Abstract
In Vibrio parahaemolyticus, scrC participates in controlling the decision to be a highly mobile swarmer cell or a more adhesive, biofilm-proficient cell type. scrC mutants display decreased swarming motility over surfaces and enhanced capsular polysaccharide production. ScrC is a cytoplasmic membrane protein that contains both GGDEF and EAL conserved protein domains. These domains have been shown in many organisms to respectively control the formation and degradation of the small signaling nucleotide cyclic dimeric GMP (c-di-GMP). The scrC gene is part of the three-gene scrABC operon. Here we report that this operon influences the cellular nucleotide pool and that c-di-GMP levels inversely modulate lateral flagellar and capsular polysaccharide gene expression. High concentrations of this nucleotide prevent swarming and promote adhesiveness. Further, we demonstrate that ScrC has intrinsic diguanylate cyclase and phosphodiesterase activities, and these activities are controlled by ScrAB. Specifically, ScrC acts to form c-di-GMP in the absence of ScrA and ScrB; whereas ScrC acts to degrade c-di-GMP in the presence of ScrA and ScrB. The scrABC operon is specifically induced by growth on a surface, and the analysis of mutant phenotypes supports a model in which the phosphodiesterase activity of ScrC plays a dominant role during surface translocation and in biofilms.
Vibrio parahaemolyticus possesses a mode of motility, called swarming, that is driven by a lateral flagellar system (laf) and is suited to rapid colonization of surfaces; the organism can also elaborate a capsular polysaccharide (cps) that enables robust biofilm formation (14, 29). How a cell varies its surface molecules to adapt and proliferate in specific environments is one key survival strategy. In V. parahaemolyticus, the scrABC locus modulates the lifestyle adaptation between swarming and sticking by participating in swarming and capsular polysaccharide gene regulation (7).
The scrABC operon encodes three proteins: ScrA, a potential pyridoxal-phosphate-dependent enzyme; ScrB, a potential extracellular solute-binding protein; and ScrC, a potential sensory protein (7). Fractionation experiments localize ScrA in the cytoplasm, ScrB in the periplasm, and ScrC with the cell membrane. The ScrC N terminus has two predicted transmembrane domains flanking a potential periplasmic region (∼300 amino acids [aa] in length). The C terminus of ScrC contains both GGDEF and EAL domains and is cytoplasmically located. These two conserved domains, named after signature amino acid motifs, are found in diguanylate cyclases and phosphodiesterases and are responsible for the formation (GGDEF) and degradation (EAL) of the nucleotide bis-3′,5′ cyclic dimeric GMP (c-di-GMP) (reviewed in reference 23). The role of c-di-GMP as a signaling molecule is being described in an expanding list of organisms (reviewed in references 13, 16, 38, and 39). Enzymes controlling the level of c-di-GMP, including proteins with GGDEF and EAL domains, as well as the phosphohydrolase-associated HD-GYP domain (16, 41), are often implicated in regulating the production of cell surface structures, such as flagella, pili, and extracellular matrixes (3, 7, 8, 11, 19, 21, 26, 37, 39, 46).
Disruption of any of the three genes in the scrABC operon reduces swarming and produces a crinkly colony morphology as the consequence of decreased laf and increased cps gene expression (7). Consistent with the mutant phenotypes, overexpression of scrABC induces the expression of laf genes and represses the transcription of cps genes. However, expression of scrC without scrAB fails to activate laf gene expression. Curiously, overproduction of ScrC without coproduction of ScrAB not only results in loss of laf expression but also causes enhanced cps transcription. Thus, ScrC appears to have two activities that are modulated by ScrAB. In many proteins with dual GGDEF and EAL domains, only one domain is catalytically active and the second domain is inactive or regulatory (35, 44). Both the GGDEF and EAL domains of ScrC seem highly conserved and hence potentially catalytically active. The conserved domain alignment tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) produced an E value of 7e-78 for the EAL domain of ScrC with 100% alignment over 240 aa. The ScrC GGDEF domain shows 94.4% alignment over 155 aa with an E value of 2e-46. A hypothesis for the opposing effects of ScrABC and ScrC is that ScrC has the capacity to both form and degrade c-di-GMP and that the enzymatic activity of the protein is influenced by ScrAB.
In this study, we investigated the effects of ScrABC/ScrC upon the cellular c-di-GMP pool, as well as upon swarming, capsular polysaccharide expression, and biofilm formation. We present genetic evidence that ScrC is a bifunctional enzyme: by itself, ScrC can synthesize c-di-GMP, whereas in the context of ScrA and ScrB, it is capable of degrading this secondary messenger. In vivo (i.e., in the context of ScrA and ScrB), ScrC appears to function as a phosphodiesterase. Consistent with its role modulating swarming and sticking, the scrABC operon was found to be expressed specifically during growth on surfaces.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this work are described in Table 1. All V. parahaemolyticus strains were derived from BB22TR (15) and were routinely grown at 30°C. Heart infusion medium (HI broth) contained 2.5% heart infusion (Difco) and 1.5% NaCl. HI swarm and nonswarm plates contained 1.4% and 2% Bacto agar (Difco), respectively. Congo red plates were prepared by adding 0.025% Congo red and 5 mM CaCl2 to 2.5% HI medium (with no added NaCl) solidified with 2% granulated agar (Difco). For biofilm experiments, the medium contained 1% HI in artificial seawater (HI ASW; 300 mM NaCl, 10 mM KCl, 50 mM MgSO4, 10 mM CaCl2) (2). Antibiotics were used at final concentrations of 10 μg/ml chloramphenicol, 25 μg/ml gentamicin, 50 μg/ml kanamycin, and 10 μg/ml tetracycline. Overexpression plasmids were induced with 2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) unless otherwise indicated.
TABLE 1.
V. parahaemolyticus strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Parent; reference |
|---|---|---|
| Strains | ||
| LM1017 | flgE313L::lux; lux fusion in lateral flagellar hook gene (translucent cell type) | BB22; 30 |
| LM5435 | LM1017/pLM2796 (scrABC+) | 7 |
| LM5674 | ΔopaR1 (translucent cell type) | 15 |
| LM5818 | cpsA1::lacZ ΔopaR1 | LM5674; 17 |
| LM5949 | flgCL3061::lacZ (Camr) ΔopaR1 | LM5674; 22 |
| LM6349 | LM1017/pLM1877 | |
| LM6453 | LM5818/pLM1877 | |
| LM6565 | ΔscrABC4::Cam flgE313L::lux | LM1017 |
| LM6567 | ΔscrABC4::Cam | LM5674 |
| LM6832 | ΔscrABC4::Cam cpsA1::lacZ ΔopaR1 | LM5818 |
| LM6837 | LM5818/pLM2796 (scrABC+) | |
| LM6933 | LM6565/pLM2449 (scrC+) | |
| LM6954 | LM5818/pLM3276 (scrCΔEAL) | |
| LM6998 | ΔscrC3::Cam flgE313L::lux | LM1017 |
| LM6999 | ΔscrC3::Cam cpsA1::lacZ ΔopaR1 | LM5818 |
| LM7001 | scrCΔEAL::Cam flgE313L::lux | LM1017 |
| LM7002 | scrCΔEAL::Cam cpsA1::lacZ ΔopaR1 | LM5818 |
| LM7012 | LM5818/pLM2449 (scrC+) | |
| LM7053 | LM1017/pLM2449 (scrC+) | |
| LM7054 | LM1017/pLM3276 (scrCΔEAL) | |
| LM7074 | LM6565/pLM1877 | |
| LM7665 | LM6565/pLM3437 (scrABCΔEAL) | |
| LM7667 | LM6565/pLM2796 (scrABC+) | |
| LM8021 | ΔscrC3::Cam ΔopaR1 | LM5674 |
| LM8022 | scrCΔEAL::Cam ΔopaR1 | LM5674 |
| LM8078 | LM1017/pLM3437 (scrABCΔEAL) | |
| LM8197 | LM5818/pLM3437 (scrABCΔEAL) | |
| LM8309 | LM1017/pLM3598 (scrABCE554A) | |
| LM8310 | LM1017/pLM3599 (scrABCD428A) | |
| LM8311 | LM5818/pLM3598 (scrABCE554A) | |
| LM8312 | LM5818/pLM3599 (scrABCD428A) | |
| LM8400 | LM1017/pLM3596 (scrCE554A) | |
| LM8401 | LM1017/pLM3597 (scrCD428A) | |
| LM8403 | LM5818/pLM3596 (scrCE554A) | |
| LM8404 | LM5818/pLM3597 (scrCD428A) | |
| LM8602 | flgCL3061::lacZ (Camr) ΔopaR1 scrC292::Tn5lux (Kanr; lux aligned) | LM5949 |
| LM8604 | flgCL3061::lacZ (Camr) ΔopaR1 scrC803::Tn5lux (Kanr; lux not aligned) | LM5949 |
| LM9000 | LM5674/pLM2641 (gfp+) | |
| LM9001 | LM6567/pLM2641 (gfp+) | |
| LM9002 | LM8022/pLM2641 (gfp+) | |
| Plasmids | ||
| pKD3 | Camr template plasmid | 12 |
| pLM1877 | Genr expression vector | 6 |
| pLM2032 | Tetr cosmid containing scrABC locus | 7 |
| pLM2449 | Genr IPTG-inducible scrC+ | pLM1877; 7 |
| pLM2641 | Genr IPTG-inducible sgGFP | pLM1877; 14 |
| pLM2796 | Genr IPTG-inducible scrABC+ | 7 |
| pLM3272 | Cosmid pLM2032 with ΔscrC3::Cam | 7 |
| pLM3273 | Cosmid pLM2032 with scrCΔEAL::Cam | LM2032 |
| pLM3276 | Genr IPTG-inducible scrCΔEAL | LM2449 |
| pLM3437 | Genr IPTG-inducible scrABCΔEAL | LM2796 |
| pLM3596 | Genr IPTG-inducible scrCE554A | LM2449 |
| pLM3597 | Genr IPTG-inducible scrCD428A | LM2449 |
| pLM3598 | Genr IPTG-inducible scrABCE554A | LM2796 |
| pLM3599 | Genr IPTG-inducible scrABCD428A | LM2796 |
Genetic and molecular techniques.
General molecular biology methods were adapted from Sambrook et al. (43). Conjugation and allelic replacement methods for V. parahaemolyticus have been previously described (45). All allelic replacements were confirmed by PCR.
Deletion/insertion mutations were made with a λ Red recombinase system in Escherichia coli (12) to introduce a chloramphenicol resistance cassette into cosmid pLM2032 for chromosomal allelic exchange or into plasmids pLM2796 and pLM2449 for overexpression studies. The ΔscrABC4 allele removed 4,379 bp of coding sequence to delete amino acids from E103 of ScrA through amino acid F753 of ScrC; the ΔscrC3 allele removed 2,153 bp to delete scrC coding sequence for Q36 through F753; the scrCΔEAL allele deleted 799 bp (including the EAL domain) to remove scrC coding sequence for L487 through F753. All deletions were replaced with a chloramphenicol resistance cassette.
Point mutations were inserted into the scrC gene by PCR site-directed mutagenesis (51). Briefly, pLM2449 (scrC+) was digested with HindIII and pLM2796 (scrABC+) was digested with PstI and NsiI. The small fragment containing part of the scrC gene from each plasmid was subcloned into the smaller pUC19 vector for the site-directed mutagenesis. The resulting plasmids were used as templates for PCR with primers that contained single substitutions in the domains of interest. Primers ScrCD428Af (GCTCGCCAGAATCGGTGGGGCTGAGTTCCTGTTGGTG) and ScrCD428Ar (CACCAACAGGAACTCAGCCCCACCGATTCTGGCGAGC) were used to insert a point mutation into the GGDEF domain at amino acid 428, changing it from aspartic acid to alanine. To inactivate the EAL domain, amino acid 554 was changed from glutamic acid to alanine with primers ScrCE554Af (GCGGCAAAATTGTTGGCGCAGCAGCATTGATGCGTTGG) and ScrCE554Ar (CCAACGCATCAATGCTGCTGCGCCAACAATTTTGCCGC). The PCR products were then inserted into HindIII-digested pLM2449 and into PstI/NsiI-digested pLM2796. DNA sequencing of the entire scrC gene was done to confirm the introduction of the specific mutations.
The lux transcriptional fusions in scrC were isolated in a Tn5lux mutagenesis of strain LM5949, which carries a lacZ reporter in a lateral flagellar gene. The mutagenesis was performed essentially as previously described (47). The scrC::lux fusion strains were identified as having a crinkly and white colony morphology on HI plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The point of transposon insertion, identified by DNA sequencing, was found to occur at nucleotides 292 and 803 in the scrC coding sequence for strains LM8602 and LM8604, respectively. The lux fusion was aligned with scrABC transcription for allele scrC292 and was in the opposite orientation for allele scrC803.
Luminescence assays.
For time course experiments with plate-grown cells, strains were pregrown on plates and suspended to an optical density at 600 nm (OD600) of 0.05 and 100-μl volumes were spread onto multiple fresh HI swarm plates (with antibiotics and IPTG, when appropriate). Plates were incubated at 30°C, and at specified times, cells were suspended in 5 ml of HI broth and OD600 and relative light unit (RLU) measurements were made. For time course experiments in liquid, overnight HI cultures were diluted to an OD600 of 0.05 into 250-ml flasks containing 25 ml of HI broth (with IPTG and antibiotics when appropriate). These cultures were incubated with shaking at 30°C with periodic sampling for OD and RLU readings. Luminescence was quantified by measuring 0.1-ml samples for 1 s in a GENios Pro 96-well plate reader (TECANResearch, Triangle Park, NC) (see Fig. 4 and 6) or by measuring samples in a TD20-202 luminometer (Turner Designs) (see Fig. 1). Dilutions were made to keep all measurements within a linear range. Luminescence is reported as specific light units (SLU; RLU per second per milliliter per unit of OD600). Light readings were taken in triplicate, and standard deviations were generally less than 10%. Each experiment was performed at least twice with similar results.
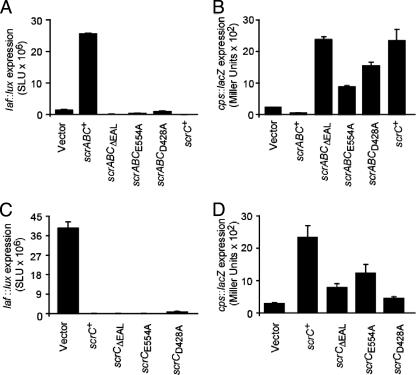
FIG. 4.
Effects of mutations on ScrABC and ScrC activities. (A) Lateral flagellar gene expression in liquid, measured in laf::lux reporter strains carrying scrABC expression plasmids. Expression plasmids were induced with IPTG, luminescence (lux) was monitored periodically throughout the growth of the cultures, and the maximal values of expression are reported as SLU. Strains: LM6349 (LM1017/vector), LM5435 (LM1017/scrABC+), LM8078 (LM1017/scrABCΔEAL), LM8309 (LM1017/scrABCE554A), LM8310 (LM1017/scrABCD428A), and LM7053 (LM1017/scrC+). (B) cpsA::lacZ gene expression measured in reporter strains carrying scrABC expression plasmids. Strains were grown on plates with IPTG and harvested at 16 h to measure β-galactosidase activity (reported as Miller units). Strains: LM6453 (LM5818/vector), LM6837 (LM5818/scrABC+), LM8197 (LM5818/scrABCΔEAL), LM8311 (LM5818/scrABCE554A), LM8312 (LM5818/scrABCD428A), and LM7012 (LM5818/scrC+). (C) laf::lux gene expression measured in reporter strains carrying scrC expression plasmids. Strains were grown on plates with IPTG and harvested periodically for lux measurements, and maximal values are reported. Strains: LM6349 (LM1017/vector), LM7053 (LM1017/scrC+), LM7054 (LM1017/scrCΔEAL), LM8400 (LM1017/scrCE554A), and LM8401 (LM1017/scrCD428A). (D) cpsA::lacZ gene expression measured in reporter strains carrying scrC expression plasmids. Strains were grown on plates with IPTG and harvested at 16 h for β-galactosidase assay. Strains: LM6453 (LM5818/vector), LM7012 (LM5818/scrC+), LM6954 (LM5818/scrCΔEAL), LM8403 (LM5818/scrCE554A), and LM8404 (LM5818/scrCD428A).
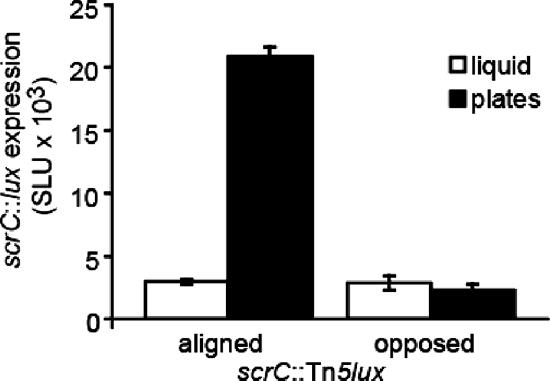
FIG. 6.
scrABC transcription is induced by growth on surfaces. Luminescence of strains carrying scrC::Tn5lux insertions was measured in triplicate periodically throughout growth on HI plates or in liquid, and maximum values are shown. The strains contained the Tn5lux reporter aligned or opposed to transcription of the scrABC operon (LM8602 and LM8604, respectively). Luminescence is reported as SLU.
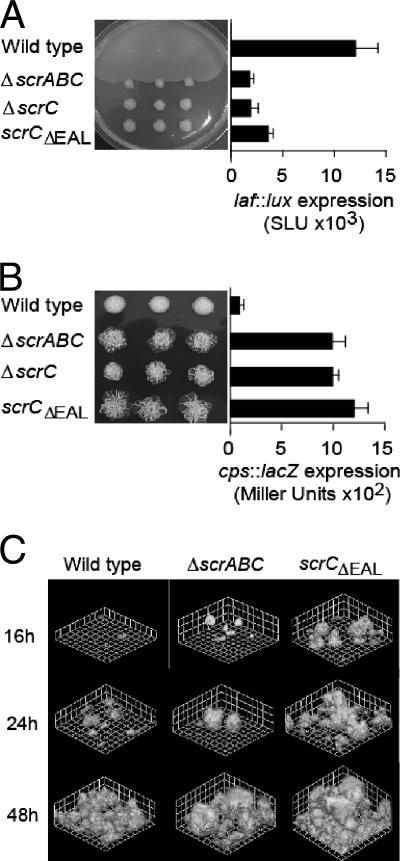
FIG. 1.
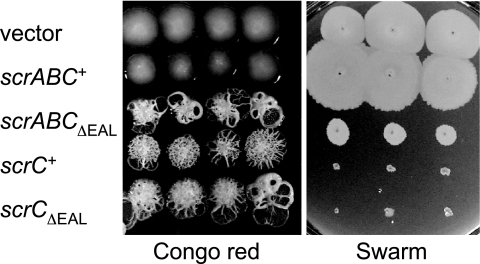
Phenotypes of scrABC mutants are determined by the EAL domain of ScrC. (A) Swarming motility of the wild-type (LM5674) and ΔscrABC (LM6567), ΔscrC (LM8021), and scrCΔEAL (LM8022) mutant strains and quantification of laf::lux transcription of the wild-type (LM1017) and ΔscrABC (LM6565), ΔscrC (LM6998), and scrCΔEAL (LM7001) mutant strains. For the swarm plate, three single colonies of each strain were inoculated onto HI swarm plates and incubated overnight. Luminescence was monitored periodically throughout the growth of the strains on the plates, and maximal values of expression are shown. Light (lux) is reported as SLU (total light units per second per milliliter per unit of OD600). (B) Colony morphology of the wild-type (LM5674) and ΔscrABC (LM6567), ΔscrC (LM8021), and scrCΔEAL (LM8022) mutant strains on Congo red medium and quantification of cpsA::lacZ transcription of the wild-type (LM5818) and ΔscrABC (LM6832), ΔscrC (LM6999), and scrCΔEAL (LM7002) mutant strains. For the Congo red plate, colonies were incubated for 1 week at room temperature. β-Galactosidase activity was measured after 16 h of growth on plates and is reported in Miller units. (C) Green fluorescent protein-labeled strains were grown in biofilm flow cell reactors, and scanning confocal laser images were acquired at the indicated times. Grid lines indicate 23.12 μm. Strains: wild type (LM9000), ΔscrABC (LM9001), and scrCΔEAL (LM9002).
β-Galactosidase assays.
For β-galactosidase assays, cells were grown as described for luminescence assays. LacZ activity was measured according to Miller (32). Cells were permeabilized by using Koch's lysis solution (36). Each experiment was performed at least twice, with measurements taken in triplicate, with similar results.
Biofilm experiments.
The flow cell system was used for microscopic examination of biofilm formation (34). The strains in these experiments contained plasmid pLM2641 encoding green fluorescent protein (14). Overnight cultures were diluted to an OD600 of 0.01 in 1% HI ASW with gentamicin and 0.5 mM IPTG. After 1 h of incubation, the flow was initiated at 0.17 ml/min. An LSM 510 confocal microscope (Carl Zeiss, Germany) was used for imaging of biofilms. The excitation wavelength used was 488 nm, and the emission wavelength was 515 ± 15 nm. After acquisition, images were processed with Volocity software v.2.6.0 (Improvision, Lexington, MA). Experiments were repeated at least three times with similar results. The image analysis program COMSTAT (18) was applied to quantify structural aspects of biofilms of six representative images per strain (two images per three experiments at the 48-h time point).
Detection of c-di-GMP.
Labeling and analysis of cellular nucleotides were performed essentially as previously described (25), by the method developed by Bochner and Ames (5). Briefly, V. parahaemolyticus strains were grown overnight in morpholinepropanesulfonic acid (MOPS) minimal medium without added phosphate (5) with 150 mM NaCl, 25 mM MgCl2, 0.4% galactose, 0.07% Casamino Acids, and 25 μg/ml gentamicin. E. coli strains were grown overnight without shaking in MOPS medium with 50 mM NaCl, 25 mM MgCl2, 0.04% glucose, 0.1% Casamino Acids, 5.2 mM arginine, and 15 μg/ml gentamicin. Cells were diluted to an OD600 of 0.05 into fresh medium containing 1 mM IPTG and 32Pi and grown at 30°C with shaking. For E. coli, 0.4% glucose was used in the labeling medium. Carrier-free 32P in an acid-free aqueous solution (Amersham Biosciences) was used at 100 μCi/ml of medium. Aliquots were removed at the indicated times, mixed with 0.1 volume of 11 N cold formic acid, and placed on ice. Extracts were applied to prewashed (0.5 M LiCl) PEI (polyethyleneimine) cellulose plates (J. T. Baker Chemical Co.). The amount of extract loaded on the chromatogram was normalized to the final OD600 (usually 3 to 5 μl/sample). Plates were then soaked in methanol and air dried. Thin-layer chromatography (TLC) solvents were adopted from previously published procedures (40, 50). Development in the first dimension was performed in 0.2 M NH4HCO3. After washing in methanol, chromatography in the second dimension was performed in 1.5 M KH2PO4, pH 3.65.
Labeling experiments were repeated at least two times, with similar results. Exposure to X-ray film was usually for 1 to 4 days, until the presence or absence of the c-di-GMP spot was clearly detectable. A control spot with an intensity similar to that of the c-di-GMP spot was chosen for normalization. Thus, the intensity of the control spot appears to change primarily because in some instances, i.e., when there was no c-di-GMP, we chose to document the longer exposure of the autoradiogram (hence resulting in a darker control spot) than in cases where there was easily detectable c-di-GMP and a shorter exposure was documented. Quantification of spot intensity was performed with the image quantification program of Molecular Dynamics (Image Quant 5.2).
RESULTS
In vivo, the EAL domain of ScrC seems essential for control of laf and cps gene expression.
Mutations in the scrABC operon were previously shown to affect both lateral flagellar (laf) and capsular polysaccharide (cps) gene expression, resulting in a severe inability to swarm and a crinkly colony morphology (7). To further dissect the roles of the proteins encoded by this locus, precise deletions were engineered to remove the entire scrABC operon, the entire scrC gene, and the C-terminal portion of the scrC gene that encodes the EAL domain. These mutations were transferred to the chromosome of the wild-type strain, as well as strains containing a laf::lux or cpsA::lacZ transcriptional reporter. All of the deletion mutants were severely disabled for swarming (Fig. 1A). Immunoblotting showed that production of lateral flagellin was also greatly decreased (data not shown). These mutations reduced laf transcription in the laf::lux reporter strains by greater than 60% (Fig. 1A). On Congo red plates, the deletion mutants formed extremely crinkly colonies compared to the smooth parental strain (Fig. 1B). This crinkliness required an intact capsular polysaccharide biosynthetic locus; prevention of Cps production upon the introduction of the cpsA::lacZ mutation into the cps biosynthetic locus converted the rough colony morphology to smooth (data not shown). The resulting reporter strains expressed cpsA::lacZ at ∼10-fold higher levels than the parental strain (Fig. 1B). These cps reporter strains also displayed severely swarming-defective phenotypes (data not shown), demonstrating that the capacity for swarming was specifically diminished and not simply the consequence of being too sticky to move effectively.
The scrABC locus had a strong effect on cps production, and thus we reasoned that scrABC might also have a significant role in biofilm formation. The ability to form biofilms was analyzed by using flow cells and confocal microscopy. Figure 1C shows that the strains containing ΔscrABC and scrCΔEAL mutations presented earlier and more robust biofilm formation compared to the wild type. At 48 h, the average thickness and maximum thickness of the wild-type biofilm were 21.6 ± 5.8 and 54.3 ± 6.4 μm, respectively, whereas these parameters were 48.5 ± 14.9 and 94.2 ± 16.9 μm for strain LM9001 (ΔscrABC) and 59.3 ± 5.5 and 89.7 ± 17.2 μm for strain LM9002 (scrCΔEAL). The ΔscrC strain produced biofilms similarly thick and precocious as those of the strains containing the ΔscrABC and scrCΔEAL mutations (data not shown). Thus, with respect to swarming, colony morphology, and biofilm formation, deletion of scrABC, scrC, or simply the portion of scrC encoding the EAL domain produced the same consequence. Although removal of the EAL domain could simply create an unstable form of the ScrC protein, these results suggested that the EAL domain of ScrC may be critical for the in vivo role of the products of the scrABC operon.
Effects of ScrABC on cellular nucleotide pools.
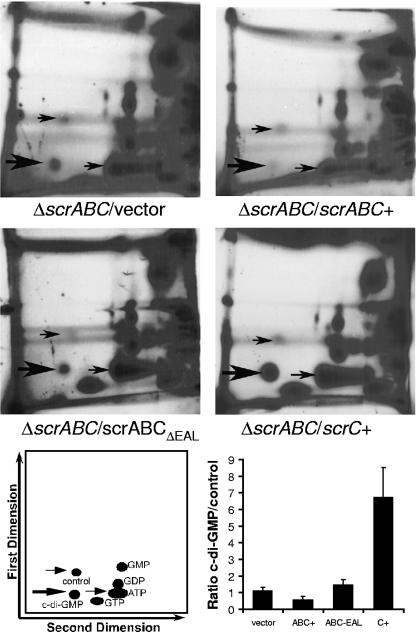
Because the EAL domain seemed to be the determining factor of ScrABC activity, we hypothesized that ScrABC might be acting as a phosphodiesterase to decrease the level of c-di-GMP. To examine this, scrABC and scrABCΔEAL were overexpressed in the ΔscrABC strain and the nucleotide pools of these cells were analyzed by two-dimensional (2D) TLC (Fig. 2). A spot, indicated by the thick arrow, that was in a position consistent with Rf values for c-di-GMP (25, 40, 50) was observed in extracts of the ΔscrABC strain containing the control vector. The average Rf values for the spot were 0.14 (± 0.02) in the first dimension and 0.31 (± 0.01) in the second dimension. The intensity of this spot decreased when scrABC, and not scrABCΔEAL or scrC, was overexpressed. Such an observation is consistent with ScrABC possessing phosphodiesterase activity and a model in which swarming is promoted by low c-di-GMP; moreover, this experiment demonstrates that the EAL domain of ScrC is critical for ScrABC activity, a result that is in keeping with the mutant phenotypes, as well as the known phosphodiesterase function, associated with other EAL domains (44, 48).
FIG. 2.
ScrABC, but not ScrABCΔEAL or ScrC, diminishes the cellular c-di-GMP level. Representative autoradiograms produced from 2D PEI cellulose TLC separations of 32P-labeled cellular nucleotides. Cultures were labeled for 12 h and normalized to the final OD600 for loading onto TLC plates. Autoradiograms were exposed for 2 to 4 days, usually until the presence or absence of the c-di-GMP spot was clearly detectable. Thick arrows indicate the spot that migrates with Rf values consistent with the mobility of c-di-GMP. Thin arrows provide orientation. Ratios of the intensity of c-di-GMP to that of the control spot are the averages of two to five independent experiments. Although the identity of the control spot is not known, it was chosen as the most appropriate normalization control because its intensity was similar to that of c-di-GMP. Strains: LM7074 (ΔscrABC/vector), LM7667 (ΔscrABC/scrABC+), LM7665 (ΔscrABC/scrABCΔEAL), and LM6933 (ΔscrABC/scrC+).
Inactivation of the EAL domain reverses ScrABC activity.
IPTG-induced expression of scrABCΔEAL not only resulted in loss of the ability of scrABC to activate swarming but also produced extremely crinkly colonies on Congo red medium and prevented swarming (Fig. 3). The gain of function by this allele suggests that deletion did not simply result in loss of enzyme activity or production of an unstable truncated product. Thus, removal of the EAL domain reversed the activity of ScrABC, converting it from promoting swarming and inhibiting capsule production to diminishing swarming and enhancing adhesiveness.
FIG. 3.
Removal of the EAL domain alters the activity of ScrABC but not ScrC. Three single colonies of each strain were grown on Congo red medium for 7 days and on swarming medium for 14 h; media also contained 2 mM IPTG and 25 μg/ml gentamicin. Strains (from top row to bottom row): LM8073 (LM5674/vector), LM8079 (LM5674/scrABC+), LM8080 (LM5674/scrABCΔEAL), LM8076 (LM5674/scrC+), and LM8081 (LM5674/scrCΔEAL).
The GGDEF and EAL domains are required for ScrABC activity.
To further analyze the activity of ScrC, we constructed scrABC expression clones containing alanine substitutions in amino acids in the signature motif demonstrated to be important for the activity of other GGDEF or EAL domain-containing proteins (35, 48). Transcription of laf and cps was examined in reporter strains containing plasmids bearing scrABCΔEAL, scrABCE554A (EAL to AAL), and scrABCD428A (GGDEF to GGAEF). When cells were grown in liquid, induction of scrABC by IPTG strongly induced laf::lux transcription whereas induction of the alanine mutant constructs caused effects similar to those caused by deletion of the EAL domain; i.e., overexpression of scrABCΔEAL, scrABCE554A, and scrABCD428A resulted in levels of luminescence similar to or lower than the levels of the control strain carrying the vector (Fig. 4A). Thus, these particular alanine substitution mutations in the EAL or the GGDEF domain caused a loss of the ability of ScrABC to enhance swarming gene expression. These data suggest that key amino acids in both the GGDEF and EAL signature motifs are critical for the phosphodiesterase activity of ScrABC.
Figure 4B shows that the loss of laf activation resulting from the impairment of the EAL domain was also accompanied by a gain of cps activation. For these experiments, cps expression was examined in strains grown on plates. Strains overexpressing scrABCΔEAL, scrABCE554A, and scrABCD428A caused increased cpsA::lacZ expression compared to the vector-containing control strain or the strain expressing scrABC. This effect on cps expression was similar to that caused by overexpression of scrC (without scrAB). Thus, removal or inactivation of the domain associated with the phosphodiesterase activity produced an altered ScrC protein that seemed insensitive (or unresponsive) to the presence of ScrA and ScrB.
The EAL domain is not essential to ScrC activity.
To further isolate the effects of these mutations, the scrC alleles were expressed by themselves (i.e., without scrAB). Figure 4C shows that induction of scrC, scrCΔEAL, and scrCE554A by IPTG strongly repressed laf gene expression compared to the vector alone, consistent with the observation that the EAL domain of ScrC is not essential for its ability to prevent swarming or enhance colony crinkliness (Fig. 3). For the experiments in Fig. 4C, laf expression was examined in strains grown on plates, i.e., conditions most appropriate for detecting down regulation of laf expression. Of the three alleles, the one containing the point mutation in the EAL domain produced the most strongly repressing form of ScrC and the allele containing the deletion produced the least active form (specifically, 12,000-fold repression of laf gene expression by scrCE554A, 2,300-fold repression by scrC+, and 620-fold repression by scrCΔEAL). We have not measured the relative stability of the mutant forms of ScrC, which could account for the differing capacities of these variants to repress laf gene expression, and so specific inferences about the consequences of the point mutation versus the deletion upon the catalytic activity cannot currently be made. However, we can conclude that whereas mutations in the EAL domain inactivate the ability of ScrABC to activate laf gene expression (Fig. 4A, which shows laf expression in liquid), these same mutations do not inactivate the ability of ScrC to repress laf gene expression (Fig. 4C, which shows laf expression on plates). Furthermore, overexpression of these alleles, either with or without scrA and scrB, results in increased cps expression (Fig. 4B and C).
The alanine substitution mutation in the GGDEF domain does not inactivate ScrC.
Alanine substitution of aspartate 428 in the GGDEF domain resulted in a form of ScrC that retained some ability to repress laf gene expression (53-fold compared to the vector control), albeit this repression was not as strong as that observed for wild-type ScrC (2,300-fold). The repressing effect on laf expression was accompanied by enhanced cps expression (Fig. 4D); in this case as well, the effect was not as strong as that caused by the wild type. Although specific conclusions about the contribution of this residue to the catalytic activity await further biochemical analysis, this genetic finding demonstrates that the D428A substitution mutation in the GGDEF domain is not essential for the activity of ScrC.
Effect of ScrC on nucleotide pools.
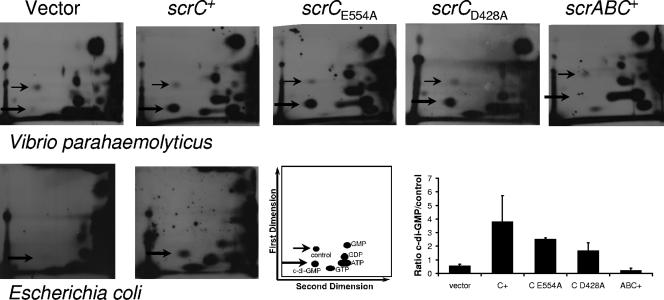
Labeling experiments were performed in the V. parahaemolyticus wild-type background and in E. coli to examine nucleotide pools (Fig. 5). In both organisms, a spot consistent with c-di-GMP was readily detected in strains expressing scrC but not in the vector-containing control strain. Furthermore, this spot could also be detected in V. parahaemolyticus strains expressing the alanine substitution scrC alleles. Thus, elevated c-di-GMP was observed in strains that showed reduced swarming (and laf gene expression) and increased colony roughness (and cps gene expression).
FIG. 5.
ScrC increases the cellular c-di-GMP level. Autoradiograms produced from 2D PEI cellulose TLC separations of 32P-labeled cellular nucleotides upon induction of scrC alleles in V. parahaemolyticus and E. coli. V. parahaemolyticus strains were labeled for 12 h, and autoradiograms were exposed for 3 to 4 days. E. coli strains were labeled for 8 h, and autoradiograms were exposed for 7 days. Thick arrows indicate spots that migrated with Rf values consistent with the reported mobility of c-di-GMP; thin arrows provide orientation and indicate the control spot used for quantitation. Ratios of the intensity of c-di-GMP to that of the control spot in V. parahaemolyticus are the average of at least two independent experiments. E. coli strains: LLM1877 (DH5α/vector) and LLM2449 (DH5α/scrC+). V. parahaemolyticus strains: LM6349 (LM1017/vector), LM7053 (LM1017/scrC+), LM8400 (LM1017/scrCE554A), LM8401 (LM1017/scrCD428A), and LM5435 (LM1017/scrABC+).
Transcription of scrABC is induced by growth on a surface.
To learn more about the cellular role of the scrABC locus, we examined its transcription by monitoring an scrC::lux fusion (Fig. 6). Tn5lux insertions were isolated in scrC on the basis of screening a transposon mutant bank (made in a laf::lacZ reporter strain) for a crinkly colony with decreased laf expression. In one mutant (LM8602), transcription of the lux reporter was aligned with the scrABC operon whereas the transposon in the second mutant (LM8604) was inserted in the opposite direction. These strains were grown in broth culture and on plates, and luminescence was monitored periodically throughout growth. The maximal light produced by LM8602 grown on plates was at least 10-fold higher than the luminescence produced when this strain was grown in liquid culture. There was no difference between plate- and liquid-grown cultures of the control strain. Microarray analysis comparing gene expression of the wild-type strain grown in liquid and on plates showed a similar fold difference in the expression of the scrABC operon (data not shown). Thus, the scrABC operon is most highly transcribed under conditions most relevant to the control of swarming and biofilm formation.
DISCUSSION
Signal transduction mechanisms are required for sensing and adaptation in bacteria; however, systems for signal integration are crucial as well. How is the most appropriate response determined when cells are confronted with multiple environmental inputs? For example, when V. parahaemolyticus encounters a surface, how does this organism process information and determine whether to differentiate into the highly mobile swarmer cell and move over that surface or to remain sessile and form a biofilm? Or, during the colonization of a surface, how does the organism assess its environment and determine when to cease surface translocation and establish a colony or when to move away from a biofilm community; i.e., in V. parahaemolyticus, what determines the cycles of swarmer cell differentiation and dedifferentiation? A network of proteins containing GGDEF or EAL (and potentially HD-GYP) motifs may participate in this task by detecting diverse environmental signals and modulating the intracellular c-di-GMP pool. Recently, the GGDEF-EAL protein of V. parahaemolyticus, ScrG, was shown to regulate swarming and sticking (25). In this study, we examined ScrC, another GGDEF-EAL-containing protein previously reported to regulate swarming and cps gene expression in V. parahaemolyticus (7). We demonstrate that ScrC, like ScrG, influences the c-di-GMP pool. ScrC is encoded by the third gene in the scrABC operon. Deletion of the region specifically encoding the EAL domain, which is associated with c-di-GMP phosphodiesterase activity (4, 10, 20, 44, 48), produced a mutant phenotype similar to that produced by deletion of the entire gene or operon, thus suggesting that the dominant in vivo role of ScrC is to degrade c-di-GMP. Such a role is also played by ScrG.
The analysis of 32P-labeled nucleotide pools is consistent with such a role assignment. A spot corresponding in mobility to c-di-GMP was readily detected in a strain bearing the ΔscrABC deletion, and the intensity of this spot decreased upon ectopic induction of scrABC and not scrABCΔEAL. Analysis of nucleotide pools also illuminated an interesting but previously unexplained observation, namely, that ScrC appeared to have two activities. Overproduction of ScrC in the presence of ScrA and ScrB induced swarming (even in liquid culture) and prevented the transcription of capsular polysaccharide biosynthetic genes. Overproduction of ScrC in the absence of ScrA and ScrB caused the completely opposite effect; i.e., swarming gene expression was prevented and cps transcription was enhanced. ScrABC caused a decreased level of c-di-GMP (Fig. 2), whereas ScrC enhanced c-di-GMP, and moreover, c-di-GMP production could also be elicited in E. coli upon the production of ScrC (Fig. 5). Thus, the two activities of ScrC could be ascribed to effects on the cellular nucleotide pool: ScrC appears to act as a diguanylate cyclase in the absence of its partners ScrA and ScrB and as a phosphodiesterase in the presence of its partners. A high concentration of c-di-GMP favors cps transcription, and a low concentration enhances lateral flagellar gene expression.
Genetic evidence further supports catalytic activities for both the EAL and GGDEF domains. Removal of the EAL domain or mutation of a key amino acid shown to be important for phosphodiesterase activity (EAL→AAL) (4, 26, 46, 50) inactivated the ability of ScrABC to induce laf and prevent cps gene expression. Removal of the EAL domain impaired the ability of ScrABC to reduce cellular c-di-GMP. Mutation of the EAL domain did not simply inactivate ScrABC but rather conferred an enhanced ability (compared to the vector control) to prevent laf and induce cps gene expression. In fact, loss of the EAL domain caused ScrABC to resemble ScrC, suggesting that interaction of ScrA and ScrB with ScrC influences the EAL domain-associated activity. Furthermore, the removal or inactivation of the EAL domain had little consequence upon the ability of ScrC to prevent laf induction, promote cps gene expression, or increase c-di-GMP, implicating the GGDEF domain in this activity. Such a result is consistent with the demonstrated ability of GGDEF domain proteins to act as diguanylate cyclases (19, 35, 42, 46).
The GGDEF signature is considered key to c-di-GMP catalysis, and mutations that alter this motif usually abolish the diguanylate cyclase activity, although mostly deletions and double substitution mutations have been studied (1, 9, 35, 46). We introduced one alanine substitution mutation into the GGDEF signature motif of ScrC (GGDEF→GGAEF; D428A). In Yersinia pestis, this particular substitution in the GGDEF-type protein HmsT inactivated the diguanylate cyclase activity (26); its substitution in ScrC diminished but did not inactivate the ability of ScrC to repress laf and induce cps. Mutations in the GGDEF signature have also been found to allosterically influence the activity of linked EAL domains. The composite GGDEF-EAL protein CC3396 of Caulobacter crescentus possesses an altered and catalytically inactive GGDEF domain, and this protein acts as a PDE. Alteration of the GGDEF motif of CC3396 was demonstrated to affect its phosphodiesterase activity (10). Similarly, we have found that the D428A substitution of ScrC affects the putative phosphodiesterase activity, as ScrABCD428A could no longer activate laf expression in liquid.
The D428A allele, with a substitution in the signature GGDEF motif, failed to produce an inactive form of ScrC, and so we conclude that this particular amino acid is not essential for the diguanylate cyclase activity. ScrCD428A seemed the least effective form of ScrC at repressing laf, activating cps, and producing c-di-GMP, suggesting some potential contribution by D428 to the catalytic activity; however, one cannot currently draw specific conclusions about the observed degree of altered function. The dissection of the contribution of particular amino acids in either the GGDEF or the EAL domain to the enzymatic activity of the protein (as well probing the influence of ScrA and ScrB on ScrC activity) will be better evaluated by using biochemical approaches that can account for protein turnover, as well as control for the influence or interaction of other GGDEF and EAL proteins and effects of nucleotide concentrations.
ScrC appears to have the capacity to both degrade and form c-di-GMP, and in this respect it resembles BphG1, a bifunctional GGDEF-EAL protein of Rhodobacter sphaeroides (49). Unlike BphG1, whose EAL domain locks down the activity of the GGDEF domain and whose EAL domain must be physically removed by cleavage to liberate the diguanylate cyclase activity of the GGDEF domain, the activity of ScrC appears to be modulated by other proteins. ScrA and ScrB modify the activity of ScrC such that the EAL domain is activated and/or the GGDEF domain is silenced. ScrB has been localized to the periplasm and resembles a solute-binding protein (pfam00497; bacterial extracellular solute-binding protein, family 3; E value = 8e-22). We propose that the activity of ScrC could hence be modulated by environmental cues—input transmitted via ligand binding to ScrB and interaction with the periplasmic domain of ScrC would modulate the activity of the cytoplasmic GGDEF-EAL domains. The role of ScrA, which is cytoplasmically located and resembles a pyridoxal-phosphate-dependent aminotransferase, is less clear, albeit complementation shows that it is an essential component in the scheme of signal transduction (7). The genes encoding ScrA, ScrB, and ScrC are organized in an operon, and thus it seems most likely that in vivo ScrC would generally be in the presence of its companions ScrA and ScrB and act as a phosphodiesterase. Environmental conditions could then influence the phosphodiesterase activity. The MbaA protein, which plays a role in biofilm formation in V. cholerae (8), has an architecture similar to that of ScrC; mbaA (VC0703) is also part of a three-gene operon. The first gene in the operon (VC0704) encodes a periplasmic protein named NspS (24), and the third gene (VC0702) encodes a conserved hypothetical protein predicted to be a nucleoside triphosphatase (33). Although not the ortholog of ScrB, the periplasmic binding protein NspS is homologous to the spermidine/putrescine-binding periplasmic protein PotD. The polyamine norspermidine was shown to influence biofilm development, and this effect requires MbaA and NspS (24). Thus, there is precedence in V. cholerae for this kind of signaling cascade whereby ligand-binding, periplasmic sensory components are linked to the activity of a GGDEF-EAL protein.
There are times during surface colonization when it must be advantageous to be the hyperflagellated swarmer cell, e.g., when initially colonizing a surface, but there are also times on a surface when it would be advantageous to be less motile and more adhesive. The scr system, which modulates c-di-GMP, seems to be central to modulating this switch between sticking and swarming (Fig. 7). Two GGDEF-EAL proteins, ScrC and ScrG, influence the transcription of lateral flagellar and cps genes. We note that they do not affect swimming motility (25). ScrC and ScrG appear to act in the same pathway, as their effects are cumulative in double-mutant strains (24). The molecular mechanism by which c-di-GMP specifically influences laf and cps transcription is not known; however, one cps-specific (and not laf) transcription factor, CpsR, has been implicated in the pathway. Loss-of-function mutants with defects in cpsR were identified as suppressor mutations of the crinkly colony morphology of the scrA mutant strain (17). The V. cholerae homolog of CpsR (VpsR) has also been implicated in c-di-GMP control of biofilm formation (28).
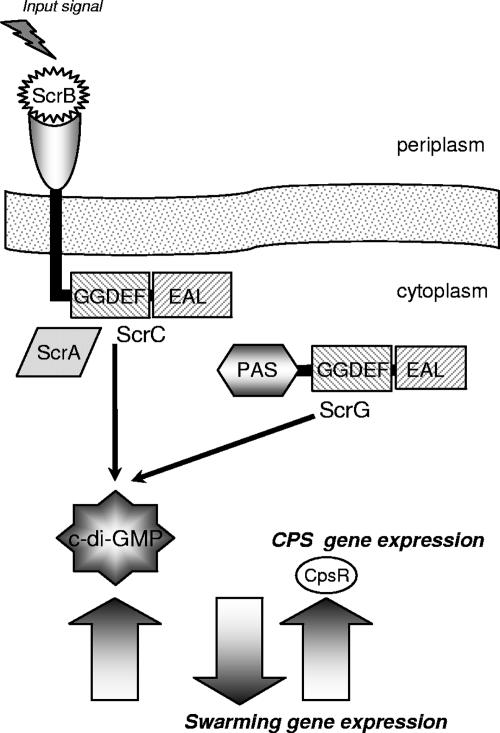
FIG. 7.
Model of the Scr c-di-GMP circuit in V. parahaemolyticus: transcriptional control of swarming and sticking. Two GGDEF-EAL proteins, ScrC and ScrG, regulate swarming and sticking gene expression by acting as phosphodiesterases to modulate the level of c-di-GMP. The phosphodiesterase activity of ScrC is controlled by interaction with ScrA, a predicted pyridoxal-phosphate-dependent aminotransferase, and ScrB, a predicted periplasmic solute-binding protein. Potentially, input signals transmitted via the periplasmic sensing domain of ScrC and the PAS domain of ScrG may moderate the activity of the output GGDEF-EAL domains. High levels of c-di-GMP promote capsular polysaccharide production and biofilm formation, and low levels of this signaling molecule promote swarming motility. The nature of the input signals and how alterations in the cellular c-di-GMP pool affect the transcription of lateral flagellar and cps genes are not known, although one Cps-specific transcriptional regulator (CpsR) acting in this circuit has been identified.
That c-di-GMP signaling participates in determining the switch between motile and sessile lifestyles is an emerging theme in other bacteria as well, e.g., in Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa (27, 31, 46). Considering that V. parahaemolyticus possesses 55 GGDEF and/or EAL proteins, a plethora of input signals may influence the level of c-di-GMP. These proteins may act to modulate a common nucleotide pool, they may act more specifically to control local concentrations of c-di-GMP, or they may act temporally, e.g., during different stages of growth (35, 46). Some of these proteins may have more important or specific roles in swarming and biofilm development than others. With respect to ScrABC, we find that its transcription is coincident with swarming; i.e., an scrC::lux reporter fusion was induced by growth on surfaces. We find that ScrA and ScrB can modify the activity of ScrC, and we suggest that environmental cues are processed in a signal transduction scheme that influences the phosphodiesterase activity of ScrC. What these precise signals are remains to be elucidated. How c-di-GMP influences gene expression also remains to be understood, and V. parahaemolyticus may be a particularly good system for studying c-di-GMP control of gene expression as the scr system has a clear and strong influence on the transcription of genes pertinent to growth on surfaces.
Acknowledgments
This work was supported by National Science Foundation grant MCB0315617.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 471695-1708. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P., L. Baumann, and M. Mandel. 1971. Taxonomy of marine bacteria: the genus Beneckea. J. Bacteriol. 107268-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 1883600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247123-130. [DOI] [PubMed] [Google Scholar]
- 5.Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 2579759-9769. [PubMed] [Google Scholar]
- 6.Boles, B. R., and L. L. McCarter. 2000. Insertional inactivation of genes encoding components of the sodium-type flagellar motor and switch of Vibrio parahaemolyticus. J. Bacteriol. 1821035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boles, B. R., and L. L. McCarter. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 1845946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 1851384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christen, B., M. Christen, R. Paul, F. Schmid, M. Folcher, P. Jenoe, M. Meuwly, and U. Jenal. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 28132015-32024. [DOI] [PubMed] [Google Scholar]
- 10.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 28030829-30837. [DOI] [PubMed] [Google Scholar]
- 11.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 1846481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dow, J. M., Y. Fouhy, J. F. Lucey, and R. P. Ryan. 2006. The HD-GYP domain, cyclic di-GMP signaling, and bacterial virulence to plants. Mol. Plant-Microbe Interact. 191378-1384. [DOI] [PubMed] [Google Scholar]
- 14.Enos-Berlage, J. L., Z. T. Guvener, C. E. Keenan, and L. L. McCarter. 2005. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 551160-1182. [DOI] [PubMed] [Google Scholar]
- 15.Enos-Berlage, J. L., and L. L. McCarter. 2000. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J. Bacteriol. 1825513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Güvener, Z. T., and L. L. McCarter. 2003. Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J. Bacteriol. 1855431-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 1462395-2407. [DOI] [PubMed] [Google Scholar]
- 19.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 10214422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hisert, K. B., M. MacCoss, M. U. Shiloh, K. H. Darwin, S. Singh, R. A. Jones, S. Ehrt, Z. Zhang, B. L. Gaffney, S. Gandotra, D. W. Holden, D. Murray, and C. Nathan. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 561234-1245. [DOI] [PubMed] [Google Scholar]
- 21.Huang, B., C. B. Whitchurch, and J. S. Mattick. 2003. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 1857068-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaques, S., and L. L. McCarter. 2006. Three new regulators of swarming in Vibrio parahaemolyticus. J. Bacteriol. 1882625-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40385-407. [DOI] [PubMed] [Google Scholar]
- 24.Karatan, E., T. R. Duncan, and P. I. Watnick. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 1877434-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, Y. K., and L. L. McCarter. 2007. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 1894094-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirillina, O., J. D. Fetherston, A. G. Bobrov, J. Abney, and R. D. Perry. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 5475-88. [DOI] [PubMed] [Google Scholar]
- 27.Kuchma, S. L., K. M. Brothers, J. H. Merritt, N. T. Liberati, F. M. Ausubel, and G. A. O'Toole. 2007. BifA, a c-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1898165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60331-348. [DOI] [PubMed] [Google Scholar]
- 29.McCarter, L. 1999. The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 151-57. [PubMed] [Google Scholar]
- 30.McCarter, L. L., and M. E. Wright. 1993. Identification of genes encoding components of the swarmer cell flagellar motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J. Bacteriol. 1753361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merritt, J. H., K. M. Brothers, S. L. Kuchma, and G. A. O'Toole. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 1898154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Ni, S., F. Forouhar, D. E. Bussiere, H. Robinson, and M. A. Kennedy. 2006. Crystal structure of VC0702 at 2.0 Å: conserved hypothetical protein from Vibrio cholerae. Proteins 63733-741. [DOI] [PubMed] [Google Scholar]
- 34.Parsek, M. R., and E. P. Greenberg. 1999. Quorum sensing signals in development of Pseudomonas aeruginosa biofilms. Methods Enzymol. 31043-55. [DOI] [PubMed] [Google Scholar]
- 35.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putnam, S. L., and A. L. Koch. 1975. Complications in the simplest cellular enzyme assay: lysis of Escherichia coli for the assay of beta-galactosidase. Anal. Biochem. 63350-360. [DOI] [PubMed] [Google Scholar]
- 37.Rashid, M. H., C. Rajanna, A. Ali, and D. K. Karaolis. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 227113-119. [DOI] [PubMed] [Google Scholar]
- 38.Römling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9218-228. [DOI] [PubMed] [Google Scholar]
- 39.Römling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57629-639. [DOI] [PubMed] [Google Scholar]
- 40.Ross, P., Y. Aloni, H. Weinhouse, D. Michaeli, P. Weinberger-Ohana, R. Mayer, and M. Benziman. 1986. Control of cellulose synthesis in Acetobacter xylinum. A unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydr. Res. 149101-117. [Google Scholar]
- 41.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 1036712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 1871792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 1874774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverman, M., R. Showalter, and L. McCarter. 1991. Genetic analysis in Vibrio. Methods Enzymol. 204515-536. [DOI] [PubMed] [Google Scholar]
- 46.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 531123-1134. [DOI] [PubMed] [Google Scholar]
- 47.Stewart, B. J., and L. L. McCarter. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 1854508-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 28033324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarutina, M., D. A. Ryjenkov, and M. Gomelsky. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 28134751-34758. [DOI] [PubMed] [Google Scholar]
- 50.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, J., and M. F. Wilkinson. 2000. Site-directed mutagenesis of large (13-kb) plasmids in a single-PCR procedure. BioTechniques 29976-978. [DOI] [PubMed] [Google Scholar]