Abstract
ADP-ribose (ADPR) is one of the main substrates of Nudix proteins. Among the eight Nudix proteins of Thermus thermophilus HB8, we previously determined the crystal structure of Ndx4, an ADPR pyrophosphatase (ADPRase). In this study we show that Ndx2 of T. thermophilus also preferentially hydrolyzes ADPR and flavin adenine dinucleotide and have determined its crystal structure. We have determined the structures of Ndx2 alone and in complex with Mg2+, with Mg2+ and AMP, and with Mg2+ and a nonhydrolyzable ADPR analogue. Although Ndx2 recognizes the AMP moiety in a manner similar to those for other ADPRases, it recognizes the terminal ribose in a distinct manner. The residues responsible for the recognition of the substrate in Ndx2 are not conserved among ADPRases. This may reflect the diversity in substrate specificity among ADPRases. Based on these results, we propose the classification of ADPRases into two types: ADPRase-I enzymes, which exhibit high specificity for ADPR; and ADPRase-II enzymes, which exhibit low specificity for ADPR. In the active site of the ternary complexes, three Mg2+ ions are coordinated to the side chains of conserved glutamate residues and water molecules. Substitution of Glu90 and Glu94 with glutamine suggests that these residues are essential for catalysis. These results suggest that ADPRase-I and ADPRase-II enzymes have nearly identical catalytic mechanisms but different mechanisms of substrate recognition.
Nudix proteins catalyze the hydrolysis of nucleoside diphosphate (NDP) linked to another moiety, x (e.g., nucleoside triphosphate [NTP], diadenosine polyphosphate [ApnA, where n = 2 to 6], NDP-sugar, NADH, and coenzyme A [CoA], etc). Nudix proteins are widely distributed in nature, including in viruses, bacteria, archaea, and eukaryotes, and share a highly conserved amino acid sequence motif called a “Nudix box” (GX5EX7RE7UXEEXGU, where U represents I, L, or V and X represents any amino acid), which forms a loop-helix-loop structure involved in catalysis (3, 7, 17). It has been proposed that the physiological function of Nudix proteins is “housecleaning” to eliminate potentially toxic nucleotide metabolites from cells and to regulate the concentrations of NDP derivatives. However, the catalytic mechanism, including substrate recognition and physiological function, remains to be elucidated for many Nudix proteins.
ADP-ribose (ADPR) is one of the main substrates of Nudix proteins in all three kingdoms. ADPR is produced enzymatically as part of the turnover of NAD, cyclic ADPR, ADP-ribosylated proteins, and poly-ADP-ribosylated proteins (37). High intracellular levels of ADPR could result in nonenzymatic ADP ribosylation and interfere with the recognition of enzymatic ADP ribosylation (11). It is likely that ADPR pyrophosphatase (ADPRase) functions to eliminate potentially deleterious ADPR. ADPRases are distributed widely throughout the three kingdoms, and many organisms have two or more ADPRases (6, 22-24, 27, 33). The specificity for ADPR over other substrates varies among the ADPRases that have been studied. Although the crystal structures of ADPRases which exhibit high specificity for ADPR have been solved (7, 12, 28, 35, 36), only one structure of ADPRases which exhibit low specificity for ADPR has been determined until now (1). Thus, much work remains to understand the structural basis underlying the substrate preference of ADPRases.
We selected Thermus thermophilus HB8 as a model organism in a project named the Whole Cell Project (34). Proteins from T. thermophilus HB8 are heat stable and suitable for physiochemical studies including, in general, X-ray crystallography. The genome has eight genes encoding Nudix proteins, designated Ndx1 to Ndx8. We previously reported that Ndx1 and Ndx3 have ApnA hydrolase activity (9, 21). On the basis of sequence similarity, Ndx2 and Ndx4 were predicted to be ADPRases. Indeed, we showed that Ndx4 catalyzes the hydrolysis of ADPR with high specificity, determined its crystal structure, and performed kinetic analysis of mutants (25, 35).
In this study, we show that Ndx2 has pyrophosphatase activity for ADPR and flavin adenine dinucleotide (FAD) and have determined the crystal structures of the free enzyme, the Mg2+-bound enzyme, and the ternary complex with Mg2+ and either AMP or a nonhydrolyzable ADPR analogue. The mutational analysis of the catalytic residues on the basis of these structures is also described. The obtained results reveal the difference in substrate recognitions between two types of ADPRases with different substrate specificities.
MATERIALS AND METHODS
Materials.
DNA-modifying enzymes, including restriction enzymes and LA Taq polymerase, were from Takara Bio Inc. Yeast extract and polypeptone were from Difco. diadenosine triphosphate (Ap3A), diadenosine tetraphosphate (Ap4A), diadenosine pentaphosphate (Ap5A), and diadenosine hexaphosphate (Ap6A), dATP, dGTP, AMP, ADPR, ADP-glucose (ADPG), GDP-mannose, UDP-glucose, FAD, NADH, CoA, and acetyl-CoA were from Sigma. 8-Oxo dGDP and CDP-choline were from Biolog. 8-Oxo dGTP was from Trilink. The ADPR analogue (α-β methylene ADPR [AMPCPR]) was synthesized by KNC Laboratories Co., Ltd. The DNA oligomers were synthesized by BEX Co. All other reagents used were of the highest grade commercially available.
Overexpression of the ndx2 gene.
Sequence analysis of the T. thermophilus HB8 genome (DDBJ/EMBL/GenBank AB107660) identified eight open reading frames (ORFs) encoding proteins containing the Nudix motif. Using this sequence information, we synthesized two primers for the amplification of the target gene (TTHA0863, project code 1256) by PCR. Amplification was carried out by standard protocols, and the amplified gene fragment was ligated into pT7Blue T-vector (Novagen, Madison, WI) by TA cloning and confirmed by sequencing. The fragment bearing the target gene from pT7Blue-ndx2 was ligated into pET-11a (Novagen) at the NdeI and BamHI sites. Escherichia coli BL21(DE3) cells transformed with the resulting plasmid were cultured at 37°C to 4 × 108 cell/ml in 1.5 liters of LB medium containing 50 μg/ml ampicillin. The cells were then incubated for 5 h in the presence of isopropyl-β-d-thiogalactopyranoside, harvested by centrifugation, and stored at −20°C.
Purification of Ndx2.
All of the following procedures were carried out at room temperature unless stated otherwise. Frozen cells (10 g) were thawed, suspended in 100 ml of buffer I (50 mM Tris-HCl [pH 8.0] and 1 mM EDTA), and disrupted by sonication on ice. The lysate was incubated at 70°C for 15 min and centrifuged (38,000 × g) for 60 min at 4°C. The resultant supernatant was loaded onto a Toyopearl SuperQ 650 M (Tosoh, Tokyo, Japan) column (bed volume, 20 ml) equilibrated with buffer I. Proteins were eluted with a linear gradient of 0 to 1 M NaCl (a total volume of 250 ml). Ammonium sulfate was added to the fractions containing the Ndx2 protein to a final concentration of 15% saturation. The protein solution was then applied to a Toyopearl Phenyl 650 M (Tosoh) column (bed volume, 20 ml) equilibrated with buffer I containing 15% saturated ammonium sulfate. The proteins were eluted with a linear gradient of ammonium sulfate from 15 to 0% saturation (a total volume of 250 ml). Fractions containing the Ndx2 protein were collected and concentrated by a Vivaspin (molecular weight cutoff, 10,000) concentrator. The concentrated solution was applied to a Superdex 75 HR 10/30 column (GE Healthcare Biosciences) equilibrated with buffer II (50 mM Tris-HCl [pH 8.0] and 100 mM KCl) and eluted with the same buffer by use of an ÄKTA explorer system (GE Healthcare Biosciences). The fractions containing the Ndx2 protein were concentrated and stored at 4°C. At each step, the fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The concentration of the purified protein was determined by using the molar absorption coefficient at 278 nm calculated according to the formula of Kuramitsu et al. (14). The N-terminal eight residues of the purified Ndx2 were confirmed by sequencing using an Applied Biosystems 473A protein sequencer.
Spectroscopic analysis.
Circular dichroism (CD) spectra in the far-UV region (200 nm to 250 nm) were obtained at 25°C with a Jasco spectropolarimeter, J-720W, by use of 5 μM enzyme in 50 mM potassium phosphate and 100 mM KCl, pH 7.5. Thermostability was investigated by recording the molar ellipticity at 222 nm from 15°C to 95°C under the same conditions as described above.
Size exclusion chromatography.
The enzyme (50 μM) was applied onto a Superdex 75 HR 10/30 column and eluted with buffer II with a flow rate of 0.5 ml/min by the ÄKTA system. The apparent molecular weight was estimated by comparing its retention time with those of molecular weight markers (Sigma).
Site-directed mutagenesis.
Site-directed mutagenesis of Glu-90, Glu-94, and Glu-136 to glutamine was performed by replacing the wild-type sequence in the pT7Blue-ndx2 plasmid with each mutated fragment. The overexpression and purification of these mutant proteins were performed in a manner similar to that used for the wild type.
Enzyme assay.
The hydrolysis of the substrate was measured by a previously described method with some modifications (9). In brief, reaction mixtures (100 μl) containing 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 200 μM substrate, and 5 μM of enzyme were incubated at 25°C. The substrates were dNTP (dATP, dGTP, and 8-oxo-dGTP) and ApnA (Ap3A, Ap4A, Ap5A, and Ap6A), NDP-sugar (ADPR, ADPG, and UDP-glucose), and others (NADH, FAD, CoA, acetyl-CoA, and CDP-choline). The reaction was stopped by adding 20 μl of 500 mM EDTA, and the protein was removed by ultrafiltration using a membrane filter (molecular weight cutoff, 5,000). A 100-μl aliquot of the filtrate was applied to a reversed-phase column (Capcell Pak C18, 4.6 × 75 mm; Shiseido) equilibrated with 50 mM Tris-HCl (pH 7.5), 5 mM tetra-n-butylammonium phosphate, and 10% methanol. Elution was performed by a gradient of 10 to 50% methanol.
To determine the steady-state kinetic parameters, the hydrolytic activities toward ADPR, ADPG, and FAD were analyzed by measuring the production of inorganic orthophosphate by use of a colorimetric assay (25). The activity for UDP-glucose and CDP-choline was also measured by a colorimetric assay. The data were fitted to the Michaelis-Menten equation, and the kinetic constant was calculated by the software Igor Pro 3.14 (Wave Metrics).
Enzyme activity in the presence of various divalent cations was measured by reversed-phase column chromatography. The reaction mixture (100 μl) contained 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM divalent cation (MgCl2, MnCl2, ZnCl2, CoCl2, NiCl2, CaCl2, or CuSO4), 3 mM FAD or ADPR, and 100 nM of enzyme.
Crystallization.
Crystallization conditions for Ndx2 were surveyed by the hanging-drop vapor diffusion method with crystal screen kits (Crystal Screen, Crystal Screen2, Crystal Screen Cryo, Crystal Screen Lite, Natrix, Index, Salt RX, and PEG/ION; Hampton Research) at 293 K. The initial protein concentration was 20 mg/ml in 20 mM Tris-HCl (pH 8.0) and 100 mM KCl, and 1 μl of the protein solution was mixed with the same volume of reservoir solution and equilibrated against the reservoir solution. After further optimization, crystals suitable for X-ray diffraction were obtained in an optimized condition containing 0.1 M MES (morpholineethanesulfonic acid) (pH 6.5), 0.16 M sodium acetate (magnesium acetate for the Mg2+ complex), 14% polyethylene glycol 8000, and 20% glycerol at 20°C. Gold derivative crystals of the Mg2+ complex were prepared by soaking the crystals in 50 mM KAu(CN)2. Cocrystallization trials with AMP and AMPCPR were set up in the same way as those of the Mg2+ complex, except for the addition of 1 μl of AMP and 1 μl of AMPCPR (final concentrations of 3.3 and 10 mM, respectively).
MAD phasing, model building, and refinement.
The gold derivative crystals were flash-cooled in a nitrogen stream (−180°C), and data were collected under cryoconditions. Three-wavelength multiwavelength anomalous diffraction (MAD) data were collected at the beamline BL26B2 at SPring-8. Diffraction images were processed with the HKL2000 program (26). Assuming the asymmetric unit to contain one subunit, the respective VM and Vsol values (19) were 2.5 Å3 Da−1 and 0.50. The structure was determined by the MAD phasing method using Solve/Resolve (30). The two sites of the gold atom were deduced from difference Patterson maps. The initial model (140 residues) was built with the aid of the amino acid sequence by use of the program Solve/Resolve, refined with the Xtalview program (10) and the CNS program (4), and subsequently refined from 50- to 2.0-Å resolution by use of phase information from the diffraction data with the program CNS. The success of model refinement was evaluated at each stage by the free R factor (Σ‖Fobs − Fcalc‖/ΣFobs) and by inspection of stereochemical parameters with the program Procheck (15). The final refinement indicated that more than 94% of the residues (expect for glycine and proline residues) fell into the most favored regions in the Ramachandran plot.
Data for the crystals of the free enzyme and complexes were collected at a wavelength of 1.0000 Å at the beamline BL44B2 at SPring-8. Diffraction images were processed with the HKL2000 program. The structures were determined by molecular replacement using an Au derivative Mg2+ complex as a starting model and refined by use of the CNS program. Figures were made with the programs CCP4 (5), Pymol (DeLano Scientific; http://www.pymol.org), and ChemDraw Ultra (CambridgeSoft Corporation, Cambridge, MA).
Atomic coordinate accession codes.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with accession codes 2YVM, 2YVN, 2YVO, and 2YVP.
RESULTS
Preparation of Ndx2.
The ndx2 gene encodes a protein of 182 amino acid residues, has a calculated molecular mass of 20.3 kDa, and has a theoretical pI of 5.4. We overexpressed Ndx2 in E. coli and purified the protein to homogeneity by using heat treatment and three column chromatography steps. Approximately 50 mg of Ndx2 was obtained from 10 g of cells.
Size exclusion chromatography was performed to investigate the oligomeric state of Ndx2. The apparent molecular mass corresponding to the peak was estimated to be 40 kDa (data not shown), which was twice the mass (20.3 kDa) calculated from the sequence. This result indicates that Ndx2 exists in a homodimeric state in solution. The far-UV (200- to 250-nm) CD spectrum suggested that Ndx2 has an α/β or an α+β fold. Ndx2 was stable up to 85°C at pH 7.5. Denaturation occurred over 85°C, and this process was irreversible (data not shown).
Enzymatic activity.
The hydrolysis activity of Ndx2 was specific for ADPR, ADPG, and FAD among the tested compounds. Relative activities, which are expressed as percentages of the activity for FAD, were 56, 22, 2, and 100 for ADPR, ADPG, NADH, and FAD, respectively. No or little activity was observed for (d)NTP, ApnA, UDP-glucose, CoA, acetyl-CoA, and CDP-choline. This specificity differs from that of Ndx4 in that Ndx4 is highly specific for ADPR and cannot hydrolyze FAD. The relative activities (kcat/Km) of Ndx4 to ADPR, ADPG, GDP-ribose, and CDP-ribose are 100, 2.9, 3.3, and 2.5, respectively (25). These results demonstrate that Ndx2 is an ADPRase with moderate specificity for ADPR. Of note is the fact that Ndx2 did not hydrolyze ApnAs. The reaction with all of these substrates generated AMP as a common product. Ribose-5′-phosphate, glucose-5′-phosphate, and flavin mononucleotide were also generated as products from ADPR, ADPG, and FAD, respectively. This type of reaction is consistent with the known features of Nudix proteins.
At neutral pH, Ndx2 exhibited stronger activity (kcat/Km) for FAD than for ADPR and ADPG (Table 1). Therefore, these results indicate that Ndx2 is a FAD pyrophosphatase (FADase) as well as an ADPRase. Ndx2 exhibited higher activity at pH 9.0 than at pH 7.5, especially for ADPR (Table 1). Such dependence of the activity on pH is also a feature of Nudix proteins.
TABLE 1.
Kinetic parameters of the Ndx2-catalyzed reactiona
| Substrate | Km (μM)b | kcat (s−1)b | kcat/Km (M−1 s−1) |
|---|---|---|---|
| ADPR | 520 (250) | 1.4 (4.9) | 2,700 (20,000) |
| ADPG | 2,200 | 0.55 | 250 |
| FAD | 300 (330) | 2.5 (5.0) | 8,300 (15,000) |
Assays were done at pH 7.5 as described in Materials and Methods. Values in parentheses represent the activity at pH 9.0.
Km and kcat were determined from a nonlinear regression analysis.
The presence of a divalent cation was essential for Ndx2 activity for ADPR and FAD. Among the divalent cations tested (each at 5 mM), Mg2+ was the most effective for inducing both ADPRase and FADase activities. The presence of Zn2+, Mn2+, Ca2+, and Co2+ ions resulted in only about 30%, 10%, 5%, and 5%, respectively, of the activity observed in the presence of Mg2+. No activity was observed without metal ions.
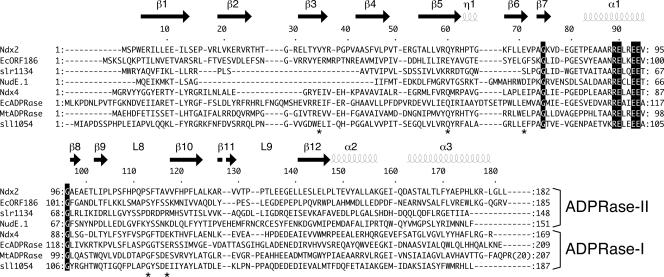
On the basis of previous research, it can be considered that many organisms have two types of ADPRase: one has high specificity for ADPR and the other has low specificity for ADPR, that is, broad substrate specificity. For example, EcADPRase and Mycobacterium tuberculosis Rv1700 (MtADPRase; NP_216216) are very specific for ADPR (6, 12). Synechocystis sp. strain PCC 6803 has five ADPRases, among which Sll1054 (NP_439968) and Slr0920 (NP_442398) hydrolyze ADPR specifically (24). Among the four ADPRases in Arabidopsis thaliana, AtNUDT10 (NP_849443) also hydrolyzes ADPR specifically (22). In this study, these enzymes are designated as ADPRase-I. The other type exhibits broad specificity for substrates including ADPR. For example, Slr1134 (NP_440605) can also hydrolyze NADH and FAD in addition to ADPR (8). E. coli ORF186 (EcORF186; NP_417856) hydrolyzes Ap3A, AP3(7-methyl)G, ADPR, diadenosine diphosphate, NADH, and FAD (23). AtNUDT2 (NP_568687), AtNUDT6 (NP_178526), and AtNUDT7 (NP_879368) also show NADH pyrophosphatase activity (22). NudE.1 from T4 bacteriophage has activity for FAD, ApnA (n = 2 to 5), and NADH (33). In this study, these enzymes are temporarily designated as ADPRase-II. In the case of T. thermophilus HB8, Ndx2, an ADPRase-II enzyme, has ADPRase and FADase activity, whereas Ndx4, an ADPRase-I enzyme, hydrolyzes ADPR with higher activity than Ndx2 and does not hydrolyze FAD (35). As mentioned above, the substrate specificity varies widely across the ADPRase-II subfamily, in contrast to what is seen for ADPRase-I. In addition, the ADPRase-II family is likely to be diverse in sequence (Fig. 1). For example, the sequence identity and similarity shared between Ndx2 and EcORF186 are 24% and 48%, respectively (Fig. 1). This classification of ADPRases is based primarily on their specificities reported in the literature and has not yet been proposed. Therefore, the structural comparison of ADPRase-I and ADPRase-II enzymes will provide clues to the molecular basis of their distinct specificities.
FIG. 1.
Alignment of Ndx2 and other ADPRases. Shown is the amino acid sequence alignment of Ndx2 and several ADPRase-I and ADPRase-II enzymes. The secondary structural elements of Ndx2 are shown above the sequence. Sequences shown here are as follows: T. thermophilus HB8 (Ndx2, Ndx4, BAC67698), E. coli (ORF186 [NP_417856], ORF209 [NP_417506]), T4 bacteriophage (NudE.1 [NP_049737]), M. tuberculosis (MtADPRase [NP_216216]), and Synechocystis sp. strain PCC 6803 (Slr1134 [NP_440605], Sll1054 [NP_439968]). Stars indicate residues that are conserved in ADPRase-I enzymes. The structural alignment for the proteins of known structure was calculated with MATRAS (13), and sequence alignment was prepared with ClustalW (31).
Several crystal structures of enzymes belonging to the ADPRase-I type have been determined: Ndx4 from T. thermophilus HB8 (35), EcADPRase from E. coli (7), MtADPRase from M. tuberculosis (12), and hNUDT5 (36) and hNUDT9 (28) from Homo sapiens. Previously, the crystal structure of EcORF186 had been the only report of the tertiary structure of an ADPRase-II (1). The structural details of EcORF186 have not been described in the published literature, however, because this structure was determined by a structural genomics project. The three-dimensional structure of Ndx2 determined in this study enables us to compare the structural details of ADPRase-II enzymes as well as those of ADPRase-I enzymes.
Overall structure of Ndx2.
The three-dimensional structure of Ndx2 was determined by X-ray crystallography as described in Materials and Methods. Crystals of the ligand-free enzyme, the Mg2+ complex, the Mg2+-AMP complex, and the Mg2+-AMPCPR complex were obtained. The structures were determined to 1.66- to 1.84-Å resolution by the MAD method using a gold derivative of the Mg2+ complex. Data collection and phasing and refinement statistics are shown in Tables 2 and 3.
TABLE 2.
Data collection and phase determination statistics using MAD method
| Data set | Value fora:
|
||
|---|---|---|---|
| Peak | Low remote | Edge | |
| Wavelength (Å) | 1.00000 | 1.03940 | 1.03985 |
| Resolution (Å) | 50-2.00 (2.07-2.00) | 50-2.00 (2.07-2.00) | 50-2.00 (2.07-2.00) |
| Space group | C2221 | C2221 | C2221 |
| Cell constants (Å) | |||
| a | 40.9 | 40.9 | 40.9 |
| b | 101.3 | 101.3 | 101.3 |
| c | 97.7 | 97.7 | 97.7 |
| α = β = γ | 90° | 90° | 90° |
| Data collection | |||
| Observed reflections | 85,118 | 88,620 | 86,170 |
| Unique reflections | 12,419 | 13,668 | 13,439 |
| Completeness (%) | 96.4 (84.7) | 97.4 (88.7) | 96.3 (83.5) |
| I/σ(I) | 20.7 (3.9) | 21.2 (4.2) | 21.1 (3.7) |
| Rmerge (%)b | 7.3 (26.2) | 7.1 (30.9) | 6.4 (28.7) |
| Phasing statistics | |||
| Phasing powerc | |||
| Acentric | 0.95 | ||
| Centric | 0.65 | ||
| Rcullisd | |||
| Acentric | 0.97 | ||
| Centric | 1.04 | ||
| Figure of merit | 0.54 | ||
Values in parentheses are for the outermost shell.
Rmerge = Σhkl ΣiIi(hkl) − I(hkl)/Σhkl ΣiIi(hkl), where Ii(hkl) is the observed intensity and I(hkl) is the averaged intensity for multiple measurements.
Phasing power = (FH/lack of closure), where FH is the calculated heavy-atom structure factor amplitude.
Rcullis = (lack of closure)/(isomorphous difference).
TABLE 3.
Data collection and refinement statistics
| Data set | Value fora:
|
|||
|---|---|---|---|---|
| Ndx2 (free) | Mg2+ (complex) | Mg2+-AMP (complex) | Mg2+-AMPCPR (complex) | |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Resolution (Å) | 50-1.84 (1.90-1.84) | 50-1.70 (1.76-1.70) | 50-1.67 (1.73-1.67) | 50-1.66 (1.72-1.66) |
| Space group | C2221 | C2221 | C2221 | C2221 |
| Cell constants (Å) | ||||
| a | 40.2 | 40.8 | 40.7 | 40.7 |
| b | 101.2 | 101.2 | 100.9 | 100.8 |
| c | 97.6 | 97.5 | 97.1 | 97.4 |
| α = β = γ | 90° | 90° | 90° | 90° |
| Data collection | ||||
| Observed reflections | 116,765 | 158,965 | 115,716 | 125,995 |
| Unique reflections | 19,698 | 22,707 | 23,468 | 23,576 |
| Completeness (%) | 99.8 (98.9) | 99.9 (99.9) | 99.1 (97.6) | 97.8 (95.8) |
| I/σ(I) | 37.2 (5.8) | 48.0 (7.0) | 48.2 (14.1) | 51.3 (14.0) |
| Rmerge (%)b | 4.5 (24.8) | 4.4 (23.9) | 3.1 (10.7) | 4.1 (9.6) |
| Refinement | ||||
| R factor (%)c | 0.224 | 0.205 | 0.180 | 0.188 |
| Rfree (%)d | 0.255 | 0.225 | 0.205 | 0.204 |
| Average B factor (Å2) | 20.7 | 19.1 | 14.3 | 16.8 |
| Modele | ||||
| Amino acid no. | 182 | 182 | 182 | 182 |
| Ligand | 2 Mg2+ | 3 Mg2+, 1 AMP | 3 Mg2+, 1 AMPCPR | |
| Water no. | 101 | 129 | 138 | 133 |
| Total atoms | 1,538 | 1,568 | 1,601 | 1,609 |
| Stereochemistryf | ||||
| RMS deviation | ||||
| Bond length (Å) | 0.008 | 0.010 | 0.010 | 0.017 (0.010) |
| Angle (°) | 1.5 | 1.6 | 1.5 | 2.8 (1.7) |
Values in parentheses are for the outermost shell; values in parentheses for RMS deviation are without AMPCPR.
Rmerge = Σhkl ΣiIi(hkl)-I(hkl)/ΣhklΣiIi(hkl), where Ii(hkl) is the observed intensity and I(hkl) is the averaged intensity for multiple measurements.
R factor = Σ‖Fobs − Fcalc‖/ΣFobs.
Rfree is monitored with 10% of the reflection data excluded from refinement.
The asymmetric unit has one subunit.
Over 90% of main-chain dihedrals fall within the “most favored regions” of the Ramachandran plot.
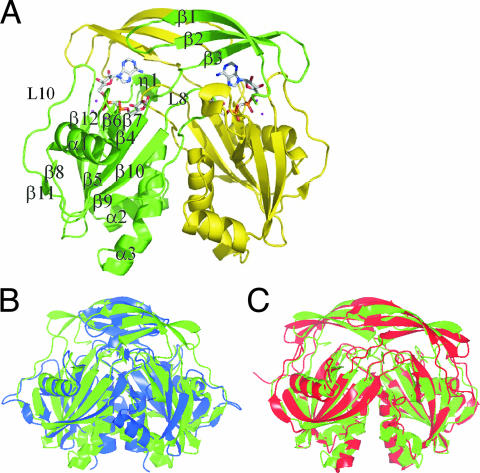
Crystals of Ndx2 contain one subunit in the asymmetric unit. By logical operation in crystallographic analysis, it is possible for Ndx2 to exist as a dimeric form as shown in Fig. 2A, in agreement with a homodimeric state in solution as indicated by size exclusion chromatography. Each subunit has 1 310-helix, 3 α-helices, and 12 β-strands and has the characteristic α-β-α sandwich fold structure of Nudix proteins. The Ndx2 homodimer is stabilized by a pair of equivalent interfaces created by swapping the N-terminal β-strands (β1 to β3) of two subunits. The Nudix box forms a loop-helix-loop structure, which includes the α1 helix, like typical Nudix proteins.
FIG. 2.
Overall structure of Ndx2. (A) The Mg2+-AMPCPR complex form. The subunits are shown in green and yellow. The Mg2+ ions are represented as magenta spheres. AMPCPR is represented in stick notation and Corey-Pauling-Kultun (CPK) color coding. (B) Overlap of the Ndx2 dimer (green) and the Ndx4 (1V8M) dimer (blue). (C) Overlap of the Ndx2 dimer (green) and the EcORF186 (1VHZ) dimer (red). The figure was produced with the CCP4MG program.
The overall structure of Ndx2 is almost identical to that of Ndx4 (35), with a root mean square (RMS) deviation of 1.78 Å for equivalent Cα atoms (Fig. 2B), although the sequence identity between them is 25%. The secondary structure elements are also almost the same: one β sheet (β6) is observed only for Ndx2. Both Ndx2 and EcORF186 are ADPRase-II enzymes, but the sequence identity is 24%. They have similar overall structures, with an RMS deviation of 2.18 Å for equivalent Cα atoms (Fig. 2C) and the same number of secondary structure elements.
Electron densities corresponding to Mg2+, AMP, and AMPCPR were identified near the Nudix box of both subunits in the respective complexes of Ndx2. Whereas the side chains of Glu136 and Glu137 in loop 10 in the free form and the Mg2+ complex are disordered, the side chain of Glu136 is well defined in the Mg2+-AMP and Mg2+-AMPCPR complexes. Except for this region, the structure of Ndx2 does not undergo a large conformational change, regardless of whether it exists as the ligand-free or ligand-bound form.
Metal-binding sites.
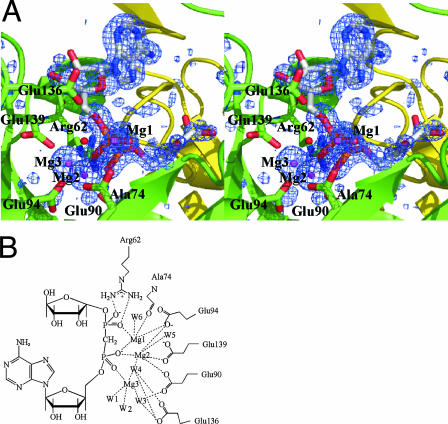
Two Mg2+ (Mg1 and Mg2) ions are found in the structures of the Mg2+ complex, whereas three Mg2+ (Mg1, Mg2, and Mg3) ions are present in the complexes with Mg2+-AMP and Mg2+-AMPCPR (Fig. 3A). Mg1 and Mg2 are found at the same position in the three complexes. Mg1, Mg2, and Mg3 are each coordinated in a nearly perfect octahedral geometry by six oxygen atoms, all about 2.1 Å from the Mg2+ ion (Fig. 3B). Mg1 is coordinated to the side chain of Glu94, the main-chain carbonyl of Ala74, and nonbridging oxygen atoms of the α-phosphate and β-phosphate of the bound nucleotide. Mg2 is coordinated to the side chains of Glu90, Glu94, and Glu139, the α-phosphate, and two water molecules, one of which is assumed to be the nucleophile water. This water molecule (W4 in Fig. 3B) is 2.92 Å away from the α-phosphorus and almost exactly in line with the scissile bond (angle, 171°), consistent with an associative mechanism. As a result, this water molecule is located in an ideal position to carry out the nucleophilic attack on the α-phosphate. In the nucleotide-bound complexes, Mg3 is coordinated to the side chain of Glu90, the α-phosphate, and four water molecules, one of which is also coordinated to Mg2.
FIG. 3.
Coordination of Mg2+ in the Mg2+-AMPCPR complex. (A) Stereo diagram of the Mg2+-binding site. Mg2+ ions and a nucleophilic water molecule are represented as magenta spheres and a red sphere, respectively. AMPCPR is represented in stick formation and Corey-Pauling-Kultun color coding. The Fo-Fc electron density map for the Mg2+ ions and AMPCPR is contoured at the 3.0-σ level. (B) Schematic representation of the Mg2+-binding site. W, water; W4 is assumed to be the nucleophilic water.
Recognition of AMP.
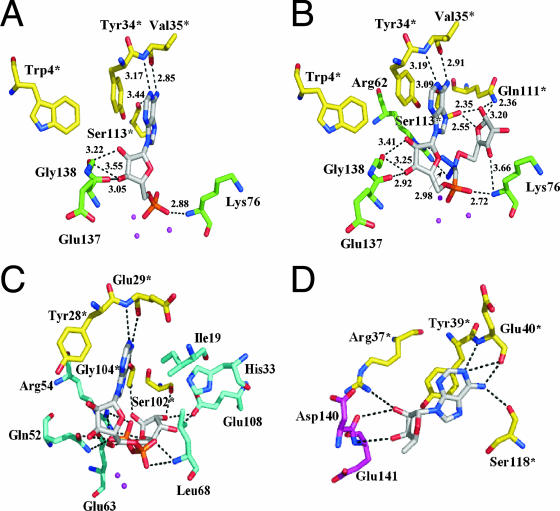
The binding pattern of AMP in the Mg2+-AMP complex structure is shown in Fig. 4A. AMP binds to the Nudix box with its adenine base in the anti conformation. The adenine ring is parallel to the side chain of Tyr34* (the asterisk represents a residue from another subunit) at a distance of 3.39 Å. Thus, it can be considered that the adenine ring is recognized by ring-stacking interactions with Tyr34. The N1 atom of the adenine moiety makes a hydrogen bond with the main-chain amide of Val35* (3.17 Å). The N6 atom of the adenine moiety makes hydrogen bonds with the main-chain carbonyls of Ser113* (3.44 Å) and Val35* (2.85 Å). The 2′-OH group of the ribose moiety makes a hydrogen bond with the main-chain amide of Gly138 (3.22 Å), whereas the 3′-OH group of the ribose moiety makes hydrogen bonds with the main-chain carbonyls of Glu137 (3.05 Å) and Gly138 (3.55 Å). One oxygen atom of the α-phosphate group makes a hydrogen bond with the main-chain amide of Lys76 (2.88 Å).
FIG. 4.
The recognition of AMP and AMPCPR. (A) The recognition site in the product complex of Ndx2 with Mg2+ and AMP. Mg2+ ions are shown as magenta spheres. AMP is shown as white lines. (B) The recognition site in the substrate analog complex with Mg2+ and AMPCPR. Mg2+ is shown as magenta spheres. AMPCPR is shown as white lines. (C) Recognition of ADPR by Ndx4. The binding residues are conserved among ADPRase-I enzymes. Gd3+ ions are shown as magenta spheres. ADPR is shown as white lines. (D) Recognition of adenosine by EcORF186. Adenosine is shown as white lines. Residue numbers followed by asterisks indicate residues of the other subunit in the dimer.
Recognition of AMPCPR.
The binding pattern of AMPCPR in the Mg2+-AMPCPR complex structure is shown in Fig. 4B. The residues involved in binding to the adenine and ribose moieties are the same as those involved in binding to AMP in the Mg2+-AMP complex, and the distances between corresponding atoms in the two complexes are also similar. The main-chain amide of Lys76 makes a hydrogen bond with one oxygen atom of the α-phosphate group (2.72 Å), similar to the binding of AMP. In addition, two oxygen atoms of the β-phosphate group make hydrogen bonds with the side chain of Arg62 (2.97 Å and 2.98 Å), which could stabilize the negative charge on the ribose-5′-phosphate. The 1′-OH group of the terminal ribose makes hydrogen bonds with the side chains of Gln111* and Ser113* (2.36 Å and 2.35 Å), and the 3′-OH group makes a hydrogen bond with the main-chain amide of Lys76 (3.66 Å). The O4 atom (ring oxygen) of the terminal ribose makes hydrogen bonds with the side chains of Gln111* and Ser113* (3.20 Å and 2.55 Å). The structure of the bound AMPCPR resembles a horseshoe. Such a conformation of the bound nucleotide sugar has been observed for the ligand-bound structures of other ADPRases. In contrast to other ADPRases, however, there is no water molecule that makes a hydrogen bond to both the N7 atom of the adenine and the β-phosphate oxygen.
Comparison with Ndx4.
The detailed mechanisms of substrate recognition are different between Ndx4 (ADPRase-I) and Ndx2 (ADPRase-II). Several residues (Glu29*, Gln52, Glu63, Gly104*, and Glu108 in Ndx4) are conserved well among members of the ADPRase-I subfamily (Fig. 1). These conserved residues are involved in the recognition of ADPR, especially the terminal ribose (Fig. 4C). The side chain of Glu63 makes hydrogen bonds with the internal oxygen of the terminal ribose (3.62 Å). The main-chain amide of Gly104* make a hydrogen bond with the terminal ribose O1′H (2.78 Å). The side chain of Glu108 makes hydrogen bonds with the terminal ribose O2′H and O3′H (2.48 and 3.23 Å). These residues are conserved among the ADPRase-I subfamily but not among the ADPRase-II subfamily (Fig. 1). Ser102* in Ndx4, corresponding to Gln111* in Ndx2 (Fig. 4C), also makes hydrogen bonds with the terminal ribose, although each residue interacts with different moieties. The residue corresponding to Ser113* is Gly104* in Ndx4. A smaller number of interactions with the terminal ribose in Ndx2 can be consistent with the low specificity of Ndx2 for ADPR compared with that of Ndx4.
Gln52 in Ndx4 is also conserved only among ADPRase-I enzymes (Fig. 1). Its side chain makes a hydrogen bond with the one oxygen of the β-phosphate group (3.27 Å) (Fig. 4C). The side chain of the corresponding residue in Ndx2, Gln60, is similarly located but too distant (4.4 Å) to interact directly with the phosphate moiety. Moreover, the glutamine residue at this position is not conserved in the ADPRase-II subfamily (Fig. 1). Another conserved residue among ADPRase-I enzymes is Glu29 in Ndx4, whose main-chain carbonyl oxygen and amide nitrogen make hydrogen bonds with the adenine N6 (3.22 Å) and N1 (3.48 Å), respectively (Fig. 4C). It should be mentioned, however, that each interaction of these conserved residues is likely to be relatively weak. Mutagenesis studies of Ndx4 have shown that the Km values of the wild-type, Q52A, E63Q, and E108Q enzymes for ADPR were 0.11, 0.24, 0.40, and 0.29 mM, respectively (25).
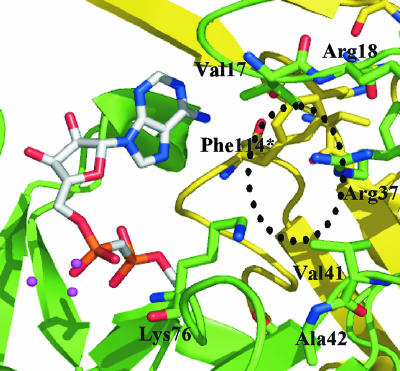
Comparison with EcORF186.
Among the residues in Ndx2 involved in the recognition of the terminal ribose, Ser113* is conserved in EcORF186 (Ser118), but Gln111* and Lys76 are not (Fig. 1). Furthermore, even Ser113* is not conserved among ADPRase-II enzymes. This may reflect the diversity in substrate specificity among ADPRase-II enzymes. In addition, in terms of recognition of the adenosine moiety, only three residues, Tyr34*, Ser113*, and Glu137, are conserved in EcORF186 (Tyr39*, Ser118*, and Glu141). In the solved structure of EcORF186 (PDB code, 1VHZ), an adenosine is bound to the active site of one subunit (Fig. 4D). These three residues are involved in the recognition of the adenosine moiety in the same way as the counterparts of Ndx2. Tyr39* forms a stacking interaction with the adenine ring, Ser118* makes a hydrogen bond with the N6 atom (3.12 Å), and Glu141 makes a hydrogen bond with the 3′-OH group of the ribose moiety (3.54 Å). However, these are not conserved among other members of the ADPRase-II family. Although the adenosine bound to EcORF186 seems to be additionally recognized by Arg37*, Glu40*, and Asp140, these residues are also not conserved. This lower conservation might be explained by the fact that the adenosine moiety is recognized by interactions with the main-chain amide and base stacking.
Mutational study of Glu90, Glu94, and Glu136.
The phosphohydrolase activity of Ndx2 mutants of three glutamic acids in the active site was examined. In EcORF209 (EcADPRase; NCBI accession no. NP_417506), Glu162 in the L9 loop has been proposed to act as a catalytic base on the basis of a structural comparison of the apoenzyme (a free form) and the Mg2+-AMPCPR complex: upon substrate binding, a major conformational change occurs in the L9 loop, which closes the active site pocket and brings Glu162 to a location that is ideal to act as the catalytic base (8). However, mutational analysis of this residue has not been performed. Here, Glu136 (corresponding to Glu162 in EcADPRase), Glu90, and Glu94 in the Nudix box of Ndx2 were mutated to glutamine. All three Ndx2 mutant proteins, the E136Q, E90Q, and E94Q mutants, were expressed and showed CD spectra similar to that of the wild-type protein. In terms of the ADPRase activity, the Km and kcat values of the E136Q mutant were 660 μM and 0.12 s−1, whereas those of the wild type were 520 μM and 1.4 s−1, respectively (Table 4). Compared with that of the wild type, the kcat was decreased by about 12-fold. In contrast, the E90Q and E94Q mutants showed marked decreases in kcat, namely, 2.2 × 10−4 s−1 and 1.4 × 10−4 s−1, respectively (Table 4). These results strongly suggest that Glu90 and Glu94 are involved in catalysis.
TABLE 4.
Kinetic constants of Ndx2 mutantsa
| Mutation | Substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| None (wild type) | ADPR | 520 | 1.4 | 2,700 |
| FAD | 300 | 2.5 | 8,300 | |
| E136Q | ADPR | 660 | 0.12 | 180 |
| FAD | 400 | 0.27 | 680 | |
| E90Q | ADPR | 820 | 0.00022 | 0.27 |
| FAD | 640 | 0.00067 | 1.0 | |
| E94Q | ADPR | 1000 | 0.00014 | 0.14 |
| FAD | 790 | 0.00017 | 0.21 |
Assays were done as described in Materials and Methods.
DISCUSSION
We propose the classification of ADPRases into two types: ADPRase-I enzymes, which exhibit high specificity for ADPR; and ADPRase-II enzymes, which exhibit low specificity for ADPR. This classification was originally based on the substrate specificity and sequence conservation. In this study, we show that Ndx2 recognizes the AMP moiety in a manner similar to ADPRase-I enzymes but recognizes the terminal ribose in a distinct manner. The residues responsible for the recognition of the substrate in Ndx2 are not conserved even among ADPRase-II enzymes. This may reflect the diversity in substrate specificity among ADPRase-II enzymes.
Glu162 located in the L9 loop of EcADPRase has been proposed to act as a catalytic base to activate a water molecule by movement of the loop. It has been reported that mutation of Glu142 in MtADPRase, corresponding to Glu162, results in a 3.2-fold increase in Km and a 300-fold decrease in kcat (20). In this study, however, the kcat values for the ADPRase and FADase activity of the E136Q mutant decreased only 12-fold and 9-fold, respectively. In contrast, the ADPRase and FADase activity of the E90Q and E94Q mutants decreased markedly, suggesting that these two glutamate residues in the Nudix box are essential for catalysis by Ndx2. The side chains of these residues coordinate to the Mg2+ ion (Mg2), which is coordinated to one water molecule (W4). The marked decreases in the activity of E90Q and E94Q might be explained if it assumed that this water molecule carries out the nucleophilic attack. This nucleophilic water may be deprotonated by Mg2 and Mg3, such that the attack on the α-phosphate and the cleavage of the P-O bond occur efficiently. The catalytic mechanism of Ndx4 is proposed to follow a pathway dependent on two metal ions (which coordinate with two glutamate residues in the Nudix box). This notion is in agreement with the experimental results for MtADPRase; that is, the contributions to kcat of Glu112 and Glu116 (corresponding to Glu90 and Glu94 of Ndx2) were found to be 103.3 and 103.0, respectively (20).
Based on the results described above, it can be concluded that ADPRase-I and ADPRase-II enzymes have nearly identical catalytic mechanisms but different mechanisms of substrate recognition. Considering that the difference of substrate recognition mechanism is important, it is reasonable to classify ADPRases into two types. Sequence conservation supports this classification. It should be mentioned here, however, that the Nudix proteins classified as ADPRase-II enzymes in this study are diverse in sequence and substrate specificity. Such diversity appears incompatible with the classification of those proteins into the same group. Further structural study might reveal a further classification of ADPRase-II enzymes into subgroups.
Zha et al. have suggested that the conserved proline residue is important to maintain the right length and position of the L8 loop, which is involved in interlocking the two subunits and in recognition of the terminal ribose (36). This proline residue is also conserved in Ndx2 (Pro112). However, several ADPRase-II enzymes, including Ndx2, hydrolyze FAD and NADH. These two compounds contain moieties larger than the terminal ribose of ADPR. By analogy to the conformation of AMPCPR bound to Ndx2, the riboflavin moiety of FAD should be situated at the bottom of the pocket. Assuming a horseshoe-like conformation of FAD, the riboflavin moiety might be positioned near the β1-β2 loop and the β9-β10 loop at the dimer interface (Fig. 5). However, the pocket does not appear to have enough room to accommodate the riboflavin moiety, especially the isoalloxazine ring, without a steric clash with the surrounding residues. To enable the pocket to accommodate FAD, a large conformational change at the dimer interface, including the L8 loop, is required. The isoalloxazine ring may be located outside the pocket by means of the flexibility of the ribitol chain. However, such a binding mode would not seem to explain the high specificity of the enzyme for FAD. Studies to obtain a crystal of the FAD-bound enzyme have not been successful so far.
FIG. 5.
Possible binding of FAD. The possible region for the riboflavin moiety is surrounded by a dashed line.
The physiological functions of many Nudix proteins, except for the MutT protein (18, 32), have not been revealed. The MutT protein functions to prevent errors in DNA replication by hydrolyzing mutagenic nucleotides such as 8-oxo-dGTP. To obtain clues to the cellular function of Ndx2 in T. thermophilus HB8, we carried out Western blotting and DNA microarray analysis to determine the amounts of Ndx2 produced (expressed) through the growth phase. These analyses showed that Ndx2 is expressed in all growth phases (data not shown), and disruption of the ndx2 gene caused no significant change of growth in rich medium (data not shown).
The fact that many organisms have multiple ADPRases highlights the physiological importance of eliminating ADPR for cell viability. On the other hand, Ndx2 showed activity for FAD. Riboflavin, FAD, and flavin mononucleotide act as coenzymes for redox enzymes called flavoproteins. These proteins catalyze a great diversity of redox reactions in oxidative phosphorylation both during cell respiration and in the intermediate metabolism of carbohydrates, amino acids, and fatty acids. The biosynthetic pathway of FAD in prokaryotes and eukaryotes has been widely characterized, whereas the degradation pathway has scarcely been studied. FADases have been found in rat liver lysosomal membrane and mitochondria (2, 29) and in human placental trophoblastic microvilli (16). This FADase activity has been proposed to play a role in the turnover of flavoproteins. However, we do not even know that these FADases are Nudix proteins, because their amino acid sequences have not been determined.
Acknowledgments
We thank Hirofumi Omori for technical assistance in DNA sequencing of the mutant enzymes in this study.
This work was supported in part by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan, and by Grant-in-Aid for Scientific Research 17770089 (to N.N.) from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Badger, J., J. M. Sauder, J. M. Adams, S. Antonysamy, K. Bain, M. G. Bergseid, S. G. Buchanan, M. D. Buchanan, Y. Batiyenko, J. A. Christopher, S. Emtage, A. Eroshkina, I. Feil, E. B. Furlong, K. S. Gajiwala, X. Gao, D. He, J. Hendle, A. Huber, K. Hoda, P. Kearins, C. Kissinger, B. Laubert, H. A. Lewis, J. Lin, K. Loomis, D. Lorimer, G. Louie, M. Maletic, C. D. Marsh, I. Miller, J. Molinari, H. J. Muller-Dieckmann, J. M. Newman, B. W. Noland, B. Pagarigan, F. Park, T. S. Peat, K. W. Post, S. Radojicic, A. Ramos, R. Romero, M. E. Rutter, W. E. Sanderson, K. D. Schwinn, J. Tresser, J. Winhoven, T. A. Wright, L. Wu, J. Xu, and T. J. Harris. 2005. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60787-796. [DOI] [PubMed] [Google Scholar]
- 2.Barile, M., C. Brizo, C. de Virgilio, S. Delfine, E. Quagliariello, and S. Passarella. 1997. Flavin adenine dinucleotide and flavin mononucleotide metabolism in rat liver. The occurrence of FAD pyrophosphatase and FMN phosphohydrolase in isolated mitochondria. Eur. J. Biochem. 249777-785. [DOI] [PubMed] [Google Scholar]
- 3.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 27125059-25062. [DOI] [PubMed] [Google Scholar]
- 4.Brünger, A. T., P. D. Adams, G. M. Clore, W. N. DeLano, P. Gros, R. W. GrosseKunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 5.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50760-763. [DOI] [PubMed] [Google Scholar]
- 6.Dunn, C. A., S. F. O'Handley, D. N. Frick, and M. J. Bessman. 1999. Studies on the ADP-ribose pyrophosphatase subfamily of the Nudix hydrolases and tentative identification of trgB, a gene associated with tellurite resistance. J. Biol. Chem. 27432318-32324. [DOI] [PubMed] [Google Scholar]
- 7.Gabelli, S. B., M. A. Bianchet, M. J. Bessman, and L. A. Amzel. 2001. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat. Struct. Biol. 8467-472. [DOI] [PubMed] [Google Scholar]
- 8.Gabelli, S. B., M. A. Bianchet, Y. Ohnishi, Y. Ichikawa, M. J. Bessman, and M. L. Amzel. 2002. Mechanism of the Escherichia coli ADP-ribose pyrophosphate, a Nudix hydrolase. Biochemistry 419279-9285. [DOI] [PubMed] [Google Scholar]
- 9.Iwai, T., S. Kuramitsu, and R. Masui. 2004. The Nudix hydrolase Ndx1 from Thermus thermophilus HB8 is a diadenosine hexaphosphate hydrolase with a novel activity. J. Biol. Chem. 27921732-21739. [DOI] [PubMed] [Google Scholar]
- 10.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47110-119. [DOI] [PubMed] [Google Scholar]
- 11.Just, I., P. Wollenberg, J. Moss, and K. Aktories. 1994. Cystein-specific ADP-ribosylation of actin. Eur. J. Biochem. 2211047-1054. [DOI] [PubMed] [Google Scholar]
- 12.Kang, L.-W., S. B. Gabelli, J. E. Cunnigham, S. F. O'Handley, and L. M. Amzel. 2003. Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis. Structure 111015-1023. [DOI] [PubMed] [Google Scholar]
- 13.Kawabata, T. 2003. MATRAS: a program for protein 3D structure comparison. Nucleic Acids Res. 313367-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuramitsu, S., K. Hiromi, H. Hayashi, Y. Morino, and H. Kagamiyama. 1990. Pre-steady-state kinetics of Escherichia coli aspartate aminotransferase catalyzed reactions and thermodynamic aspects of its substrate specificity. Biochemistry 295469-5476. [DOI] [PubMed] [Google Scholar]
- 15.Laskowski, R. A., M. W. McArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26283-291. [Google Scholar]
- 16.Lee, R. S.-F., and H. C. Ford. 1988. 5′-Nucleotidase of human placental trophoblastic microvilli possesses cobalt-stimulated FAD pyrophosphatase activity. J. Biol. Chem. 26314878-14883. [PubMed] [Google Scholar]
- 17.Lin, J., C. Abeygunawardana, D. N. Frick, M. J. Bessman, and A. S. Mildvan. 1997. Solution structure of the quaternary MutT-M2+-AMPCPP-M2+ complex and mechanism of its pyrophosphohydrolase action. Biochemistry 361199-1211. [DOI] [PubMed] [Google Scholar]
- 18.Maki, H., and M. Sekiguchi. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355273-275. [DOI] [PubMed] [Google Scholar]
- 19.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33491-497. [DOI] [PubMed] [Google Scholar]
- 20.Mildvan, A. S., Z. Xia, H. F. Azurmendi, V. Saraswat, P. M. Legler, M. A. Massiah, S. B. Gabelli, M. A. Bianchet, L.-W. Kang, and L. M. Amzel. 2005. Structures and mechanisms of Nudix hydrolases. Arch. Biochem. Biophys. 433129-143. [DOI] [PubMed] [Google Scholar]
- 21.Nishikubo, T., N. Nakagawa, S. Kuramitsu, and R. Masui. 2005. Improved heterologous gene expression in Escherichia coli by optimization of the AT-content of codons immediately downstream of the initiation codon. J. Biotechnol. 120341-346. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa, T., Y. Ueda, K. Yoshimura, and S. Shigeoka. 2005. Comprehensive analysis of cytosolic Nudix hydrolases in Arabidopsis thaliana. J. Biol. Chem. 28025277-25283. [DOI] [PubMed] [Google Scholar]
- 23.O'Handley, S. F., D. N. Frick, C. A. Dunn, and M. J. Bessman. 1998. Orf186 represents a new member of the Nudix hydrolases, active on adenosine(5′)triphospho(5′)adenosine, ADP-ribose, and NADH. J. Biol. Chem. 2733192-3197. [DOI] [PubMed] [Google Scholar]
- 24.Okuda, K., H. Hayashi, and Y. Nishiyama. 2005. Systematic characterization of the ADP-ribose pyrophosphatase family in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 1874984-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooga, T., S. Yoshiba, N. Nakagawa, S. Kuramitsu, and R. Masui. 2005. Molecular mechanism of the Thermus thermophilus ADP-ribose pyrophosphatase from mutational and kinetic studies. Biochemistry 449320-9329. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276307-326. [DOI] [PubMed] [Google Scholar]
- 27.Sheikh, S., S. F. O'Handley, C. A. Dunn, and M. J. Bessman. 1998. Identification and characterization of the Nudix hydrolase from the archaeon, Methanococcus jannaschii, as a highly specific ADP-ribose pyrophosphatase. J. Biol. Chem. 27320924-20928. [DOI] [PubMed] [Google Scholar]
- 28.Shen, B. W., A.-L. Perraud, A. Scharenberg, and B. L. Stoddard. 2003. The crystal structure and mutational analysis of human NUDT9. J. Mol. Biol. 332385-398. [DOI] [PubMed] [Google Scholar]
- 29.Shin, H. J., and W. K. Lim. 1996. A rat liver lysosomal membrane flavin-adenine dinucleotide phosphohydrolase. J. Biochem. Mol. Biol. 29253-260. [DOI] [PubMed] [Google Scholar]
- 30.Terwilliger, T. 2004. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 1149-52. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuzuki, T., A. Egashira, H. Igarashi, T. Iwakuma, Y. Nakatsuru, Y. Tominaga, H. Kawate, K. Nakao, K. Nakamura, F. Ide, S. Kura, Y. Nakabeppu, M. Katsuki, T. Ishikawa, and M. Sekiguchi. 2001. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc. Natl. Acad. Sci. USA 9811456-11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, W., P. Gauss, J. Shen, C. A. Dunn, and M. J. Bessman. 2002. The gene e.1 (nudE.1) of T4 bacteriophage designates a new member of the Nudix hydrolase superfamily active on flavin adenine dinucleotide, adenosine 5′-triphospho-5′-adenosine, and ADP-ribose. J. Biol. Chem. 27723181-23185. [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama, S., H. Hirota, T. Kigawa, T. Yabuki, M. Shirouzu, T. Terada, Y. Ito, Y. Matsuo, Y. Kuroda, Y. Nishimura, Y. Kyogoku, K. Miki, R. Masui, and S. Kuramitsu. 2000. Structural genomics projects in Japan. Nat. Struct. Biol. 7943-945. [DOI] [PubMed] [Google Scholar]
- 35.Yoshiba, S., T. Ooga, N. Nakagawa, T. Shibata, Y. Inoue, S. Yokoyama, S. Kuramitsu, and R. Masui. 2004. Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal. J. Biol. Chem. 27937163-37174. [DOI] [PubMed] [Google Scholar]
- 36.Zha, M., C. Zhong, Y. Peng, H. Hu, and J. Ding. 2006. Crystal structures of human NUDT5 reveal insights into the structural basis of the substrate specificity. J. Mol. Biol. 3641021-1033. [DOI] [PubMed] [Google Scholar]
- 37.Ziegler, M. 2000. New functions of a long-known molecule. Emerging roles of NAD in cellular signaling. Eur. J. Biochem. 2671550-1564. [DOI] [PubMed] [Google Scholar]