Abstract
Prokaryotic secretion relies on proteins that are widely conserved, including NTPases and secretins, and on proteins that are system specific. The Tad secretion system in Aggregatibacter actinomycetemcomitans is dedicated to the assembly and export of Flp pili, which are needed for tight adherence. Consistent with predictions that RcpA forms the multimeric outer membrane secretion channel (secretin) of the Flp pilus biogenesis apparatus, we observed the RcpA protein in multimers that were stable in the presence of detergent and found that rcpA and its closely related homologs form a novel and distinct subfamily within a well-supported gene phylogeny of the entire secretin gene superfamily. We also found that rcpA-like genes were always linked to Aggregatibacter rcpB- or Caulobacter cpaD-like genes. Using antisera, we determined the localization and gross abundances of conserved (RcpA and TadC) and unique (RcpB, RcpC, and TadD) Tad proteins. The three Rcp proteins (RcpA, RcpB, and RcpC) and TadD, a putative lipoprotein, localized to the bacterial outer membrane. RcpA, RcpC, and TadD were also found in the inner membrane, while TadC localized exclusively to the inner membrane. The RcpA secretin was necessary for wild-type abundances of RcpB and RcpC, and TadC was required for normal levels of all three Rcp proteins. TadC abundance defects were observed in rcpA and rcpC mutants. TadD production was essential for wild-type RcpA and RcpB abundances, and RcpA did not multimerize or localize to the outer membrane without the expression of TadD. These data indicate that membrane proteins TadC and TadD may influence the assembly, transport, and/or function of individual outer membrane Rcp proteins.
Aggregatibacter (formerly Actinobacillus) actinomycetemcomitans is a gram-negative, capnophilic coccobacillus, principally known as the etiologic agent of localized aggressive periodontitis (16, 25, 39). A. actinomycetemcomitans is occasionally able to colonize sites outside of the oral cavity to produce other infections, and it is one of the so-called HACEK organisms, gram-negative bacteria that cause approximately 3% of infective endocarditis cases (20). Clinical isolates of A. actinomycetemcomitans adhere to surfaces nonspecifically and form remarkably strong biofilms (14, 15, 29). Insertional mutagenesis studies have shown that genes of the tad (tight adherence) locus are necessary for the nonspecific adherence of the bacterium to surfaces, as well as for the phenotypes of rough colony morphology, autoaggregation, and production of type IVb Flp pili (29, 31, 42, 45). Genetic and biochemical analyses have indicated that 13 tad gene products are involved (3, 29, 31, 42, 45, 66); only the flp-2 product is not (42).
tad loci have been identified in over half of the sequenced bacteria and in all of the archaeal genomes that have been completed (45, 52). Phylogenetic evidence strongly indicates that many bacterial species have acquired the tad genes from foreign sources, and because of its apparent propensity for horizontal transfer, the tad locus has also been named the widespread colonization island (44, 45). The tad locus has been implicated in the pathogenesis of several bacterial diseases. A. actinomycetemcomitans strains with inactivated flp-1 or tadA fail to colonize tooth surfaces and cause characteristic bone loss, reminiscent of localized aggressive periodontitis, in a rat model of that disease (59). A tadA mutant of the human pathogen Haemophilus ducreyi is avirulent in a skin pustule test (62). During in vivo assays to identify virulence genes, tadD was identified as likely to be critical for virulence in both the animal pathogen Pasteurella multocida and the fish-colonizing bacterium Yersinia ruckeri (12, 17). Thus, the study of tad loci and the molecular functions of Tad proteins is predicted to produce valuable insights about the life styles of diverse and disease-causing prokaryotes.
Many tad locus proteins (Flp1, TadV, RcpA, TadA, TadB, TadC, TadE, and TadF) exhibit similarity to proteins that participate in type II secretion (T2S), type IV pilus (T4P) assembly, type IV secretion, and type III secretion (T3S), but some of the Tad proteins (RcpB, RcpC, TadZ, TadD, and TadG) are not significantly similar to known proteins. The Tad proteins are predicted to assemble to form a macromolecular structure for the assembly and secretion of Flp pili (67). Although the tad genes are widespread, the architecture and operation of the Tad secretion system have not been elucidated.
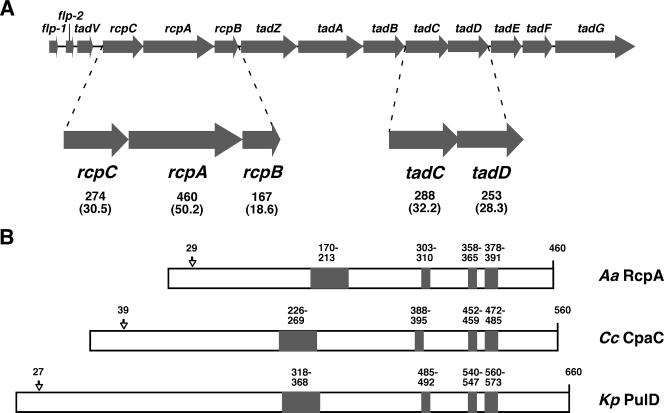
Rough-colony proteins RcpA and RcpB were discovered in an outer membrane fraction from adherent, rough-colony-forming A. actinomycetemcomitans cells, and three open reading frames, called rcpCAB, were found by nucleotide sequencing (23). We mapped the rcp genes to the tad locus (see Fig. 1A) (29). As in other genes of the tad locus, transposon insertions in rcpA and rcpC abolish the nonspecific adherence and associated phenotypes, including Flp-fibril secretion, and these defects can be restored by genetic complementation (45). Our attempts to inactivate rcpB in adherent strains failed to produce viable transformants, and we determined that rcpB is essential for viability in the context of an intact tad locus (42).
FIG. 1.
tad locus and conserved secretin sequences. (A) The tad locus is composed of 14 genes that are transcribed in the same direction (22). The number of amino acid residues for the full-length protein products of rcpCAB and tadCD are shown by the top numbers under the genes. The sizes (in kilodaltons) of the proteins are specified by the numbers in parentheses directly below. The nucleotide sequence was from the strain CU1000N. (B) Conservation in secretins. Conserved regions A to D in A. actinomycetemcomitans (Aa) RcpA, C. crescentus (Cc) CpaC, and K. pneumoniae (Kp) PulD are shaded. The residue numbers spanned by each region are denoted. The alignment of the proteins is based on regions C and D, which are spaced equally in all three proteins. Arrows indicate predicted signal sequence cleavage sites.
Based on its sequence and its presence in the outer membrane, the RcpA protein is expected to form the channel through which Flp pili of the Tad secretion system traverse the outer membrane (23). The RcpA homolog CpaC has been localized to the pole of the environmental organism Caulobacter crescentus, whose tad locus has been named cpa for Caulobacter pilus assembly (60). Both CpaC and RcpA contain C-terminal domains that are conserved among known secretin proteins (see Fig. 1B), including the proteins PulD of Klebsiella spp., pIV of filamentous phage, and PilQ of Neisseria gonorrhoeae (19, 53, 60). The evolutionary relationship of the RcpA protein to other secretins and its ability to function as a secretin are not well-characterized.
Although rcpB is probably essential for the production of Flp pili, an outer membrane function for RcpB cannot be inferred from its predicted primary sequence. Obvious homologs of rcpB are found exclusively in tad loci of members of the Pasteurellaceae family (30). The cpaD gene, located in the cpa locus in a position analogous to that of rcpB (i.e., between homologs of rcpA and tadZ), may be a relative of rcpB despite our inability to detect any sequence similarity, and cpaD is known to be required for pilus biogenesis in C. crescentus (60).
Sequence predictions indicate that RcpC and TadD may localize to the bacterial outer membrane. The RcpC protein does not have a strongly predicted function. tadD is absent from the tad loci of gram-positive bacteria, as is true for the rcpAB genes, which encode proteins known to localize to the outer membrane (67). The protein sequence of TadD contains tetratricopeptide repeat (TPR) domains and a predicted site for N-terminal cleavage by signal peptidase II. These sequence features are common among a class of known outer membrane lipoproteins, called docking proteins for their known role in positioning secretin proteins at the outer membrane (2, 6, 40). Finally, TadC displays similarity to PilC-like proteins (41) and is likely to have multiple inner membrane-spanning segments, which presents the likelihood that this protein may be required for the stability of other members of the Tad system, potentially as an inner membrane scaffold. The localization and characterization of these proteins in A. actinomycetemcomitans have not been reported.
In this study, we examined properties of known and predicted membrane proteins of the Tad secretion system. We have compared the RcpA amino acid sequence to sequences of other secretins to demonstrate the evolutionary relationship of RcpA within the protein superfamily and among tad loci-containing organisms. We also generated antisera specific to Rcp and Tad proteins to demonstrate evidence of RcpA multimer formation and to show the localization patterns of several Tad proteins and the Tad-related requirements for their presence.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. The addition of “N” to a strain designation denotes a spontaneous nalidixic acid-resistant derivative. Two derivatives of rough clinical isolates, CU1000N (serotype f) (29) and DF2200N (serotype a) (32), were used as wild-type strains. A. actinomycetemcomitans strains were grown and stored in A. actinomycetemcomitans growth medium (AAGM) broth or agar, each with 0.75% glucose (wt/vol) and 0.4% sodium bicarbonate (wt/vol), as previously described (65). Chloramphenicol (2 μg/ml) and nalidixic acid (20 μg/ml) were used to supplement AAGM agar plates or broth, as necessary.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| A. actinomycetemcomitans | ||
| Aa0727 | Aa1577 with pJMM108; Kmr Nalr Cmr | Morrison and Figurski, unpublished |
| Aa0886 | CU1000N tadZ::IS903φkan; Kmr Nalr | 45 |
| Aa1005 | CU1000N; spontaneous Nalr mutant of rough clinical isolate CU1000, serotype f; Nalr | 29 |
| Aa1332 | CU1000N tadB::IS903φkan; Kmr Nalr | 29 |
| Aa1347 | CU1000N tadE::IS903φkan; Kmr Nalr | 29 |
| Aa1354 | CU1000N rcpA::IS903φkan; Kmr Nalr | 45 |
| Aa1359 | CU1000N tadC::IS903φkan; Kmr Nalr | 29 |
| Aa1360 | CU1000N tadA::IS903φkan; Kmr Nalr | 29 |
| Aa1361 | CU1000N rcpC::IS903φkan; Kmr Nalr | 45 |
| Aa1511 | CU1000N tadF::IS903φkan; Kmr Nalr | 29 |
| Aa1561 | CU1000N tadG::IS903φkan; Kmr Nalr | 29 |
| Aa1577 | CU1000N tadD::IS903φkan; Kmr Nalr | 29 |
| Aa1758 | Aa1577 with pJAK17; Kmr Nalr Cmr | This study |
| Aa1759 | Aa1577 with pPP40; Kmr Nalr Cmr | This study |
| Aa3074 | DF2200N tadV::mini-Tn5Km; Kmr Nalr | 42 |
| JK1010 | CU1000N flp-1::IS903φkan; Kmr Nalr | 31 |
| MB1228 | DF2200N; spontaneous Nalr mutant of rough clinical isolate DF2200, serotype a; Nalr | 32 |
| E. coli | ||
| BL21(DE3) | F−dcm ompT hsdSB(rB− mB−) gal (λDE3) | Novagen |
| SC1031 | BL21(DE3) with pSAC117; Kmr | This study |
| SC1032 | BL21(DE3) with pSAC118; Kmr | This study |
| SC1041 | BL21(DE3) with pPP40; Cmr | This study |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) deoR ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG (φ80lacZΔM15) | Invitrogen |
| Plasmids | ||
| pBP100 | RcpB in pCR2.1-TOPO | This study |
| pBP111 | RcpC in pCR2.1-TOPO | This study |
| pCR2.1-TOPO | TA cloning vector; Apr Kmr | Invitrogen |
| pET30a | Cloning vector used to produce N-terminal fusions to six-His tag; Kmr | Novagen |
| pJAK17 | Broad-host-range mobilizable IncQ vector derived from pMMB67; carries tac promoter; CmrlacIq | Kornacki, unpublished |
| pJMM108 | tadC upstream region-tadD-T7 coding sequence in pJAK17; Cmr | Morrison and Figurski, unpublished |
| pPP40 | rcpA from CU1000N in pJAK17; Cmr | 45 |
| pSAC117 | rcpC from CU1000N in pET30a with 5′ BamHI and 3′ EcoRI ends; Kmr | This study |
| pSAC118 | rcpB from CU1000N in pET30a with 5′ BamHI and 3′ Sall ends; Kmr | This study |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Nalr, nalidixic acid resistance; Strr, streptomycin resistance.
Escherichia coli Top10 (Invitrogen) was used for all subcloning procedures and served as a conjugative donor for the mobilization of broad-host-range IncQ expression vectors into A. actinomycetemcomitans. As described previously (21, 65), an oriT-defective mutant of the IncP RK2 plasmid, pRK21761, which does not self-transfer efficiently, was used to mobilize IncQ vectors, including pJAK17 (J. A. Kornacki, unpublished data). E. coli strains were grown on Luria-Bertani (LB) (56) agar plates or in LB broth at 37°C, and kanamycin (50 μg/ml) or chloramphenicol (50 μg/ml) was used for the appropriate selection of plasmids. Electroporation or CaCl2 transformation of E. coli with recombinant plasmids was done as described previously (7). For the expression of all proteins in E. coli strain BL21(DE3), LB was supplemented with the Overnight Express autoinduction system (Novagen) and the cells were grown at 30°C.
DNA manipulations.
All A. actinomycetemcomitans gene sequences were amplified from CU1000N genomic DNA that was extracted using the DNeasy tissue kit (QIAGEN). PCR was done using the high-fidelity DNA polymerase TripleMaster Taq (Eppendorf) and primers that were purchased from Sigma Genosys. Restriction endonucleases and T4 DNA ligase were used according to the instructions of the manufacturer (New England Biolabs). After agarose gel electrophoresis, the extraction of digested plasmid or amplified DNA was done using either the QIAex II kit (QIAGEN) or SpinX tubes (Corning). Extracted PCR products were ligated into the pCR2.1-TOPO TA cloning vector (Invitrogen). Plasmid preparations were made with the QIAprep spin miniprep kit (QIAGEN), and all cloned PCR products were confirmed by nucleotide sequencing at the Columbia University DNA sequencing facility by using an Applied Biosystems 3100 capillary sequencer.
Protein sequence analysis.
Within the MacVector 7.2 software (Accelrys Inc.), the ClustalW algorithm was used to produce amino acid alignments. The NCBI Entrez (http://www.ncbi.nlm.nih.gov) sequences used were those of A. actinomycetemcomitans RcpA (AAN75208), C. crescentus CpaC (AAF40192), and Klebsiella pneumoniae PulD (AAA25126.2). PSORT (http://www.psort.org) (18) and CELLO (http://cello.life.nctu.edu.tw/) (70) were used to make predictions of protein features and subcellular localization in gram-negative bacteria.
Database sequence collection and phylogenetic analysis.
Sequences of the following proteins were obtained from the NCBI website (http://www.ncbi.nlm.nih.gov) and used for searching for the secretin superfamily: RcpA from A. actinomycetemcomitans, CpaC from C. crescentus, PilQ from Pseudomonas aeruginosa, PulD from Klebsiella pneumoniae, InvG from Salmonella enterica, SpiA from S. enterica, BfpB from E. coli, TcpC from Vibrio cholerae, HrcC from Erwinia amylovora, and pIV from filamentous phage from E. coli. Each sequence was used in a separate BLAST search (1) of GenBank and unfinished genome sequences available in the NCBI database, and the results from each search were compiled. Default settings of the BLAST program were used, except that we did not use a low-complexity filter, as this option often excluded clearly conserved regions from similarity calculations. In all, a nonredundant data set of 186 amino acid sequences was used, each of which significantly exceeded BLAST default criteria.
Protein sequences were aligned using CLUSTAL X 1.63 (64), gap parameters were varied in three independent alignments, and the three resulting matrices were concatenated for analysis: gap/change ratios for each alignment were 4, 7, and 10. This method is called elision and can be used to account for the possibility that certain portions of the alignment are incorrect (68). The alignment is available upon request.
We did parsimony analysis using the heuristic “ratchet” method (38), implemented with the aid of PAUPRat (Sikes & Lewis, Storrs, CT) in conjunction with PAUP. Two hundred replicates of the ratchet method were done. In each iteration, 15% of the characters were upweighted using tree-branch reconnection (TBR). Only one tree at each step was saved. TBR with the MulTrees option was done on the resulting trees. All characters and state transformations were given equal weight, and columns with gaps were retained as phylogenetically informative characters (43).
To calculate confidence in the trees, we generated bootstrap values and Bremer decay indices using the program AutoDecay with PAUP (10). Ten TBR replicates at each node in the phylogeny were done to obtain the Bremer index. Because Bremer indices were calculated using the elided data set of three independent alignments, each value was divided by three and rounded to the nearest 10th. We did 100 bootstrap replicates in PAUP, with one iteration of random addition followed by TBR, limiting the search time to 1 h for each replicate. Bootstrap values were retained if they were consistent with the 50% majority consensus tree.
Generation of antisera.
RcpC and RcpB recombinant proteins were constructed as follows. The full-length rcpC and rcpB genes were amplified using the primer sets RcpCBamHIup (5′-GGATCCATGAATTACAGAACGCTCTTG-3′) and RcpCEcoRIdwn (5′-GAATTCTTATTGACCTCTTAATTTTCTGATGAA-3′) and RcpBBamHIup (5′-GGATCCATGAGAAAATTAGTTATTACGGCCTC-3′) and RcpBSalIdwn (5′GTCGACTTAATACTTCAATTGAACACGCTGATT-3′), respectively, and then cloned into pCR2.1-TOPO (Invitrogen) to generate pBP111 and pBP100. The RcpC coding sequence was excised with BamHI and EcoRI, and the fragment encoding RcpB was removed with BamHI and SalI. Each of these fragments was then ligated into the expression vector pET30a (Novagen), proximal to a sequence encoding a His6 tag, producing pSAC117 and pSAC118.
In E. coli, His-tagged RcpB and RcpC recombinant proteins were expressed from pSAC117 and pSAC118, whereas the native (full-length) RcpA was expressed from pPP40 (45). Autoinduced cells were treated as described in the protocol for the Bugbuster protein extraction reagent (Novagen), with the addition of 100 mM NaCl and 5 mM MgCl2 to either Bugbuster or the equivalent B-PER II solution (Pierce) to increase protein stability.
Following lysis, the resuspension of insoluble material with 6 M urea solubilized recombinant RcpB and RcpC, each of which was purified on a column (Pierce) by using the His-Bind kit (Novagen) with the addition of 6 M urea to all buffers. Purified proteins were sent to Invitrogen for the immunization of rabbits.
For purification and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of RcpA, insoluble material was resuspended in Laemmli buffer (35) and boiled after Bugbuster lysis. The abundant protein band was removed in gel slices and sent for the production of antisera in rabbits (at Cocalico Biologicals, Reamstown, PA).
When necessary, the adsorption of nonspecific antibodies with acetone powders was used to reduce the background (24).
The TadC antiserum was generated using the TadC peptide comprising amino acids 40 to 58 (ERPKDQDSDEVAKNKSKQQ) conjugated to the keyhole limpet hemocyanin carrier protein via an N-terminal cysteine residue. The resulting protein was used to generate specific antisera in rabbits at Zymed Laboratories, Inc. The antisera were enriched with TadC peptide-specific antibodies by affinity purification at Zymed Laboratories, Inc.
Preparation of soluble and membrane fractions.
The fractionation of A. actinomycetemcomitans strains by differential detergent solubilization was done essentially as described before (3, 23), with a few modifications. Briefly, adherent A. actinomycetemcomitans cells were grown in 750-ml tissue culture flasks (BD Falcon), scraped, and pelleted by centrifugation. Nonadherent cells were grown in and harvested from 50-ml conical tubes (BD Falcon). For lysis, the cell pellets of adherent or nonadherent cultures were first frozen and then resuspended in 10 mM HEPES (pH 7.5) with 50 mM NaCl and incubated with 0.1 mg of lysozyme (Sigma)/ml on ice prior to physical disruption by sonication. Sonicates were cleared twice by centrifugation at 7,000 × g for 10 min. Subsequent centrifugations were carried out in a type 70.1 Ti Beckman ultracentrifuge rotor at 105,000 × g for 1 h. The soluble fraction contained cytoplasmic and, presumably, periplasmic proteins. The pellet, comprising bacterial membranes, was resuspended in buffer containing 0.5% Sarkosyl (N-laurylsarcosine; Sigma), and the suspension was rocked for 30 min at room temperature. The Sarkosyl-soluble fraction contained inner membrane proteins, whereas the pellet, resuspended in a 500-μl solution of 10 mM HEPES (pH 7.5) with 50 mM NaCl, 1% Sarkosyl, and 10 mM EDTA, consisted of outer membrane proteins.
For experiments using the strain Aa1759, fractions were prepared from twice the volume of overnight cultures.
Immunoblot analysis.
A. actinomycetemcomitans strains were grown in 10 ml of AAGM broth for 17 h. Cultures were centrifuged to harvest bacterial cells, and the pellets were resuspended and boiled for 5 min in standard Laemmli loading buffer (35) containing 2% SDS and 5% β-mercaptoethanol. Equal amounts of protein from each strain were resolved by SDS-PAGE and analyzed by immunoblotting with anti-RcpA (1:10,000), anti-RcpB (1:2,500), anti-RcpC (1:10,000), or anti-TadC (1:1,000) antiserum. An anti-rabbit immunoglobulin G antibody conjugated to horseradish peroxidase (HRP; Pierce) was used as a secondary antibody at 1:50,000. We were unable to raise a suitable antiserum for TadD, but for the detection of TadD, we used a functional, tagged version of TadD (TadD-T7) expressed from pJMM108 (J. M. Morrison and D. H. Figurski, unpublished data) and anti-T7 epitope tag antibody (1:5,000; Novagen), along with HRP-conjugated anti-mouse immunoglobulin G secondary antibody at a dilution of 1:50,000 (Pierce). Femto Western chemiluminescence reagent (Pierce) was used as a substrate for HRP.
To determine subcellular localizations by immunoblotting, ethanol precipitation was used to concentrate inner membrane fractions. Protein samples from soluble and membrane fractions were prepared by adding 4× Laemmli loading buffer and were treated as described above.
Multimerization samples were run on 8% polyacrylamide gels after resuspended pellets were heated at 37, 65, or 100°C for 5 min. For the efficient detection of these samples by immunoblotting, it was necessary to use modified electroblot buffer prepared with 10% methanol (71) and to transfer proteins onto nitrocellulose overnight at 30 V.
For experiments using the strain Aa1759, multimerization samples were prepared from twice the volume of overnight cultures.
RESULTS
rcpA-like secretin genes are phylogenetically distinct.
Based on strong scores in BLAST searches and apparently conserved sequence features (Fig. 1B) (23), rcpA is a member of the secretin gene superfamily. To examine further the relationship of rcpA and its close homologs to other members of the secretin gene family, we undertook a rigorous phylogenetic analysis of the entire superfamily. One hundred eighty-six amino acid sequences were identified, all from organisms in the domain Bacteria. No significant homologs in members of the Archaea or Eukarya or in other organisms with a single lipid membrane were identified, strengthening the idea that secretins are dedicated exclusively to trafficking proteins across the outer membrane.
The secretin superfamily tree was tentatively rooted with the monophyletic clade representing secretins involved in the macromolecular process of T3S (e.g., the product of sctC), and secretin genes were divided into two major families (Fig. 2). The sctC family, labeled based on the inclusive nomenclature of Hueck (26), includes genes involved in T3S and flagellar biosynthesis. The second family, gspD, is based on the inclusive nomenclature of Pugsley and Possot (48, 49), and the gspD/T2S subfamilies were named using the first described representative of each clade based on our literature review (5, 33, 54, 55, 61).
FIG. 2.
Phylogeny of secretin gene superfamily. We have divided the secretin gene superfamily into two primary families (sctC and gspD), whose names are based on proposals for inclusive nomenclature schemes. T3S secretin genes and genes for secretins associated with flagellar biosynthesis are labeled sctC, for secretion and cellular translocation, as proposed by Hueck (26). Genes involved in T2S, T4P biosynthesis, and Tad secretion are collectively labeled gspD, for general secretion pathway (48, 49). Subfamilies are named for the first described representative of the clade based on our literature review (see the text). The rcpA subfamily, along with its closest relatives, is shown enlarged. Within the rcpA subfamily, a white box indicates rcpA genes that were identified as being adjacent to a putative homolog of cpaD without other tad locus genes found in the near vicinity. All other members of the rcpA subfamily were found with closely linked tad locus homologs. Numbers on branches indicate Bremer support values (designated with a “d” by convention) and bootstrap values (a dot indicates a bootstrap value of 100). An expanded superfamily tree with named secretin proteins, a list of secretin genes used, and Bremer and bootstrap indices are available as supplemental figures (see the supplemental material). Complete names of organisms listed are as follows: Pasteurella multocida, Haemophilus ducreyi, Yersinia enterocolitica, Yersinia pestis, Agrobacterium tumefaciens, Sinorhizobium meliloti, Mesorhizobium loti, Caulobacter crescentus, Rhodopseudomonas palustris, Magnetospirillum magnetotacticum, Chlorobium limicola, Chlorobium tepidum, Ralstonia metallidurans, Ralstonia solanacearum, Desulfovibrio vulgaris, Bradyrhizobium japonicum, Sinorhizobium fredii, Burkholderia xenovorans, Bordetella pertussis, Acidithiobacillus ferrooxidans, Pseudomonas fluorescens, Pseudomonas syringae, Pseudomonas aeruginosa, Deinococcus radiodurans, Thermus thermophilus, and Thermotoga maritima.
rcpA-like genes form a monophyletic clade distinct from genes for secretins for classical T2S (pulD), T4P biogenesis (pilQ), filamentous phage extrusion (gene IV), and T3S (sctC). The rcpA subfamily is most closely related to a small clade of putative secretin genes from the Ralstonia solanacearum chromosome (RSc2303, accession no. NP_520424) and megaplasmid (RSp0143, accession no. NP_521704 and RSp0474, accession no. NP_522035) (Fig. 2), which are not classified as part of a tad locus based on the phylogenetic relationships of nearby genes. Interestingly, a small clade of genes from the Rhizobiaceae (e.g., rhcC2) is included within the rcpA subfamily, but these genes are not found closely linked to tad loci or to any other definable loci for transport systems (Fig. 2). However, in all cases, these genes are found adjacent to a gene with strong similarity to cpaD, a gene that is found in the same position as rcpB in many tad loci. Indeed, to our knowledge, all genes in the rcpA subfamily are adjacent to a clear homolog of either cpaD or rcpB.
Genes for secretins from bacteria outside of the proteobacteria, such as Thermotoga maritima, Deinococcus radiodurans, Synechocystis spp., Aquifex aeolicus, and Fusobacterium nucleatum, tended to be found at basal (early-branching) positions, suggesting ancient divergences of these genes—reflective of the ancient divergence of the genomes of these bacteria from the proteobacterial lineage. One small clade of secretins from these early-branching organisms is the sister group of the clade composed of the rcpA subfamily and the R. solanacearum secretin genes discussed above. This phylogenetic pattern indicates that the rcpA secretin subfamily has a very ancient origin.
RcpA forms multimers that are stable in the presence of detergent.
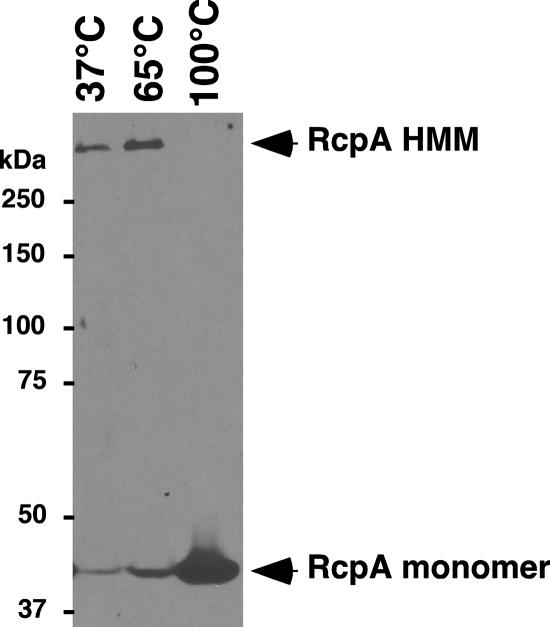
Known secretin proteins form multimers of 12 to 14 subunits (2, 53, 63) that run as high-molecular-weight species on polyacrylamide gels and resist dissociation by detergent and heat (36, 37). Whole-cell extracts of A. actinomycetemcomitans were heated at three different temperatures and examined for multimers by immunoblot analysis with antiserum specific to the RcpA protein. At 37 and 65°C, we detected a band migrating above 250 kDa (Fig. 3). The SDS-resistant high-molecular-weight multimer (HMM) in these samples was accompanied by a minor band at 46 kDa, the correct molecular mass for the monomeric form of RcpA. At 100°C, we detected the lower RcpA band as the dominant species, and no HMM was detected (Fig. 3). That the HMM was relatively detergent resistant at the lower temperatures, but not after boiling, may indicate the existence of RcpA predominantly as an assembled multimer in the bacterium. The 37 and 65°C HMM-containing samples displayed only monomers upon boiling and electrophoresis (data not shown).
FIG. 3.
Multimerization of RcpA. Whole-cell extracts of A. actinomycetemcomitans CU1000N (wild type) were heated at 37, 65, or 100°C for 5 min in standard loading buffer and separated by SDS-PAGE through an 8% polyacrylamide gel, as described in the text. Immunoblotting was done with anti-RcpA antiserum. Arrows indicate the positions of the RcpA monomer and the HMM.
The same banding pattern of RcpA was observed with the derivatives of other clinical isolates of A. actinomycetemcomitans: DF2200N (serotype a), HK1651N (serotype b), and IDH781N (serotype d) (data not shown). In addition, we have found and sequenced a tad locus from Aggregatibacter (Haemophilus) aphrophilus (34, 39, 45; C. Sheth, C. Mott, A. Soczewska, P. J. Planet, and D. H. Figurski, unpublished data), and we were able to detect both the RcpA HMM and the monomer by probing A. aphrophilus cells with the antiserum specific for A. actinomycetemcomitans RcpA (data not shown). Together, these data provide the first biochemical evidence that the RcpA proteins in tad locus-containing organisms participate in a higher-ordered structure.
Localization of Rcp and Tad proteins to the bacterial membranes.
The rcpCAB genes are predicted to code for proteins of 274, 460, and 167 residues, respectively, while the tadCD genes are likely to produce 288- and 253-amino-acid proteins (Fig. 1A). Predicted signal sequence cleavage sites of CU1000N RcpA (residue 29) and RcpB (residue 23) would produce proteins of approximately 46.9 and 16.4 kDa, respectively (18). Haase et al. (23) reported the migration of RcpA at 43 kDa and RcpB at 20 kDa in outer membrane fractions. We successfully generated polyclonal antisera to each of the three Rcp proteins and to the TadC protein. We also made two attempts to generate antiserum specific to TadD, but neither antiserum was able to detect the native protein in A. actinomycetemcomitans (data not shown).
The subcellular fractionation of A. actinomycetemcomitans was achieved by ultracentrifugation and selective solubilization of the inner membrane with the anionic detergent Sarkosyl. Previously, we used this method to demonstrate the localization pattern of the recombinant TadA-T7 protein (3). In addition, using this method with TadA-specific antiserum, we have since confirmed that native TadA localizes to the soluble and inner membrane fractions (data not shown).
Immunoblotting of subcellular fractions for RcpA, RcpB, and RcpC revealed that none of the Rcp proteins were present in detectable amounts in the buffer-soluble fraction, which consisted of cytoplasmic and, probably, periplasmic proteins (Fig. 4, lane S). RcpA and RcpC were observed in the inner membrane (Fig. 4, lane IM). All three Rcp proteins, including RcpB, were abundant in the Sarkosyl-insoluble outer membrane of A. actinomycetemcomitans (Fig. 4, lane OM). The TadC protein was not present in the soluble or outer membrane fractions but was contained exclusively in the inner membrane fraction (Fig. 4).
FIG. 4.
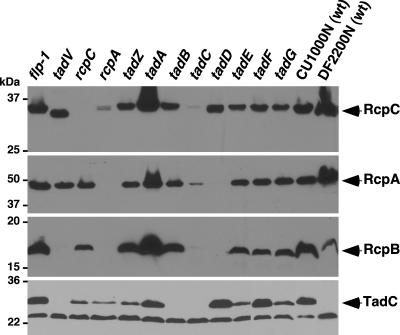
Localization of Rcp proteins. Subcellular fractions of CU1000N (wild type [wt]) and Aa0727, the tadD mutant strain with added TadD-T7 (via pJMM108), were prepared by differential detergent solubilization of the inner membrane with 0.5% Sarkosyl, as described in the text. Immunoblots were probed with antiserum specific to RcpC, RcpA, RcpB, or TadC or to the T7 epitope tag. Arrows indicate the positions of RcpC, RcpA, RcpB, TadC, and TadD-T7. S, soluble fraction; IM, inner membrane; OM, outer membrane.
We used a plasmid construct, pJMM108, to determine the localization of TadD by expressing TadD-T7 in the A. actinomycetemcomitans CU1000N-derived tadD mutant. The open reading frames of tadC and tadD overlap, and previous plasmid constructs used to complement the tadD mutation included tadC for its Shine-Dalgarno sequence (29). In contrast, the pJMM108 plasmid (J. M. Morrison and D. H. Figurski, unpublished) provides a functional copy of tadD that initiates protein synthesis from a small region upstream of tadC, without the tadC open reading frame. This construct (tadC upstream region-tadD-T7 coding sequence) also includes a sequence encoding a T7 tag at the 3′ end of tadD so that the tagged protein (TadD-T7) can be visualized with anti-T7 epitope tag antiserum. The protein is functional because it complements the tadD mutation for the adherence-related phenotypes (J. M. Morrison and D. H. Figurski, unpublished). Immunoblots of the subcellular fractions prepared from the tadD strain complemented with TadD-T7 showed the presence of the tagged protein in the inner and outer membranes (Fig. 4). We were unable to detect TadD-T7 in the soluble fraction (Fig. 4), indicating that TadD does not remain in the cytoplasmic interior and is probably not in the periplasm either.
As expected from previous work (23), these results confirm the presence of RcpA and RcpB in the outer membrane. They also provide the first evidence for the localization of RcpC in both membranes and reveal that some RcpA protein may be observed in the inner membrane fraction. Additionally, TadC was localized to the inner membrane, and TadD-T7 was found in both of the bacterial membranes.
Other Tad proteins influence Rcp protein abundance.
The association of tad locus proteins into a complex may be necessary for the operation of the Tad secretion system (45, 67). We reasoned that other proteins produced from the tad locus might influence the expression, abundance, and/or stability of the Rcp proteins. We compared the levels of production of the RcpA, RcpB, and RcpC proteins within nonpolar tad mutants (29, 31, 42, 45) to the levels produced by the parental, wild-type strains CU1000N and DF2200N. Our analysis did not include a mutant form of flp-2, as flp-2 is not required for the production of Flp pili or for adherence, autoaggregation, and rough-colony formation by A. actinomycetemcomitans (42). We also did not include an rcpB mutant, since rcpB appears to be indispensable for the survival of otherwise tad+ bacteria (42).
When immunoblots were probed with the RcpC-specific antiserum, RcpC was found as an abundant band in wild-type bacteria whereas no band was detected in the rcpC mutant, as expected (Fig. 5). We found that the abundance of the RcpC protein in the rcpA mutant was greatly reduced and that RcpC was nearly absent from the tadC mutant. The abundance of RcpC in other tad mutants was not significantly affected, although it was observed that the protein in the tadV strain migrated slightly faster than the RcpC proteins in the other strains (Fig. 5). Because the tadV strain was made in a DF2200N background (42), we compared its RcpC banding pattern to that of the DF2200N wild type. As the protein in the tadV strain, RcpC in the DF2200N wild type migrated faster than the RcpC proteins in the other strains analyzed. Therefore, the different migration is likely a property of the background strain.
FIG. 5.
Abundances of Rcp proteins in tad mutant strains. Whole-cell extracts of nonpolar (complementable) tad mutant strains and of wild-type (wt) strains CU1000N and DF2200N were prepared and analyzed by immunoblotting with anti-RcpC, anti-RcpA, anti-RcpB, or anti-TadC antiserum. The stronger signals from the tadA mutant strain were reproducible, despite equivalent protein concentrations (at A280) in the extracts of the other strains. Arrows indicate the positions of RcpC, RcpA, RcpB, and TadC. In the TadC blot, the bottom band is nonspecific (see the text).
The RcpA protein was absent in the rcpA mutant, and it was abundant in both wild-type strains (Fig. 5). In most other mutant strains, the intensity of the RcpA band was comparable to that of the band observed in wild-type A. actinomycetemcomitans. However, RcpA was visible only as a faint band in a tadC mutant and not at all in a tadD mutant (Fig. 5). When more-concentrated tadD samples were subjected to immunoblot analysis, RcpA could be detected, but only at very low levels (data not shown). This result indicates a severe defect in the production and/or stability of RcpA in the absence of TadD.
The anti-RcpB antiserum detected an abundance of native RcpB in wild-type CU1000N and in many of the CU1000N tad mutants, but RcpB was almost undetectable in the rcpA, tadC, and tadD mutant strains (Fig. 5). The anti-RcpB antiserum displayed poor recognition of the RcpB protein in both the DF2200N wild type and the tadV strain, which was made from DF2200N (Fig. 5). All of our antisera were generated using protein antigens from CU1000N. In contrast to RcpB, the RcpA and RcpC proteins were detected by their specific antisera in whole-cell extracts prepared from both CU1000N and DF2200N.
Because TadC appeared to be critical for the abundances of all three Rcp proteins, we tested whether or not the abundance of TadC was altered in the tad locus mutants, including rcpA and rcpC. Our TadC-specific antiserum recognized the native protein at the predicted size in CU1000N, as well as a lower, nonspecific band that was present in all of the strains we examined (Fig. 5). TadC was not detected in the tadC mutant, as expected. We found that in the absence of TadB, TadC abundance was severely diminished. TadC production and/or stability in several other mutants, including the rcpA, rcpC, tadZ, tadE, and tadG mutant strains, was less than that in the wild type (Fig. 5). The CU1000N TadC-specific antiserum failed to recognize its cognate protein in the DF2200N wild-type and tadV strains; therefore, as for RcpB, we were unable to determine the abundance of TadC in a tadV mutant.
In summary, we found defects in the production and/or stability of all three of the Rcp proteins in the tadC mutant and reciprocally diminished abundance of the TadC protein in rcpC- and rcpA-deficient strains. Additionally, RcpA and RcpB were nearly undetectable in the tadD mutant strain, and TadC was difficult to detect in the tadB mutant. RcpB and RcpC were detected at diminished levels in the rcpA mutant strain.
Importance of outer membrane protein TadD.
The near absence of RcpA in the tadD mutant was dramatic. PSORT analysis (18) of the predicted CU1000N TadD protein sequence indicated that TadD may be a lipoprotein cleaved by signal peptidase II and localized to the outer membrane. Our results showed the localization of a tagged version of TadD in the outer membrane, though it was also detected in the inner membrane. Since some secretin proteins require an outer membrane lipoprotein for stabilization or membrane insertion (2), we considered that TadD may be an outer membrane docking protein for RcpA in the Tad system and that alterations in RcpA protein distribution may occur in its absence.
We did a complementation analysis to ascertain whether the production of TadD could restore the abundance of RcpA to the wild-type level. Immunoblots of whole-cell extracts showed that, whereas RcpA was severely diminished in the tadD mutant, the addition of TadD-T7 returned the abundance of RcpA to an intensity comparable to that in wild-type CU1000N (Fig. 6A). We also examined the production of RcpA in a tadD (plus RcpA) strain, which is the tadD mutant strain containing pPP40, a tac-mediated, RcpA-producing plasmid that complements an rcpA mutant (45). The tadD mutant strain contained only the chromosomal copy of rcpA, and although RcpA was detected in protein extracts of the tadD (plus RcpA) strain as a faint band (Fig. 6A), it seemed that additional copies of rcpA were not sufficient to restore wild-type levels of RcpA to the tadD mutant. We also observed that the addition of RcpA (vial pPP40) to the tadD mutant failed to restore adherence or rough colony morphology (data not shown).
FIG. 6.
TadD and its effects on RcpA. (A) Whole-cell extracts of the following strains were prepared and immunoblotted using anti-RcpA antiserum: lane 1, Aa1577 (tadD mutant); lane 2, Aa1758 (tadD mutant with empty vector); lane 3, Aa1759 (tadD mutant with RcpA); lane 4, Aa0727 (tadD mutant with TadD-T7); lane 5, CU1000N (wild type). (B) Whole-cell extracts of Aa1759 [tadD (+ RcpA)] were heated at 37, 65, or 100°C for 5 min in standard loading buffer and separated by SDS-PAGE through an 8% polyacrylamide gel. Immunoblotting was done with anti-RcpA antiserum. (C) Subcellular fractions of Aa1759 were prepared by differential detergent solubilization of the inner membrane with 0.5% Sarkosyl, and immunoblots were probed with anti-RcpA antiserum. S, soluble fraction; IM, inner membrane; and OM, outer membrane. Arrows indicate the positions of RcpA. In panel B, the RcpA HMM is indicated.
We used the tadD (plus RcpA) strain to observe the oligomerization of RcpA in this mutant. Without TadD protein production, RcpA was no longer detected as an HMM but was present only in its monomeric form (Fig. 6B). In the tadD mutant, the RcpA protein was also no longer detected in the outer membrane but was present predominantly in the inner membrane fraction and was detected at low levels in the cytoplasm (Fig. 6C). RcpA did not exhibit this outer membrane defect in either the tadC or the rcpC mutant (data not shown), indicating that the mislocalization of RcpA in the tadD (plus RcpA) strain was because of the absence of TadD. Complementation of the tadD mutation restored both the localization pattern and the multimerization of RcpA (data not shown).
In summary, RcpA abundance could be fully restored by the complementation of the tadD mutation with TadD-T7. We did not detect RcpA at wild-type intensity without TadD production. In the absence of TadD, the outer membrane localization and oligomerization of RcpA were abolished.
DISCUSSION
The secretin family of outer membrane proteins form oligomeric channels through which fully folded and assembled secretion substrates exit the cell. RcpA has been assumed to operate as the secretin for the Flp pilus secretion system because of (i) the similarity of the sequence of its C terminus to those of members of the secretin superfamily, (ii) the detection of this protein in A. actinomycetemcomitans outer membranes (23), and (iii) the genetic requirement for a functional rcpA gene for Flp pilus production (45). Here, we have shown that the RcpA protein of A. actinomycetemcomitans exhibits an evolutionary relationship with known secretins, and we have shown patterns of membrane localization and multimerization typical of well-studied secretins.
Our parsimony-based phylogenetic analysis of 186 genes, which included measurements of nodal support, is the largest secretin gene superfamily phylogeny to date (69). We found that the rcpA gene and its close homologs form a distinct, major monophyletic subfamily with an ancient divergence from other well-known secretin genes. The rcpA subfamily groups with other secretin genes from T2S systems, which is consistent with the functional similarities reported here and with the numerous similarities between T2S systems and the Tad secretion system (67). Indeed, we have proposed that the Tad secretion system be classified as a T2S system (44, 67). The ancient divergence of the rcpA family indicates that these functional similarities are very strongly selected and conserved.
Association with tad locus genes and participation in Flp pilus assembly are the exclusive properties of rcpA subfamily genes. In all cases, genes of the rcpA subfamily are found in close proximity to at least some other tad locus genes, and such associations are not seen outside the rcpA clade, indicating that the rcpA subfamily is representative of a historical event in which the ancestral rcpA secretin gene became associated with the tad locus. The sister group of the rcpA subfamily, which is composed of chromosomal and plasmid-borne secretin genes from R. solanacearum, is associated with secretion genes only distantly related to tad locus genes. This observation indicates that the initial event that created the rcpA lineage may have been the addition of a secretin gene from another secretion system to a preexisting tad locus. It is possible that this was a plasmid-mediated event because several of the closest relatives in R. solanacearum are found on megaplasmids.
The secretin N terminus is proposed to reside at least partially in the periplasm, where it may interact with other secretory proteins or physically gate the opening of its own channel (13, 57). In our alignment (Fig. 1B), the N terminus of RcpA was noticeably shorter than those of the proteins CpaC, its close homolog, and PulD. The relative shortness of the N terminus of RcpA may argue against a self-gating function, and another protein in the Tad system may be required. Additionally, our observation that the overexpression of rcpA is toxic in both A. actinomycetemcomitans and E. coli (data not shown) may be interpreted as an inability of RcpA to provide a gate on its own, resulting in detrimental effects such as membrane leakiness.
Mounting evidence leads us to suggest that RcpB may be required in the Tad secretion system as a gating protein. (i) The necessity for rcpB in tad+ bacteria implies that something about the composition of an rcpB-deficient Tad system causes toxicity to the point of lethality. (ii) RcpB is in the outer membrane and has been observed in cross-linked homomultimers (data not shown), much like RcpA. (iii) RcpB abundance is dependent on the presence of the RcpA secretin, and like the secretin, RcpB was less abundant and no longer localized to the outer membrane in the tadD mutant (data not shown). Finally, (iv) our phylogenetic analysis offers support for the requirement of an interacting protein encoded by a gene with sequence similarity to rcpB or to cpaD, which immediately follows every rcpA subfamily gene we examined.
Protein abundance patterns have been used successfully to deduce protein interactions within transport systems (4, 27, 50, 51, 58). It is possible that the loss of stabilizing physical interactions in tad mutant strains may account for some of the abundance defects we observed. Consequently, an abundance defect for a noncognate protein in a tad mutant may represent the degradation of that protein in the absence of interaction with the missing gene product. Reciprocal abundance relationships have been observed for TadC-RcpC and TadC-RcpA, predicting that there may be stabilizing or complex-related contacts occurring. In addition, all three Rcp proteins were less abundant in a tadC mutant than in the wild type, RcpA and RcpB were nearly absent in a tadD mutant, and RcpB and RcpC seemed to depend on the presence of RcpA.
We found TadC to be a 32-kDa inner membrane protein. In homology searches, both A. actinomycetemcomitans TadB and TadC show similarity to PilC-like proteins, which are integral inner membrane members of T2S systems (41). TadB and TadC may associate to form a heteromultimer that allows the passage of other Tad components across the inner membrane (8). In support of a possible association, we found that TadC protein levels in the tadB mutant were severely depleted, while many other mutants exhibited wild-type levels. We have not been able to examine the expression profile or localization of TadB because we were not able to make an antiserum that recognizes the native protein in A. actinomycetemcomitans, and attempts to detect a T7-tagged fusion were not successful.
The RcpC/CpaB homologs, including those in H. ducreyi, P. multocida, C. crescentus (CpaB), and Pseudomonas spp. (CpaB) contain two β-clip motifs (28). While β-clip folds were previously known to function in fish antifreeze proteins, in sialic acid synthase, and in a range of other enzymes, it has been proposed that in RcpC/CpaB proteins they may function by binding carbohydrate moieties of peptidoglycan to assemble structures like Flp pili or flagella. Our finding (66) that the Flp1 protein migrates faster in an rcpC mutant than in the wild-type strain or in the other tad mutants indicates that the RcpC protein may be involved in the known glycosylation of Flp1, and this study has revealed that the RcpC protein may function in association with one or both bacterial membranes.
Our immunoblots showed RcpC, RcpA, and TadD-T7 in both the outer membrane fraction and the Sarkosyl-soluble inner membrane. The detection of these proteins in the inner membrane may be a manifestation of their trafficking to the outer membrane or of their extraction resulting from interactions at the outer face of the inner membrane, as has been suggested previously for other systems (50, 55). Contamination of fractions is not a factor (unpublished results). Evidence of outer membrane proteins in the cytoplasmic membrane was also found in localization studies of the secretins PulD and TcpC (4, 9), and evidence of intermembrane interactions for many secretin proteins has been supported biochemically (11, 27, 46, 47, 51).
Outer membrane lipoproteins have been implicated in the assembly of secretin oligomers, in outer membrane insertion of secretins, and in the general stabilization of assembled secretin complexes. Monomeric RcpA in the tadD mutant was difficult to detect, even when extra copies of rcpA were provided in trans. One possibility is that without TadD there is severe destabilization and probably near-complete degradation of the RcpA protein. Although we cannot yet separate the effects, our multimerization and localization results indicate that TadD may mediate the association of RcpA subunits and act as an assembly factor for the insertion of RcpA into the outer membrane. The docking proteins PilW and Tgl, both lipoproteins, do this for their secretins (6, 40). They each contain TPR sequences that are thought to be important for interactions during secretin complex assembly (40). The TadD sequence contains a 34-residue TPR domain that may mediate an interaction with the RcpA protein.
Through localization, protein stability, and additional analyses, this study has started to uncover potential functions of membrane-bound Tad proteins in Flp pilus biogenesis and likely relationships to other proteins within the secreton. Since its component proteins seem to be interdependent for stability, the Tad secretion system probably forms a complex that spans both membranes, and this complex may include an outer membrane module. It is clear that the continued study of these proteins will help to define the mechanism by which the Tad secreton operates.
Supplementary Material
Acknowledgments
We thank Dan Fine for his help, interest, and support; Rob DeSalle for advice and guidance in phylogenetic analysis and gene taxonomy; Neil Sarkar for computer support and applications for sequence retrieval; Patrick Viollier for CpaC antiserum; and members of the Figurski lab for helpful discussions. We are grateful to Juliet Morrison for constructing pJMM108 and Aa0727. D.H.F. also appreciates the help of Saul Silverstein and Aaron Mitchell.
This work was supported by grants from the U.S. National Institutes of Health: DE014713 (to D.H.F.) and GM062351 (to R. DeSalle, P.J.P., and D.H.F.). Partial support was provided by training grant 5T32AI007161 (to S.A.C.) and funds from the Kirschstein-NRSA Individual Fellowship Program (to B.A.P.).
Footnotes
Published ahead of print on 30 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayan, N., I. Guilvout, and A. P. Pugsley. 2006. Secretins take shape. Mol. Microbiol. 601-4. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee, M. K., S. C. Kachlany, D. H. Fine, and D. H. Figurski. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 1835927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bose, N., and R. K. Taylor. 2005. Identification of a TcpC-TcpQ outer membrane complex involved in the biogenesis of the toxin-coregulated pilus of Vibrio cholerae. J. Bacteriol. 1872225-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brissette, J. L., and M. Russel. 1990. Secretion and membrane integration of a filamentous phage-encoded morphogenetic protein. J. Mol. Biol. 211565-580. [DOI] [PubMed] [Google Scholar]
- 6.Carbonnelle, E., S. Helaine, L. Prouvensier, X. Nassif, and V. Pelicic. 2005. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 5554-64. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 692110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bentzmann, S., M. Aurouze, G. Ball, and A. Filloux. 2006. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 1884851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.d'Enfert, C., I. Reyss, C. Wandersman, and A. P. Pugsley. 1989. Protein secretion by gram-negative bacteria. Characterization of two membrane proteins required for pullulanase secretion by Escherichia coli K-12. J. Biol. Chem. 26417462-17468. [PubMed] [Google Scholar]
- 10.Eriksson, T. 1997. AutoDecay, version 2.9.9. Botaniska institutionen, Stockholm University, Stockholm, Sweden.
- 11.Feng, J. N., P. Model, and M. Russel. 1999. A trans-envelope protein complex needed for filamentous phage assembly and export. Mol. Microbiol. 34745-755. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez, L., I. Marquez, and J. A. Guijarro. 2004. Identification of specific in vivo-induced (ivi) genes in Yersinia ruckeri and analysis of ruckerbactin, a catecholate siderophore iron acquisition system. Appl. Environ Microbiol. 705199-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filloux, A. 2004. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694163-179. [DOI] [PubMed] [Google Scholar]
- 14.Fine, D. H., D. Furgang, J. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 441063-1076. [DOI] [PubMed] [Google Scholar]
- 15.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 1451335-1347. [DOI] [PubMed] [Google Scholar]
- 16.Fine, D. H., J. B. Kaplan, S. C. Kachlany, and H. C. Schreiner. 2006. How we got attached to Actinobacillus actinomycetemcomitans: a model for infectious diseases. Periodontol. 2000 42114-157. [DOI] [PubMed] [Google Scholar]
- 17.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 2925-38. [DOI] [PubMed] [Google Scholar]
- 18.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. Brinkman. 2005. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21617-623. [DOI] [PubMed] [Google Scholar]
- 19.Genin, S., and C. A. Boucher. 1994. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet. 243112-118. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg, M. H., and J. Katz. 2006. Infective endocarditis caused by fastidious oro-pharyngeal HACEK micro-organisms. J. Oral Maxillofac. Surg. 64969-971. [DOI] [PubMed] [Google Scholar]
- 21.Goncharoff, P., J. K. Yip, H. Wang, H. C. Schreiner, J. A. Pai, D. Furgang, R. H. Stevens, D. H. Figurski, and D. H. Fine. 1993. Conjugal transfer of broad-host-range incompatibility group P and Q plasmids from Escherichia coli to Actinobacillus actinomycetemcomitans. Infect. Immun. 613544-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haase, E. M., J. O. Stream, and F. A. Scannapieco. 2003. Transcriptional analysis of the 5′ terminus of the flp fimbrial gene cluster from Actinobacillus actinomycetemcomitans. Microbiology 149205-215. [DOI] [PubMed] [Google Scholar]
- 23.Haase, E. M., J. L. Zmuda, and F. A. Scannapieco. 1999. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 672901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Henderson, B., S. P. Nair, J. M. Ward, and M. Wilson. 2003. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annu. Rev. Microbiol. 5729-55. [DOI] [PubMed] [Google Scholar]
- 26.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang, J., D. Bieber, S. W. Ramer, C. Y. Wu, and G. K. Schoolnik. 2003. Structural and topographical studies of the type IV bundle-forming pilus assembly complex of enteropathogenic Escherichia coli. J. Bacteriol. 1856695-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer, L. M., and L. Aravind. 2004. The emergence of catalytic and structural diversity within the beta-clip fold. Proteins 55977-991. [DOI] [PubMed] [Google Scholar]
- 29.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 1826169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, and D. H. Figurski. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9429-437. [DOI] [PubMed] [Google Scholar]
- 31.Kachlany, S. C., P. J. Planet, R. Desalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40542-554. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan, J. B., H. C. Schreiner, D. Furgang, and D. H. Fine. 2002. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. J. Clin. Microbiol. 401181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kazmierczak, B. I., D. L. Mielke, M. Russel, and P. Model. 1994. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J. Mol. Biol. 238187-198. [DOI] [PubMed] [Google Scholar]
- 34.Kilian, M. 1976. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 939-62. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 36.Linderoth, N. A., P. Model, and M. Russel. 1996. Essential role of a sodium dodecyl sulfate-resistant protein IV multimer in assembly-export of filamentous phage. J. Bacteriol. 1781962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newhall, W. J., C. E. Wilde III, W. D. Sawyer, and R. A. Haak. 1980. High-molecular-weight antigenic protein complex in the outer membrane of Neisseria gonorrhoeae. Infect. Immun. 27475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nixon, K. C. 1999. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 15407-414. [DOI] [PubMed] [Google Scholar]
- 39.Norskov-Lauritsen, N., and M. Kilian. 2006. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int. J. Syst. Evol. Microbiol. 562135-2146. [DOI] [PubMed] [Google Scholar]
- 40.Nudleman, E., D. Wall, and D. Kaiser. 2006. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 6016-29. [DOI] [PubMed] [Google Scholar]
- 41.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 1493051-3072. [DOI] [PubMed] [Google Scholar]
- 42.Perez, B. A., P. J. Planet, S. C. Kachlany, M. Tomich, D. H. Fine, and D. H. Figurski. 2006. Genetic analysis of the requirement for flp-2, tadV, and rcpB in Actinobacillus actinomycetemcomitans biofilm formation. J. Bacteriol. 1886361-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips, A., D. Janies, and W. Wheeler. 2000. Multiple sequence alignment in phylogenetic analysis. Mol. Phylogenet. Evol. 16317-330. [DOI] [PubMed] [Google Scholar]
- 44.Planet, P. J., D. H. Figurski, and R. DeSalle. 2006. Function, evolution, and classification of macromolecular transport systems, p. 189-219. In H. S. Seifert and V. J. DiRita (ed.), Evolution of microbial pathogens. ASM Press, Washington, DC.
- 45.Planet, P. J., S. C. Kachlany, D. H. Fine, R. DeSalle, and D. H. Figurski. 2003. The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat. Genet. 34193-198. [DOI] [PubMed] [Google Scholar]
- 46.Possot, O. M., M. Gerard-Vincent, and A. P. Pugsley. 1999. Membrane association and multimerization of secreton component PulC. J. Bacteriol. 1814004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Possot, O. M., G. Vignon, N. Bomchil, F. Ebel, and A. P. Pugsley. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol. 1822142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 5750-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pugsley, A. P., and O. Possot. 1993. The general secretory pathway of Klebsiella oxytoca: no evidence for relocalization or assembly of pilin-like PulG protein into a multiprotein complex. Mol. Microbiol. 10665-674. [DOI] [PubMed] [Google Scholar]
- 50.Ramer, S. W., D. Bieber, and G. K. Schoolnik. 1996. BfpB, an outer membrane lipoprotein required for the biogenesis of bundle-forming pili in enteropathogenic Escherichia coli. J. Bacteriol. 1786555-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramer, S. W., G. K. Schoolnik, C. Y. Wu, J. Hwang, S. A. Schmidt, and D. Bieber. 2002. The type IV pilus assembly complex: biogenic interactions among the bundle-forming pilus proteins of enteropathogenic Escherichia coli. J. Bacteriol. 1843457-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenfeld, J. A., I. N. Sarkar, P. J. Planet, D. H. Figurski, and R. DeSalle. 2004. ORFcurator: molecular curation of genes and gene clusters in prokaryotic organisms. Bioinformatics 203462-3465. [DOI] [PubMed] [Google Scholar]
- 53.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279485-499. [DOI] [PubMed] [Google Scholar]
- 54.Russel, M. 1994. Mutants at conserved positions in gene IV, a gene required for assembly and secretion of filamentous phages. Mol. Microbiol. 14357-369. [DOI] [PubMed] [Google Scholar]
- 55.Russel, M., and B. Kazmierczak. 1993. Analysis of the structure and subcellular location of filamentous phage pIV. J. Bacteriol. 1753998-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 57.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40271-283. [DOI] [PubMed] [Google Scholar]
- 58.Schmidt, S. A., D. Bieber, S. W. Ramer, J. Hwang, C. Y. Wu, and G. Schoolnik. 2001. Structure-function analysis of BfpB, a secretin-like protein encoded by the bundle-forming-pilus operon of enteropathogenic Escherichia coli. J. Bacteriol. 1834848-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schreiner, H. C., K. Sinatra, J. B. Kaplan, D. Furgang, S. C. Kachlany, P. J. Planet, B. A. Perez, D. H. Figurski, and D. H. Fine. 2003. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. USA 1007295-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 193223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, G. P. 1988. Filamentous phage assembly: morphogenetically defective mutants that do not kill the host. Virology 167156-165. [DOI] [PubMed] [Google Scholar]
- 62.Spinola, S. M., K. R. Fortney, B. P. Katz, J. L. Latimer, J. R. Mock, M. Vakevainen, and E. J. Hansen. 2003. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect. Immun. 717178-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thanassi, D. G. 2002. Ushers and secretins: channels for the secretion of folded proteins across the bacterial outer membrane. J. Mol. Microbiol. Biotechnol. 411-20. [PubMed] [Google Scholar]
- 64.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1997. CLUSTAL X multiple sequence alignment program, version 1.63. European Molecular Biology Organization, Hamburg, Germany.
- 65.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 1817298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomich, M., D. H. Fine, and D. H. Figurski. 2006. The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J. Bacteriol. 1886899-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomich, M., P. J. Planet, and D. H. Figurski. 2007. The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5363-375. [DOI] [PubMed] [Google Scholar]
- 68.Wheeler, W. C., J. Gatesy, and R. DeSalle. 1995. Elision: a method for accomodating multiple molecular sequence alignments with alignment-ambiguous sites. Mol. Phylogenet. Evol. 41-9. [DOI] [PubMed] [Google Scholar]
- 69.Yen, M. R., C. R. Peabody, S. M. Partovi, Y. Zhai, Y. H. Tseng, and M. H. Saier. 2002. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim. Biophys. Acta 15626-31. [DOI] [PubMed] [Google Scholar]
- 70.Yu, C. S., C. J. Lin, and J. K. Hwang. 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 131402-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao, S., D. M. Tobiason, M. Hu, H. S. Seifert, and R. A. Nicholas. 2005. The penC mutation conferring antibiotic resistance in Neisseria gonorrhoeae arises from a mutation in the PilQ secretin that interferes with multimer stability. Mol. Microbiol. 571238-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.