Abstract
Carbon dioxide occupies a central position in the physiology of Helicobacter pylori owing to its capnophilic nature, the large amounts of carbon dioxide produced by urease-mediated urea hydrolysis, and the constant bicarbonate supply in the stomach. Carbonic anhydrases (CA) catalyze the interconversion of carbon dioxide and bicarbonate and are involved in functions such as CO2 transport or trapping and pH homeostasis. H. pylori encodes a periplasmic α-CA (α-CA-HP) and a cytoplasmic β-CA (β-CA-HP). Single CA inactivation and double CA inactivation were obtained for five genetic backgrounds, indicating that H. pylori CA are not essential for growth in vitro. Bicarbonate-carbon dioxide exchange rates were measured by nuclear magnetic resonance spectroscopy using lysates of parental strains and CA mutants. Only the mutants defective in the α-CA-HP enzyme showed strongly reduced exchange rates. In H. pylori, urease activity is essential for acid resistance in the gastric environment. Urease activity measured using crude cell extracts was not modified by the absence of CA. With intact CA mutant cells incubated in acidic conditions (pH 2.2) in the presence of urea there was a delay in the increase in the pH of the incubation medium, a phenotype most pronounced in the absence of H. pylori α-CA. This correlated with a delay in acid activation of the urease as measured by slower ammonia production in whole cells. The role of CA in vivo was examined using the mouse model of infection with two mouse-adapted H. pylori strains, SS1 and X47-2AL. Compared to colonization by the wild-type strain, colonization by X47-2AL single and double CA mutants was strongly reduced. Colonization by SS1 CA mutants was not significantly different from colonization by wild-type strain SS1. However, when mice were infected by SS1 Δ(β-CA-HP) or by a SS1 double CA mutant, the inflammation scores of the mouse gastric mucosa were strongly reduced. In conclusion, CA contribute to the urease-dependent response to acidity of H. pylori and are required for high-grade inflammation and efficient colonization by some strains.
Carbonic anhydrases (CA) (carbonate dehydratase; EC 4.2.1.1) are zinc-containing metalloenzymes that catalyze the interconversion of carbon dioxide and bicarbonate: CO2 + H2O ↔ H+ + HCO3−.
CA are present in most living organisms and have been classified in five phylogenetically unrelated categories (30, 57, 64): (i) α-CA, present in mammals and about 30 Eubacteria and not present in Archaea; (ii) β-CA, present in invertebrates, photosynthetic eukaryotes, Eubacteria, and Archaea; (iii) γ-CA, present in Archaea and Eubacteria; (iv) δ-CA of the marine diatom Thalassiosira weissflogii (TWCA1) and the coccolithoporid alga Emiliana huxleyi (58); and, (v) ɛ-CA of chemolithoautotrophic bacteria and some cyanobacteria.
The reactions catalyzed by CA are involved in various biochemical processes (56), including: (i) transport of CO2 or HCO3−, in some cases in antiport with other compounds, such as chloride in mammals (55); (ii) trapping of CO2 or HCO3− to maintain their intracellular levels in order to optimize carboxylation reactions by enzymes, such as pyruvate carboxylase, or CO2 fixation by ribulose 1,5-bisphosphate carboxylase-oxygenase in autotrophic organisms; and (iii) degradation of other substrates, such as degradation of esters by α-CA or degradation of carbamate by some β-CA. In animals, CA play a major role in metabolic and physiological processes, such as acid-base equilibrium (pKa of CO2/HCO3−, 6.1), ion transport, or oncogenesis (15). The roles of CA in eukaryotes have been well documented, but less is known about their function in prokaryotes despite their wide distribution; often two or more different CA are present in a single microbial species. Notably, a predicted β-CA gene (mig5) of Salmonella enterica serovar Typhimurium is up-regulated when this bacterium is in macrophages, and the protein encoded by this gene is involved in adaptation to mouse colonization (65). Escherichia coli and Ralstonia eutropha contain a β-CA encoded by the can gene (previously yadF in E. coli) which is essential for growth at ambient CO2 concentrations under standard conditions (33, 38).
The bacterium Helicobacter pylori colonizes the gastric mucosa of almost one-half the world's population, and its persistence is associated with chronic gastritis, which, depending on environmental factors, can evolve into peptic ulcer disease or neoplasia (23). H. pylori is a microaerophile which grows under atmospheres with low O2 partial pressures and, as a capnophile, requires a CO2-enriched atmosphere (5 to 10% CO2). Thus, carbon dioxide appears to be an important component of the physiology of H. pylori, and it may also have a role in the pathogenicity of this organism since the urease of the bacterium produces large amounts of CO2 by degradation of urea (42). Two predicted CA have been identified by analysis of the sequenced genomes of H. pylori strains 26695 (62) and J99 (3). The open reading frame (ORF) designated HP0004 in strain 26695 encodes a β-CA-type enzyme (β-CA-HP) which shares 30% identity with E. coli CynT (28) and 48.5% identity with its close homologue, the predicted β-CA of Helicobacter hepaticus (60). H. pylori is one of the few bacteria that possesses an α-CA-type enzyme (α-CA-HP), which is encoded by ORF HP1186 in strain 26695. This enzyme shares 24 and 28% identical residues with the α-CA of Neisseria gonorrhoeae (16) and the predicted α-CA of H. hepaticus (60), respectively. Comparison of the published sequences of the α-CA-HP genes of strains 26695 and J99 initially suggested that strain 26695 encoded an inactive truncated form of α-CA. This was contradicted by Chirica et al. (18), who identified two errors in the sequence of the 26695 HP1186 gene. These authors characterized in vitro the enzymatic activity of a recombinant α-CA-HP expressed in E. coli (18). This α-CA-HP possesses a characteristic N-terminal signal peptide sequence predicted to be cleaved between residues 18 and 19 (7) and no “cytoplasmic retention signal” as proposed by Marcus et al. (37), a structure known to be restricted to eukaryotes. While the known prokaryotic α-CA are periplasmic, data on the actual localization of the α-CA in H. pylori are inconsistent. α-CA-HP has been reported to be secreted into the extracellular milieu (8), to be at the cell surface of H. pylori (50), possibly in association with the outer membrane (18), or to be in the periplasm in association with the inner membrane (37). In contrast, β-CA-HP is intracellular (18). Interestingly, Campylobacter jejuni (47) and Wolinella succinogenes (4), which are phylogenetically close to H. pylori, both possess a β-CA and no α-CA. The two CA of H. pylori have attracted considerable interest since these enzymes were inhibited in vitro by several sulfonamide and sulfamate inhibitors and therefore have been proposed to be alternative therapeutic targets for the management of patients infected by drug-resistant H. pylori (44, 45). Strikingly, data showed previously that CA inhibitors enhanced the healing of peptic ulcers (48). The first pilot study testing the CA inhibitor acetazolamide on volunteers with active H. pylori infection did not reveal any significant effect on colonization (52). Despite this interest, no study on the role of the CA of H. pylori during colonization of animal models has been reported until now.
Regarding the function of the CA in H. pylori, participation of these enzymes in the capnophily of this organism has been proposed previously but has never been demonstrated (31). A role for α-CA of H. pylori in acid acclimation has been shown (37), while another group concluded that β-CA, but not α-CA, was involved in acid resistance (59). To investigate the role of H. pylori CA more systematically, we constructed single and double CA mutants in five different H. pylori genetic backgrounds, measured for the first time the contribution of each CA to bicarbonate-carbon dioxide exchange rates in this bacterium, and studied the roles of these enzymes in H. pylori physiology. Finally, H. pylori strains SS1 and X47-2AL, which have been reported to have different gastric tropisms, were used to evaluate the impact of CA deficiency on the capacity of H. pylori to colonize the mouse stomach and to generate inflammation of the gastric mucosa.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain MC1061 (13) grown at 37°C on solid or liquid Luria-Bertani medium (40) was used as a host for preparation of the plasmids employed to transform H. pylori. The antibiotics used for selection of recombinant E. coli strains were kanamycin (20 μg ml−1), spectinomycin (100 μg ml−1), and gentamicin (30 μg ml−1). The H. pylori strains employed in this study were SS1 (36), X47-2AL (22), 26695 (62), J99 (3), and N6 (24), as well as CA mutants of these strains (Table 1). H. pylori was grown on blood agar base 2 (Oxoid) plates supplemented with 10% defibrinated horse blood and with an antibiotic-fungicide mixture consisting of vancomycin (12.5 μg ml−1), polymyxin B (0.31 μg ml−1), trimethoprim (6.25 μg ml−1), and amphotericin B (2.5 μg ml−1). Plates were incubated at 37°C for 24 to 48 h at 37°C under microaerobic conditions (7% O2, 10% CO2; Campigen gas pack; Oxoid). To determine growth kinetics, plate-grown H. pylori strains were inoculated at an initial optical density at 600 nm (OD600) of 0.1 into liquid brain heart infusion (BHI) (Oxoid) or brucella broth (Difco) supplemented with 0.2% β-cyclodextrin or 10% fetal calf serum (FCS) and with the antibiotic-fungicide mixture. When mentioned specifically below, filter-sterilized NaHCO3 was added to the liquid medium at concentrations of 10 to 30 mM, or filter-sterilized NiCl2 was added at a concentration of 250 μM. Growth under moderately acidic conditions was assessed in BHI liquid medium containing 0.2% β-cyclodextrin, whose initial pH was adjusted to 5 with HCl.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Reference |
|---|---|---|
| E. coli MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (Strr) hsdR2(rK− mK+) mcrA mcrB1 | 13 |
| H. pylori strains | ||
| 26695 | Parent wild-type strain | 62 |
| X47-2AL | Parent wild-type strain | 22 |
| SS1 | Parent wild-type strain | 36 |
| N6 | Parent wild-type strain | 24 |
| J99 | Parent wild-type strain | 3 |
| 26695 Δ(β-CA-HP) | 26695 HP0004ΩaphA-3 | This study |
| X47-2AL Δ(β-CA-HP) | X47-2AL HP0004ΩaphA-3 | This study |
| SS1 Δ(β-CA-HP) | SS1 HP0004ΩaphA-3 | This study |
| N6 Δ(β-CA-HP) | N6 HP0004ΩaphA-3 | This study |
| J99 Δ(β-CA-HP) | J99 HP0004ΩaphA-3 | This study |
| 26695 Δ(α-CA-HP) | 26695 HP1186Ωaac(3)-VI | This study |
| X47-2AL Δ(α-CA-HP) | X47-2AL HP1186Ωaac(3)-VI | This study |
| SS1 Δ(α-CA-HP) | SS1 HP1186Ωaac(3)-VI | This study |
| N6 Δ(α-CA-HP) | N6 HP1186Ωaac(3)-VI | This study |
| J99 Δ(α-CA-HP) | J99 HP1186Ωaac(3)-VI | This study |
| 26695 Δ(α+β-CA-HP) | 26695 HP0004ΩaphA-3 HP1186Ωaac(3)-VI | This study |
| X47-2AL Δ(α+β-CA-HP) | X47-2AL HP0004ΩaphA-3 HP1186Ωaac(3)-VI | This study |
| SS1 Δ(α+β-CA-HP) | SS1 HP0004ΩaphA-3 HP1186Ωaac(3)-VI | This study |
| N6 Δ(α+β-CA-HP) | N6 HP0004ΩaphA-3 HP1186Ωaac(3)-VI | This study |
| J99 Δ(α+β-CA-HP) | J99 HP0004ΩaphA-3 HP1186Ωaac(3)-VI | This study |
| Plasmids | ||
| pILL485 | pILL570-1 suicide plasmid containing HP0004ΩaphA-3 | This study |
| pILL486 | pILL570-1 suicide plasmid containing HP1186Ωaac(3)-VI | This study |
Molecular techniques, PCR, and sequencing.
Standard procedures were used for small-scale plasmid preparation, endonuclease digestion, ligation, agarose gel electrophoresis, and elution of DNA fragments from agarose gels (51). Midi or maxi Qiagen columns were employed for large-scale plasmid preparation. A QiaAmp DNA extraction kit (Qiagen) was used to extract chromosomal DNA from H. pylori. PCR was carried out according to the manufacturer's recommendations using a Taq DNA polymerase kit (Amersham). The procedure comprised an initial 2-min denaturation step at 94°C, followed by 30 cycles of 30 s at 94°C, 1 min at 55°C, and 1 min at 72°C. PCR were used to define the genetic organization upstream and downstream of HP0004 and HP1186. The following primer pairs were used: for HP0004, IA5S/H117 and IA3S/H120; and for HP1186, XIIIA11S/H124 and XIIIB1R/H125 (Table 2 and Fig. 1). The expected allelic exchange events in the mutants were checked by performing PCR with primer pairs H119/H17, H116/H50, and H116/H119 for HP0004 and with primer pairs H123/H121 (or H154/H121 for SS1 strains), H126/H122, and H123/H126 for HP1186 (Table 2 and Fig. 1).
TABLE 2.
Oligonucleotides used in this study
| Target gene | Oligonucleotide | Sequence (5′ to 3′)a |
|---|---|---|
| HP0004 | H116 | CCATCGATGAGTGAAAGCGTTTTTAGGAG |
| H117 | GGAATTCCCGCAAGCCCCACAATCGC | |
| H119 | AACTGCAGGAAGTTTTCATGACTTTTCC | |
| H120 | CGGGATCCCCTTACATTGCAAACTGG | |
| IA5S | CUACUACUATTAACAATCCTTTAAAAGCTC | |
| IA3S | CUACUACUATTCCTTTAAAATAAATTTTGG | |
| HP1186 | H123 | CCATCGATCGCTTTAGCGCTTACGGC |
| H124 | CGGGATCCAACACATAGTCATGCCC | |
| H125 | CGGGATCCGTGCATTTCCACGCCCCTATG | |
| H126 | AACTGCAGCTAATCACAAACCATGCCACC | |
| H154 | CGCAGCGGTATTTTTCGATC | |
| XIIIA11S | CUACUACUACTCTTTATGTTTTTTTAAACT | |
| XIIIB1R | CAUCAUCAUAATATGACTATATACACTACA | |
| aphA-3 | H17 | TTTGACTTACTGGGGATCAAGCCTG |
| H50 | CCGGTGATATTCTCATTTTAGCC | |
| aac(3)-IV | H121 | GGCTTTTCGCCATTCGTATTGC |
| H122 | GACGTTGGAGGGGCAAGGTCG |
Restriction sites are underlined.
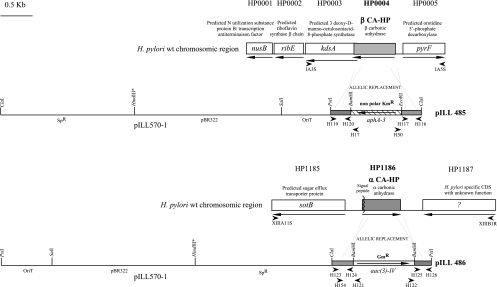
FIG. 1.
Insertional mutations of the HP0004 and HP1186 genes in H. pylori strains. Restriction maps of pILL485 and pILL486 are shown. For each construction, the vector is represented by a thin line. Genes are represented by boxes with arrows indicating the direction of transcription. SpR indicates the gene conferring resistance to spectinomycin. The aac(3)-IV gene confers resistance to gentamicin, and the aphA-3 gene confers resistance to kanamycin. The chromosomal organization of the resulting mutant strains is shown with the adjacent genes. Specific primers, indicated by small arrowheads, were used to verify the constructions and the genetic organization of the HP0004 and HP1186 regions in different H. pylori strains. wt, wild type.
Insertion of the nonpolar kanamycin cassette in HP0004 was verified by sequencing with an ABI 310 automated DNA sequencer (Perkin-Elmer) as previously described (11).
Construction of H. pylori mutants.
Plasmids were constructed to generate the H. pylori mutants shown in Table 1. Plasmid pILL485 (Fig. 1) was derived from the vector pILL570-1 (19) by inserting the nonpolar aphA-3 kanamycin resistance cassette (54) between BamHI and EcoRI sites generated after cloning of a 318-bp fragment corresponding to the HP0004 5′ end and a 304-bp fragment corresponding to the HP0004 3′ end by amplification with primer pairs H116/H117 and H119/H120, respectively (Table 2). Primer H120 contained a ribosome binding site necessary to generate a nonpolar insertion. Plasmid pILL486 (Fig. 1) was also a derivative of the pILL570-1 vector (19), in which an apramycin/gentamicin resistance cassette [aac(3)-IV (10)] was inserted at a unique BamHI restriction site generated after cloning of a 320-bp fragment corresponding to the HP1186 5′ end and a 288-bp fragment corresponding to the HP1186 3′ end by amplification with primer pairs H123/H124 and H126/H125, respectively (Table 2). The H. pylori mutants were obtained by natural transformation as previously described (11) with approximately 2 μg of plasmid DNA (prepared with maxi Qiagen). Clones that had undergone allelic exchange were selected after 4 to 5 days of growth on plates containing 20 μg ml−1 kanamycin for pILL485 or 5 μg ml−1 gentamicin for pILL486. Double mutants were obtained by transformation of the Δ(α-CA-HP) mutant by pILL485 and selection with both gentamicin and kanamycin.
Measurement of exchange rates by 13C-NMR spectroscopy assays.
Bacterial lysates were prepared from H. pylori strains J99, SS1, and X47-2AL, the corresponding Δ(α-CA-HP) and Δ(β-CA-HP) single mutants, and the Δ(α+β-CA-HP) double mutants grown on plates as described above. Cultures were subcultured every 28 to 32 h and incubated under microaerobic conditions (5% CO2, 10% O2, 80% N2) with 95% relative humidity at 37°C. Cells were harvested after 24 h in 0.9% (wt/vol) NaCl, the purity was checked by phase-contrast microscopy, and the preparations were tested for urease and catalase activities. Cell suspensions were centrifuged at 17,000 × g for 8 min at 6°C. Each pellet was washed twice in NaCl. Cells were lysed by freezing them in liquid nitrogen and thawing them at room temperature three times. The exchange rates of CA were determined at 37°C using H. pylori whole-cell lysates suspended in a buffer containing HEPES (85 mM, pH 7.5), Tris-HCl (16 mM, pH 8.5), NaCl (50 mM), and KCl (13 mM) and placed in 5-mm tubes (Wilmad, Buena, NJ). The reaction was initiated by adding H13CO3− at a final concentration of 100 mM. A column of mineral oil was added on top of each sample to prevent 13CO2 from diffusing away. Enzyme activities were determined using 13C nuclear magnetic resonance (13C-NMR) spectroscopy and using the selective inversion magnetization transfer technique (34, 35). The intensities of the bicarbonate and CO2 resonances were measured after the bicarbonate resonance was inverted and partial relaxation/exchange was allowed at various times. Information about the exchange rates for the two metabolites was present in the magnetization exchanged between the nuclei of both compounds. One-dimensional 13C-NMR spectra were acquired at 150.93 MHz using a Bruker DMX-600 spectrometer equipped with a dual 13C/1H probe operating in the Fourier transform mode with quadrature detection. The following instrument parameters were used: spectral width, 17,361 Hz; number of time domain points, 32,000; acquisition time, 0.943 s; pulse angle, 90° (5.4 μs); and relaxation delay, >5 times the spin-lattice relaxation time (typically 120 s). Exponential multiplication with line broadening of 1.5 Hz was applied prior to Fourier transformation. For the inversion transfer experiments, the selective 180° pulse was a 1,000-point IBURP2 pulse that was 9.04 ms long and was applied to the bicarbonate 13C nuclei on resonance. Typically, between 18 and 22 experiments were performed, and the exchange delays were between 10 ms and 52 s, with successive delays that were 1.5 times the previous delay. The order of collection of the different time points was randomized. Rates of bicarbonate exchange were obtained by using the CIFIT program (5). The relaxation rate of CO2 was not fitted into the calculations but was kept constant at 0.4 to 0.1 s−1 scaled with the relaxation rate of the bicarbonate. The exact value used for the CO2 relaxation rate has a negligible effect on the calculated rate constant since there is always a large excess of the bicarbonate. The errors were expressed as standard deviations.
In vitro assays of survival to acidity in the absence of urea.
Strains SS1 and X47-2AL were grown in BHI liquid medium as described above. Bacteria were harvested either after one generation time (4 h) or in early stationary phase (24 h). Bacterial cells were washed once in phosphate-buffered saline (PBS) (pH 7.4) (Roche), the concentration was adjusted to 2 × 108 bacteria ml−1, and the bacteria were incubated in PBS adjusted to pH 7 or 3. After 15 min of incubation at room temperature under aerobic conditions, the surviving bacteria were enumerated by serial dilution on blood agar plates.
Measurement of urease activity and kinetics of ammonia production by H. pylori strains.
Urease activities were measured using crude extracts prepared by sonication as previously described (20, 53). One unit of urease activity was defined as the amount of enzyme required to hydrolyze 1 μmol of urea (producing 2 μmol of ammonia) per min per mg of total protein. The amount of ammonia released was determined from a standard curve. The protein concentration was determined with a commercial version of the Bradford assay (Sigma Chemical Co.) using bovine serum albumin as a standard.
For measurement of ammonia production, bacteria were washed once in PBS, and the concentration was adjusted to 2 × 108 bacteria ml−1 in 5 ml PBS at an initial pH of 2.2 with 5 or 10 mM urea. PBS was chosen because it mimics the buffering capacities of the stomach (low at an acidic pH and higher at a neutral pH) and allows direct and rapid estimation of urea consumption by measurement of the pH. After 15 or 30 min of incubation at room temperature, the pH of a bacterial suspension filtrate was determined. After 5, 15, and 30 min and 1 h, 0.5-ml aliquots were centrifuged to pellet bacteria, and the supernatant was saved on ice. Ten microliters was used to measure the extracellular ammonia concentration with a specific enzymatic assay based on glutamate dehydrogenase activity (Sigma), as previously described (11).
Mouse model for colonization.
H. pylori SS1 and X47-2AL parental or mutant strains were grown on blood agar plates, harvested after 24 h, and suspended in peptone broth. The bacterial concentrations were adjusted to 108 cells ml−1, and the numbers of CFU per milliliter were subsequently determined. Aliquots (100 μl) of a bacterial suspension were administered orogastrically to seven female NMRI mice (4 weeks old; provided by Iffa-Credo) per strain as described previously (25). In each experiment, five mice were inoculated with peptone broth as a negative control (not shown). Mice were sacrificed 4 weeks after inoculation. The stomach was removed from each mouse and cut longitudinally along the greater curvature, and the forestomach was removed. The antrum and the corpus of the remaining stomach were separated using macroscopic criteria as described by Akada et al. (2). For histology, a longitudinal segment approximately 5 mm wide was removed. Quantitative cultures of H. pylori were grown separately for the antrum and the corpus of each sample on blood agar plates with the usual antibiotic-fungicide mixture in the presence of bacitracin (200 μg ml−1) plus nalidixic acid (10 μg ml−1), as described previously (25). These cultures were grown (i) without additional antibiotics for the mice infected with the parental strains or on plates supplemented with (ii) 20 μg ml−1 kanamycin for mice infected with the Δ(β-CA-HP) mutant, (iii) 5 μg ml−1 gentamicin for mice infected with the Δ(α-CA-HP) mutant, or (iv) both kanamycin and gentamicin for mice infected with the Δ(α+β-CA-HP) double mutant. Data confirmed that the presence of one or two antibiotics did not affect the numbers of CFU of the mutant strains.
Histology.
Gastric tissue samples were taken from uninfected and infected mice, and for each sample the antrum was separated from the corpus. Tissue sections were fixed in formalin and embedded in paraffin wax. Serial 4-μm sections were cut and stained with hematoxylin and eosin, Giemsa stain, periodic acid-Schiff stain, and alcian blue using standard procedures (63). Tissue sections were examined in order to (i) verify the quality of the macroscopically based separation of the antrum and corpus of each sample and (ii) evaluate blindly the gastric lesions. The severity of gastritis in all groups of mice was evaluated semiquantitatively by examining the recruitment of polymorphonuclear (PMN) cells, lymphocytes, and plasma cells as previously described for the mouse model (21, 63). The grading procedure involved evaluation of the recruitment of inflammatory cells (PMN and mononuclear cells) and also the changes in the architecture of the mucosa. The morphological changes in both the antrum and the corpus were classified into four groups: none, mild, moderate, or severe.
Statistical analyses.
The Wilcoxon signed-rank test was used to compare H. pylori density and inflammation in the corpus and antrum (paired samples) in every mouse. The Mann-Whitney U test was used to analyze the differences between the groups of mice infected with the different CA mutants and the corresponding groups of mice infected with the wild-type strains (unpaired samples). P values of <0.05 were considered significant.
RESULTS
Construction and growth phenotypes of single and double CA mutants of H. pylori.
In strain 26695 (62), the HP0004 gene encoding β-CA-HP is followed immediately by genes encoding proteins having unrelated functions, with which it might form an operon (Fig. 1). In contrast, the HP1186 gene encoding α-CA-HP seems to be monocistronic in strain 26695 (Fig. 1). Similar gene organizations have been found in strain J99 (3). PCR was used to examine the ORF flanking these two genes in three different H. pylori strains, SS1 (36), X47-2AL (22), and N6 (24). In these three strains, the regions around HP0004 showed a syntenic genetic organization with that of strains 26695 and J99. The regions surrounding HP1186 in strains 26695, SS1, X47-2AL, and J99 have similar genetic organizations. In strain N6, the region downstream of HP1186 could not be amplified with the primers employed for the other four strains, suggesting either that there is microdiversity or that the genetic organization is different from that of strain 26695 (Fig. 1).
To avoid potential polar effects on the expression of genes downstream from HP0004, this gene was inactivated with a nonpolar kanamycin cassette (54) employing plasmid pILL485 (Fig. 1). To inactivate α-CA-HP, plasmid pILL486 was employed; in this plasmid the HP1186 gene is interrupted by the aac(3)-IV cassette, conferring gentamicin resistance (10) (Fig. 1). Single and double CA mutants could be generated in the H. pylori SS1, X47-2AL, 26695, J99, and N6 backgrounds with comparable transformation efficiencies for single and double mutants, indicating that the CA, individually or together, were not essential for in vitro growth of H. pylori.
Growth of the CA-HP single and double mutant strains in BHI liquid medium supplemented with β-cyclodextrin was monitored for 48 h. No significant differences between the 26695, X47-2AL, and N6 parental strains and their isogenic mutants were observed. Only the SS1 CA mutants had a significantly reduced growth rate. The generation times in exponential phase were as follows: 1.8 h for wild-type strain SS1, 2.6 h for the SS1 Δ(CA-HP) single mutants, and 3 h for the SS1 Δ(α+β CA-HP) mutant. In addition, a premature decline was observed after 24 h of growth of the single and double SS1 CA mutants. For these strains, the OD600 of the cultures correlated with the number of viable bacteria (1 OD600 = 109 bacteria ml−1) in exponential (4 h) or early stationary (24 h) growth phase. Addition of nickel (250 μM), of bicarbonate at a physiological concentration (10 or 30 mM), or of CO2 (30%) did not affect the growth phenotype (data not shown). Higher concentrations of bicarbonate, like those used in the study of Stähler et al. (59) (50 and 75 mM), were not tested as they are much greater than the concentrations encountered by H. pylori in the stomach and should cause cell toxicity due to excessive alkalinization of the medium.
Similar results were obtained when β-cyclodextrin was replaced by FCS or when brucella broth was used instead of BHI broth.
CA activities in H. pylori parental and mutant strains.
The 13C-NMR method was used to measure CA exchange rates in the complex milieu of H. pylori lysates.
To establish the validity of the test, bicarbonate-to-carbon dioxide chemical exchange rates were measured for samples with various chemical compositions. In aqueous HEPES-Tris buffer at initial bicarbonate concentrations ranging from 25 to 100 mM and with 0.5 mg ml−1 bovine serum albumin, the rates were 11 × 10−3 ± 3 × 10−3 and 12 × 10−3 ± 2 × 10−3 s−1, respectively. At protein concentrations between 0.1 and 0.4 mg ml−1, similar background exchange rates (11 × 10−3 ± 2 × 10−3 s−1) were obtained with heat-denatured lysates of H. pylori strains J99 and X47-2AL suspended in the buffer. Thus, the selective inversion magnetization transfer technique yielded low basal rates of bicarbonate-to-carbon dioxide chemical exchange (11 × 10−3 ± 3 × 10−3 s−1) and was suitable for measuring enzyme exchange rates in H. pylori lysate suspensions.
The bicarbonate exchange rates determined for lysates of strains J99, SS1, and X47-2AL and their CA-deficient mutants in suspensions adjusted to obtain a protein content of 0.2 mg ml−1 are shown in Table 3. Similar rates (within experimental error) were obtained for wild-type strains and mutants lacking the β-CA-HP encoded by the HP0004 gene. Weak activities greater than the background exchange rate were obtained for mutants lacking α-CA-HP. Surprisingly, the exchange rates obtained for the double mutants lacking both enzymes were significantly higher than the rates obtained for the α-CA-HP single mutants but were only one-half the rates obtained for the corresponding parental strains.
TABLE 3.
Bicarbonate exchange rates at 37°C for H. pylori lysates suspended in HEPES-Tris buffer
| Strain | Rate (10−3 s−1)a
|
|||
|---|---|---|---|---|
| Wild type | Δ(α-CA-HP) | Δ(β-CA-HP) | Δ(α+β-CA-HP) | |
| J99 | 88 ± 14 | 17 ± 3 | 85 ± 11 | 42 ± 6 |
| SS1 | 78 ± 12 | 18 ± 3 | 70 ± 10 | 34 ± 5 |
| X47-2AL | 93 ± 14 | 14 ± 2 | 88 ± 10 | 36 ± 5 |
Rates are given as measurements without subtraction of the background chemical exchange rate, which under the conditions used was estimated to be 11 × 10−3 ± 3 × 10−3 s−1. The initial [13C]bicarbonate concentration was 100 mM. The values are means ± standard deviations.
Resistance to acidity of H. pylori parental and CA mutant strains.
The resistance to acidity of the SS1 and X47-2AL parental strains and double CA mutants was determined in the absence of urea by counting viable bacteria after 15 min of incubation in PBS at pH 3 and comparing the number of surviving bacteria to the number of surviving bacteria incubated at pH 7. Wild-type and mutant strains showed similar resistance to such a stress.
The role of each CA in the urease-dependent response to acidity was also examined. First, the urease activities of crude extracts of the parental strains and single and double CA mutants were measured. Significant differences were observed in the basal urease activity levels between the H. pylori parental strains (SS1, 9 IU; N6, 11 IU; X47-2AL, 5.3 IU; 26695, 2.2 IU), but the enzyme activities of the parental strains and their isogenic CA mutants did not differ significantly.
Strains SS1, N6, X47-2AL, and 26695 and their CA mutants were incubated at pH 2.2 with 5 or 10 mM urea, and the changes in the extracellular medium pH, reflecting urea hydrolysis, were determined after 15 and 30 min (Table 4). Independent of the genetic background, the external pH was significantly lower for the mutants defective in α-CA-HP and in both α-CA-HP and β-CA-HP than for the corresponding parental strains. In addition, lower values were observed with strain SS1 in the absence of β-CA-HP. The time and the urea concentration at which the effect was maximal depended on the strain; the phenotype was more pronounced for strains N6 and SS1 than for strains X47-2AL and 26695. This may be related to the higher urease activity found in strains N6 and SS1 (see below).
TABLE 4.
Extracellular pH values of the medium after 15 or 30 min of exposure of H. pylori strains in PBS at an initial pH of 2.2 in the presence of 5 or 10 mM urea
| Strain | pH valuea
|
|||
|---|---|---|---|---|
| 15 min and 5 mM urea | 30 min and 5 mM urea | 15 min and 10 mM urea | 30 min and 10 mM urea | |
| SS1 | 3.94 (0.85) | 5.26 (0.42) | 6.21 (0.13) | 6.40 (0.08) |
| SS1 Δ(β-CA-HP) | 2.75 (0.14) | 4.41 (1.00) | 2.97 (0.34) | 6.32 (0.25) |
| SS1 Δ(α-CA-HP) | 2.56 (0.10) | 3.12 (0.39) | 2.57 (0.01) | 4.94 (0.84) |
| SS1 Δ(α+β-CA-HP) | 2.61 (0.25) | 3.24 (0.91) | 2.53 (0.03) | 4.41 (1.17) |
| N6 | 5.04 (0.26) | 5.61 (0.43) | 6.37 (0.06) | 6.60 (0.12) |
| N6 Δ(β-CA-HP) | 4.24 (1.51) | 5.63 (0.31) | 6.28 (0.16) | 6.55 (0.12) |
| N6 Δ(α-CA-HP) | 2.92 (0.12) | 5.46 (0.23) | 4.69 (1.47) | 6.58 (0.11) |
| N6 Δ(α+β-CA-HP) | 2.93 (0.33) | 4.50 (0.50) | 3.92 (1.39) | 6.44 (0.10) |
| X47-2AL | 2.58 (0.10) | 2.86 (0.28) | 2.62 (0.17) | 5.75 (0.32) |
| X47-2AL Δ(β-CA-HP) | 2.43 (0.10) | 2.86 (0.44) | 2.52 (0.12) | 5.70 (0.26) |
| X47-2AL Δ(α-CA-HP) | 2.39 (0.04) | 2.58 (0.07) | 2.42 (0.05) | 2.98 (0.46) |
| X47-2AL Δ(α+β-CA-HP) | 2.41 (0.01) | 2.52 (0.17) | 2.47 (0.04) | 3.18 (0.76) |
| 26695 | 2.57 (0.07) | 3.78 (0.24) | 2.68 (0.24) | 6.01 (0.17) |
| 26695 Δ(β-CA-HP) | 2.42 (0.06) | 3.33 (0.49) | 2.48 (0.10) | 5.25 (1.45) |
| 26695 Δ(α-CA-HP) | 2.39 (0.10) | 2.68 (0.06) | 2.47 (0.06) | 2.89 (0.24) |
| 26695 Δ(α+β-CA-HP) | 2.42 (0.09) | 2.70 (0.21) | 2.51 (0.03) | 3.11 (0.50) |
Values significantly different from the wild-type value are indicated by bold type (P < 0.05, Student t test). The data are results from at least three independent experiments. The values in parentheses are standard deviations.
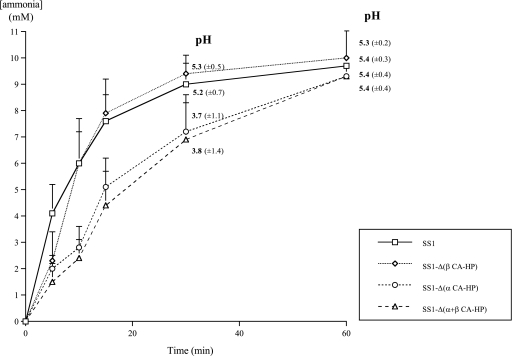
The extracellular pH and ammonia concentration were measured as a function of time for suspensions of wild-type SS1 and CA mutant cells incubated in the presence of 5 mM urea. A delay in the increase in pH and in ammonia production was observed for the CA mutants with an inactivated HP1186 gene encoding α-CA-HP (Fig. 2). Finally, the survival after exposure to an acidic medium supplemented with urea was tested for strains 26695, X47-2AL, and SS1 and their single and double CA mutants. The protocol described by Marcus et al. (37) with 108 bacteria/ml was used, but the medium used was brucella broth (Difco) because, in contrast to the observations of Marcus et al., we found that a wild-type strain did not survive incubation at pH 2 with 10 mM urea in BHI medium (Oxoid). Surviving bacteria were enumerated after incubation at pH 7 or at pH 2 in the presence of 10 mM urea. No difference was observed between the survival of the wild-type strains and the survival of the mutants at pH 7 or at pH 2 with urea (100% survival) even when the incubation time was extended from 30 min to 2 h. Thus, these test conditions did not reveal a role of the CA during in vitro exposure to acidity, clearly showing the limits of performing such experiments in poorly defined complex media and the importance of the analysis of the role of the enzymes during in vivo colonization.
FIG. 2.
Extracellular ammonia production by the H. pylori SS1 parental strain and CA single and double mutants incubated in PBS with 5 mM urea at an initial pH of 2.2. The pH values of the medium and the standard deviations for incubation times of 30 min and 1 h are indicated. The results are representative of at least four independent experiments. Standard deviations for ammonia production are indicated by error bars.
Colonization of mouse gastric mucosa by CA mutant strains.
The mouse model of colonization by H. pylori was used to evaluate the role of the two CA-HP enzymes in vivo and their impact on gastric colonization tropism. Mice were infected with two different strains, SS1 (36) and X47-2AL (22), which in some mouse lines (C57BL/6J and BALB/cJ NMRI) have been reported to colonize preferentially the antrum and the corpus (a more acidic region of the stomach), respectively (2). One month after orogastric inoculation, the colonization loads of each parental H. pylori strain and its single and double CA mutants were measured separately using the gastric antrum and corpus of Swiss outbred mice (NMRI), a model of infection that is well established and has been validated (25). In addition, histological analysis was performed for every portion of the stomach (see below).
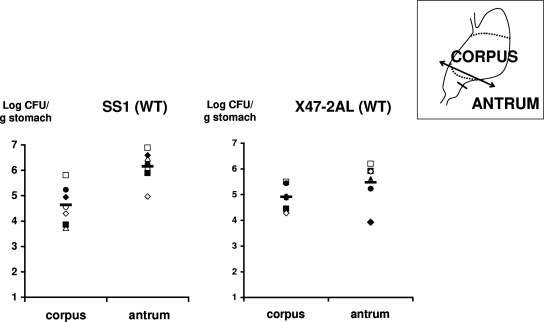
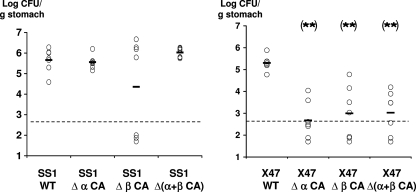
In the NMRI mouse line, the wild-type SS1 strain preferentially colonized the antrum, while the two stomach regions were colonized equally by the wild-type X47-2AL strain (Fig. 3). The relative distribution of each single and double CA mutant in the two gastric regions was similar to that of the corresponding parental strain (data not shown). Most interestingly, the colonization by the X47-2AL single and double CA mutants was strongly diminished compared to the colonization by the parental strain; this result was reproduced in four independent experiments, including the experiment whose results are shown in Fig. 4. The number of mice infected with the mutants was lower, and the gastric colonization loads were smaller (Fig. 4). In contrast, no statistically significant changes in colonization loads were observed with the SS1 single and double CA mutants, although three mice were not colonized by the SS1 Δ(β-CA-HP) mutant (Fig. 4).
FIG. 3.
Distribution of the H. pylori SS1 and X47-2AL parental strains in regions of the stomach of NMRI Swiss mice. Each symbol indicates the H. pylori colonization titer in the corpus or antrum of a single mouse. The horizontal bars indicate the geometric mean for each group of data. The results for the single and double H. pylori CA mutants were similar to those for the corresponding wild-type strains (WT) shown. The colonization density of the SS1 strain was statistically significantly higher in the antrum (P = 0.018, Wilcoxon signed-rank test).
FIG. 4.
Effects of deficiency of one or both CA-HP enzymes on mouse gastric colonization by H. pylori strains SS1 and X47-2AL. The bacterial strains used to inoculate mice are indicated on the x axis. The detection limit of the method is indicated by the dashed horizontal lines. Each point corresponds to the gastric colonization load (antrum plus corpus) for one mouse, and the horizontal bars indicate the geometric mean for each group of mice. Statistically significant differences (using the Mann-Whitney test) between mutant and parental strains are indicated by two asterisks (P < 0.01). WT, wild type.
Inflammation of the mouse gastric mucosa upon infection by H. pylori strains SS1 and X47-2AL and their CA mutants.
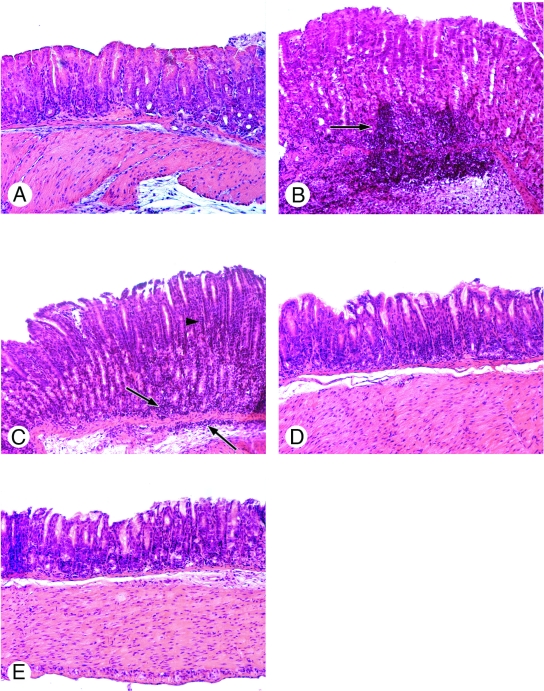
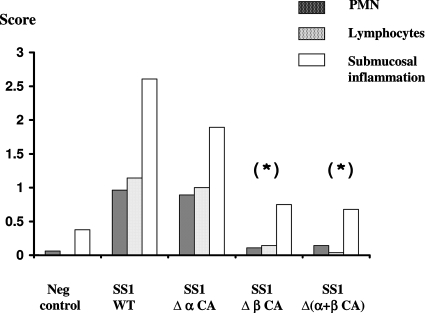
To obtain better insight into the in vivo behavior of the H. pylori CA mutants, inflammation scores were determined for every gastric section. The gastritis scores included evaluation of the recruitment of inflammatory cells (PMN cells and lymphocytes) and changes in the architecture of the mucosa on a scale from 0 to 4, according to the system proposed by Eaton et al. (21) (Fig. 5 and 6). For mice infected with strain X47-2AL, the inflammation scores were very low (0.5 to 1), and there were no statistically significant differences between infected and noninfected mice (data not shown). In contrast, the inflammation scores were higher (1 to 2.5) after infection with the SS1 wild-type strain or with SS1 Δ(α-CA-HP), revealing that there was acute diffuse gastritis (presence of PMN cells) in the mucosal and submucosal regions of these mice compared to those of uninfected animals (Fig. 5B, 5C, and 6).
FIG. 5.
Histological analysis of the antrum mucosa of mice infected by the SS1 wild-type strain and the single and double CA mutants: representative stained (hematoxylin and eosin) sections of (A) normal gastric mucosa from a naive mouse, (B) gastric mucosa from a mouse infected by the SS1 wild-type strain, (C) gastric mucosa from a mouse infected by the SS1 Δ(α-CA-HP) mutant, (D) gastric mucosa from a mouse infected by the SS1 Δ(β-CA-HP) mutant, and (E) gastric mucosa from a mouse infected by the SS1 Δ(α+β-CA-HP) mutant. The arrows indicate infiltrates. Magnification, ×100.
FIG. 6.
Inflammation of the mucosa of mice infected by the SS1 wild-type strain and the single and double CA mutants. The intensity of the lesions on gastric sections from every mouse infected by the SS1 wild-type strain (WT) and single and double mutants and from the negative controls (Neg control) was evaluated semiquantitatively using two inflammation scoring criteria. Infiltrates of PMN and lymphocytes were scored as follows: 0, no infiltrate; 1, mild multifocal infiltration; 2, mild widespread infiltration; 3, mild widespread and moderate multifocal infiltration; and 4, moderate widespread infiltration. The submucosal inflammation was scored as follows: 1, mild multifocal; 2, mild widespread; and 3, moderate multifocal. Mean values of the scores are shown. Statistically significant differences (determined using the Mann-Whitney test) between the scores for mutant strains and the parental strain scores are indicated by an asterisk (P < 0.05).
For the infected animals, the mean scores for the antrum and the mean scores for the corpus were not significantly different. Interestingly, inflammation was significantly diminished (P < 0.05) in mice infected with SS1 Δ(β-CA) (Fig. 5D and 6) or the double mutant SS1 Δ(α+β CA-HP) (Fig. 5E and 6), and there was mild recruitment of lymphocytes without PMN cells, an effect observed in both the antrum and the corpus.
DISCUSSION
Carbon dioxide and bicarbonate have important roles in H. pylori physiology manifested by the dependence of the bacterium on CO2 for growth in vitro (capnophily) and supported by the abundance of these metabolites in its ecological niche, the human stomach, owing to urease activity and gastric epithelial cell secretion. A chemotactic response of H. pylori to bicarbonate through a methyl-accepting chemotaxis receptor protein has been reported (14, 41). Thus, CA which catalyze the interconversion of carbon dioxide and bicarbonate were expected to be important in the metabolism of H. pylori, and the roles of the two CA in the physiology of this organism were investigated. Mutants defective in one or both CA were constructed in five different H. pylori genetic backgrounds, demonstrating that the enzymes were not essential in vitro. These data were in agreement with those reported by Chirica et al. (18), who observed that H. pylori strains 26695 and J99 can grow in the presence of the CA inhibitor acetazolamide at a concentration of 5 μM. In addition, a double CA mutant of strain 26695 was obtained by Stähler et al. (59), but the possibility of a polar effect of the cassette that these workers used to disrupt HP0004, encoding β-CA, could not be excluded.
CA activities in H. pylori lysates of three strains and their single and double CA mutants were measured by 13C-NMR spectroscopy (Table 3). The measured activities indicated that under the experimental conditions used, almost all of the CA activity of H. pylori was provided by α-CA-HP. Chirica et al. found that the exchange rate of purified α-CA-HP from strain 26695 was 240 × 10−3 ± 10 × 10−3 s−1 at pH 8.9 (17). The rates measured in the present study at pH 7.5 were approximately threefold lower. This difference is reasonable considering that the activity of the purified enzyme decreased with pH and was reduced to approximately 60% at pH 7.5 (17) and that the activity of the CA may be modulated in complex mixtures, such as whole-cell lysates.
Surprisingly, the CA activity of the mutants deficient in both H. pylori CA was higher than that of the corresponding Δ(α-CA-HP) single mutant. It is possible that the absence of both CA induced compensatory changes in the activity and/or expression of other enzymes involved in bicarbonate metabolism. Uncharacterized suppressors of an E. coli CA mutant (Δcan, ΔcynT) were obtained by screening for its recovered ability to grow in air (38). Compensatory metabolic changes might involve enzymes similar to members of the γ-CA group reported in E. coli (38) or zinc-containing enzymes which ordinarily do not catalyze bicarbonate/carbon dioxide exchange but which have esterase activities and can catalyze hydrolysis or closely related transfer reactions.
The fact that some CA-defective mutants exhibited only reduced stomach colonization suggested that at least in strain X47-2AL, both CA-HP enzymes were expressed and active in vivo. The CA activities measured were compatible with the hypothesis that the expression of the enzymes and/or their activities are subject to regulation. In many organisms, CA are expressed at low basal levels and induced under specific conditions, e.g., diminished pCO2 in Neisseria spp. (43), the presence of cyanate in E. coli (28), or enhanced or decreased pO2 in yeast (26) and Rhodopseudomonas palustris (49), respectively. In H. pylori, the amounts of α- and β-CA-HP measured by Western blotting in 2-day-old H. pylori cultures with 0.1 or 10% CO2 were constant, suggesting that the expression of the two CA genes is not regulated by pCO2 (18). Inconsistent results for acid regulation of the expression of the HP1186 gene encoding α-CA-HP showed repression at low pH (39) or up-regulation (68). A more recent study implicated the ArsRS acid-responsive two-component system in the acid-induced expression of the HP1186 gene (67). In a global analysis of the genes regulated during growth in moderately acidic conditions, no significant modifications in HP1186 or HP0004 gene expression were measured (12). The disparity in the results could be due to variation in CA expression depending on the various strains and acidic conditions used in these studies. This interpretation is supported by the observation that in H. pylori strains 26695 and ATCC 43504 expression of the HP1186 gene relies on two distinct promoters: TSP1, which is subject to pH-dependent regulation by ArsR, and TSP2, which may be responsible for the basal transcription level of this gene (67).
The molecular basis of the capnophily of H. pylori is not understood, but it has been proposed that enzymes with low affinity for CO2, such as the acetyl coenzyme A carboxylase (9), require elevated partial pressures of this gas. The role of the two CA-HP enzymes on H. pylori growth was investigated using the five genetic backgrounds to circumvent potential strain-dependent effects. Only the SS1 CA mutants showed a significant and reproducible growth defect, indicating that other factors which depend on the genetic background were implicated in this phenotype. Supplementation with HCO3−, CO2, Ni2+, or FCS did not restore a wild-type growth phenotype to the SS1 CA mutants. In microorganisms, inactivation of CA often leads to growth defects (26, 28, 29), and β-CA have a crucial role in preventing bicarbonate depletion for metabolic carboxylation reactions (for example, in E. coli during cyanate degradation [28, 32] or in Neisseria sicca incubated at low pCO2 [1]). E. coli or R. eutropha strains deficient in the constitutive β-CA encoded by can (previously yadF in E. coli) are unable to grow at atmospheric partial pressures of CO2. In E. coli, this defect leads to a capnophile-type phenotype since it can be compensated for by addition of 2% CO2 (38), suggesting that under the conditions in which the E. coli CA is essential, the rate of an uncatalyzed hydration reaction converting CO2 into bicarbonate is too low to meet the demand for bicarbonate.
The roles of CA-HP enzymes in the response of H. pylori to acidity and in the regulation of urease activity were investigated. In vitro tests of acid shock resistance in the presence or absence of urea revealed that the absence of one or both CA-HP enzymes had no effect on viability. A role for CA in maintaining urease activity under nickel-limiting conditions has been described previously (59). However, our results and those of Marcus et al. (37) convincingly show that CA has no effect on in vitro urease activity under standard conditions. This is in contrast to the results of Stähler et al., who observed a reduction in urease activity in both CA mutants studied (59). Surprisingly, we noticed that the basal urease activity measured by Strähler et al. for the 26695 wild-type strain was about fivefold lower than the activity obtained by others and by us (see Results) for the same strain and that this level was at the limit of sensitivity of the urease assay. Therefore, we suggest that the data of Stähler et al. (59) reveal an effect of CA on in vitro urease activity that is apparent only under nonoptimal urease activity conditions and thus difficult to interpret.
In contrast, measurements obtained with whole cells revealed that CA-HP enzymes are involved in the urease-dependent response of H. pylori to acidic pH. In five different genetic backgrounds, inactivation of the CA-HP enzymes resulted in delays in the increase in the pH of the acidic medium during incubation in the presence of urea; little or no effect was observed in the absence of β-CA-HP, and significant effects were observed in the absence α-CA-HP (Table 4). CA inactivation resulted in a phenotype with delayed production of ammonia from urea, and the level depended on the strain tested, the urea concentration, and the exposure time. The delay in ammonia production by the mutants lacking α-CA-HP (Fig. 3) correlated with the significantly lower CA exchange rates observed for these mutants (Table 3). Thus, the data suggested that α-CA-HP regulated the urea-dependent response to acidity of H. pylori. This result is consistent with a study showing that H. pylori strain ATCC 43504, which was deficient in α-CA activity due to either deletion of the HP1186 gene or addition of the CA inhibitor acetazolamide, presented an impaired capacity to survive acidic pH exposure in the presence of urea (37).
Together, these data supported the hypothesis that there is a link between CO2, CA, and resistance to acidity in microorganisms, as suggested by other reports. In S. enterica serovar Typhimurium, a putative CA gene (mig5) is induced during survival in the acidic environment inside macrophages. The mig5 gene is regulated by the PhoP/PhoQ two-component system (65) involved in the pH response in enterobacteria (6). In addition, the synthesis of inducible decarboxylases, which is the main response of enterobacteria to acidity, is induced by a lower pCO2 (61).
Several hypotheses for the mechanism responsible for the delayed ammonia production by H. pylori CA mutants were considered. The first hypothesis is that CO2 is needed for in vitro nickel incorporation into mature active urease (46) and might be limiting in the CA mutants. This hypothesis was discarded because the urease activities measured for H. pylori crude extracts were not significantly lower than those measured for the CA mutants. The second hypothesis is that the potential intracellular accumulation of bicarbonate in the CA mutants may generate product inhibition effects on urease activity. This hypothesis was rejected because addition of high concentrations of bicarbonate (up to 100 mM) in the in vitro assays did not impair urease activity. Finally, considering that CA are involved in proton capture and CO2 production, it was possible that under extracellular acidic conditions the H. pylori CA mutants had a decreased capacity to buffer the periplasmic and/or intracellular pH. This hypothesis was consistent with data of Marcus et al. (37), who, using pH-sensitive fluorescent probes, showed that at a low pH the increase in the cytoplasmic and periplasmic pH with urea was abolished in the absence of α-CA-HP activity.
The mechanism proposed is that the lack of CA activity results in a transiently lower cellular pH, which indirectly leads to decreased urea hydrolysis and consequently to delayed ammonia production. Supporting this view are the observation that changes in the intracellular pH could modify urease enzymatic activity (optimum pH, between 7.5 and 8.2), as suggested for Yersinia enterocolitica (69), and the observation that in H. pylori the specific urea channel, UreI, opens when the extracellular pH falls below 5 (11, 66). Alternatively, changes in the periplasmic pH could influence UreI activation and thus urea availability, resulting in a delayed increase in urease activity and ammonia production.
The role of CA-HP enzymes in colonization of the mouse gastric mucosa was investigated using two H. pylori mouse-adapted strains, SS1 and X47-2AL, which have been reported to colonize preferentially the antrum and the corpus, respectively, in some mouse lines (2). In the NMRI Swiss mouse strain, the antrum tropism of strain SS1 was reproduced, while strain X47-2AL was distributed equally in the two regions. The absence of CA did not affect these distributions. The capacities of the SS1 CA mutants to colonize mice were not significantly affected, in contrast to the capacities of the strain X47-2AL α- and β-CA mutants, which were severely affected. Despite the weak activity that was attributed to β-CA by in vitro measurement (Table 3), the colonization data suggest that the HP0004 gene encoding β-CA is expressed under in vivo conditions in strain X47-2AL. A significant difference between these two strains is their basal urease activities, 10 IU for SS1 and 5 IU for X47-2AL, which correlated with the greater resistance of SS1 to growth on acidic medium plates (2). The observation that strain X47-2AL mutants have an impaired colonization capacity could be explained by the CA mutants lowering the “in vivo” resistance to acidity to a level below that compatible with efficient colonization of the acidic environment of the stomach.
Although the SS1 mutants did not exhibit a deficiency in colonization or modified tropism, the inflammation caused by the Δ(β-CA-HP) and double CA mutants was strongly reduced. This decrease in inflammation involved both poor infiltration by inflammatory cells and diminished modification of the mucosal structure. However, a complete understanding of this new relationship between CA activity and inflammation requires additional studies. A possible hypothesis is that in order to incorporate enough carbon dioxide, these mutants need to colonize regions of the mucus layer that are farther from the epithelial cells than the regions colonized by the wild-type strain, generating at those sites a less intense inflammatory response. Consistent with this proposition is the fact that the bicarbonate/carbon dioxide ratio (which is strongly pH dependent) is not uniform in the mucus layer, with higher carbon dioxide concentrations in the more acidic regions that are distant from the epithelial cells. Interestingly, antibodies directed against H. pylori CA have been proposed to have a molecular mimicry role in the postulated link between gastric infection by H. pylori and autoimmune pancreatitis (27).
The results of this study revealed a role for the α-CA-HP in the urease-dependent response to exposure to low pH and the involvement of both CA-HP enzymes in the adaptation of H. pylori to the gastric environment, thus opening new perspectives on the biological roles of CA in H. pylori and in bacteria in general.
Acknowledgments
We thank I. G. Boneca and S. Skouloubris for helpful discussions and critical reading of the manuscript.
We thank the Australian Research Council for its support. K. Stingl was supported by postdoctoral fellowships from the German Academic Exchange Service (DAAD) and the Fondation pour la Recherche Médicale (FRM).
Editor: V. J. DiRita
Footnotes
Published ahead of print on 19 November 2007.
REFERENCES
- 1.Adler, L., J. Brundell, S. O. Falkbring, and P. O. Nyman. 1972. Carbonic anhydrase from Neisseria sicca strain 6021. I. Bacterial growth and purification of the enzyme. Biochim. Biophys. Acta 284298-310. [DOI] [PubMed] [Google Scholar]
- 2.Akada, J. K., K. Ogura, D. Dailidiene, G. Dailide, J. M. Cheverud, and D. E. Berg. 2003. Helicobacter pylori tissue tropism: mouse-colonizing strains can target different gastric niches. Microbiology 1491901-1909. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., L. S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397176-180. [DOI] [PubMed] [Google Scholar]
- 4.Baar, C., M. Eppinger, G. Raddatz, J. Simon, C. Lanz, O. Klimmek, R. Nandakumar, R. Gross, A. Rosinus, H. Keller, P. Jagtap, B. Linke, F. Meyer, H. Lederer, and S. C. Schuster. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. USA 10011690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bain, A. D., and J. A. Cramer. 1996. Slow chemical exchange in an eight-coordinated bicentered ruthenium complex studied by one-dimensional methods. Data fitting and error analysis. J. Magn. Reson. 118A21-27. [Google Scholar]
- 6.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147173-180. [DOI] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 8.Bumann, D., S. Aksu, M. Wendland, K. Janek, U. Zimmy-Arndt, N. Sabarth, T. F. Meyer, and P. R. Jungblut. 2002. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect. Immun. 703396-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns, B., S. Hazell, and G. L. Mendz. 1995. Acetyl-CoA carboxylase activity in Helicobacter pylori and the requirement of increased CO2 for growth. Microbiology 1413113-3118. [DOI] [PubMed] [Google Scholar]
- 10.Bury-Moné, S., S. Skouloubris, C. Dauga, J.-M. Thiberge, D. Dailidiene, D. E. Berg, A. Labigne, and H. De Reuse. 2003. Presence of active aliphatic amidases in Helicobacter species able to colonize the stomach. Infect. Immun. 715613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bury-Moné, S., S. Skouloubris, A. Labigne, and H. De Reuse. 2001. The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Mol. Microbiol. 421021-1034. [DOI] [PubMed] [Google Scholar]
- 12.Bury-Moné, S., J.-M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53623-638. [DOI] [PubMed] [Google Scholar]
- 13.Casadaban, M., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusions and cloning in E. coli. J. Mol. Biol. 138179-207. [DOI] [PubMed] [Google Scholar]
- 14.Cerda, O., A. Rivas, and H. Toledo. 2003. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol. Lett. 224175-181. [DOI] [PubMed] [Google Scholar]
- 15.Chegwidden, W. R., N. D. Carter, and Y. H. Edwards. 2000. Introduction to the carbonic anhydrases, p. 12-28. In W. R. Chegwidden, N. D. Carter, and M. R. Edwards (ed.), The carbonic anhydrases: new horizons, vol. 90. Birkhaüser Verlag, Basel, Switzerland. [Google Scholar]
- 16.Chirica, L. C., B. Elleby, B. H. Jonsson, and S. Lindskorg. 1997. The complete sequence, expression in Escherichia coli, purification and some properties of carbonic anhydrase from Neisseria gonorrhoeae. Eur. J. Biochem. 244755-760. [DOI] [PubMed] [Google Scholar]
- 17.Chirica, L. C., B. Elleby, and S. Lindskorg. 2001. Cloning, expression and some properties of α-carbonic anhydrases from Helicobacter pylori. Biochim. Biophys. Acta 154455-63. [DOI] [PubMed] [Google Scholar]
- 18.Chirica, L. C., C. Petersson, M. Hurtig, B. H. Jonsson, T. Borén, and S. Lindskorg. 2002. Expression and localization of α- and β-carbonic anhydrases in Helicobacter pylori. Biochim. Biophys. Acta 1601192-199. [DOI] [PubMed] [Google Scholar]
- 19.Colland, F., J.-C. Rain, P. Gounon, A. Labigne, P. Legrain, and H. De Reuse. 2001. Identification of the Helicobacter pylori anti-σ28 factor. Mol. Microbiol. 41477-487. [DOI] [PubMed] [Google Scholar]
- 20.Cussac, V., R. L. Ferrero, and A. Labigne. 1992. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 1742466-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eaton, K. A., M. J. Radin, and S. Krakowa. 1995. An animal model of gastric ulcer due to bacterial gastritis in mice. Vet. Pathol. 32489-497. [DOI] [PubMed] [Google Scholar]
- 22.Ermak, T. H., P. J. Giannasca, R. Nichols, G. A. Myers, J. Nedrud, R. Weltzin, C. K. Lee, H. Kleanthous, and T. P. Monath. 1998. Immunization of mice with urease vaccine affords protection against Helicobacter pylori infection in the absence of antibodies and is mediated by MHC class II-restricted response. J. Exp. Med. 1882277-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst, P. B., and B. D. Gold. 2000. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54615-640. [DOI] [PubMed] [Google Scholar]
- 24.Ferrero, R. L., V. Cussac, P. Courcoux, and A. Labigne. 1992. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J. Bacteriol. 1744212-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrero, R. L., J.-M. Thiberge, M. Huerre, and A. Labigne. 1998. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect. Immun. 661349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gotz, R., A. Gnann, and F. K. Zimmermann. 1999. Deletion of the carbonic anhydrase-like gene NCE103 of the yeast Saccharomyces cerevisiae causes an oxygen-sensitive growth defect. Yeast 15855-864. [DOI] [PubMed] [Google Scholar]
- 27.Guarneri, F., C. Guarneri, and S. Benvenga. 2005. Helicobacter pylori and autoimmune pancreatitis: role of carbonic anhydrase via molecular mimicry? J. Cell. Mol. Med. 9741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guilloton, M. B., J. J. Korte, A. F. Lamblin, J. A. Fuchs, and P. M. Anderson. 1992. Carbonic anhydrase in Escherichia coli. A product of the cyn operon. J. Biol. Chem. 2673731-3734. [PubMed] [Google Scholar]
- 29.Hashimoto, M., and J. Kato. 2003. Indispensability of the Escherichia coli carbonic anhydrases YadF and CynT in cell proliferation at a low CO2 partial pressure. Biosci. Biotechnol. Biochem. 67919-922. [DOI] [PubMed] [Google Scholar]
- 30.Hewett-Emmett, D. 2000. Evolution and distribution of the carbonic anhydrase gene families, p. 29-76. In W. R. Chegwidden, N. D. Carter, and M. R. Edwards (ed.), The carbonic anhydrases: new horizons, vol. 90. Birkhaüser Verlag, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 31.Kelly, D. J., N. J. Hughes, and R. K. Poole. 2001. Microaerobic physiology: aerobic respiration, anaerobic respiration, and carbon dioxide metabolism, p. 113-124. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC. [PubMed]
- 32.Kozliak, E. I., J. A. Fuchs, M. B. Guilloton, and P. M. Anderson. 1995. Role of bicarbonate/CO2 in the inhibition of Escherichia coli growth by cyanate. J. Bacteriol. 1773213-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusian, B., D. Sueltemeyer, and B. Bowien. 2002. Carbonic anhydrase is essential for growth of Ralstonia eutropha at ambient CO2 concentrations. J. Bacteriol. 1845018-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Led, J. J., H. Gesmar, and F. Abildgaard. 1989. Applicability of magnetization transfer nuclear magnetic resonance to study chemical exchange reactions. Methods Enzymol. 176311-329. [DOI] [PubMed] [Google Scholar]
- 35.Led, J. J., E. Neesgaard, and J. T. Johansen. 1982. Carbon dioxide hydration activity and metal-substrate distances of manganese(II) human carbonic anhydrase B determined by 13C magnetization-transfer NMR. FEBS Lett. 14774-80. [DOI] [PubMed] [Google Scholar]
- 36.Lee, A., J. O'Rourke, M. Corazon De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 1121386-1397. [DOI] [PubMed] [Google Scholar]
- 37.Marcus, E. A., A. P. Moshfegh, G. Sachs, and D. R. Scott. 2005. The periplasmic α-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J. Bacteriol. 187729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merlin, C., M. Masters, S. McAteer, and A. Coulson. 2003. Why is carbonic anhydrase essential to Escherichia coli? J. Bacteriol. 1856415-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 713529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Mizote, T., H. Yoshiyama, and T. Nakazawa. 1997. Urease-independent chemotaxis responses of Helicobacter pylori to urea, urease inhibitor, and sodium bicarbonate. Infect. Immun. 651519-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mobley, H. L. T., G. L. Mendz, and S. L. Hazell (ed.). 2001. Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC. [PubMed]
- 43.Nafi, B. M., R. J. Miles, L. O. Butler, N. D. Carter, C. Kelly, and S. Jeffery. 1990. Expression of carbonic anhydrase in Neisseriae and other heterotrophic bacteria. J. Med. Microbiol. 321-7. [DOI] [PubMed] [Google Scholar]
- 44.Nishimori, I., T. Minakuchi, T. Kohsaki, S. Onishi, H. Takeuchi, D. Vullo, A. Scozzafava, and C. T. Supuran. 2007. Carbonic anhydrase inhibitors: the beta-carbonic anhydrase from Helicobacter pylori is a new target for sulfonamide and sulfamate inhibitors. Bioorg. Med. Chem. Lett. 173585-3594. [DOI] [PubMed] [Google Scholar]
- 45.Nishimori, I., T. Minakuchi, K. Morimoto, S. Sano, S. Onishi, H. Takeuchi, D. Vullo, S. Andrea, and C. T. Supuran. 2006. Carbonic anhydrase inhibitors: DNA cloning and inhibition studies of the alpha-carbonic anhydrase from Helicobacter pylori, a new target for developing sulfonamide and sulfamate gastric drugs. J. Med. Chem. 492117-2126. [DOI] [PubMed] [Google Scholar]
- 46.Park, I. S., and R. P. Hausinger. 1995. Requirement of carbon dioxide for in vitro assembly of the urease nickel metallocenter. Science 2671156-1158. [DOI] [PubMed] [Google Scholar]
- 47.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. Quail, M. A. Rajandream, K. M. Rutherford, A. VanVliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403665-668. [DOI] [PubMed] [Google Scholar]
- 48.Puscas, D., I. Paun, R. Ursea, N. Lenghel, and C. A. Dinga. 1975. Carbonic anhydrase inhibitors in the treatment of gastro-duodenal ulcer. Gastroenterol. Endocrinol. Nutr. 3093-100. (In Spanish.) [PubMed] [Google Scholar]
- 49.Puskas, L. G., M. Inui, K. Zahn, and H. Yukawa. 2000. A periplasmic, α-type carbonic anhydrase from Rhodopseudomonas palustris is essential for bicarbonate uptake. Microbiology 1462957-2966. [DOI] [PubMed] [Google Scholar]
- 50.Sabarth, N., S. Lamer, U. Zimny-Arndt, P. R. Jungblut, T. F. Meyer, and D. Bumann. 2002. Identification of surface proteins of Helicobacter pylori by selective biotinylation, affinity purification and two-dimensional gel electrophoresis. J. Biol. Chem. 27727896-27902. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 52.Shahidzadeh, R., A. Openkun, A. Shiotani, and D. Y. Graham. 2005. Effect of the carbonic anhydrase inhibitor, acetazolamide, on Helicobacter pylori infection in vivo: a pilot study. Helicobacter 10136-138. [DOI] [PubMed] [Google Scholar]
- 53.Skouloubris, S., A. Labigne, and H. De Reuse. 1997. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol. Microbiol. 25989-998. [DOI] [PubMed] [Google Scholar]
- 54.Skouloubris, S., J. M. Thiberge, A. Labigne, and H. De Reuse. 1998. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 664517-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sly, W. S., and P. Y. Hu. 1995. Human carbonic anhydrases and carbonic anhydrases deficiencies. Annu. Rev. Biochem. 64375-401. [DOI] [PubMed] [Google Scholar]
- 56.Smith, K. S., and J. G. Ferry. 2000. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24335-366. [DOI] [PubMed] [Google Scholar]
- 57.So, A. K.-C., G. S. Espie, E. B. Williams, J. M. Shively, S. Heinhorst, and G. C. Cannon. 2004. A novel evolutionary lineage of carbonic anhydrase (ɛ class) is a component of the carboxysome shell. J. Bacteriol. 186623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soto, A. R., H. Zheng, D. Shoemaker, J. Rodriguez, B. A. Read, and T. M. Wahlund. 2006. Identification and preliminary characterization of two cDNAs encoding unique carbonic anhydrases from the marine alga Emiliana huxleyi. Appl. Environ. Microbiol. 725500-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stähler, F. N., L. Ganter, K. Lederer, M. Kist, and S. Bereswill. 2005. Mutational analysis of the Helicobacter pylori carbonic anhydrases. FEMS Immunol. Med. Microbiol. 44183-189. [DOI] [PubMed] [Google Scholar]
- 60.Suerbaum, S., C. Josenhans, T. Sterzenbach, B. Drescher, P. Brandt, M. Bell, M. Droge, B. Fartmann, H. P. Fischer, Z. Ge, A. Horster, R. Holland, K. Klein, J. Konig, L. Macko, G. L. Mendz, G. Nyakatura, D. B. Schauer, Z. Shen, J. Weber, M. Frosch, and J. G. Fox. 2003. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 1007901-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takayama, M., T. Ohyama, K. Igarashi, and H. Kobayashi. 1994. Escherichia coli cad operon functions as a supplier of carbon dioxide. Mol. Microbiol. 11913-918. [DOI] [PubMed] [Google Scholar]
- 62.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Globek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fuji, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388539-547. [DOI] [PubMed] [Google Scholar]
- 63.Touati, E., V. Michel, J.-M. Thiberge, N. Wuscher, M. Huerre, and A. Labigne. 2003. Chronic Helicobacter pylori infections induce gastric mutations in mice. Gastroenterology 1241408-1419. [DOI] [PubMed] [Google Scholar]
- 64.Tripp, B. C., K. S. Smith, and J. G. Ferry. 2001. Carbonic anhydrase: new insights for an ancient enzyme. J. Biol. Chem. 27648615-48618. [DOI] [PubMed] [Google Scholar]
- 65.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 2772007-2011. [DOI] [PubMed] [Google Scholar]
- 66.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287482-485. [DOI] [PubMed] [Google Scholar]
- 67.Wen, Y., J. Feng, D. R. Scott, E. A. Marcus, and G. Sachs. 2007. The HP0165-HP0166 two-component system (ArsRS) regulates acid-induced expression of HP1186 α-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. J. Bacteriol. 1892426-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 715921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young, G. M., D. Amid, and V. L. Miller. 1996. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 1786487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]