aspects of plague. Proc. Natl. Acad. Sci. USA 978778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darby, C., S. L. Ananth, L. Tan, and B. J. Hinnebusch. 2005. Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect. Immun. 737236-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417243-244. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 1844601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelthaler, D. M., B. J. Hinnebusch, C. M. Rittner, and K. L. Gage. 2000. Quantitative competitive PCR as a technique for exploring flea-Yersina pestis dynamics. Am. J. Trop. Med. Hyg. 62552-560. [DOI] [PubMed] [Google Scholar]
- 13.Hanski, C., M. Naumann, A. Grutzkau, G. Pluschke, B. Friedrich, H. Hahn, and E. O. Riecken. 1991. Humoral and cellular defense against intestinal murine infection with Yersinia enterocolitica. Infect. Immun. 591106-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heffernan, E. J., S. Reed, J. Hackett, J. Fierer, C. Roudier, and D. Guiney. 1992. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella typhimurium virulence plasmid gene rck. J. Clin. Investig. 90953-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heffernan, E. J., L. Wu, J. Louie, S. Okamoto, J. Fierer, and D. G. Guiney. 1994. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect. Immun. 625183-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinnebusch, B. J. 2005. The evolution of flea-borne transmission in Yersinia pestis. Curr. Issues Mol. Biol. 7197-212. [PubMed] [Google Scholar]
- 17.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273367-370. [DOI] [PubMed] [Google Scholar]
- 18.Jones, S. M., K. F. Griffin, I. Hodgson, and E. D. Williamson. 2003. Protective efficacy of a fully recombinant plague vaccine in the guinea pig. Vaccine 213912-3918. [DOI] [PubMed] [Google Scholar]
- 19.Kolodziejek, A. M., D. J. Sinclair, K. S. Seo, D. R. Schnider, C. F. Deobald, H. N. Rohde, A. K. Viall, S. S. Minnich, C. J. Hovde, S. A. Minnich, and G. A. Bohach. 2007. Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology 1532941-2951. [DOI] [PubMed] [Google Scholar]
- 20.Krushkal, J., C. Kemper, and I. Gigli. 1998. Ancient origin of human complement factor H. J. Mol. Evol. 47625-630. [DOI] [PubMed] [Google Scholar]
- 21.Lindler, L. E., G. V. Plano, V. Burland, G. F. Mayhew, and F. R. Blattner. 1998. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect. Immun. 665731-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorange, E. A., B. L. Race, F. Sebbane, and B. Joseph Hinnebusch. 2005. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J. Infect. Dis. 1911907-1912. [DOI] [PubMed] [Google Scholar]
- 23.Marcus, S., D. W. Esplin, and D. M. Donaldson. 1954. Lack of bactericidal effect of mouse serum on a number of common microorganisms. Science 119877. [DOI] [PubMed] [Google Scholar]
- 24.Mecsas, J., R. Welch, J. W. Erickson, and C. A. Gross. 1995. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J. Bacteriol. 177799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, V. L., K. B. Beer, G. Heusipp, B. M. Young, and M. R. Wachtel. 2001. Identification of regions of Ail required for the invasion and serum resistance phenotypes. Mol. Microbiol. 411053-1062. [DOI] [PubMed] [Google Scholar]
- 26.Miller, V. L., J. B. Bliska, and S. Falkow. 1990. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J. Bacteriol. 1721062-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 561242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nonaka, M., and F. Yoshizaki. 2004. Primitive complement system of invertebrates. Immunol. Rev. 198203-215. [DOI] [PubMed] [Google Scholar]
- 29.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413523-527. [DOI] [PubMed] [Google Scholar]
- 30.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 1035-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry, R. D., S. C. Straley, J. D. Fetherston, D. J. Rose, J. Gregor, and F. R. Blattner. 1998. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect. Immun. 664611-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierson, D. E. 1994. Mutations affecting lipopolysaccharide enhance ail-mediated entry of Yersinia enterocolitica into mammalian cells. J. Bacteriol. 1764043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plano, G. V., and S. C. Straley. 1995. Mutations in yscC, yscD, and yscG prevent high-level expression and secretion of V antigen and Yops in Yersinia pestis. J. Bacteriol. 1773843-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porat, R., W. R. McCabe, and R. R. Brubaker. 1995. Lipopolysaccharide-associated resistance to killing of yersiniae by complement. J. Endotoxin Res. 291-97. [Google Scholar]
- 35.Protsenko, O. A., P. I. Anisimov, O. T. Mozharov, N. P. Konnov, and A. Popov Iu. 1983. Detection and characterization of the plasmids of the plague microbe which determine the synthesis of pesticin I, fraction I antigen and “mouse” toxin exotoxin. Genetika 191081-1090. (In Russian.) [PubMed] [Google Scholar]
- 36.Pulkkinen, W. S., and S. I. Miller. 1991. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J. Bacteriol. 17386-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikkema, D. J., and R. R. Brubaker. 1987. Resistance to pesticin, storage of iron, and invasion of HeLa cells by yersiniae. Infect. Immun. 55572-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Styer, K. L., G. W. Hopkins, S. S. Bartra, G. V. Plano, R. Frothingham, and A. Aballay. 2005. Yersinia pestis kills Caenorhabditis elegans by a biofilm-independent process that involves novel virulence factors. EMBO Rep. 6992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Une, T., and R. R. Brubaker. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect. Immun. 43895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viboud, G. I., and J. B. Bliska. 2004. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 5969-89. [DOI] [PubMed] [Google Scholar]
- 42.Vogt, J., and G. E. Schulz. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure 71301-1309. [DOI] [PubMed] [Google Scholar]
- 43.Wachtel, M. R., and V. L. Miller. 1995. In vitro and in vivo characterization of an ail mutant of Yersinia enterocolitica. Infect. Immun. 632541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, Y., J. J. Merriam, J. P. Mueller, and R. R. Isberg. 1996. The psa locus is responsible for thermoinducible binding of Yersinia pseudotuberculosis to cultured cells. Infect. Immun. 642483-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]