Abstract
Uropathogenic Escherichia coli strain J96 carries multiple determinants for fimbrial adhesins. The regulatory protein PapB of P fimbriae has previously been implicated in potential coregulatory events. The focB gene of the F1C fimbria determinant is highly homologous to papB; the translated sequences share 81% identity. In this study we investigated the role of PapB and FocB in regulation of the F1C fimbriae. By using gel mobility shift assays, we showed that FocB binds to sequences in both the pap and foc operons in a somewhat different manner than PapB. The results of both in vitro cross-linking and in vivo oligomerization tests indicated that FocB could function in an oligomeric fashion. Furthermore, our results suggest that PapB and FocB can form heterodimers and that these complexes can repress expression of the foc operon. The effect of FocB on expression of type 1 fimbriae was also tested. Taken together, the results that we present expand our knowledge about a regulatory network for different adhesin gene systems in uropathogenic E. coli and suggest a hierarchy for expression of the fimbrial adhesins.
Fimbriae are adhesive organelles of paramount importance for successful bacterial recognition and colonization of specific host tissues. The fimbriae on Escherichia coli strains associated with urinary tract infections can be differentiated into three categories on the basis of their hemagglutinating abilities. The first category encompasses fimbriae mediating d-mannose-resistant agglutination of human erythrocytes, e.g., P and S fimbriae (25, 27, 44). The second group consists of the type 1 fimbriae and mediates d-mannose-sensitive agglutination of various erythrocytes, e.g., guinea pig erythrocytes (10, 43). To the third class belong fimbriae that do not possess any known hemagglutinating characteristics, notably F1C fimbriae (23, 50, 51). Although F1C fimbriae do not hemagglutinate, they do contribute to the adhesive properties of uropathogenic E. coli strains. It has been suggested that F1C fimbriae mediate specific adherence to the collecting ducts and the distal tubules of the human kidney (51). The F1C fimbriae bind to glycosphingolipids (2, 19).
The foc gene cluster involved in the synthesis of F1C fimbriae is highly homologous to the sfa gene cluster encoding S-fimbria adhesins (37, 38). The sfr gene cluster is another member of this highly homologous family (39). The structure of the F1C fimbriae, like the structure of S, P, and type 1 fimbriae, appears to be complex, as these fimbriae consist of major and minor subunits (1, 20, 28, 29, 32, 42). The focA gene encodes the major fimbrial subunit, while focG and focH encode minor fimbrial subunits; the focC and focD genes are required to encode a chaperone protein and molecular usher (41). It has been reported previously that the two-component export systems of type 1 and F1C fimbriae are interchangeable and that the minor fimbrial structural elements of these two fimbrial systems can be exchanged, resulting in hybrid organelles with changed receptor specificity (21, 22, 24). However, the regulatory proteins of the foc operon have remained largely uncharacterized.
A particular E. coli strain may have at least four different fimbrial antigens that are potentially involved in binding to urinary tract tissue (10, 26, 36, 49). In uropathogenic strain J96, three different adhesin gene clusters, pap, prs, and fim, have been described (18, 30), and it is now known that there is also a fourth cluster, the foc gene cluster. The F1C fimbriae are associated with uropathogenic O4 and O6 strains of E. coli that commonly express P fimbriae (35, 40). Binding of type 1C-fimbriated bacteria to collecting ducts may increase the adhesiveness of E. coli carrying this fimbrial type, since strains with P fimbriae adhere to collecting ducts only weakly (34).
PapB is a fimbrial regulator of the pap operon, and it belongs to a family of proteins which are involved in regulation of the production of different fimbrial adhesins. In vitro binding studies have shown that the PapB protein interacts with three different sequences (sites 1, 2, and 3), two within the papI-papB intercistronic region and one within the papB coding sequence (11). Site 1 appeared to be the preferential binding site, and sites 2 and 3 appeared to be low-affinity sites. PapB has an autoregulatory role, and the expression of P fimbriae is repressed when the expression of PapB is high, presumably by binding of PapB to low-affinity sites 2 and 3 (11). PapB has been shown to bind in an oligomeric fashion to 9-bp sequence motifs that contain T/A triplets at site 1 (53). The amino acid residues crucial for DNA binding and oligomerization are almost completely conserved in the whole PapB family of proteins (55), suggesting that they are conserved structurally and functionally.
We have shown previously that introduction of PapB into E. coli K-12 can repress expression of the α-d-mannose-sensitive type 1 fimbriae by affecting the FimB and FimE recombinases (54). That PapB may regulate type 1 fimbria expression was recently shown to be true at wild-type expression levels of PapB in several clinical uropathogenic isolates (16). Further studies of the effect of several different PapB paralogues on the phase variation of type 1 fimbriae showed that this effect was due to certain amino acids in the C-terminal part of the protein, which are present only in a subset of the PapB homologous proteins (17). Thus, in E. coli, only PapB and the fimbrial regulator of S fimbriae, SfaB, were able to repress fim expression and required both a previously identified amino acid important for DNA binding and specific amino acids at the carboxy terminus. In a recent study it was shown that expression of type 1 fimbriae also could repress P fimbria expression (45); however, the mechanism for this regulation is not known yet. In the present study, the role of a papB homologous gene in the F1C fimbria determinant, focB, was analyzed. We found that the FocB protein binds DNA in an oligomeric fashion, and our evidence suggests a potential interaction between PapB and FocB. Our findings suggest that the two proteins could interact to form heterooligomers that can be involved in the regulation of E. coli fimbrial biogenesis. To our knowledge, our description of contact between PapB and FocB is the first report of an interaction between different PapB homologous proteins. Since the fimbriae recognize receptors that may represent different tissue domains, coordination of expression between the different fimbrial gene clusters might help the bacterial population to rapidly adhere to and colonize different surfaces of the human urinary tract. This may be particularly important in the case of P and type 1C fimbriae, which often occur in the same strains (52).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains used in this study are shown in Table 1. The media used included L broth (LB); L agar consisted of LB containing 1.5% agar. Liquid cultures were grown at the indicated temperature. The antibiotics used were carbenicillin (50 μg/ml), chloramphenicol (10 μg/ml), and tetracycline (10 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| J96 | Uropathogenic isolate | 18 |
| SAL1 | J96 ΔfocB | This study |
| HB101 | F−lacY1 recA13 | 6 |
| JM109 | recA1 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | 56 |
| JH607 | λ1120sPs | 4 |
| AAEC198A | fimB+E+fimA-lacZYA | 5 |
| AAEC374A | fimBE fimA-lacZYA | 5 |
| Plasmids | ||
| pHMG15 | pap (I+, B1) A:lacZ, Cbr | 3 |
| pHMG1 | pap (I+, B+) A:lacZ, Cbr | 15 |
| pHMG63 | pACYC184 carrying the lacIq repressor gene | This study |
| pACYC184 | Cloning vector, Tcr | 7 |
| pBB1 | focI,B cloned into pACYC184, Tcr | This study |
| pQE30 | Expression vector, Cbr | Qiagen Inc. |
| pYN89 | pQE30 His-FocB construct | This study |
| pBSN50 | Cosmid clone containing the foc gene cluster from J96 | 30 |
| pYN21 | Wild-type papB gene | 55 |
| pYN22 | Mutant papB gene (R61A) | 55 |
| pYN24 | Mutant papB gene (C65A) | 55 |
| pYN41 | Mutant papB gene (L55A,V56A) | 55 |
| pYN46 | Truncated papB gene (amino acids 1 to 80) | 55 |
| pBF21 | Wild-type cI | 46 |
| pBF22 | cI (1-159) | 46 |
| pYN69 | cI (Δ57-152)-papB (wild type) | 55 |
| pYN51 | cI (1-159)-papB (wild type) | 55 |
| pYN108 | cI (1-159)-focB (wild type) | This study |
Cb, carbenicillin; Tc, tetracycline.
DNA techniques and construction of plasmids.
Plasmid isolation, gel electrophoresis, transformation, amplification of DNA by PCR, and DNA labeling were performed by standard procedures. Restriction endonuclease digestion and DNA ligation reactions were performed under conditions recommended by the manufacturers (Boehringer Mannheim and New England Biolabs Inc.). A previously described cosmid cloning library constructed from E. coli uropathogenic strain J96 DNA was used to screen clones with the fimbrial adhesin gene sequence (30). Southern blotting of DNA showed that three EcoRI fragments hybridized to either papI- or papB-specific probes (12). The sizes of the two larger fragments (15.5 and 21 kb) corresponded to the sizes of the previously cloned and characterized pap and prs gene clusters, respectively (18, 30). A cosmid designated pBSN50 was found to hybridize to probes from the pap regulatory region, but it was different from the clones containing the pap or prs gene clusters. Further analysis showed that pBSN50 carried a determinant for F1C fimbriae. After enzyme-linked immunosorbent assays were performed with polyclonal antisera to F1C pili, it was concluded that the cosmid and subclones containing this third EcoRI fragment carried a determinant that produced F1C pili in E. coli K-12 (data not shown). Plasmid pBB1 was constructed by cloning the 4.8-kb EcoRI fragment from cosmid clone pBSN50 into the EcoRI site of pACYC184 (7). Plasmid pQE30 provides high-level expression of proteins containing a six-His affinity tag in E. coli. The DNA fragment containing the wild-type focB gene was obtained by PCR amplification with primers foc2 and foc3 (Table 2). Cosmid clone pBSN50 from J96 containing the foc DNA was used as a template. The fragment was then digested with BamHI-HindIII and cloned into pQE30 to produce pYN89. Plasmids pHMG1 (15), pHMG15 (3), pBF21 and pBF22 (46), and pYN51 and pYN69 (55) have been described previously. A DNA fragment containing the wild-type focB gene obtained by PCR amplification with primers foc4 and foc5 (Table 2) was digested with EcoRV-HindIII and then cloned into plasmid pBF21 to produce plasmid pYN108. Plasmid pHMG63 was constructed by cloning a 1.1-kb EcoRI fragment of lacIq into the EcoRI site of pACYC184. Strain SAL1 was created by deleting part of the focB locus in strain J96 by using lambda Red-mediated recombination of linear DNA fragments as described previously (9, 33). The deletion was designed to remove all but the first two codons of the gene, and it was confirmed by DNA sequence analysis.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| foc2 | GCC CGA ATT TCG GAT CCA TGG CAC AGC ATG AAG TTA TTA C |
| foc3 | GCC CCT GCA GAA GCT TTT ATT ACA GGG AAG CAG CTT CAG C |
| foc4 | GCC CAA GCT TTA TGG CAC AGC ATG AAG TTA T |
| foc5 | GCC CGA TAT CTT ATT ACA GGG AAG CAG CTT C |
| 740 | CCC AAG CTT AAT CCG TTA CCG CCA GCG CCT |
| 1140 | CCC AAG CTT TAA ACG ATC TTT TAA CCC ACA AAA C |
| 1012 | CGG ACT TTC TTT TCA CAG AAA AA |
| 1116 | GTT TTG TGG GTT AAA AGA TCG TTA |
| 1290 | GTC CTT TAC GCA AAA ACG ACT T |
| type1C no.1 | TGC CTG GTA ATC CGT TAC CGC |
| type1C no.7 | TTG ACG ATC TTT TGA TCT GTA |
| type1C no.3 | TCC TCT CTA TAT AAG CAA AAG G |
| type1C no.6 | TTT TTC AAT GGT AAG GAA CGT |
| type1C no.10 | AAG CAT CCC CTC CCC GGG TAA |
Expression and purification of the FocB protein.
Strain JM109/pYN89 was grown in LB at 37°C, and protein expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration, 1 mM) at the logarithmic phase. His-tagged FocB protein was purified using a Ni-nitrilotriacetic acid column as previously described for PapB (55). According to a sodium dodecyl sulfate (SDS)-polyacrylamide gel analysis, the molecular mass of FocB was approximately 12 kDa. The FocB protein could be recognized by anti-PapB polyclonal antibody based on results of immunoblot analyses, but, as shown below (see Results), the antibody reaction activity was much lower than that of PapB.
SDS-PAGE and immunoblot analysis.
For determination of the FocA and PapB levels in the cells, bacteria were grown at 37°C in LB (with or without IPTG) to mid-log phase. Cell extracts were obtained by suspending bacterial pellets in sample buffer (62.5 mM Tris [pH 6.8], 2% SDS, 5% glycerol, 0.25% β-mercaptoethanol, 0.1% bromophenol blue). The samples were boiled for 3 to 5 min and loaded onto an SDS-polyacrylamide gel electrophoresis (PAGE) (15% acrylamide) gel. Western blotting was performed using standard procedures essentially as described previously (8). Polyclonal antisera with antibodies recognizing PapB or FocA were obtained from immunization of rabbits with purified proteins as described previously (40, 55). The PapB antibody was further purified using the procedure described by Taraseviciene et al. (47). Anti-His antibodies (Qiagen) were also used for detection of His-tagged PapB or FocB proteins. The protein bands were detected using ECL+ (Amersham Biosciences) and were quantified using the Quantity One Fluor-S MultiImager system (Bio-Rad).
In vivo test of cooperative DNA binding.
An in vivo assay for testing cooperative DNA binding activity was employed as described previously for the PapB protein (55). This assay is based on hybrid gene construction with the bacteriophage lambda cI repressor gene. It relies on the ability of the oligomerization-proficient protein domain to functionally replace the C-terminal domain of the phage repressor protein and thereby confer activity to the N-terminal DNA binding domain (57). Plasmids encoding different domains or hybrids were introduced into strain JH607, which can be used as a reporter system for cooperative binding of repressor proteins to adjacent operators (4). Different receptor activities can be distinguished by measuring the chloramphenicol sensitivity and/or β-galactosidase activity of the bacterial cells.
β-Galactosidase assay.
To measure the β-galactosidase specific activity, we used the method described by Miller (31). All data below are average values obtained from at least three separate experiments.
Chloramphenicol sensitivity test.
Chloramphenicol sensitivity was tested by plating dilutions from a fresh overnight culture on LB plates containing chloramphenicol at concentrations of 0, 10, 25, 50, 100, 150, and 200 μg/ml (57). Sensitivity was expressed as the chloramphenicol concentration at which the plating efficiency of the cells fell below 50%.
Analysis of protein-DNA interactions.
Gel mobility shift assays to detect protein-DNA interactions were performed as previously described (13, 14). DNA fragments containing the PapB binding site of the pap or foc operon (13, 53) were obtained by end labeling purified PCR products with [γ-32P]ATP and T4 polynucleotide kinase. For the pap operon, oligonucleotides 740 and 1140 giving DNA fragment pap III (Fig. 1A), oligonucleotides 740 and 1012 giving DNA fragment pap I, and oligonucleotides 1116 and 1290 giving DNA fragment pap II were used as the primers (Table 2), while plasmid pHMG1 (15) was used as the template for PCR amplification. For the foc operon, oligonucleotides type1C no.1 and type1C no.7 giving DNA fragment foc III (Fig. 1A), oligonucleotides type1C no.3 and type1C no.7 giving DNA fragment foc I, and oligonucleotides type1C no.6 and type1C no.10 giving DNA fragment foc II were used as the primers (Table 2), while plasmid pBSN50 was used as the template. The purified FocB protein was mixed (final volume, 10 μl) with 32P-end-labeled DNA fragments (5,000 to 10,000 cpm) in the presence of 0.5 μg poly(dI-dC) and 50 mM KCl in buffer B (25 mM HEPES [pH 7.5], 0.1 mM EDTA, 5 mM dithiothreitol, 10% glycerol). The reaction mixtures were incubated at 25°C for 15 min and then immediately loaded onto an 8% polyacrylamide-bisacrylamide (37.5:1) gel for electrophoresis.
FIG. 1.
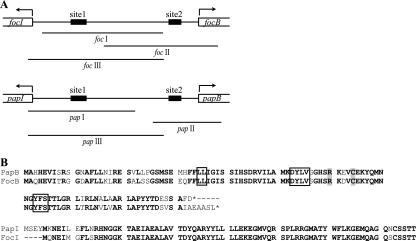
Regulatory regions of the pap and foc operons. (A) Schematic diagram of the foc and pap regulatory regions, showing binding sites 1 and 2 for the regulatory proteins FocB and PapB. Sequences corresponding to PCR fragments used in the experiments are designated foc I to foc III and pap I to pap III, respectively. (B) Amino acid sequence comparison of the FocB and PapB proteins and the PapI and FocI proteins in E. coli J96. Conserved amino acids in PapB and FocB important for oligomerization and DNA binding are enclosed in boxes and are shaded, respectively (55).
Cross-linking.
In vitro cross-linking with purified proteins was carried out as described by Ueguchi et al. (48). About 500 ng of purified wild-type PapB (53) and/or His-FocB was incubated at room temperature in 200 μl of the cross-linking buffer with or without dimethyl suberimidate (DMS) (1 mg/ml). Proteins were precipitated from the reaction mixtures with trichloroacetic acid and then analyzed by SDS-PAGE. Protein bands were detected by immunoblotting with purified polyclonal anti-PapB antiserum.
Bacterial samples for cross-linking of protein complexes formed in vivo were obtained by growing JM109/pYN89 and JM109/pYN89+pYN21 expressing His-FocB and His-FocB plus wild-type PapB, respectively, from the plasmid clones. Protein expression was induced by addition of IPTG (final concentration, 0.1 mM) at the logarithmic phase. The bacteria were harvested by centrifugation, and after washing the cells were frozen, thawed, and resuspended in a buffer (500 mM NaCl, 20 mM Tris-HCl, 1 mM imidazole). The cells were lysed by treatment with glass beads in a Mini-Beadbeater (BioSpec Products, Inc.), and the lysate obtained was used in the cross-linking reaction. The lysates of the two bacterial strains were incubated with or without 25 μM glutaraldehyde for 5 min at 37°C. The reaction was terminated by adding 1 M Tris-HCl (pH 8.0). The results were analyzed by SDS-PAGE and immunoblotting using anti-His tag antibodies (Qiagen) as described above.
AFM imaging.
For atomic force microscopy (AFM) imaging of DNA-protein complexes, the samples were prepared essentially as described previously (53). Imaging of bacteria and fimbriae was done using whole-cell samples prepared from overnight cultures grown on L agar plates at 37°C. The cells were resuspended in 50 μl MilliQ water, and 5 μl was placed on mica, air dried, and desiccated for 1 h. The samples were imaged in air using a Nanoscope IIIa AFM (Digital Instruments) in tapping mode. The scan parameters were as follows: scan rate, 1.001 Hz; drive frequency, 318 kHz; and scan size, 10 μm. Images were presented in amplitude mode. Several regions of each sample were scanned to confirm consistency.
Nucleotide sequence accession number.
Data resulting from the DNA sequence analysis of the regulatory region, including the focB and focI genes, have been deposited in the EMBL nucleotide sequence database under accession no. AM887937.
RESULTS AND DISCUSSION
Analysis of the regulatory region in the F1C pilus determinant from strain J96.
In a cosmid clone bank of E. coli uropathogenic isolate J96, a clone (pBSN50) including the foc gene cluster was identified. By performing a DNA sequence analysis of a subclone of this F1C pilus determinant from J96, we localized the regulatory region, which includes genes and sequences corresponding to the papB-papI region in the pap gene cluster (Fig. 1A). We identified a papB homologous gene that could encode a 109-amino-acid protein (which we designated FocB) that shared 81% identity and 88% similarity with PapB (Fig. 1B). The C-terminal part of the putative protein contains five additional amino acids compared with PapB. The gene was designated focB in accordance with the nomenclature for the other F1C fimbria genes characterized by Riegman et al. (41). The coding sequence of the papI homologous gene showed 88% identity at the protein level to PapI, and the protein was designated FocI (Fig. 1B).
PapB is a transcriptional regulator of the pap operon (3, 13, 53, 54). We utilized a series of papB mutant clones in order to test the effect of PapB on expression of the foc operon in vivo. E. coli K-12 strain HB101 harboring the foc+ cosmid pBSN50 was transformed with plasmids expressing either the wild-type (pYN21) or mutant papB allele (55). The mutant PapB variants were defective either in DNA binding (pYN22 and pYN24) or in protein oligomerization (pYN41 and pYN46). The expression of the major foc fimbrial subunit protein FocA was determined by Western blotting. Introduction of wild-type PapB or the DNA binding-deficient PapB protein resulted in complete repression of FocA (Fig. 2A, lanes 2 to 4). The dominant negative feature of these mutants (pYN22 and pYN24) suggested that there was an interaction between the PapB and FocB proteins. Introduction of the oligomerization-deficient PapB mutant constructs (pYN41 and pYN46) did not cause strong repression, but there was only a rather modest effect on the expression of FocA (Fig. 2A, lanes 5 and 6). The difference in effects of the pYN41 and pYN46 derivatives was presumed to be due to the relatively high levels of the PapB mutant protein expressed from pYN41, assuming that it retained weak oligomerization activity. In conclusion, the repression by DNA binding mutant PapB indicated that there might be formation of heterooligomeric complexes of PapB and FocB and that PapB thereby titrated out the activating FocB oligomers. Also, oligomerization-defective PapB had a limited effect on the expression of FocA. To further determine how the fimbriation of the bacteria was affected by PapB, bacterial cells were examined by AFM. Clear fimbriation was observed in strain HB101/pBSN50 carrying the foc gene cluster in the absence of PapB (Fig. 2B, panel 1), whereas there was a lack of fimbriae on the bacteria expressing either wild-type PapB (Fig. 2B, panel 2) or DNA binding-defective PapB (Fig. 2B, panel 3) in trans. We concluded from these results that the pap and foc regulatory proteins may interact and that, in particular, the PapB protein can repress foc operon expression. Figure 2C shows a summary of how the action in the focB regulatory region of the FocB and PapB proteins may be envisioned.
FIG. 2.
Effect of PapB on expression of foc. (A) Western blot analysis of expression of PapB and the FocA fimbrial subunit proteins in strain HB101 harboring cosmid pBSN50 encoding the whole foc determinant and plasmids expressing wild-type (wt) (lane 2), DNA binding-negative (lanes 3 to 4), and oligomerization-negative (lanes 5 to 6) PapB protein variants. (B) AFM micrographs showing F1C fimbriation of HB101/pBSN50 cells (left panel) and the lack of fimbriation in the same strain overexpressing wild-type PapB (middle panel) or DNA binding-deficient PapB (right panel). The scanned area was 10 by 10 μm. (C) Schematic diagram of how the FocB and PapB proteins could act in the focB regulatory region to affect foc expression. Open ellipses represent the FocB protein, and filled ellipses represent the PapB protein. Filled ellipses with a black dot and filled ellipses with a triangle represent DNA binding mutant and oligomerization-deficient PapB, respectively. Open triangles represent the FocA protein.
DNA binding properties of the FocB and PapB proteins.
Purified FocB and PapB proteins were obtained (see Materials and Methods) in order to study how they might interact with the regulatory DNA sequences of the pap and foc operons. We used DNA fragments corresponding to the region (Fig. 1A, fragments foc III and pap III) where PapB has been shown to bind to a 52-bp sequence in the pap DNA (53). The results of gel mobility shift assays indicated that FocB could bind to DNA from both the foc and the pap operons (Fig. 3A). The foc DNA was shifted at a somewhat lower FocB concentration, indicating that the binding was more efficient than the binding of the pap DNA (1.6 μM compared to 3.4 μM) (Fig. 3A, lanes 4 and 11). A DNA fragment corresponding to fragment pap III, but with a 51-bp deletion of site 1, was used as a negative control for the specificity of the FocB binding to the pap DNA (Fig. 3A, lanes 13 to 16). Only at the highest protein concentration was there some binding to this DNA, but no distinct bands were observed (lane 16). FocB binding to the foc operon was also visualized at single-molecule level by AFM imaging. Measurement of the length of the DNA (in nm) and the position of the protein binding on the DNA (Fig. 3B and C) indicated that the FocB protein bound to the DNA in an oligomeric fashion. Footprint analysis of the pap regulatory region showed protection from DNase I cleavage by FocB both at site 1 and in the predicted site 2 region (data not shown).
FIG. 3.
In vitro DNA binding by FocB. (A) Gel mobility shift assays of purified FocB protein with DNA fragments foc III (left panel) and pap III (middle panel). The panel on the right shows the results of tests of FocB binding to a DNA fragment corresponding to fragment pap III, in which the 51-bp region including site 1 had been deleted. The concentration of FocB in each reaction mixture is indicated at the top. (B) Representative AFM micrographs showing binding of FocB to DNA fragment foc I (panel a, only DNA; panels b to d, DNA incubated with 3.2 μM FocB). The area of the DNA that was occupied by the FocB protein is indicated by arrows and bars. The scanned area was 145 by 120 nm. (C) Schematic diagram showing the calculated binding region of FocB for the foc I DNA fragment, based on measurements from AFM images. The estimated length of the 401-bp DNA fragment was 136 ± 8 nm (corresponding to 387 ± 23 bp; n = 15).
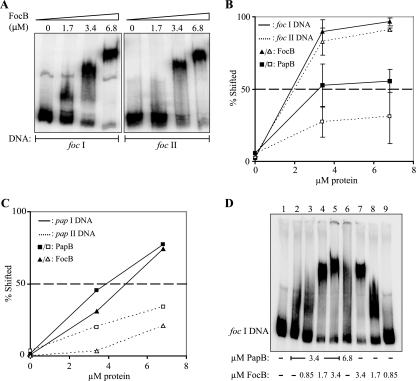
In order to further assess the similarities or differences in the binding capacity of FocB for the different sequences in the regulatory region of the foc operon, we carried out a series of gel shift assays with different DNA fragments. Figure 4A shows the binding of FocB to DNA fragments corresponding to the upstream regulatory region of the foc gene cluster and including binding site 1 (fragment foc I) or site 2 (fragment foc II). With the gradually increasing amounts of FocB protein added, the DNA appeared increasingly in the form of higher-order complexes (Fig. 4A, left panel). For example, the bands seen with 1.7 μM presumably represent faster-migrating complexes in which the protein occupied only part of the site 1 DNA region. The bands seen with the highest concentration represent oligomer complexes with fully occupied site 1 DNA. The results were consistent with the findings obtained by single-molecule analysis by AFM (full occupation of site 1 sequences at a FocB concentration of approximately 3 μM) (Fig. 3B). We carried out similar studies with the corresponding DNA fragments from both the pap gene cluster (fragments pap I and pap II [Fig. 1A]) and the foc gene cluster (fragments foc I and foc II [Fig. 1A]) and with both the FocB and PapB proteins in each case. Our estimates of the binding capacities of the two proteins for the foc DNA, as quantified by monitoring the percentage of DNA shifted, are shown in Fig. 4B. FocB showed stronger binding than PapB to both the foc I and the foc II DNA targets. The 50% levels of shifted DNA were obtained at approximately 1.8 μM FocB protein. With PapB, a 50% level of shifted fragment foc I DNA required 3.4 μM (Fig. 4B). The results of quantification of the binding capacities of PapB and FocB for the pap DNA are shown in Fig. 4C. Higher concentrations of both PapB and FocB were required for shifts of the pap I DNA, and a 50% shift was obtained with approximately 4 μM (Fig. 4C).
FIG. 4.
In vitro DNA binding of FocB and PapB to foc and pap DNA. (A) Gel mobility shift assay of purified FocB protein with DNA fragments foc I and foc II (see Fig. 1A). The concentration of FocB used in each reaction mixture is indicated at the top. (B) Graph showing the gel shift of fragments foc I (solid lines) and foc II (dotted lines) in response to different concentrations of FocB (triangles) or PapB (squares). The 50% level of shifted DNA is indicated by a dashed line. A 50% level of shifted foc II DNA required up to 10 μM PapB (data not shown). (C) Graph showing the gel shift of fragments pap I (solid lines) and pap II (dotted lines) in response to different concentrations of PapB (squares) or FocB (triangles). The 50% level of shifted DNA is indicated by a dashed line. PapB gave a 50% shift of the fragment pap II DNA at 12 μM, whereas a 50% shift required >15 μM FocB (data not shown). (D) Gel mobility shift assay analysis of a combination of purified PapB and FocB at different concentrations with DNA fragment foc I. The concentration of FocB increased from 0.8 to 3.4 μM in lanes 3 to 5 and decreased from 3.4 to 0.8 μM in lanes 7 to 9. The concentrations of PapB were 3.4 μM in lanes 2 to 5 and 6.8 μM in lane 6.
The indication from in vivo tests described above that FocB and PapB may interact for regulation of foc operon expression (Fig. 2) raised the question of whether heterooligomeric complexes have DNA binding properties different from those of the corresponding homooligomers. A series of gel mobility shift assays using different combinations of the PapB and FocB proteins was therefore carried out with fragment foc I DNA (Fig. 4D). We found that mixing of PapB (3.4 μM) and FocB (1.7 μM) resulted in a distinct shift (Fig. 4D, lane 4). In contrast, considerably less-distinct shifts were observed when either 3.4 or 6.8 μM PapB alone was added (lanes 2 and 6). Thus, addition of the FocB protein appeared to increase the PapB binding to the foc DNA. These results indicate that heteromeric PapB/FocB complexes may be formed and alter the DNA interactions.
Formation of oligomeric and heteromeric PapB and FocB complexes.
To test the in vivo dimerization/oligomerization capability of FocB, we constructed plasmid pYN108 encoding a chimeric protein consisting of the N-terminal DNA binding domain of the λ cI repressor and the wild-type FocB molecule. Plasmid pYN108 was introduced by transformation into strain JH607 (= λ 112OsPs), which can function as a reporter for cooperative binding by cI repressor proteins to two adjacent operators. Thus, cooperative binding to both operator sites increases the efficiency of repression for the downstream reporter genes cat and lacZ. By exchanging the C-terminal domain of the λ cI repressor, which confers the dimerization ability of the protein, for the FocB protein, the ability of FocB to dimerize could be monitored. The FocB protein could replace to a large extent the dimerization function of the C-terminal domain of the cI repressor, and the resulting chimera substantially repressed expression of the two reporter genes. The FocB-cI chimera (pYN108) reduced the β-galactosidase activity to less than one-half the control activity, and it reduced expression of chloramphenicol acetyltransferase to a level that resulted in clearly enhanced chloramphenicol sensitivity (Table 3). These results strongly support the suggestion that FocB can form dimers and oligomers in vivo.
TABLE 3.
In vivo test of cooperative DNA binding
| Plasmid | Proteina | Cm sensitivity (μg/ml)b | β-Galactosidase activityc
|
|
|---|---|---|---|---|
| Miller units | Relative value | |||
| pBF22 | cI (1-159) | 150 | 1,078.0 | 1.0 |
| pBF21 | cI (wild type) | 10 | 65.6 | 0.06 |
| pYN69 | cI (Δ57-152)-PapB (wild type) | 150 | 1,081.2 | 1.0 |
| pYN51 | cI (1-159)-PapB (wild type) | 25 | 322.8 | 0.30 |
| pYN108 | cI (1-159)-FocB (wild type) | 50 | 506.6 | 0.47 |
The proteins that were tested included the wild-type and the N-terminal DNA binding domain of λ cI repressor, chimeric proteins composed of the DNA binding domain of cI, and wild-type PapB and FocB.
Chloramphenicol sensitivity was tested as described in the text. The values are the concentrations at which the plating efficiency of the cells fell below 50%.
β-Galactosidase activity was measured as described by Miller (31). The value for strain JH607/pBF22, which produced the N-terminal DNA binding domain of the cI repressor, was defined as 1.0 for comparison.
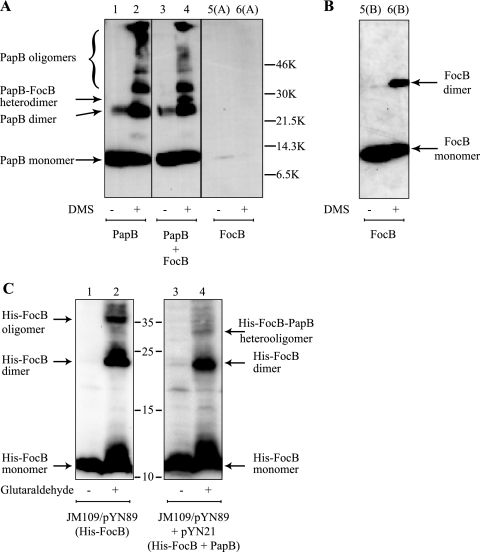
Next, we tested if we could detect a direct interaction between the FocB and PapB proteins. Purified wild-type PapB and FocB were incubated separately or together in the absence or presence of the cross-linker DMS. We used an anti-PapB antiserum and immunoblot analysis for detection of protein complexes. The in vitro cross-linking assay showed that PapB could form dimers and even larger oligomers in the presence of DMS (Fig. 5A), which is consistent with our previous data (55). Furthermore, when wild-type PapB and FocB were incubated together with DMS, one extra band was observed at a molecular size larger than that of the PapB dimer but somewhat smaller than that of the FocB dimer (Fig. 5A, lane 4). Our explanation for this band is that it represents heterodimers formed between wild-type PapB and FocB. While the anti-PapB polyclonal antiserum readily allowed detection of PapB, it was rather poor at recognizing the FocB protein. The band corresponding to the FocB dimer was detected only after prolonged exposure of the ECL developed blot [Fig. 5B, lanes 5(B) and 6(B)]. Furthermore, we performed cross-linking experiments with whole-cell lysates of strains JM109/pYN89 and JM109/pYN89+pYN21 expressing His-FocB and His-FocB plus wild-type PapB from the plasmids. For this purpose we used glutaraldehyde as a cross-linking agent, and the His-tagged protein was detected by anti-His immunoblot analysis; the results are shown in Fig. 5C. With the strain expressing only the His-FocB protein there were bands after cross-linking that represented both dimers and larger oligomers (Fig. 5C, lane 2). Similarly, in the sample from the strain expressing both the His-FocB and the PapB proteins we observed, after cross-linking, bands representing the His-FocB dimers and oligomers. In addition, there was a band representing a protein complex with a somewhat smaller molecular size than the His-FocB oligomer, and we suggest that this band corresponds to a heteromeric complex of His-FocB and PapB (Fig. 5C, lane 4). In the presence of the PapB protein, the His-FocB oligomer forms appeared to be a little less abundant in this analysis. This could have been because these complexes were a little less abundant or because they were less clearly detectable by the anti-His antibody. One possible explanation for the relatively weak band observed for the heteromers is that the His tag, to which the antibody was directed, was somehow concealed by the PapB protein when the two proteins formed heteromers. Taken together, the results suggest that FocB could function like PapB as an oligomer in DNA binding, affecting transcriptional regulation of pilus biogenesis, and that these two proteins could interact with each other and form heterooligomers.
FIG. 5.
Cross-linking of PapB and FocB proteins. (A) Chemical cross-linking with DMS of wild-type PapB and His-FocB. Protein bands were detected by immunoblotting with anti-PapB antisera. The presumed positions of the different PapB and FocB derivatives are indicated, as are the positions of molecular size markers. (B) Detection of FocB protein in lanes 5(A) and 6(A) after prolonged exposure. (C) Cross-linking with glutaraldehyde and detection of protein complexes formed in vivo by His-FocB and PapB in strain JM109. The protein bands were detected by immunoblotting using anti-His tag antiserum. The positions of molecular mass markers (in kDa) and the different forms of His-FocB are indicated.
Effect of PapB on FocA expression in clinical isolate J96.
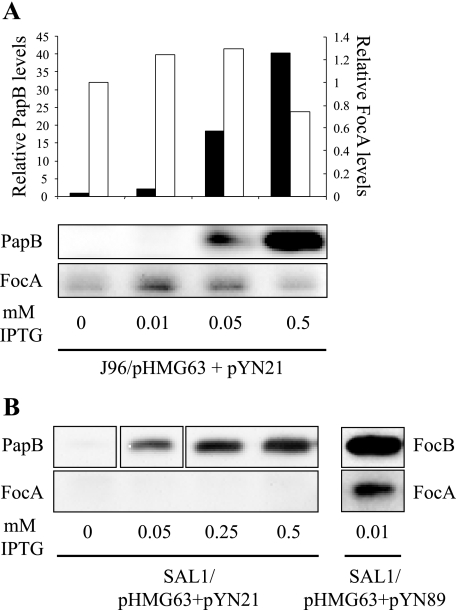
In order to further assess the importance of PapB/FocB heteromer formation in vivo, a plasmid containing a wild-type copy of papB under control of a tac promoter (pYN21) was introduced by transformation into clinical isolate J96 together with a plasmid encoding a lacIq repressor (pHMG63). Transcription of papB was induced gradually by addition of IPTG, and the expression of PapB and FocA was monitored by Western immunoblotting. In wild-type strain J96, an increase in the amount of PapB expression caused repression of FocA (Fig. 6A). Subsequently, the same experimental setup was used with a focB deletion mutant of J96 (strain SAL1). This strain was negative for FocA expression, but the phenotype could be complemented by introducing a copy of focB in trans (Fig. 6B). As anticipated and in accordance with our other results, PapB was not able to complement and restore expression of FocA in strain SAL1, thus leaving focA completely silent (Fig. 6B). The in vivo experiments carried out with clinical isolate J96 and its isogenic ΔfocB variant SAL1 further substantiated the role of PapB/FocB heteromer formation in regulation of the foc operon. When levels of PapB expression in J96 increased, the expression of FocA decreased, suggesting that PapB sequesters FocB.
FIG. 6.
In vivo effect of PapB on expression of FocA in J96. (A) Western blot analysis of expression of FocA and PapB in J96 transformed with IPTG-inducible plasmid pYN21 harboring papB and pHMG63 encoding the lacIq repressor. The concentrations of IPTG used for induction are indicated at the bottom. (B) Western blot analysis of the effect of induced levels of PapB or FocB on FocA expression as monitored with strains SAL1/pHMG63+pYN21 and SAL1/pHMG63/pYN89.
The in vitro studies of the DNA binding of PapB to the foc regulatory region showed that PapB was less efficient than FocB in binding to the same region (Fig. 4B), and there was apparently better binding to the foc DNA by the combination of PapB and FocB than by PapB alone (Fig. 4D). Our results suggest that PapB could act as a repressor of the foc operon both by sequestering FocB and by PapB/FocB heteromeric binding to the regulatory region and thereby blocking transcription. This is also consistent with the finding that PapB was unable to transcomplement the ΔfocB mutant derivative of J96 (Fig. 6B).
Effects of FocB on pap and fim expression.
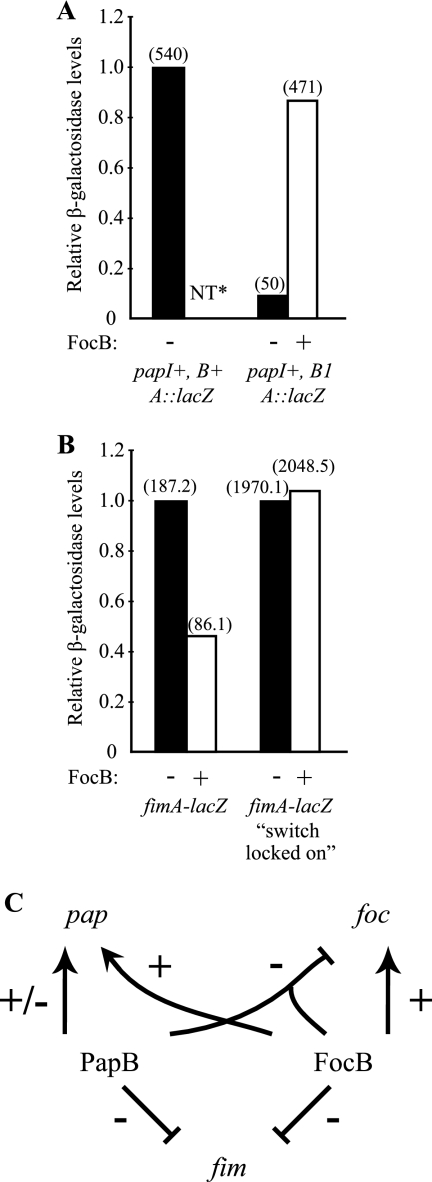
We used a strain containing the papI+ papB1 papA-lacZ fusion plasmid pHMG15 to test if the focB gene in trans could influence pap expression and substitute for papB. In this derivative the papB gene was disrupted by insertional mutation, and the level of expression of pHMG15 was 10-fold lower than that of a similar construct (pHMG1) carrying an intact papB+ gene (Fig. 7A). When the focB+ plasmid pBB1 was present in trans together with pHMG15, the activity was restored to 90% of the activity obtained with the strain carrying the papB+ derivative pHMG1. The results indicated that the focB+ gene could complement a mutation in the papB gene to restore papA transcription.
FIG. 7.
Effect of FocB on pap and fim expression. The effect of FocB on pap and fim expression was examined by analyzing expression of plasmid pBB1 (focB+) or the vector control pACYC184 in different papA-lacZ (A) or fimA-lacZ (B) fusion strains. β-Galactosidase activity was measured as described by Miller (31). The relative β-galactosidase activity level obtained with the coresident pACYC184 vector control was defined as 1.0 for each strain. The absolute values are indicated in parentheses. NT, not tested. (C) Summary of the observed regulatory cross talk between the pap, foc, and fim operons in uropathogenic isolate J96.
In addition to the foc and pap determinants strain J96 carries the gene cluster (fim) for type 1 fimbriae, and previous studies showed that PapB can reduce the expression of this cluster (16, 17, 54). Therefore, the effect of FocB on fim expression was also studied, and plasmid pBB1 was introduced into two fimA-lacZYA transcriptional fusion strains, AAEC198A (fimB+E+) and AAEC374A (fimBE). No effect of FocB was detected in the case of phase “locked-on” strain AAEC374A. However, in strain AAEC198A, the β-galactosidase level was reduced to less than one-half the control level (46%) when the focB clone was present (Fig. 7B). This result indicated that focB altered fim phase switching frequencies, and the findings are similar to those obtained with PapB (54). We concluded that FocB can inhibit type 1 fimbria expression by influencing phase switching, and our results suggest that FocB can play a role similar to that of PapB in the regulatory cross talk with the fim determinant.
In summary, the in vitro and in vivo data suggest that there is both a kind of hierarchy and homeostasis in the regulatory cross talk of the pap, foc, and fim operons (Fig. 7C). Whereas FocB alone could activate pap expression, there was no stimulation of foc expression by PapB in the absence of FocB. In the presence of FocB there was a PapB effect on foc expression, presumably due to the formation of heterooligomeric PapB/FocB complexes, eventually resulting in reduced levels of foc expression. Either PapB or FocB appeared to cause reduced expression of the genes for type 1 fimbriae. These findings indicate that there is intricate cross talk between different adhesin gene clusters and that the different PapB-like regulators may form heterodimeric/oligomeric complexes involved in the coordination of fimbrial biogenesis.
Acknowledgments
We thank Kristina Forsman-Semb for her contribution to the early phase of this work, Nicola Holden for critically reading the manuscript, and Monica Persson for skillful technical assistance.
This work was supported by grants from the Swedish Research Council, the Göran Gustafsson Foundation for Research in Natural Science and Medicine, the Swedish Foundation for International Cooperation in Research and Higher Education (STINT), the International Graduate College (IGK 587/2), and the EU FP6 EuroPathoGenomics Network of Excellence and was performed in part at the Umeå Centre for Microbial Research (UCMR).
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Abraham, S. N., J. D. Goguen, D. Sun, P. Klemm, and E. H. Beachey. 1987. Identification of two ancillary subunits of Escherichia coli type 1 fimbriae by using antibodies against synthetic oligopeptides of fim gene products. J. Bacteriol. 1695530-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed, F., B. Alsen, N. Roche, J. Angstrom, A. von Euler, M. E. Breimer, B. Westerlund-Wikstrom, S. Teneberg, and A. Richter-Dahlfors. 2002. Identification of target tissue glycosphingolipid receptors for uropathogenic, F1C-fimbriated Escherichia coli and its role in mucosal inflammation. J. Biol. Chem. 27718198-18205. [DOI] [PubMed] [Google Scholar]
- 3.Baga, M., M. Goransson, S. Normark, and B. E. Uhlin. 1985. Transcriptional activation of a pap pilus virulence operon from uropathogenic Escherichia coli. EMBO J. 43887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckett, D., D. S. Burz, G. K. Ackers, and R. T. Sauer. 1993. Isolation of lambda repressor mutants with defects in cooperative operator binding. Biochemistry 329073-9079. [DOI] [PubMed] [Google Scholar]
- 5.Blomfield, I. C., P. J. Calie, K. J. Eberhardt, M. S. McClain, and B. I. Eisenstein. 1993. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 17527-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41459-472. [DOI] [PubMed] [Google Scholar]
- 7.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1341141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dagberg, B., and B. E. Uhlin. 1992. Regulation of virulence-associated plasmid genes in enteroinvasive Escherichia coli. J. Bacteriol. 1747606-7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duguid, J. P., and D. C Old. 1980. Bacterial adherence. Chapman & Hall, London, United Kingdom.
- 11.Forsman, K., M. Goransson, and B. E. Uhlin. 1989. Autoregulation and multiple DNA interactions by a transcriptional regulatory protein in E. coli pili biogenesis. EMBO J. 81271-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goransson, M., K. Forsman, and B. E. Uhlin. 1988. Functional and structural homology among regulatory cistrons of pili-adhesin determinants in Escherichia coli. Mol. Gen. Genet. 212412-417. [DOI] [PubMed] [Google Scholar]
- 13.Goransson, M., K. Forsman, and B. E. Uhlin. 1989. Regulatory genes in the thermoregulation of Escherichia coli pili gene transcription. Genes Dev. 3123-130. [DOI] [PubMed] [Google Scholar]
- 14.Goransson, M., P. Forsman, P. Nilsson, and B. E. Uhlin. 1989. Upstream activating sequences that are shared by two divergently transcribed operons mediate cAMP-CRP regulation of pilus-adhesin in Escherichia coli. Mol. Microbiol. 31557-1565. [DOI] [PubMed] [Google Scholar]
- 15.Goransson, M., and B. E. Uhlin. 1984. Environmental temperature regulates transcription of a virulence pili operon in E. coli. EMBO J. 32885-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holden, N. J., M. Totsika, E. Mahler, A. J. Roe, K. Catherwood, K. Lindner, U. Dobrindt, and D. L. Gally. 2006. Demonstration of regulatory cross-talk between P fimbriae and type 1 fimbriae in uropathogenic Escherichia coli. Microbiology 1521143-1153. [DOI] [PubMed] [Google Scholar]
- 17.Holden, N. J., B. E. Uhlin, and D. L. Gally. 2001. PapB paralogues and their effect on the phase variation of type 1 fimbriae in Escherichia coli. Mol. Microbiol. 42319-330. [DOI] [PubMed] [Google Scholar]
- 18.Hull, R. A., R. E. Gill, P. Hsu, B. H. Minshew, and S. Falkow. 1981. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun. 33933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan, A. S., B. Kniep, T. A. Oelschlaeger, I. Van Die, T. Korhonen, and J. Hacker. 2000. Receptor structure for F1C fimbriae of uropathogenic Escherichia coli. Infect. Immun. 683541-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klemm, P., and G. Christiansen. 1987. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol. Gen. Genet. 208439-445. [DOI] [PubMed] [Google Scholar]
- 21.Klemm, P., G. Christiansen, B. Kreft, R. Marre, and H. Bergmans. 1994. Reciprocal exchange of minor components of type 1 and F1C fimbriae results in hybrid organelles with changed receptor specificities. J. Bacteriol. 1762227-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klemm, P., B. J. Jorgensen, B. Kreft, and G. Christiansen. 1995. The export systems of type 1 and F1C fimbriae are interchangeable but work in parental pairs. J. Bacteriol. 177621-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klemm, P., I. Orskov, and F. Orskov. 1982. F7 and type 1-like fimbriae from three Escherichia coli strains isolated from urinary tract infections: protein chemical and immunological aspects. Infect. Immun. 36462-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudsen, T. B., and P. Klemm. 1998. Probing the receptor recognition site of the FimH adhesin by fimbriae-displayed FimH-FocH hybrids. Microbiology 1441919-1929. [DOI] [PubMed] [Google Scholar]
- 25.Korhonen, T. K., V. Vaisanen-Rhen, M. Rhen, A. Pere, J. Parkkinen, and J. Finne. 1984. Escherichia coli fimbriae recognizing sialyl galactosides. J. Bacteriol. 159762-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korhonen, T. K., V. Vaisanen, H. Saxen, H. Hultberg, and S. B. Svenson. 1982. P-antigen-recognizing fimbriae from human uropathogenic Escherichia coli strains. Infect. Immun. 37286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korhonen, T. K., M. V. Valtonen, J. Parkkinen, V. Vaisanen-Rhen, J. Finne, F. Orskov, I. Orskov, S. B. Svenson, and P. H. Makela. 1985. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindberg, F., B. Lund, L. Johansson, and S. Normark. 1987. Localization of the receptor-binding protein adhesin at the tip of the bacterial pilus. Nature 32884-87. [DOI] [PubMed] [Google Scholar]
- 29.Lindberg, F., B. Lund, and S. Normark. 1986. Gene products specifying adhesion of uropathogenic Escherichia coli are minor components of pili. Proc. Natl. Acad. Sci. USA 831891-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lund, B., B. I. Marklund, N. Stromberg, F. Lindberg, K. A. Karlsson, and S. Normark. 1988. Uropathogenic Escherichia coli can express serologically identical pili of different receptor binding specificities. Mol. Microbiol. 2255-263. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Moch, T., H. Hoschutzky, J. Hacker, K. D. Kroncke, and K. Jann. 1987. Isolation and characterization of the alpha-sialyl-beta-2,3-galactosyl-specific adhesin from fimbriated Escherichia coli. Proc. Natl. Acad. Sci. USA 843462-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowicki, B., H. Holthofer, T. Saraneva, M. Rhen, V. Vaisanen-Rhen, and T. K. Korhonen. 1986. Location of adhesion sites for P-fimbriated and for 075X-positive Escherichia coli in the human kidney. Microb. Pathog. 1169-180. [DOI] [PubMed] [Google Scholar]
- 35.Orskov, I., and F. Orskov. 1983. Serology of Escherichia coli fimbriae. Prog. Allergy 3380-105. [PubMed] [Google Scholar]
- 36.Orskov, I., F. Orskov, and A. Birch-Andersen. 1980. Comparison of Escherichia coli fimbrial antigen F7 with type 1 fimbriae. Infect. Immun. 27657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott, M., H. Hoschutzky, K. Jann, I. Van Die, and J. Hacker. 1988. Gene clusters for S fimbrial adhesin (sfa) and F1C fimbriae (foc) of Escherichia coli: comparative aspects of structure and function. J. Bacteriol. 1703983-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott, M., T. Schmoll, W. Goebel, I. Van Die, and J. Hacker. 1987. Comparison of the genetic determinant coding for the S-fimbrial adhesin (sfa) of Escherichia coli to other chromosomally encoded fimbrial determinants. Infect. Immun. 551940-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawelzik, M., J. Heesemann, J. Hacker, and W. Opferkuch. 1988. Cloning and characterization of a new type of fimbria (S/F1C-related fimbria) expressed by an Escherichia coli O75:K1:H7 blood culture isolate. Infect. Immun. 562918-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pere, A., M. Leinonen, V. Vaisanen-Rhen, M. Rhen, and T. K. Korhonen. 1985. Occurrence of type-1C fimbriae on Escherichia coli strains isolated from human extraintestinal infections. J. Gen. Microbiol. 1311705-1711. [DOI] [PubMed] [Google Scholar]
- 41.Riegman, N., R. Kusters, H. Van Veggel, H. Bergmans, P. Van Bergen en Henegouwen, J. Hacker, and I. Van Die. 1990. F1C fimbriae of a uropathogenic Escherichia coli strain: genetic and functional organization of the foc gene cluster and identification of minor subunits. J. Bacteriol. 1721114-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riegman, N., I. van Die, J. Leunissen, W. Hoekstra, and H. Bergmans. 1988. Biogenesis of F7(1) and F7(2) fimbriae of uropathogenic Escherichia coli: influence of the FsoF and FstFG proteins and localization of the Fso/FstE protein. Mol. Microbiol. 273-80. [PubMed] [Google Scholar]
- 43.Salit, I. E., and E. C. Gotschlich. 1977. Type I Escherichia coli pili: characterization of binding to monkey kidney cells. J. Exp. Med. 1461182-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmoll, T., H. Hoschutzky, J. Morschhauser, F. Lottspeich, K. Jann, and J. Hacker. 1989. Analysis of genes coding for the sialic acid-binding adhesin and two other minor fimbrial subunits of the S-fimbrial adhesin determinant of Escherichia coli. Mol. Microbiol. 31735-1744. [DOI] [PubMed] [Google Scholar]
- 45.Snyder, J. A., B. J. Haugen, C. V. Lockatell, N. Maroncle, E. C. Hagan, D. E. Johnson, R. A. Welch, and H. L. Mobley. 2005. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 737588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spurio, R., M. Falconi, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 161795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taraseviciene, L., S. Naureckiene, and B. E. Uhlin. 1994. Immunoaffinity purification of the Escherichia coli rne gene product. Evidence that the rne gene encodes the processing endoribonuclease RNase E. J. Biol. Chem. 26912167-12172. [PubMed] [Google Scholar]
- 48.Ueguchi, C., C. Seto, T. Suzuki, and T. Mizuno. 1997. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 274145-151. [DOI] [PubMed] [Google Scholar]
- 49.Vaisanen-Rhen, V., J. Elo, E. Vaisanen, A. Siitonen, I. Orskov, F. Orskov, S. B. Svenson, P. H. Makela, and T. K. Korhonen. 1984. P-fimbriated clones among uropathogenic Escherichia coli strains. Infect. Immun. 43149-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Die, I., B. van Geffen, W. Hoekstra, and H. Bergmans. 1985. Type 1C fimbriae of a uropathogenic Escherichia coli strain: cloning and characterization of the genes involved in the expression of the 1C antigen and nucleotide sequence of the subunit gene. Gene 34187-196. [DOI] [PubMed] [Google Scholar]
- 51.Virkola, R., B. Westerlund, H. Holthofer, J. Parkkinen, M. Kekomaki, and T. K. Korhonen. 1988. Binding characteristics of Escherichia coli adhesins in human urinary bladder. Infect. Immun. 562615-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia, Y., K. Forsman, J. Jass, and B. E. Uhlin. 1998. Oligomeric interaction of the PapB transcriptional regulator with the upstream activating region of pili adhesin gene promoters in Escherichia coli. Mol. Microbiol. 30513-523. [DOI] [PubMed] [Google Scholar]
- 54.Xia, Y., D. Gally, K. Forsman-Semb, and B. E. Uhlin. 2000. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J. 191450-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia, Y., and B. E. Uhlin. 1999. Mutational analysis of the PapB transcriptional regulator in Escherichia coli. Regions important for DNA binding and oligomerization. J. Biol. Chem. 27419723-19730. [DOI] [PubMed] [Google Scholar]
- 56.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 57.Zeng, X., and J. C. Hu. 1997. Detection of tetramerization domains in vivo by cooperative DNA binding to tandem lambda operator sites. Gene 185245-249. [DOI] [PubMed] [Google Scholar]