Abstract
The general stress response mediated by the sigma factor RpoS is important for survival of bacteria in adverse environments. A mutant unable to produce RpoS was constructed using the diazotrophic bacterium Azotobacter vinelandii strain UW. Under nondesiccating, solid-medium growth conditions the wild type was culturable for 16.5 years, while the rpoS mutant remained viable for only 10 months. The rpoS mutant exhibited reduced survival compared to the wild type following hydrogen peroxide stress, and stationary phase cells were killed rapidly by 15 mM H2O2. Three catalases (Kat1, Kat2, and Kat3) were expressed in the wild type under the conditions used. Kat2 was expressed in exponential phase during shake flask growth and could be induced under highly aerated conditions in all growth phases, suggesting that there was induction by reactive oxygen intermediates. Kat3 was possibly an isoform of Kat2. In contrast, Kat1 was expressed in an RpoS-dependent manner during the mid-exponential to late stationary phases. RpoS expression did not occur exclusively in stationary phase but was influenced by changes in carbon and nitrogen source availability. There was 26- to 28-fold induction of the RpoS protein during acetate-to-glucose and ammonium-to-N2 diauxic shifts. Following recovery of growth on the alternative carbon or nitrogen source, RpoS protein concentrations declined rapidly to a basal level. However, rpoS mRNA levels did not correlate directly to RpoS levels, suggesting that there was posttranscriptional regulation. Evidence obtained using the RpoS-dependent reporter Kat1 suggested that there is regulation of the RNAP:RpoS holoenzyme at the level of complex formation or activity.
The stress and starvation response is primarily regulated by the alternative sigma factor, variably designated σ38, σS, or RpoS, in many gram-negative organisms. Expression of this factor during stationary phase has been shown to regulate (positively or negatively) nearly 360 genes or 10% of the annotated genome of Escherichia coli (32), while the homologue in Pseudomonas aeruginosa regulates 772 genes or 14% of the genome (38). RpoS has been shown to regulate the production of secondary metabolites and various pathogenicity determinants in pseudomonads (14, 43). In addition to stationary-phase expression, RpoS induction is also observed when E. coli exhausts specific nutrients during growth in minimal medium (35). The magnitude of RpoS induction is the same (∼4-fold) whether the stress is a terminal starvation event, such as glucose exhaustion (35), or a transitional starvation event during a diauxic shift from glucose to lactose (10). Following the stress event, the concentration of RpoS decreases to a basal level, whether starvation is alleviated or not (10, 35). Although equivalent studies have been done during nitrogen exhaustion, no RpoS studies have been done during nitrogen diauxie in a diazotroph.
Catalase, peroxidase, and superoxide dismutase are used by aerobes in all kingdoms to degrade reactive oxygen intermediates (ROIs), such as •O2− and H2O2, that are produced during respiration. During periods of stress or starvation, many organisms supplement their constitutively expressed “housekeeping” catalase with an inducible enzyme. For example, P. aeruginosa modulates the expression of a single housekeeping catalase (KatA) in response to various growth conditions (5), while a second catalase (KatB) is induced under oxygen stress conditions by the regulator OxyR (27). In other organisms supplemental expression of an RpoS-dependent catalase occurs in stationary phase (7).
In this study we examined the role of RpoS in the long-term survival of Azotobacter vinelandii under starvation conditions and ROI stress conditions. RpoS and rpoS mRNA levels were measured by quantitative Western and Northern analyses. Although these methods are laborious, they were preferable to the use of lacZ fusions, as a careful analysis of previously described transcriptional (44) and translational (42) chimeric reporter systems indicated that they exhibit different stabilities than the native rpoS products (25). To determine if RpoS was functional during exponential growth, we used the stationary-phase catalase Kat1 (identified in this study) as a naturally occurring reporter of RpoS activity. RpoS appears to be functional during nutrient shifts, but the regulation of RpoS accumulation and activity in the cells is complex.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The A. vinelandii strains used were the non-alginate-producing strain UW (= ATCC 13705) and the rpoS mutant strain UWS, which was constructed in this study. Strains were grown in Burk's complete medium (BBGN) composed of Burk's buffer salts (BB) supplemented with 18.0 μM ferric citrate, 55.5 mM glucose, and 15 mM ammonium acetate, using the growth conditions described previously (29). In the ammonium augmentation experiments, 100 ml of 7 mM (NH4)2SO4 (pH 7.0) was added to 2.5 liters of BBGN at a rate of 5 ml min−1. BBG medium contained Burk's buffer supplemented with 18.0 μM ferriccitrate and 55.5 mM glucose. Kanamycin was added at a final concentration of 10 μg ml−1 to both solid and liquid media, as appropriate.
Shake flask cultures of A. vinelandii were inoculated from 3-day-old BBGN agar slants to obtain an initial optical density at 600 nm (OD600) of 0.030 to 0.040 and were grown with agitation at 200 rpm on a rotary shaker at 28°C. Large batch cultures were grown in a 2.5-liter BioFlowIII bioreactor (New Brunswick Scientific) that was inoculated using a liquid seed culture (see above). The inoculum was no more than 4% (vol/vol), and the culture was aerated with air (1 volume/min) using a constant impeller speed of 200 rpm. Dissolved oxygen (dO2) was monitored electronically, and samples were removed manually at the times indicated in the Results.
Quantification of cell fractions and nutrients.
Glucose and ammonium concentrations in the culture fluid were determined by the Trinder assay (Sigma Diagnostics) and the sodium nitroprusside method (41), respectively. The acetate concentration was determined by gas chromatography (Hewlett-Packard 5890) using a DB-FFAP megabore column (Agilent). All experiments were repeated at least twice, and the values reported below are means of duplicate or triplicate assays.
Hydrogen peroxide survival assay.
Bacteria used for hydrogen peroxide treatment were grown in shake flask cultures to the desired growth phase, removed from the culture medium by centrifugation at 3,000 × g for 5 min (Sorval RC2-B) at room temperature, and resuspended in BB to a final OD600 of 1.0. A sample obtained at time zero was immediately plated on BBGN to determine the number of CFU, and hydrogen peroxide (final concentration, 15 mM) was added to the remaining culture. Then the culture was incubated with shaking.
Molecular biology procedures.
A. vinelandii chromosomal DNA was cut by XhoI and cloned into pBluescript (Stratagene). The chromosomal fragment containing the rpoS gene was detected by colony hybridization (36), using an rpoS probe prepared by PCR using primers WJP58 and WJP59 (see Fig. S1 in the supplemental material). The recombinant plasmid containing rpoS was designated pRPOS, and the sequence of this fragment was determined using a DYEnamic ET kit (Molecular Dynamics). A 960-bp kanamycin resistance cassette was excised from p34S-Kmr (9) with SmaI and was ligated into the NruI site of pRPOS (see Fig. S1 in the supplemental material) to generate pRPOS::Km. This plasmid was transformed into A. vinelandii (30), and homologous recombination produced the kanamycin-resistant rpoS mutant UWS. Insertion of the Kmr cassette into rpoS was confirmed by PCR and sequencing.
Production of His-RpoS protein and antiserum.
The rpoS gene in pRPOS was amplified by PCR using primer WJP106 (see Fig. S1 in the supplemental material) and the M13-20 universal primer and was cloned into pRSETa (Invitrogen) after digestion with HindIII and NcoI. The ligated product, pRSET-rpoS, was transformed into E. coli DH5α. The His-RpoS protein was overexpressed, solubilized, and purified with Ni-nitrilotriacetic acid-agarose (Qiagen), followed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and electroelution (Tyler Research Instruments model EE-04) using a 12- to 14-kDa-cutoff membrane under conditions recommended by the manufacturer. The resultant His-RpoS protein was used to raise polyclonal antiserum in rabbits (Department of Biological Sciences Animal Services Unit).
Preparation of cell lysates.
Azotobacter cell lysate was prepared by the bead beating method of Benov and Al-Ibraheem (3). E. coli lysate was prepared by sonication (Braun Sonic 2000). Soluble protein concentrations were determined as described previously (22).
Zymographic catalase assay.
The proteins in the cell lysate were separated using 7.5 or 10% nondenaturing PAGE (Hoefer Scientific SE500) as indicated in the Results. Catalases were detected by the activity staining method of Clare et al. (8).
Quantification of RpoS protein by Western analysis.
Proteins were separated by 10% SDS-PAGE using a large-format Hoefer Scientific SE500 or Daltsix (Amersham) gel system. Proteins were transferred to Immun-Blot polyvinylidene difluoride membranes (Bio-Rad) by wet-cell electrotransfer (TE Transfor; Hoefer Scientific). RpoS protein was identified using polyclonal anti-His-RpoS antiserum, followed by IRDye800-conjugated donkey anti-rabbit antibody (Rockland Immunochemicals). Detection and quantification were done using an Odyssey infrared imaging system (Li-Cor) at a resolution of 84 μm.
The quantification method was validated as described by Jishage and Ishihama (16). The amount of RpoS present in 25 μg of strain UW cell lysate harvested at 48 h was arbitrarily designated 100 fluorescence units, and the amount of RpoS present in the same amount of lysate from strain UWS at 48 h was arbitrarily designated 0 fluorescence units. The relative fluorescence was linear (R2 = 0.996) from 1 to 1,500 relative fluorescence units (RFU).
Determination of RpoS stability.
A culture (35 ml) was removed aseptically from the bioreactor and placed in sterile 125-ml shake flasks. A 5-ml sample was immediately removed at time zero, and tetracycline (final concentration, 1.7 mM) was added to the remaining culture, which was incubated at 28°C with shaking at 250 rpm. Samples were pelleted by centrifugation at 1,000 × g with a model CL clinical centrifuge (International Equipment Co.) for exactly 2 min at 4°C and flash frozen in liquid N2. Samples were lysed and subjected to Western analysis.
Northern analysis.
The hot phenol method (37) was used to purify RNA from A. vinelandii. RNA (5 μg) was separated on 4% formaldehyde agarose gels and transferred to Highbond N+ (GE Healthcare) membranes. The rpoS template was created by PCR amplification using primers WJP188 (5′-TGGGAATTCAGGGGACGACAACGATGGCTG-3′) and WJP189 (5′-CTCGAGCTCTTTTCACTGGAACAGCGCATCG-3′). An [α-32P]dCTP-labeled rpoS probe was produced from the template by random priming (Roche) and was hybridized overnight at 65°C. Membranes were washed at 68°C in buffer B (0.5× SSPE, 0.1% SDS) (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), exposed to a phosphor screen (Molecular Dynamics) overnight, and imaged using a PhophorImager (Molecular Dynamics). The relative rpoS mRNA signal was normalized against a 16S rRNA internal control. The probe for 16S rRNA was produced by [γ-32P]dATP end labeling (Roche) oligonucleotide WJP16S (5′-CCGTCAATTCATTTGAGTTT-3′). The 16S rRNA probe was hybridized to the same membrane overnight at 50°C, washed several times with buffer B at 51°C, and imaged after a 15-min exposure.
Nucleotide sequence accession number.
The sequence of the rpoS gene of A. vinelandii and the surrounding genes has been deposited in the GenBank database under accession number AF421351.
RESULTS
rpoS gene of A. vinelandii.
The rpoS gene of A. vinelandii was isolated from chromosomal DNA, and surrounding gene sequences were identified (see Fig. S1 in the supplemental material). The order of the genes was later confirmed by whole-genome sequencing (http://azotobacter.org).
The rpoS gene of A. vinelandii is 1,005 bp long and encodes a 334-amino-acid protein with a predicted molecular mass of 38,306 Da. The A. vinelandii RpoS open reading frame is 87% identical to that of P. aeruginosa (protein accession number NP_252312). Sigma factor regions 2.4 and 2.5, responsible for recognition of the −10 and extended −10 promoter sequences, were 100% identical, while region 4.2, which is responsible for recognition of the −35 promoter sequences, shared 96% identity (13, 33, 40) (see Fig. S2 in the supplemental material).
RpoS is required during starvation survival.
The rpoS gene in pRPOS was disrupted with a Kmr cassette and was used to produce the rpoS mutant UWS by homologous gene replacement. Wild-type strain UW and mutant strain UWS had identical growth rates and growth curves in BBGN. The long-term survival of strains UW and UWS was tested on slants of solid BBGN. Strain UW stored in the dark at room temperature (21°C) was culturable on BBGN after 16.5 years, whereas UWS stored under identical conditions remained culturable for only 10 months. However, neither strain was viable if the medium became dry (28; this study).
The strains were also grown for 2 days in shake flask cultures until glucose became limiting, and then cell survival was examined for an additional 8 days (see Fig. S3 in the supplemental material). The viability of strain UW cells had decreased 60.5% by the eighth day of starvation. In contrast, the viability of strain UWS decreased 99.5% over the same period of time.
Expression of an RpoS-dependent catalase affects survival under oxidative stress conditions.
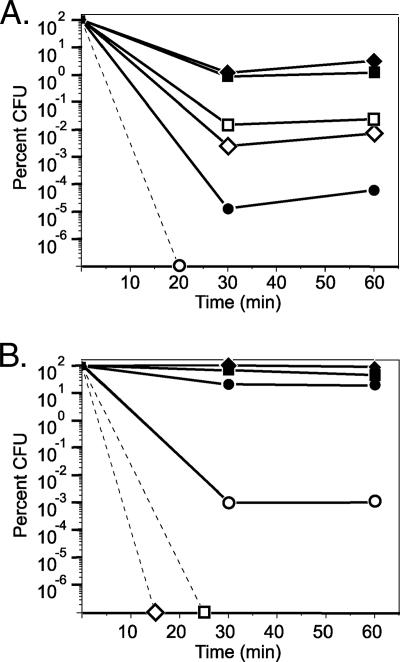
Strains UW and UWS were grown from early exponential phase (7 h) to late stationary phase (72 h) and treated with 15 mM H2O2 to determine their sensitivities to ROI stress. The survival of strain UW was worst during exponential phase (maximum, 3.2% survival) (Fig. 1A) and best during stationary phase (20 to 93% survival) (Fig. 1B). In contrast, the survival of strain UWS was worst during stationary phase (maximum, 0.001%) (Fig. 1B) and best during mid- to late exponential phase (0.007 to 0.024%) (Fig. 1A). This suggested that catalase activity was expressed in an RpoS-dependent manner.
FIG. 1.
Percent survival of strains UW (filled symbols) and UWS (open symbols) following treatment with 15 mM H2O2. The cultures were normalized to an OD600 of 1.0 in BB, and the levels of survival were determined for (A) 7-h (circles), 14-h (diamonds), and 24-h (squares) cultures before and after peroxide treatment and for (B) 36-h (circles), 48-h (diamonds), and 72-h (squares) cultures before and after peroxide treatment. Dashed lines indicate that no CFU were recovered following 30 min of peroxide treatment.
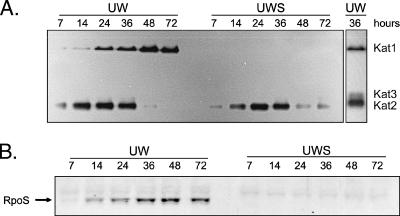
To determine if multiple catalases were expressed by A. vinelandii, cell lysates were analyzed for the presence of catalase activity after nondenaturing PAGE. Strain UW had two bands of catalase activity, designated Kat1 and Kat2, when it was assayed zymographically for 5 min (Fig. 2A); a third faint band of catalase activity (Kat3) was detected after 15 min (Fig. 2A, right panel). Kat2 was expressed in early exponential phase but not in 48- to 72-h stationary-phase cells. Conversely, Kat1 was not expressed in early exponential phase, but the concentration of this catalase increased in later phases. The absence of Kat1 in the rpoS mutant indicated that this catalase was expressed in an RpoS-dependent manner. The mutant exhibited wild-type expression of Kat2 during exponential growth but continued to produce low levels of Kat2 in late stationary phase (Fig. 2A).
FIG. 2.
Catalase expression during 72 h of growth of A. vinelandii. (A) In situ catalase zymographs of 50 μg of cell lysate from cultures of strains UW and UWS, separated by nondenaturing PAGE. Two strong bands (Kat1 and Kat2) were detected after 5 min of exposure to diaminobenzidine. Longer exposure times allowed detection of an additional catalase band, Kat3. (B) The same lysates (20 μg) were separated by denaturing PAGE and analyzed for RpoS by Western analysis. The sample times are indicated above the lanes. These cultures also were used as the inocula for the experiment whose results are shown in Fig. 1.
The apparent anomaly of expression of an RpoS-dependent catalase during exponential growth was explained by Western analysis (Fig. 2B). RpoS was detected in strain UW in mid-exponential phase, and the maximum levels were reached by 36 to 72 h, which coincided with the pattern of Kat1 expression. RpoS was not detected in strain UWS at any time (Fig. 2B).
RpoS expression during an acetate-to-glucose diauxic shift.
The expression of an RpoS-dependent catalase and of RpoS itself during exponential growth was unexpected, as a previous study (6) reported that A. vinelandii rpoS mRNA expression occurred only in late stationary phase. To determine what event might be responsible for RpoS induction during exponential growth, we grew strain UW in batch culture in a bioreactor and then monitored the RpoS concentration and nutrient availability for 48 h.
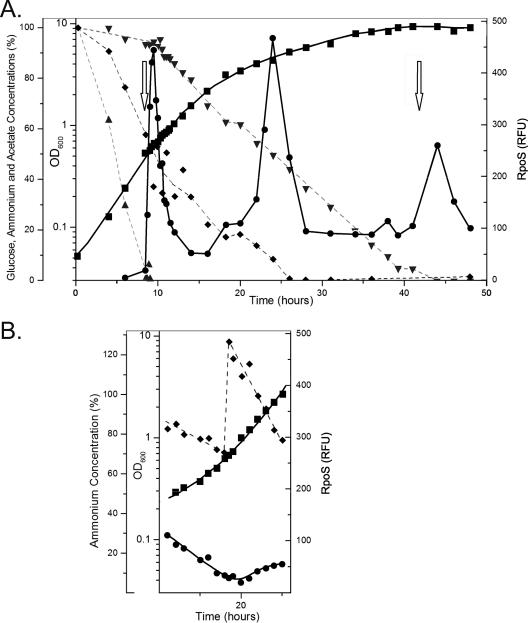
Strain UW used acetate in BBGN preferentially during early exponential phase, with a mean generation time of 2.48 h (Fig. 3A). During this time the dO2 levels in the bioreactor decreased, indicative of active metabolism. The acetate concentration decreased rapidly, while glucose was consumed at a very low rate, typical of the carbon source preference of A. vinelandii (11). A rapid increase in the dO2 concentration at 8.6 h signaled the cessation of metabolism due to acetate depletion (Fig. 3A). Rapid metabolism of the nonpreferred carbon source glucose began at 9.5 h and continued into stationary phase.
FIG. 3.
Growth of strain UW in BBGN in a 2.5-liter bioreactor. Growth was determined optically (OD600) (▪). The acetate (▴), ammonium (♦), and glucose (▾) concentrations are expressed as percentages of their initial concentrations. The relative amount of RpoS protein was determined by quantitative Western analysis (•). Periods of oxygen saturation, indicative of metabolic cessation, are indicated by arrows. (A) Data from a representative batch culture grown for 48 h. (B) Data from an ammonium-fed culture to which (NH4)2SO4 was added immediately after 18 h. Panel B is aligned directly below the equivalent times in panel A.
Western analysis of cell lysates showed that very low quantities of RpoS (0 to 17 RFU) were present during early exponential growth. The RpoS concentration increased within 9 min after acetate depletion and reached 443 RFU within 1 h, which was 26-fold induction. The RpoS concentration declined to a basal level (51 RFU) by 14 h (Fig. 4A), coincident with the recovery of metabolic activity on glucose. A. vinelandii resumed growth on glucose with a reduced generation time, 3.8 h.
FIG. 4.
Analysis of selected time points for the bioreactor culture in Fig. 3A to determine RpoS and catalase activities. (A) Western analysis of RpoS from 25 μg of cell lysate separated by SDS-PAGE. (B) Catalase zymography of 50 μg of cell lysate separated by nondenaturing PAGE. The sample times (in hours) are indicated above the lanes.
RpoS expression during an ammonium-to-N2 diauxic shift.
At 18 h the culture growth rate decreased (generation time, 11.3 h), and the RpoS levels increased 28-fold (relative to the levels in early exponential phase) by 24 h (Fig. 4A). Although the ammonium was not completely depleted by 18 h, it is possible that the cells experienced nitrogen limitation, since there was a significant decrease in the growth rate. The RpoS concentration declined rapidly after 24 h to a new basal level, 92 RFU.
RpoS is not regulated by oxygen limitation.
At 24 h the dO2 level of the medium was below 1% saturation, indicating that oxygen was being consumed as fast as it could be added to the culture. To determine if the observed RpoS response was due to nitrogen limitation or oxygen limitation, an ammonium augmentation experiment was performed (Fig. 3B). A culture was incubated in BBGN for 18 h, at which time there was still excess glucose, ammonium, and oxygen. Ammonium sulfate was added so that there was excess ammonium for the duration of the experiment. The RpoS levels did not increase even though the dO2 levels dropped below 1% saturation by 20 h. From this result we concluded that the production of RpoS starting at 18 h, as shown in Fig. 3A, was due to ammonium limitation rather than oxygen limitation.
RpoS expression during nitrogen fixation and glucose depletion.
After ammonium depletion, the cells fixed nitrogen from 27 to 40 h, using glucose as the carbon and energy source. Since a significant amount of energy is required for nitrogen fixation (11), it seemed likely that the organism would undergo a stress response during this period. The growth rate was greatly decreased (generation time, 31 h), but the RpoS concentration changed very little during this period (92 to 95 RFU).
Glucose was depleted at approximately 42 h, and there was a coincident increase in the dO2 level, indicating that active metabolism ceased (Fig. 3A). The RpoS concentrations increased and returned to the basal levels by 48 h. It is likely that the cells switched to metabolism of endogenous polyhydroxybutyrate reserves in preparation for encystment (28), although this was not measured in these experiments.
Catalase expression during growth on BBGN.
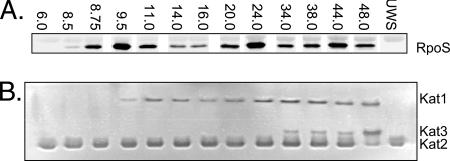
To determine if the RpoS produced by A. vinelandii during the metabolic shifts was transcriptionally active, we used Kat1 activity as a reporter of RpoS-dependent transcriptional activity. The first appearance of Kat1 (Fig. 4B) coincided with the induction of RpoS (Fig. 4A) during the acetate-to-glucose diauxic shift at 9.5 h. Kat1 activity continued to increase until 14 h, although the RpoS levels decreased hours before this. Similarly, an increase in Kat1 activity occurred at 24 h, coincident with the ammonium-to-N2 diauxic shift, but high levels of Kat1 activity persisted for an additional 14 h despite a fourfold decrease in the RpoS level in the same time frame. Although the catalase staining method is not strictly quantitative, comparison of the relative catalase (Fig. 4A) and RpoS (Fig. 4B) band strengths at these time points indicated that there is a nonlinear relationship between the RpoS concentration and Kat1 activity in A. vinelandii.
Contrary to what was observed in the shake flask study (Fig. 2A), Kat2 expression occurred throughout the bioreactor experiment, and a third catalase (Kat3) became more evident in stationary phase (34 to 48 h) (Fig. 4B).
RpoS stability in various growth conditions.
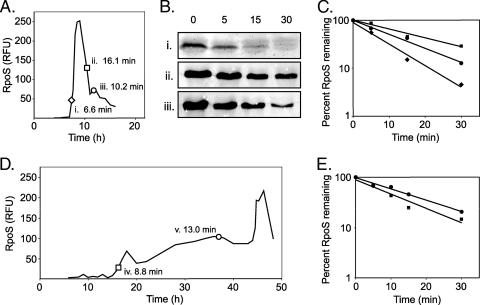
The stability of the RpoS protein was determined by antibiotic treatment, followed by Western analysis. The total amount of RpoS produced by a culture during the acetate-to-glucose diauxic shift and the specific times at which samples were taken for half-life determination are shown in Fig. 5A. Cultures were treated with tetracycline for up to 30 min, lysed, and analyzed by Western analysis (Fig. 5B). A graphic representation of RpoS stability is shown in Fig. 5C. During the acetate-to-glucose diauxic shift, half-lives of 6.6 to 16.1 min were observed at the time points indicated in Fig. 5A.
FIG. 5.
Determination of the half-life of RpoS during bioreactor growth of A. vinelandii in BBGN and BBG medium. (A) Relative RpoS expression during an acetate-to-glucose diauxic shift. Samples were removed during acetate limitation (i), diauxic lag (ii), and growth recovery on glucose (iii) and then treated with tetracycline to stop translational initiation. (B) Western analysis of RpoS following tetracycline addition. (C) Relative amount of RpoS following treatment plotted as a percentage of the RpoS present prior to tetracycline addition. The calculated half lives are indicated in panel A. (D and E) RpoS expression during growth on BBG medium. Samples were removed at two time points (iv and v) and treated with tetracycline. The relative amount of RpoS following treatment was plotted as a percentage of the RpoS present prior to tetracycline addition (E), and the calculated half-lives are indicated in panel D.
RpoS expression and stability during growth on BBG medium under identical aeration, mixing, and temperature conditions were also determined (Fig. 5D). It is interesting that the RpoS levels were low for the first 15 h of growth but then steadily increased over the next 30 h, an observation that directly contradicts the observations of Castañeda et al. (6), which suggested that rpoS expression occurred strictly during stationary phase. Half-lives of 8.8 to 13.0 min were determined for different time points (Fig. 5D), and the RpoS concentrations are plotted in Fig. 5E.
rpoS mRNA expression and RpoS expression are asynchronous.
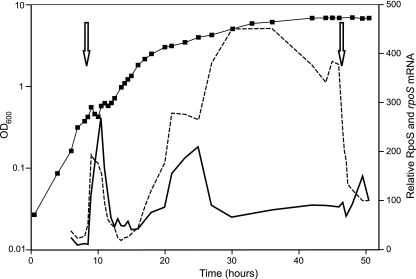
Both the relative RpoS protein and rpoS mRNA levels were monitored during growth on BBGN in a bioreactor batch culture (Fig. 6). During the initial acetate-to-glucose diauxic shift, the rpoS mRNA levels increased just before the RpoS levels increased and decreased slightly before the RpoS levels decreased. During ammonium limitation (16 h), rpoS mRNA induction also preceded RpoS accumulation. However, while the RpoS levels decreased during nitrogen-fixing growth (Fig. 3A), the rpoS mRNA levels continued to increase, reaching maximal induction (16-fold) by 30 h. The rpoS transcript levels remained elevated for an additional 17 h, until glucose became limiting. Following glucose limitation the rpoS mRNA levels decreased dramatically, while the RpoS levels increased slightly in the same time frame.
FIG. 6.
Northern and Western analyses of the products rpoS mRNA (dashed line) and RpoS protein (solid line) from A. vinelandii UW cultures grown on BBGN. Bacterial growth was determined spectrophotometrically at 600 nm (▪). Periods of oxygen saturation, indicative of metabolic cessation, are indicated by arrows.
DISCUSSION
RpoS and stationary-phase survival.
The stationary-phase sigma factor RpoS is needed for long-term survival of A. vinelandii. The vegetative growth of the wild-type strain and the vegetative growth of the rpoS strain were essentially identical, but the stationary-phase survival characteristics were different. It is tempting to say that wild-type strain UW formed cysts, while strain UWS did not. Old cultures of strain UWS contained no cystlike structures when they were viewed with an electron microscope (data not shown), nor does strain UW form fully “mature” cysts (28). The RpoS mutant survived as vegetative cells for about 10 months, even though the polyhydroxybutyrate reserves of wild-type cells are expected to last only 2.5 months (1). However, the wild type was able to survive for at least 16.5 years, suggesting that RpoS is necessary for the development of a dormant cell, independent of complete cyst formation.
RpoS is also required for the survival of stationary-phase A. vinelandii exposed to hydrogen peroxide. Five putative catalase genes are predicted to be present in the A. vinelandii genome, but only three genes were observed under the conditions used. It is interesting that despite the high respiratory rate of A. vinelandii, this organism has a relatively low tolerance for exogenous H2O2 compared to members of the closely related Pseudomonadaceae (14, 18). Mounting evidence suggests that catalases are produced primarily to deal with exogenous peroxide threats, while endogenously created peroxides are removed mainly by peroxidases (31). Many pathogenic and strongly rhizosphere-associated pseudomonads produce large quantities and, in some cases, several types of catalases (19, 20) for protection from host defenses and interspecies competition. As a free-living organism, A. vinelandii may not have developed this degree of ROI defense since it is less likely to encounter high levels of exogenous hydrogen peroxide. The A. vinelandii survival in the presence of hydrogen peroxide was best during stationary phase, which correlated to both RpoS expression and elevated activities of the RpoS-dependent catalase. The decreased survival of strain UWS in stationary phase can be largely attributed to its failure to express Kat1.
In contrast to most bacteria described to date, which either supplement a constitutively expressed catalase with additional catalase species (19, 26) or up-regulate the housekeeping catalase during stationary phase (5), A. vinelandii appears to undergo complete catalase switching from Kat2 in exponential phase to Kat1 in stationary phase. We also did not observe expression of a phase-specific catalase during or after the transition to nitrogen-fixing conditions. However, the increased production of Kat2 and Kat3 with higher aeration does suggest that these enzymes are ROI inducible, as described previously for many other catalases (7, 27). The reciprocal production of Kat3 at the expense of Kat2 suggests that Kat3 may be an isoform of Kat2, as observed previously with KatG (15).
Regulation of RpoS expression.
The expression of RpoS in A. vinelandii during the acetate-to-glucose diauxic shift was similar to that described in E. coli during a glucose-to-lactose shift (10), except that the magnitude of induction was much greater in this study. The rpoS mRNA and protein concentrations rose and fell synchronously, indicating that RpoS accumulation is regulated primarily at the transcriptional level during the acetate-to-glucose diauxic shift. Since A. vinelandii is able to fix nitrogen, it was used to examine the general stress response during a nitrogen diauxic shift and subsequent nitrogen-fixing growth. Up to 28-fold induction of RpoS and 16-fold induction of rpoS mRNA occurred during the nitrogen diauxic shift. Thus, it appears that A. vinelandii accumulates similar amounts of RpoS whether it experiences carbon or nitrogen limitation. However, during diauxic recovery on N2 the RpoS concentrations decreased to basal levels even though the mRNA levels continued to increase. Because RpoS stability did not change significantly in A. vinelandii (see below) and has not been observed to do so in other pseudomonads (4), the most probable interpretation of the RpoS/rpoS mRNA asynchrony is that a significant degree of posttranscriptional regulation occurs in A. vinelandii under certain nutrient-limiting growth conditions. Equivalent modulation of translation is mediated primarily by small RNA species in E. coli (12), but no such species have been reported for A. vinelandii to date. Alternatively, the RpoS/rpoS mRNA asynchrony may be due to sequestration of large amounts of the transcript in a protected but translationally inactive form by chaperones, such as Hfq or ribosomal protein S1 (39).
The RpoS level in E. coli is regulated by a dedicated proteolytic circuit involving the recognition factor SprE/RssB (34). RssB binds to RpoS at the critical residue K173 and then transfers it to the protease ClpXP for degradation (2). This residue and surrounding regions 2.4 and 2.5 of E. coli RpoS are conserved (96% identity) in A. vinelandii, despite an overall protein identity of only 76% (see Fig. S2 in the supplemental material). No rssB homologue was observed in the A. vinelandii genome, despite the presence of a possible rssA homologue (accession number ZP_00418183). Whereas RssB-directed regulation in E. coli modulates RpoS stability nearly 38-fold (the half-lives range from 4.1 to 154 min) during carbon limitation (25), the stability changed a mere 1.4- to 2.4-fold during the nitrogen- and carbon-limiting events tested with A. vinelandii. Therefore, it appears that no RssB-like proteolytic regulatory mechanism exists in A. vinelandii. Consequently, the conservation of RpoS regions 2.4 and 2.5 in these divergent organisms is most likely driven by recognition requirements for the −10 promoter sequence.
The efficiency of RNA polymerase:RpoS holoenzyme formation and subsequent transcriptional activity was assessed using the natural reporter Kat1. Considering that RpoS has the lowest affinity for RNA polymerase of all the E. coli sigma factors (23), it would be expected that the high level of induction of RpoS during the various diauxic shifts would result in increased RNA polymerase:RpoS formation and resultant activity (24) and thus greater expression of the reporter Kat1. Although Kat1 activity was observed only during periods of RpoS expression, this activity was not directly proportional to RpoS levels. For example, the Kat1 levels were similar during peak (24 h) and basal (34, 38, and 48 h) (Fig. 6) RpoS expression. Similar phenomena have been observed for E. coli, where effector molecules such as ppGpp and trehalose modulate the formation or activity of the RpoS holoenzyme (17, 21). From these observations we concluded that the RpoS of A. vinelandii is regulated at the transcriptional, posttranscriptional, and formation/activity levels.
Supplementary Material
Acknowledgments
We express our gratitude to Sonya Kujat-Choy, who made mutant strain UWS, purified the His-tagged RpoS protein, and made the RpoS antibody. We thank Anthony Cornish, George Owttrim, and Ryan Will for helpful comments regarding the manuscript. We also sincerely thank Trevor Samis and Angela Scott for helpful discussions regarding the results.
This research was funded by the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 30 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aladegbami, S. L., J. C. Tsai, and G. R. Vela. 1979. Adenilate energy charge of Azotobacter vinelandii during encystment. Curr. Microbiol. 2327-329. [Google Scholar]
- 2.Becker, G., E. Klauck, and R. Hengge-Aronis. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. USA 966439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benov, L., and J. Al-Ibraheem. 2002. Disrupting Escherichia coli: a comparison of methods. J. Biochem. Mol. Biol. 35428-431. [DOI] [PubMed] [Google Scholar]
- 4.Bertani, I., M. Ševo, M. Kojic, and V. Venturi. 2003. Role of GacA, LasI, RhlI, Ppk, PsrA, Vfr and ClpXP in the regulation of the stationary-phase sigma factor rpoS/RpoS in Pseudomonas. Arch. Microbiol. 180264-271. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. M., M. L. Howell, M. L. Vasil, A. J. Anderson, and D. J. Hassett. 1995. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J. Bacteriol. 1776536-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castañeda, M., J. Sánchez, S. Moreno, C. Núñez, and G. Espín. 2001. The global regulators GacA and σS form part of a cascade that controls alginate production in Azotobacter vinelandii. J. Bacteriol. 1836787-6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chelikani, P., I. Fita, and P. C. Loewen. 2004. Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61192-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clare, D. A., M. N. Duong, D. Darr, F. Archibald, and I. Fridovich. 1984. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal. Biochem. 140532-537. [DOI] [PubMed] [Google Scholar]
- 9.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 642710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, D., A. Teich, P. Neubauer, and R. Hengge-Aronis. 1998. The general stress sigma factor σS of Escherichia coli is induced during diauxic shift from glucose to lactose. J. Bacteriol. 1806203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George, S. E., C. J. Costenbader, and T. Melton. 1985. Diauxic growth in Azotobacter vinelandii. J. Bacteriol. 164866-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58303-328. [DOI] [PubMed] [Google Scholar]
- 13.Gross, C. A., C. Chan, A. Dombroski, T. Gruber, M. Sharp, J. Tupy, and B. Young. 1998. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 63141-155. [DOI] [PubMed] [Google Scholar]
- 14.Heeb, S., C. Valverde, C. Gigot-Bonnefoy, and D. Haas. 2005. Role of the stress sigma factor RpoS in GacA/RsmA-controlled secondary metabolism and resistance to oxidative stress in Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 243251-258. [DOI] [PubMed] [Google Scholar]
- 15.Hillar, A., and P. C. Loewen. 1995. Comparison of isoniazid oxidation catalyzed by bacterial catalase-peroxidases and horseradish peroxidase. Arch. Biochem. Biophys. 323438-446. [DOI] [PubMed] [Google Scholar]
- 16.Jishage, M., and A. Ishihama. 1995. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of σ70 and σ38. J. Bacteriol. 1776832-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 161260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jørgensen, F., M. Bally, V. Chapon-Herve, G. Michel, A. Lazdunski, P. Williams, and G. S. A. B. Stewart. 1999. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145835-844. [DOI] [PubMed] [Google Scholar]
- 19.Kang, B. R., B. H. Cho, A. J. Anderson, and Y. C. Kim. 2004. The global regulator GacS of a biocontrol bacterium Pseudomonas chlororaphis O6 regulates transcription from the rpoS gene encoding a stationary-phase sigma factor and affects survival in oxidative stress. Gene 325137-143. [DOI] [PubMed] [Google Scholar]
- 20.Klotz, M. G., and S. W. Hutcheson. 1992. Multiple periplasmic catalases in phytopathogenic strains of Pseudomonas syringae. Appl. Environ. Microbiol. 582468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusano, S., and A. Ishihama. 1997. Functional interaction of Escherichia coli RNA polymerase with inorganic polyphosphate. Genes Cells 2433-441. [DOI] [PubMed] [Google Scholar]
- 22.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 23.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli σ subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 283497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik, S., K. Zalenskaya, and A. Goldfarb. 1987. Competition between sigma factors for core RNA polymerase. Nucleic Acids Res. 158521-8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel, M. J., and T. J. Silhavy. 2005. Starvation for different nutrients in Escherichia coli results in differential modulation of RpoS levels and stability. J. Bacteriol. 187434-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, C. D., Y. C. Kim, and A. J. Anderson. 1997. Cloning and mutational analysis of the gene for the stationary-phase inducible catalase (catC) from Pseudomonas putida. J. Bacteriol. 1795241-5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochsner, U. A., M. L. Vasil, E. Alsabbagh, K. Parvatiyar, and D. J. Hassett. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 1824533-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Page, W. J. 1983. Formation of cystlike structures by iron-limited Azotobacter vinelandii strain UW during prolonged storage. Can. J. Microbiol. 291110-1118. [Google Scholar]
- 29.Page, W. J., A. Tindale, M. Chandra, and E. Kwon. 2001. Alginate formation in Azotobacter vinelandii UWD during stationary phase and the turnover of poly-beta-hydroxybutyrate. Microbiology 147483-490. [DOI] [PubMed] [Google Scholar]
- 30.Page, W. J., and M. von Tigerstrom. 1978. Induction of transformation competence in Azotobacter vinelandii iron-limited cultures. Can. J. Microbiol. 241590-1594. [DOI] [PubMed] [Google Scholar]
- 31.Park, S., X. You, and J. A. Imlay. 2005. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx− mutants of Escherichia coli. Proc. Natl. Acad. Sci. USA 1029317-9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272580-591. [DOI] [PubMed] [Google Scholar]
- 33.Ponnambalam, S., C. Webster, A. Bingham, and S. Busby. 1986. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal −35 region sequences. J. Biol. Chem. 26116043-16048. [PubMed] [Google Scholar]
- 34.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. USA 932488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruteanu, M., and R. Hengge-Aronis. 2002. The cellular level of the recognition factor RssB is rate-limiting for σS proteolysis: implications for RssB regulation and signal transduction in σS turnover in Escherichia coli. Mol. Microbiol. 451701-1714. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sandercock, J. R., and L. S. Frost. 1998. Analysis of the major domains of the F fertility inhibition protein, FinO. Mol. Gen. Genet. 259622-629. [DOI] [PubMed] [Google Scholar]
- 38.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51973-985. [DOI] [PubMed] [Google Scholar]
- 39.Ševo, M., E. Buratti, and V. Venturi. 2004. Ribosomal protein S1 specifically binds to the 5′ untranslated region of the Pseudomonas aeruginosa stationary phase sigma factor rpoS mRNA in the logarithmic phase of growth. J. Bacteriol. 1864903-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegele, D. A., J. C. Hu, W. A. Walter, and C. A. Gross. 1989. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 206591-603. [DOI] [PubMed] [Google Scholar]
- 41.Strickland, J. D. H., and T. R. Parsons. 1972. A practical handbook of sea water analysis. Fish. Res. Bd. Can. Bull. 16771-89. [Google Scholar]
- 42.Stüdemann, A., M. Noirclerc-Savoye, E. Klauck, G. Becker, D. Schneider, and R. Hengge. 2003. Sequential recognition of two distinct sites in σS by the proteolytic targeting factor RssB and ClpX. EMBO J. 224111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suh, S.-J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. H. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 1813890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zgurskaya, H. I., M. Keyhan, and A. Matin. 1997. The σS level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol. Microbiol. 24643-651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.