Abstract
We describe a transcriptomic study of the effect of hha and ydgT mutations in Salmonella enterica serovar Typhimurium. A large number of genes showing altered expression are located in AT-rich horizontally acquired DNA sequences. Many of these genes have also been reported to be targets for H-NS. As Hha and YdgT interact with H-NS, our findings strongly suggest that Hha and/or YdgT must form complexes with H-NS when they silence these DNA regions.
The genomes of all members of genera belonging to the family Enterobacteriaceae contain at least one copy of a gene that encodes a member of the Hha-YmoA family of proteins (23, 24). These low-molecular-mass proteins show high degrees of similarity, and all of them have been identified as modulators of the expression of virulence factors (7, 30). Hha regulates, among other genes, expression of Escherichia coli α-hemolysin and esp operons (23, 29, 31, 36), the Salmonella enterica serovar Typhimurium hilA modulator of Salmonella pathogenicity island 1 (SPI1) (14, 32), and virulence genes in SPI2 (37). The Hha paralogue YdgT contributes to modulation of virulence genes in S. enterica serovar Typhimurium SPI2 (6, 37). The YmoA protein regulates the expression of several Yersinia virulence factors, as well as the RovA transcriptional activator (7, 13, 27). The Hha-YmoA proteins are encoded exclusively in the genomes of members of the family Enterobacteriaceae and in conjugative plasmids isolated from these organisms (24). Cells in which Hha has been depleted exhibit phenotypic properties similar to those of cells lacking the nucleoid-associated protein (NAP) H-NS, and it has been proposed that Hha-like proteins represent a new class of NAPs (27).
Several lines of evidence have shown that proteins belonging to the Hha family interact with members of the H-NS family to modulate gene expression (13, 16, 17, 31, 32, 33). Hha-like proteins mimic the H-NS oligomerization domain (23, 30), and interaction of Hha with H-NS increases the repressive ability of H-NS (25, 31). H-NS is the most extensively studied example of a NAP that has a role as an environmentally dependent modulator of gene expression (11, 35). H-NS is considered a transcriptional repressor, playing a relevant role in silencing xenogeneic DNA (12, 22, 28, 34).
It remains to be determined whether the set of H-NS-regulated genes coincides with the set of Hha-regulated genes or whether the latter is simply a subset of the former. A genome-wide analysis of the modulatory role of Hha-like proteins has not been performed. Here we describe a transcriptomic study of the effect of depletion of Hha-like proteins in S. enterica serovar Typhimurium and provide evidence that one of the main targets for Hha and/or its paralogue YdgT are the genes in horizontally acquired DNA sequences that are silenced by H-NS.
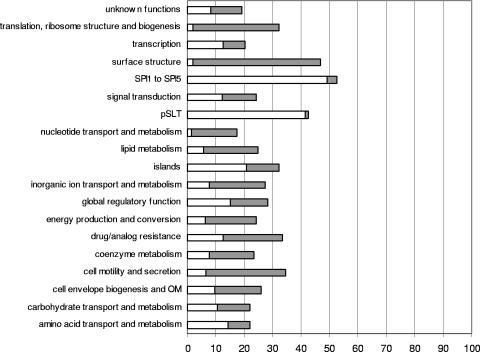
The genome of Salmonella contains, in addition to hha, a copy of the ydgT gene, which codes for an Hha paralogue, YdgT. When cells grow under nonstress conditions, YdgT does not significantly contribute to modulation. Nevertheless, this protein is overexpressed in hha mutants, which attenuates the hha phenotype (33). To prevent attenuation, we used a double hha ydgT mutant (strain SV5015HY). This deletion mutant, obtained by gene replacement, was constructed as described by Datsenko and Wanner (9). To obtain the ydgT mutant, the antibiotic resistance of plasmid pKD4 (kanamycin) was amplified using primers YDGTKAM1 and YDGTKAM2. To obtain the hha mutant, the antibiotic resistance of plasmid pKD3 (chloramphenicol) was amplified using primers HHAP1 and HHAP2. Both constructions were verified by using primers YDGTSA and YDGTSA2, primers HHA3 and HHA5, and primers corresponding to pKD3 (c1/c2) and pKD4 sequences (k1/k2) (primer sequences are described in Table S1 in the supplemental material). Strain SV5015HY (hha ydgT) was obtained by P22 HT transduction of the hha deletion into the ydgT strain (Table 1). Transcriptomic analyses were performed by using a Salmonella microarray that contained 6,119 probes corresponding to 5,116 open reading frames, 21 rRNAs, 86 tRNAs, 51 small RNAs, and 845 intergenic regions. The methods used for RNA extraction, retrotranscription, labeling, hybridization, microarray scanning, and data analysis will be described elsewhere (J. F. Mariscotti and F. García del Portillo, submitted for publication). Compared to the wild-type (wt) strain, strain SV5015HY showed altered expression of about 1,000 genes. The mRNA levels of 471 genes were >2-fold higher in the hha ydgT mutant, indicating that Hha and YdgT repress gene expression in the wt strain. The mRNA levels of 504 genes were >2-fold lower in the mutant strain, indicating that Hha and YdgT activate gene expression in the wt strain (see Tables S1 and S2 in the supplemental material). Up-regulation predominated in genes belonging to several functional categories, including genomic islands (SPI1 to SPI5 and other genes related to horizontally acquired DNA regions), and the pSLT plasmid. In contrast, down-regulation predominated in genes belonging to the surface structure, cell motility-secretion, and translation functional categories (Fig. 1).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmida | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| SV5015 | SL1344 His+ | J. Casadesús |
| SV3081 | LT2/pSLT− | 39 |
| SV5015HY | Δhha::Cm Δydgt::Km | This study |
| SV5015HY-2 | Δhha Δydgt | This study |
| SV4478 | LT2 ΔfinO::Km | 3 |
| SV5015-finO | SV5015/pSLT ΔfinO::Km | This study |
| SV5015HY-2-finO | SV5015HY-2/pSLT ΔfinO::Km | This study |
| Plasmids | ||
| pLG338-30 | oripSC101; Apr | 8 |
| pUBM22 | pBR322 hha Apr | 29 |
| pKD46 | Red helper plasmid, Apr | 9 |
| pKD3 | Template plasmid, Cmr | 9 |
| pKD4 | Template plasmid, Kmr | 9 |
| pCP20 | FLP helper plasmid, Apr Cmr | 5 |
The S. enterica serovar Typhimurium strains were derived from SV5015, a His+ derivative of strain SL1344 (19).
FIG. 1.
Changes in expression of genes belonging to functional groups and in pathogenicity islands (26, 38). The bars indicate the percentages of genes belonging to each group that show altered expression in SV5015HY cells. The gray bars indicate the proportions of genes that are down-regulated (M < 0) and the open bars indicate the proportions of genes that are up-regulated (M > 0) for each group (where M is the fold change log2 ratio). OM, outer membrane.
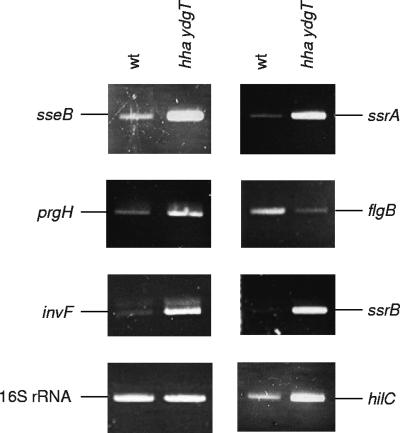
Most of the genes in SPI1 to SPI5 (93.22% of the genes) were predominantly overexpressed in strain SV5015HY (Table 2). To confirm the transcriptomic results obtained, we used reverse transcription (RT)-PCR to analyze the mRNA levels of some of the genes that were differentially expressed. Ready-to-Go RT-PCR beads (Amersham Biosciences) and primer pairs shown in Table S1 in the supplemental material were used. The genes selected were prgH (which encodes a component of the needle complex of the type III secretion apparatus of SPI1), hilC and invF (which encode invasion regulatory proteins of SPI1), sseB (which encodes a component of the translocation machinery of SPI2), and ssrA/ssrB (which encodes a two-component regulatory system of SPI2). RT-PCR analysis confirmed overexpression of all these genes in strain SV5015HY (Fig. 2).
TABLE 2.
Hha-YdgT-dependent expression of SPI, flagellar, and pSLT genes identified by transcriptomic analysis
| Genea | hha ydgT/wt expression ratio | Protein function |
|---|---|---|
| SPI5 | ||
| pipB | 4.18 | Secreted effector protein |
| pipC | 3.08 | Pathogenicity island-encoded protein C |
| sopB | 14.37 | Secreted effector protein |
| SPI2 | ||
| STM1381 | 0.30 | Putative cytoplasmic protein |
| STM1382 | 3.46 | Putative regulatory protein |
| STM1390 | 3.28 | Putative regulatory protein |
| ssrB | 5.26 | Transcriptional activator |
| ssrA | 8.85 | Sensor kinase |
| ssaE | 4.91 | Secretion system effector |
| sseB | 12.04 | Translocation machinery component |
| sscA | 5.24 | Secretion system chaperone |
| sseC | 9.88 | Translocation machinery component |
| sseD | 7.16 | Translocation machinery component |
| sseE | 3.19 | Secreted effector protein |
| sscB | 4.44 | Secretion system chaperone |
| ssaH | 3.82 | Type III secretion system apparatus protein |
| ssaI | 3.72 | Type III secretion system apparatus protein |
| ssaJ | 3.38 | Needle complex inner membrane lipoprotein |
| ssaK | 2.89 | Type III secretion system apparatus protein |
| ssaV | 2.49 | Type III secretion system apparatus protein |
| ssaO | 3.72 | Type III secretion system apparatus protein |
| ssaT | 0.34 | Type III secretion system apparatus protein |
| SPI1 | ||
| avrA | 7.73 | Secreted effector protein |
| sprB | 13.36 | Transcriptional regulator |
| hilC | 2.55 | Invasion regulatory protein |
| orgC | 4.69 | Putative cytoplasmic protein |
| orgB | 16.80 | Needle complex export protein |
| prgK | 6.34 | Needle complex inner membrane lipoprotein |
| prgJ | 8.17 | Needle complex minor subunit |
| prgI | 2.45 | Needle complex major subunit |
| prgH | 12.77 | Needle complex inner membrane protein |
| hilA | 2.78 | Invasion protein transcriptional activator |
| sptP | 3.52 | Protein tyrosine phosphatase/GTPase activating protein |
| sicP | 13.74 | Secretion chaperone |
| iacP | 3.48 | Acyl carrier protein |
| sipA | 3.59 | Secreted effector protein |
| sipD | 28.15 | Translocation machinery component |
| sipC | 19.90 | Translocation machinery component |
| sipB | 14.17 | Translocation machinery component |
| spaS | 5.22 | Type III secretion protein |
| spaR | 2.45 | Needle complex export protein |
| invJ | 13.18 | Needle length control protein |
| invI | 13.18 | Needle complex assembly protein |
| invB | 6.04 | Secretion chaperone |
| invA | 3.47 | Needle complex export protein |
| invF | 3.39 | Invasion regulatory protein |
| invH | 4.03 | Needle complex outer membrane lipoprotein precursor |
| STM2913 | 0.37 | Putative permease |
| SPI3 | ||
| slsA | 4.41 | Putative inner membrane protein |
| cigR | 3.71 | Putative inner membrane protein |
| mgtC | 4.18 | Mg2+ transport protein |
| STM3767 | 0.28 | Putative cytoplasmic protein |
| STM3770 | 4.21 | Putative phosphotransferase system enzyme IIC |
| STM3774 | 2.52 | Putative inner membrane protein |
| STM3782 | 3.81 | Putative phosphotransferase system galactitol-specific enzyme IIC |
| Flagellar genes | ||
| flgN | 0.16 | Putative FlgK/FlgL export chaperone |
| flgM | 0.22 | Anti-FliA factor |
| flgB | 0.16 | Flagellar basal body rod protein |
| flgC | 0.21 | Flagellar basal body rod protein |
| flgD | 0.13 | Flagellar basal body rod modification protein |
| flgE | 0.24 | Flagellar hook protein |
| flgF | 0.16 | Cell-proximal portion of basal body rod |
| flgH | 0.17 | Flagellar L-ring protein precursor |
| flgI | 0.37 | Flagellar P-ring protein precursor |
| flgJ | 0.29 | Flagellar biosynthesis protein |
| flgK | 0.22 | Flagellar hook-associated protein |
| cheB | 0.16 | Chemotaxis-specific methylesterase |
| cheW | 0.12 | Chemotaxis docking protein |
| fliD | 0.41 | Flagellar hook-associated protein |
| fliS | 0.41 | Flagellar protein FliS |
| fliT | 0.33 | Possible FliD export chaperone |
| fliF | 0.20 | Flagellar M-ring protein |
| fliH | 0.16 | Flagellar assembly protein |
| fliN | 0.30 | Flagellar motor switch protein |
| fliO | 0.22 | Flagellar biosynthetic protein |
| fljB | 0.14 | Flagellar biosynthesis protein |
| pSLT genes | ||
| rcK | 11.04 | Resistance to complement killing |
| srgA | 5.58 | Putative thiol-disulfide isomerase or thioredoxin |
| orf7 | 8.08 | Putative bacterial regulatory protein |
| pefI | 6.94 | Putative bacterial regulatory protein |
| orf6 | 9.58 | Putative outer membrane protein |
| orf5 | 8.08 | Putative outer membrane protein |
| pefA | 4.29 | Major fimbrial subunit |
| repA2 | 4.08 | DNA replication protein |
| PSLT025 | 0.46 | Putative cytoplasmic protein |
| PSLT045 | 3.81 | Putative resolvase |
| PSLT046 | 11.00 | Putative carbonic anhydrase |
| tlpA | 2.89 | Alpha-helical coiled-coil protein |
| psiB | 4.98 | Plasmid SOS inhibition |
| psiA | 4.24 | Plasmid SOS inhibition |
| traA | 4.84 | Pilus subunit |
| traL | 3.62 | Pilus assembly protein |
| traK | 8.34 | Pilus assembly protein |
| traP | 6.96 | Conjugative transfer protein |
| traC | 3.28 | ATP-binding protein |
| trbI | 2.55 | Pilus assembly protein |
| traU | 5.30 | Pilus assembly protein |
| trbC | 3.82 | Pilus assembly protein |
| traF | 5.05 | Pilus assembly protein |
| trbB | 3.57 | Conjugative transfer protein |
| traG | 10.89 | Mating pair stabilization and pilus assembly protein |
| traS | 4.41 | Entry exclusion protein |
| traT | 3.90 | Surface exclusion protein |
| PSLT107 | 2.75 | Putative cytoplasmic protein |
| finO | 2.61 | FinP binding protein |
| traJ | 2.55 | Conjugative transfer regulation |
| traM | 3.30 | Conjugative transfer mating signal |
| traN | 3.01 | Conjugative transfer aggregate stability |
| traQ | 2.29 | Conjugative transfer fimbrial synthesis |
Bold type indicates the genes containing binding sites for H-NS as described by Navarre et al. (28).
FIG. 2.
RT-PCR analysis of transcription of the prgH, hilC, invF, sseB, ssrA, and ssrB genes from strains SV5015 (wt) and SV5015HY (hha ydgT). 16S rRNA was used as a control to confirm that equivalent quantities of templates were loaded.
To examine the effect of deregulation of virulence genes in the SPIs, we used the competitive index (2). Equivalent numbers of cells from strains SV5015 and SV5015HY were combined and used to inoculate an animal host (BALB/c mice) (input). Bacteria were recovered after 48 h from the spleen and liver (output), and the competitive index was determined. The competitive indexes were 0.08 ± 0.05 and 0.05 ± 0.03 in spleen and liver homogenates, respectively. As previously reported for S. enterica serovar Typhimurium strain SL1344 (6, 37 ), the hha and ydgT alleles are responsible for an attenuated virulence phenotype.
Several (42.52%) of the putative open reading frames in the virulence plasmid pSLT exhibited altered expression in strain SV5015HY. Most of them (97%), including those in the tra operon, were overexpressed (Table 2). To further examine the effect of the hha and ydgT mutations on conjugation of plasmid pSLT, mating experiments were performed using a conjugation-derepressed derivative of plasmid pSLT, finO::Km (4). The antibiotic resistance cassettes associated with hha (chloramphenicol) and ydgT (kanamycin) mutations in strain SV5015HY were first deleted. To do this, we used FLP recombinase of plasmid pCP20 (9) and obtained strain SV5015HY-2. P22 HT transduction was used to transfer the finO::Km allele from plasmid pSLT of SV4478 into SV5015 and SV5015HY-2, generating strains SV5015-finO and SV5015HY-2-finO, respectively. The recipient used in the mating experiments was strain SV3081, a pSLT-cured strain, which was transformed with plasmid pLG338-30 to confer ampicillin resistance for selection of transconjugants. The frequency of plasmid transfer was calculated per donor bacterium. The frequency of pSLT FinO− plasmid transfer was 10-fold higher when strain SV5015HY-2 was used as the donor than when the wt strain was used as the donor (1.75 × 10−4 and 1.84 × 10−5, respectively). These results show that the Hha-YdgT proteins participate in the regulation of plasmid pSLT conjugation. The participation of Hha-like proteins in the modulation of conjugative plasmid transfer has also been reported previously for the IncH1 plasmid R27 (15).
These results show that Hha-like proteins negatively modulate the expression of horizontally acquired genes in enteric bacteria, either directly or indirectly. This observation coincides with data for target genes previously described for Hha-like proteins (23). Hha-YdgT may negatively modulate gene expression either directly or indirectly. A good example of this is the virulence genes of SPI1; Hha modulates the master regulator, the hilA gene (14, 20).
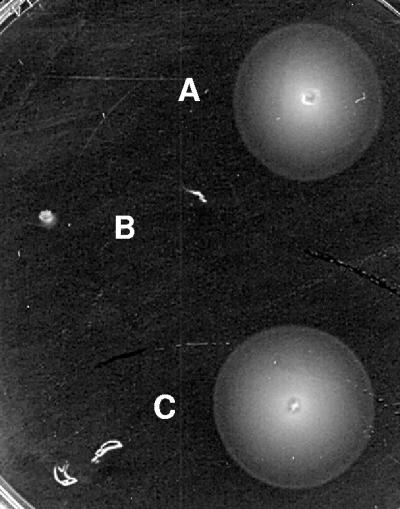
Genes showing reduced expression in strain SV5015HY belong to several functional categories. The decreased expression of many of these genes may be due to an indirect effect of the double mutation on cell physiology (most likely the genes in the translation, ribosome structure, and biogenesis functional categories) or on specific transcriptional repressors. Remarkably, one significant set of genes showing reduced expression includes many genes involved in flagellar biogenesis (Table 2). We also used RT-PCR to confirm deregulation of flgB, which encodes a flagellar basal body rod protein (Fig. 2). We also compared strains SV5015 and SV5015HY in motility agar plates (18). Strain SV5015HY formed a very small halo (Fig. 3). This phenotype was complemented by expressing a plasmid that contains hha (pUBM22). Neither hha nor ydgT single mutants exhibited the drastic effect on motility shown by the hha ydgT double mutant (data not shown). Hence, the lower expression of several flagellar genes in strain SV5015HY resulted in reduced motility. The doubling time in LB medium of SV5015HY was 40% lower than that of the wt strain. The decrease in the growth rate may be explained by the down-regulation of the genes involved in translation, ribosomes, and biogenesis, as shown in the transcriptomic analysis (Fig. 1). Nevertheless, the difference in the doubling times of the strains analyzed is not sufficient to explain the difference in the reduced halos formed in motility plates.
FIG. 3.
Growth of strains SV5015 (wt) (colony A), SV5015HY (hha ydgT) (colony B), and SV5015HY(pUBM22) (colony C) on a motility agar plate.
Hha-like proteins form nucleoprotein complexes with H-NS (13, 15, 23, 30, 33). Recent results presented independently by different groups have shown that this NAP silences the expression of horizontally acquired DNA under nonpermissive conditions (22, 28, 34). Hence, a significant set of genes that are silenced by H-NS should also be silenced by Hha-YdgT. To confirm this, we compared the reported H-NS binding sites in the Salmonella genome (28) and our transcriptomic data. The results (Table 2) clearly support the hypothesis that Hha and YdgT interact with H-NS to favor silencing of xenogeneic DNA. Although these results were obtained with two different Salmonella strains (SV5015 and 14028) and the experimental approaches differed, 87% of SPI genes were repressed by Hha-YdgT and are reported to contain binding sites for H-NS. The coincidence is also high for other SV5015HY up-regulated genes on other genomic islands (34%) and for the pSLT plasmid (46%). It has been suggested that H-NS is not an effective silencer at all binding sites for some horizontally transferred sequences (28). This suggestion is supported by our finding that Hha-like proteins also participate in silencing xenogeneic DNA. Coregulation of genes in SPIs by H-NS and Hha-YdgT can be also inferred from two recent independent reports. The Hha and YdgT proteins repress expression of SPI2 virulence genes (37). Furthermore, H-NS down-regulates SPI2 expression, in competition with the two-component activator system SsrA-SsrB (40). The results reported here show that the Hha and YdgT proteins also repress ssrA and ssrB expression, thereby supporting the hypothesis that the Hha/H-NS complex modulates SPI2 gene expression.
For many other functional categories, the coincidence between Hha-regulated genes and H-NS-regulated genes was low. This finding can probably be attributed to H-NS modulating housekeeping functions in the absence of Hha-YdgT or to indirect effects of the double hha ydgT mutation on cell physiology. Some well-characterized examples of H-NS-modulated operons are bgl and proU (10, 21). We have been unable to show that Hha-YdgT modulates these operons (unpublished results). We therefore suggest that the regulatory regions of the H-NS target genes include two categories: genes for which H-NS requires the formation of heteromeric complexes with Hha-like proteins in order to achieve efficient repression and genes repressed by H-NS homooligomers. Many of the H-NS-modulated genes that are located in horizontally acquired DNA, among others, belong to the first category.
The mechanism by which H-NS activates gene expression is not well characterized. This is also the case for Hha-YdgT. Nevertheless, we also found coincidences between genes down-regulated in strain SV5015HY and genes down-regulated in hns mutants. Genes related to flagellar biogenesis are positively regulated by H-NS in E. coli or Salmonella (1, 28). Our transcriptomic analysis showed that the Hha and YdgT proteins also positively regulate flagellar genes. Again, these findings point to participation of Hha-like proteins in some of the cellular processes that require H-NS.
Microarray data accession number.
The complete data set has been deposited under accession number E-MEXP-1303 at htpp:/www.ebi.ac.uk/arrayexpress.
Acknowledgments
We thank J. Casadesús for providing bacterial strains and for advice on conjugation experiments.
This work was supported by grants from the Ministerio de Ciencia y Tecnología (grants GEN2003-20234-C06-06 and BIO2004-02747 to A.J. and GEN2003-20234-C06-01 to F.G.-D.P.) and from the Generalitat de Catalunya (grant 2005SGR00635).
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bertin, P., E. H. Lee, P. Lejeune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 1765537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beuzón, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 31345-1352. [DOI] [PubMed] [Google Scholar]
- 3.Camacho, E., and J. Casadesús. 2005. Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol. Microbiol. 571700-1718. [DOI] [PubMed] [Google Scholar]
- 4.Camacho, E. M., and J. Casadesús. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by a leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 441589-1598. [DOI] [PubMed] [Google Scholar]
- 5.Cheperanov, P. P., and W. W. Wackernagel. 1995. Gene disruption in Escherichia coli: Tcr and Kmr cassetes with the option of Flp-catalyzed excision of the antibiotic resistance determinant. Gene 1582-14. [DOI] [PubMed] [Google Scholar]
- 6.Coombes, B. K., M. E. Wickham, M. J. Lowden, N. F. Brown, and B. B. Finlay. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contectual control of virulence during typhoid. Proc. Natl. Acad. Sci. USA 10217460-17465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis, G., C. Sluiters, I. Delor, D. Gelb, K. Kaniga, C. Lambert de Rouvroit, M. P. Sory, J. C. Vanooteghem, and T. Michiels. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 51023-1034. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, T. P., R. C. Montelaro, and E. P. Greenberg. 1993. Lentivirus envelope sequences and proviral genomes are stabilized in Escherichia coli when cloned in low-copy-number plasmid vector. Gene 993-98. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K12. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dole, S., V. Nagarajavel, and K. Schnetz. 2004. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 52589-600. [DOI] [PubMed] [Google Scholar]
- 11.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2391-400. [DOI] [PubMed] [Google Scholar]
- 12.Dorman, C. J. 2007. H-NS, the genome sentinel. Nat. Rev. Microbiol. 5157-161. [DOI] [PubMed] [Google Scholar]
- 13.Ellison, D. W., and V. L. Miller. 2006. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J. Bacteriol. 1885101-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 1836620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forns, N., R. C. Baños, C. Balsalobre, A. Juárez, and C. Madrid. 2005. Temperature-dependent conjugative transfer of R27: role of chromosome- and plasmid-encoded Hha and H-NS proteins. J. Bacteriol. 1873950-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García, J., T. N. Cordeiro, J. M. Nieto, I. Pons, A. Juárez, and M. Pons. 2005. Interaction between the bacterial nucleoid-associated proteins Hha and H-NS involves a conformational change of Hha. Biochem. J. 388755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García, J., C. Madrid, A. Juárez, and M. Pons. 2006. New roles for key residues in helices H1 and H2 of the Escherichia coli H-NS N-terminal domain: H-NS dimer stabilization and Hha binding. J. Mol. Biol. 359679-689. [DOI] [PubMed] [Google Scholar]
- 18.Gillen, K. L., and K. T. Hughes. 1991. Negative regulatory loci coupling flagellar synthesis to flagellar assembly in Salmonella typhimurium. J. Bacteriol. 1732301-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella enterica are non-virulent and effective as live vaccines. Nature 291238-239. [DOI] [PubMed] [Google Scholar]
- 20.Jones, B. D. 2005. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 43110-117. [PubMed] [Google Scholar]
- 21.Jordi, B. J., and C. F. Higgins. 2000. The downstream regulatory element of the proU operon of Salmonella typhimurium inhibits open complex formation by RNA polymerase at a distance. J. Biol. Chem. 27512123-12128. [DOI] [PubMed] [Google Scholar]
- 22.Lucchini, S., G. Rowley, M. D. Goldberg, D. Hurd, M. Harrison, and J. C. Hinton. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madrid, C., C. Balsalobre, J. García, and A. Juárez. 2007. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol. Microbiol. 637-14. [DOI] [PubMed] [Google Scholar]
- 24.Madrid, C., J. García, M. Pons, and A. Juárez. 2007. Molecular evolution of the H-NS protein: interaction with Hha-like proteins is restricted to Enterobacteriaceae. J. Bacteriol. 189265-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madrid, C., J. M. Nieto, S. Paytubí, F. Falconi, C. O. Gualerzi, and A. Juárez. 2002. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 1845058-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413852-856. [DOI] [PubMed] [Google Scholar]
- 27.Mikulskis, A. V., and G. Cornelis. 1994. A new class of proteins regulating gene expression in enterobacteria. Mol. Microbiol. 1177-86. [DOI] [PubMed] [Google Scholar]
- 28.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313236-238. [DOI] [PubMed] [Google Scholar]
- 29.Nieto, J. M., M. Carmona, S. Bolland, Y. Jubete, F. de la Cruz, and A. Juárez. 1991. The hha gene modulates hemolysin expression in Escherichia coli. Mol. Microbiol. 51285-1293. [DOI] [PubMed] [Google Scholar]
- 30.Nieto, J. M., C. Madrid, E. Miquelay, J. L. Parra, S. Rodríguez, and A. Juárez. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nieto, J. M., C. Madrid, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and A. Juárez. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 263349-358. [DOI] [PubMed] [Google Scholar]
- 32.Olekhnovich, I. N., and R. J. Kadner. 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357373-386. [DOI] [PubMed] [Google Scholar]
- 33.Paytubi, S., C. Madrid, N. Forns, J. M. Nieto, C. Balsalobre, B. E. Uhlin, and A. Juárez. 2004. YdgT, the Hha paralogue in Escherichia coli, forms heteromeric complexes with H-NS and StpA. Mol. Microbiol. 54251-263. [DOI] [PubMed] [Google Scholar]
- 34.Pflum, M. K. 2006. H-NS gives invading DNA the silent treatment. Nat. Chem. Biol. 2400-401. [DOI] [PubMed] [Google Scholar]
- 35.Rimsky, S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7109-114. [DOI] [PubMed] [Google Scholar]
- 36.Sharma, V. K., and R. L. J. Zuerner. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 1867290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silphaduang, U., M. Mascarenhas, M. Karmali, and B. K. Coombes. 2007. Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J. Bacteriol. 1893669-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonina. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2922-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torreblanca, J., S. Marqués, and J. Casadesús. 1999. Synthesis of finP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics 15231-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walthers, D., R. K. Carroll, W. W. Navarre, S. J. Libby, F. C. Fang, and L. J. Kenney. 2007. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol. Microbiol. 65477-493. [DOI] [PubMed] [Google Scholar]