Abstract
RNA polymerase is a central macromolecular machine controlling the flow of information from genotype to phenotype, and insights into global transcriptional regulation can be gained by studying mutational perturbations in the enzyme. Mutations in the RNA polymerase β subunit gene rpoB causing resistance to rifampin (Rifr) in Bacillus subtilis were previously shown to lead to alterations in the expression of a number of global phenotypes known to be under transcriptional control, such as growth, competence for transformation, sporulation, and germination (H. Maughan, B. Galeano, and W. L. Nicholson, J. Bacteriol. 186:2481-2486, 2004). To better understand the global effects of rpoB mutations on metabolism, wild-type and 11 distinct congenic Rifr mutant strains of B. subtilis were tested for utilization of 95 substrates by use of Biolog GP2 MicroPlates. A number of alterations of substrate utilization patterns were observed in the Rifr mutants, including the utilization of novel substrates previously unknown in B. subtilis, such as gentiobiose, β-methyl-d-glucoside, and d-psicose. The results indicate that combining global metabolic profiling with mutations in RNA polymerase provides a system-wide approach for uncovering previously unknown metabolic capabilities and further understanding global transcriptional control circuitry in B. subtilis.
A central goal of the genomics-bioinformatics era is to understand at a global level how cells coordinate the conversion of their genomic information into phenotype. From this perspective, macromolecular machines such as the replication, transcription, and translation complexes can be viewed as central control devices that optimize the flow of genetic information to the various cellular tasks needed for sensing and responding to environmental cues. It has been well established for the gram-positive spore-forming bacterium Bacillus subtilis that growth and metabolism, as well as several developmental events (e.g., extracellular enzyme synthesis, motility, chemotaxis, competence, sporulation, spore resistance properties, germination, and outgrowth), are controlled in large part at the transcriptional level (26, 27, 28). Because the RNA polymerase transcription complex contacts every promoter in the B. subtilis genome, single amino acid substitutions in critical portions of the enzyme can lead to global changes in gene transcription and hence in physiology and metabolism.
One such critical target resides on the β subunit of RNA polymerase in the binding site for the antibiotic rifampin (Rif). Rifampin resistance (Rifr) results from mutations in the rpoB gene leading to specific amino acid alterations in β (12; reviewed in reference 37). Evidence connecting RNA polymerase, Rifr, and global regulation of growth and development in B. subtilis has been accumulating. (i) Rothstein et al. (31) first isolated a Rifr B. subtilis mutant exhibiting a temperature-sensitive sporulation defect; the mutation responsible, rfm2103, was later identified as causing the amino acid change H482Y in cluster I of β (7). (ii) Rifr mutations mapping to cluster I (Q469K, Q469R, and H482Y) affected the efficiency of rho-dependent transcription termination (10). (iii) Rifr mutations in cluster I, especially S487L, were reported to enhance postexponential production of the extracellular enzymes amylase and protease (S. T. Jørgensen, unpublished data; see also reference 9a). (iv) We observed that four Rifr mutations in cluster I (Q469R, H482R, H482Y, and S487L) caused both locus-specific and allele-specific alterations in the expression of global transcriptional regulons controlling growth, competence, sporulation, and germination (19). Collectively these observations suggest that alteration of single amino acids in the Rif-binding pocket of β results in fundamental alterations in the global interactions of RNA polymerase with promoters and transcriptional regulators, leading to profound changes in global phenotypes.
Using B. subtilis as a model organism, we have been exploring the connections linking environmental selective pressure, mutagenesis, altered phenotype, and fitness during evolution (18). We found that during long-term laboratory evolution of B. subtilis the rate of spontaneous mutation to Rifr increased in evolving cultures by 1 to 2 orders of magnitude over the ancestral mutation rate (18), and we have noted that differing cellular environments can alter the spectrum of spontaneous mutations to Rifr (21, 22). Thus, different selective environments can lead to different amino acid substitutions in the RNA polymerase β subunit, which in turn alter global transcriptional patterns, which in turn affect phenotype and evolutionary fitness.
One of the goals of our research is to understand how Rifr mutations might activate new metabolic capabilities that enhance the evolutionary fitness of spore-forming bacteria in novel selective environments. In a previous communication we reported that four rpoB mutations to Rifr (Q469R, H482R, H482Y, and S487L) caused global changes in B. subtilis growth, sporulation, germination, and competence for DNA-mediated transformation (19). Because these global phenotypes are controlled largely at the level of transcription, we reasoned that mutations located in the Rif binding pocket of the β subunit may alter the RNA polymerase structure and hence its regulated interaction with specific promoters. However, it is difficult to identify specific individual promoter-RNA polymerase interactions leading to changes in gross phenotypes, such as growth rate or sporulation, which result from the concerted action of hundreds of genes. Therefore, with a more comprehensive collection of rpoB mutations in hand, we undertook a larger-scale search for specific phenotypes altered in Rifr strains that may be more amenable to individual genetic analysis. We have isolated more than 100 independent Rifr mutants of B. subtilis and have collected a total of 11 distinct rpoB mutations, one in the N cluster (V135F) and 10 in cluster I (Q469K/L/R, H482D/P/R/Y, and S487F/L/Y) (A. E. Perkins and W. L. Nicholson, unpublished data). In order to search for novel phenotypes on a large scale, we report in this communication the use of high-throughput metabolic profiling in a 96-well format using Biolog GP2 MicroPlates coupled with the kinetic monitoring feature of the Omnilog automated plate reader (3-5). Using this format we were able to uncover a number of new phenotypes exhibited by various rpoB mutants that were previously not apparent in the congenic “wild-type” (wt) laboratory strain of B. subtilis. This technique has the potential for better understanding the ecology and evolution of B. subtilis, for identifying previously unknown or unannotated biochemical pathways in the B. subtilis genome, and for further understanding transcriptional regulation mechanisms of known metabolic pathways.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Spontaneously arising mutations of rpoB conferring a Rifr phenotype were derived from vegetative cells or spores of B. subtilis strain 168. Details of their isolation and characterization have either been reported previously (22) or will be described in detail elsewhere (Perkins and Nicholson, unpublished). The rpoB mutations S487F and S487Y were isolated from spores of B. subtilis exposed to ultrahigh vacuum (21) and were kindly provided to us by Nobuo Munakata. To assure that the phenotypes observed in this study resulted directly from the rpoB alleles tested, a set of congenic strains was constructed. All alleles were PCR amplified from their respective Rifr mutants as described previously (22) and transferred by transformation (8, 22) into the common host background of B. subtilis strain MH5636, a generous gift from Marion Hulett. Strain MH5636 (trpC2 pheA1 Cmr) carries an engineered rpoC gene expressing a six-histidine-tagged version of the RNA polymerase β′ subunit, which does not interfere with normal RNA polymerase function (19, 29; Perkins and Nicholson, unpublished). In all constructs, the presence of the correct rpoB allele after its transformation into MH5636 was confirmed by PCR amplification and sequencing as described previously (22). (In this work, strain MH5636 is referred to as the wt with respect to RNA polymerase, and all congenic mutant strains are referred to by the amino acid substitution they carry in the β subunit).
Media used were Luria-Bertani (LB) medium (20) and Spizizen minimal medium (SMM) (33) in which glucose was replaced with d-ribose. All media were either used in liquid form or solidified with a final concentration of 1.5% agar as needed. SMM contained (final concentration) tryptophan (50 μg/ml), phenylalanine (50 μg/ml), and various concentrations of d-ribose (see Results). Antibiotics used were (final concentration) chloramphenicol (Cm) (5 μg/ml) and Rif (50 μg/ml). Liquid cultures were grown at 37°C with vigorous aeration, and growth was monitored using either a Klett-Summerson photometer fitted with a no. 66 (red) filter or a spectrophometer set at 660 nm.
Biolog phenotype profiling.
Overnight cultures of each strain were grown in 2 ml of LB broth (wt) or LB plus Rif (rpoB mutants), and the next day the overnight cultures were diluted 100-fold into 20 ml of LB. Cultures were incubated at 37°C until mid-logarithmic phase (80 to 90 Klett units). Cells were harvested by centrifugation (10,000 × g, 10 min, 24°C), and the cell pellets were resuspended in 1 ml of phosphate-buffered saline buffer (10 mM potassium phosphate, pH 7.2, 150 mM NaCl) (23). Cells were diluted into GN/GP inoculating fluid (Biolog, Hayward, CA), and the optical density of the inoculating suspension was adjusted to 28% transmittance by use of a turbidimeter (Biolog model 21907). Cell suspensions were inoculated (150 μl of suspension per well) into 96-well Biolog gram-positive (GP2) MicroPlates; the complete listing of the 95 substrates in these plates is available at www.biolog.com/pdf/GP2b_Brochure.pdf. Plates were placed in an Omnilog phenotype microarray instrument (Biolog) programmed to run at 30°C for 24 h with absorbance readings taken every 15 min. Each strain was assayed in duplicate plates. Data were exported into Microsoft Excel and Kaleidograph (version 3.6.2; Synergy Software, Reading, PA) for further analysis.
Statistical analyses.
All assays were performed in at least duplicate. Basic statistical parameters and analyses of variance (ANOVA) were performed using commercial statistical software (Kaleidagraph). Differences with P values of <0.05 were considered statistically significant.
RESULTS AND DISCUSSION
All Rifr mutant strains and their congenic wt counterpart were inoculated into duplicate Biolog GP2 MicroPlates and incubated in an Omnilog automated plate reader to discern their substrate utilization patterns. In order to ascertain a baseline for phenotype scoring, the data from all 12 strains in negative control well A01 (water) were analyzed. It was noted that at no time did any of the 12 strains exhibit more than 50 arbitrary absorbance units (U); thus, it was decided that less than 50 U constituted a negative signal. Using this criterion, we determined that the wt strain and all 11 Rifr mutants tested were unable to utilize 63 out of the 95 substrates included in GP2 MicroPlates (Table 1).
TABLE 1.
Substrates in GP2 MicroPlates not utilized (<50 U) by the wt and Rifr mutants of B. subtilis
| Substrate |
|---|
| α-Cyclodextrin |
| β-Cyclodextrin |
| Inulin |
| Mannan |
| Tween-40 |
| Tween-80 |
| N-Acetyl-d-glucosamine |
| N-Acetyl-β-d-glucosamine |
| d-Arabitol |
| l-Fucose |
| d-Galactose |
| d-Galacturonic acid |
| d-Gluconic acid |
| m-Inositol |
| α-d-Lactose |
| Lactulose |
| Maltose |
| Maltotriose |
| d-Melezitose |
| d-Melibiose |
| α-methyl-d-Galactoside |
| β-methyl-d-Galactoside |
| α-methyl-d-Glucose |
| α-methyl-d-Mannoside |
| d-Raffinose |
| l-Rhamnose |
| Sedoheptulosan |
| d-Sorbitol |
| Stachyose |
| d-Tagatose |
| Turanose |
| Xylitol |
| Acetic acid |
| α-Hydroxybutyrate |
| β-Hydroxybutyrate |
| γ-Hydroxybutyrate |
| p-Hydroxyphenylacetate |
| α-Ketoglutarate |
| α-Ketovaleric acid |
| Lactamide |
| d-Lactic acid |
| l-Lactic acid |
| d-Malic acid |
| Succinic acid methyl ester |
| Propionic acid |
| Succinamic acid |
| Succinic acid |
| N-Acetyl glutamic acid |
| l-Alaninamide |
| d-Alanine |
| l-Alanyl-glycine |
| Glycyl-l-glutamic acid |
| l-Pyroglutamic acid |
| Putrescine |
| 2,3-Butanediol |
| Adenosine |
| 2′-Deoxy-adenosine |
| Adenosine-5′-monophosphate |
| Thymidine-5′-monophosphate |
| Uridine-5′-monophosphate |
| d-Fructose-6-phosphate |
| Glucose-1-phosphate |
| Glucose-6-phosphate |
We noted that the 32 substrates utilized by the wt strain could be divided further into two broad categories. Thirteen substrates were strongly utilized (>100 U), and these included a number of readily metabolized sugars (arabinose, fructose, glucose, mannose, ribose, sucrose, trehalose, and xylose), dextrin, the aromatic glycoside salicin, pyruvic acid, and the nucleosides inosine and uridine (Table 2). The remaining 19 substrates we classified as weakly utilized by wt (50 to 100 U); these included a number of sugar polymers, rare sugars, sugar alcohols, amino acids, and the nucleoside thymidine (Table 3).
TABLE 2.
Substrate utilization by Rifr mutants: substrates utilized strongly by the wt (>100 U)
| Substrate | Maximum absorbance value (avg ± SD)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt | V135F | Q469K | Q469L | Q469R | H482D | H482P | H482R | H482Y | S487F | S487L | S487Y | |
| A04 Dextrin | 133 ± 3.5 | 156 ± 5.7 | 75 ± 16 | 161 ± 16 | 111 ± 18 | 94 ± 12 | 194 ± 5.7 | 150 ± 3.5 | 159 ± 6.4 | 97 ± 6.4 | 128 ± 3.5 | 156 ± 5.7 |
| B01 l-Arabinose | 106 ± 6.4 | 30 ± 3.5 | 42 ± 4.9 | 38 ± 3.5 | 33 ± 2.1 | 43 ± 4.9 | 48 ± 4.2 | 41 ± 4.2 | 40 ± 0.7 | 35 ± 7.8 | 96 ± 6.4 | 34 ± 8.5 |
| B05 d-Fructose | 149 ± 28 | 158 ± 13 | 141 ± 4.9 | 148 ± 1.4 | 205 ± 60 | 138 ± 4.9 | 196 ± 73 | 147 ± 12 | 153 ± 1.4 | 188 ± 25 | 168 ± 9.9 | 138 ± 3.5 |
| B11 α-d-Glucose | 195 ± 9.9 | 200 ± 19 | 204 ± 12 | 197 ± 0.7 | 261 ± 27 | 172 ± 21 | 263 ± 31 | 180 ± 9.2 | 161 ± 3.5 | 188 ± 1.4 | 162 ± 6.4 | 198 ± 2.1 |
| C06 d-Mannose | 165 ± 14 | 178 ± 3.5 | 172 ± 2.8 | 174 ± 30 | 180 ± 28 | 170 ± 21 | 180 ± 0 | 205 ± 71 | 153 ± 1.4 | 163 ± 25 | 169 ± 1.4 | 180 ± 35 |
| D07 d-Ribose | 218 ± 2.8 | 217 ± 13 | 203 ± 2.8 | 225 ± 22 | 42 ± 14 | 38 ± 3.5 | 250 ± 33 | 203 ± 2.8 | 194 ± 2.8 | 41 ± 0 | 189 ± 2.8 | 196 ± 1.4 |
| D08 Salicin | 114 ± 9.2 | 193 ± 71 | 174 ± 0.7 | 152 ± 7.8 | 153 ± 9.9 | 117 ± 10.6 | 142 ± 0 | 134 ± 3.5 | 121 ± 0 | 122 ± 0.7 | 132 ± 3.5 | 149 ± 2.8 |
| D12 Sucrose | 187 ± 30 | 193 ± 12 | 204 ± 0.7 | 208 ± 7.8 | 253 ± 46 | 187 ± 36 | 214 ± 25 | 187 ± 25 | 181 ± 26 | 181 ± 27 | 92 ± 88 | 197 ± 28 |
| E02 d-Trehalose | 165 ± 6.4 | 195 ± 17 | 208 ± 2.8 | 242 ± 4.2 | 222 ± 21 | 193 ± 1.4 | 249 ± 40 | 192 ± 18 | 218 ± 60 | 186 ± 8.5 | 167 ± 5.7 | 209 ± 2.8 |
| E05 d-Xylose | 146 ± 16.3 | 60 ± 3.5 | 46 ± 0.7 | 50 ± 14 | 34 ± 1.4 | 24 ± 0.7 | 68 ± 2.8 | 56 ± 6.4 | 61 ± 0.7 | 37 ± 6.4 | 125 ± 8.5 | 44 ± 14 |
| F09 Pyruvic acid | 114 ± 9.9 | 134 ± 1.4 | 143 ± 0.7 | 117 ± 9.2 | 16 ± 8.5 | 17 ± 9.2 | 136 ± 21 | 106 ± 1.4 | 100 ± 4.9 | 28 ± 9.9 | 176 ± 8.5 | 113 ± 3.5 |
| H03 Inosine | 105 ± 5.7 | 77 ± 17 | 71 ± 42 | 86 ± 15 | 50 ± 4.5 | 57 ± 7.1 | 123 ± 3.5 | 67 ± 28 | 88 ± 9.9 | 42 ± 23 | 115 ± 0.7 | 90 ± 13 |
| H05 Uridine | 100 ± 9.2 | 108 ± 4.9 | 76 ± 21 | 98 ± 8.5 | 39 ± 11 | 31 ± 3.5 | 116 ± 36 | 94 ± 4.9 | 94 ± 2.1 | 28 ± 4.9 | 141 ± 7.8 | 65 ± 4.2 |
Data are expressed as averages ±standard deviations of maximum absorbance values obtained in duplicate runs. Values in boldface are significantly higher than values for the wt by ANOVA (P < 0.05). Values in italics are significantly lower than values for the wt by ANOVA (P < 0.05).
TABLE 3.
Substrate utilization by Rifr mutants: substrates utilized weakly by the wt (50 to 100 U)
| Substrate | Maximum absorbance value (avg ± SD)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt | V135F | Q469K | Q469L | Q469R | H482D | H482P | H482R | H482Y | S487F | S487L | S487Y | |
| A05 Glycogen | 61 ± 11 | 45 ± 3.5 | 37 ± 23 | 39 ± 11 | 23 ± 14 | 17 ± 6.4 | 59 ± 6.4 | 37 ± 5.7 | 55 ± 8.5 | 29 ± 7.8 | 85 ± 1.4 | 33 ± 1.4 |
| A12 Amygdalin | 50 ± 26 | 87 ± 23 | 76 ± 28 | 79 ± 28 | 67 ± 33 | 65 ± 29 | 64 ± 16 | 67 ± 3.5 | 78 ± 19 | 70 ± 15 | 62 ± 4.9 | 93 ± 25 |
| B03 Arbutin | 90 ± 3.5 | 97 ± 31 | 138 ± 39 | 136 ± 4.2 | 116 ± 9.9 | 93 ± 5.7 | 126 ± 4.2 | 94 ± 37 | 82 ± 30 | 78 ± 37 | 86 ± 1.4 | 148 ± 11 |
| B04 d-Cellobiose | 91 ± 14 | 144 ± 13 | 112 ± 2.1 | 147 ± 1.4 | 76 ± 0 | 57 ± 3.5 | 145 ± 18 | 95 ± 0.7 | 115 ± 32 | 74 ± 15 | 96 ± 4.2 | 138 ± 3.5 |
| B09 Gentiobiose | 67 ± 3.5 | 142 ± 5.7 | 141 ± 4.2 | 156 ± 2.8 | 41 ± 22 | 74 ± 11 | 108 ± 29 | 53 ± 11 | 43 ± 3.5 | 31 ± 11 | 80 ± 2.1 | 149 ± 16 |
| C05 d-Mannitol | 92 ± 13 | 100 ± 7.8 | 142 ± 18 | 138 ± 42 | 53 ± 0 | 82 ± 9.2 | 125 ± 31 | 106 ± 7.8 | 160 ± 1.4 | 85 ± 2.8 | 101 ± 2.8 | 76 ± 14 |
| C11 3-methyl-d-Glucose | 70 ± 0 | 110 ± 35 | 75 ± 15 | 106 ± 16 | 48 ± 4.2 | 62 ± 30 | 153 ± 32 | 73 ± 3.5 | 139 ± 49 | 60 ± 21 | 122 ± 23 | 73 ± 3.5 |
| D01 β-methyl-d-Glucoside | 83 ± 23 | 206 ± 4.2 | 231 ± 6.3 | 217 ± 12 | 115 ± 1.4 | 129 ± 5.7 | 198 ± 0.7 | 200 ± 2.1 | 176 ± 25 | 160 ± 2.8 | 92 ± 2.1 | 208 ± 4.9 |
| D03 Palatinose | 85 ± 28 | 50 ± 7.1 | 47 ± 23 | 42 ± 4.9 | 33 ± 7.8 | 20 ± 7.1 | 67 ± 4.9 | 31 ± 0.7 | 35 ± 6.4 | 38 ± 25 | 81 ± 3.5 | 44 ± 19 |
| D04 d-Psicose | 82 ± 2.8 | 132 ± 2.1 | 130 ± 4.2 | 123 ± 7.8 | 141 ± 9.2 | 106 ± 1.4 | 163 ± 13 | 130 ± 23 | 154 ± 49 | 95 ± 4.9 | 125 ± 35 | 125 ± 13 |
| F05 l-Malic acid | 69 ± 9.2 | 75 ± 2.8 | 76 ± 4.2 | 75 ± 9.2 | 46 ± 6.4 | 36 ± 5.7 | 103 ± 39 | 84 ± 4.2 | 101 ± 34 | 37 ± 7.1 | 119 ± 16 | 48 ± 6.4 |
| F06 Pyruvic acid methyl ester | 81 ± 8.5 | 121 ± 1.4 | 100 ± 7.1 | 108 ± 18 | 23 ± 2.8 | 14 ± 16 | 125 ± 0 | 108 ± 4.9 | 112 ± 7.1 | 29 ± 2.1 | 96 ± 3.5 | 92 ± 26 |
| G03 l-Alanine | 68 ± 3.5 | 52 ± 15 | 30 ± 18 | 43 ± 18 | 32 ± 2.8 | 20 ± 2.8 | 108 ± 44 | 47 ± 23 | 61 ± 15 | 19 ± 2.8 | 109 ± 11 | 49 ± 16 |
| G05 l-Asparagine | 91 ± 22 | 76 ± 16 | 108 ± 3.5 | 93 ± 17 | 69 ± 14 | 72 ± 15 | 91 ± 13 | 74 ± 27 | 105 ± 35 | 70 ± 28 | 190 ± 0.7 | 65 ± 1.4 |
| G06 l-Glutamic acid | 60 ± 7.1 | 59 ± 5.7 | 76 ± 16 | 59 ± 6.4 | 52 ± 12 | 37 ± 9.2 | 86 ± 5.7 | 76 ± 5.7 | 61 ± 4.2 | 36 ± 2.8 | 133 ± 15 | 57 ± 2.1 |
| G09 l-Serine | 69 ± 9.2 | 64 ± 2.1 | 55 ± 4.2 | 54 ± 1.4 | 20 ± 1.4 | 9 ± 1.4 | 71 ± 8.4 | 68 ± 3.5 | 80 ± 0.7 | 20 ± 4.9 | 110 ± 7.1 | 55 ± 3.5 |
| G12 Glycerol | 83 ± 18 | 81 ± 3.5 | 82 ± 1.4 | 87 ± 7.8 | 45 ± 7.1 | 54 ± 5.7 | 117 ± 1.4 | 83 ± 21 | 82 ± 2.8 | 47 ± 4.9 | 74 ± 16 | 73 ± 11 |
| H04 Thymidine | 61 ± 1.4 | 56 ± 15 | 39 ± 5.7 | 55 ± 7.1 | 33 ± 2.8 | 42 ± 27 | 83 ± 27 | 51 ± 3.5 | 47 ± 6.4 | 33 ± 15 | 77 ± 6.4 | 66 ± 11 |
| H12 d-l-α-Glycerol phosphate | 65 ± 0 | 71 ± 8.5 | 62 ± 7.1 | 66 ± 7.8 | 45 ± 4.2 | 37 ± 9.2 | 73 ± 7.1 | 44 ± 11 | 56 ± 6.4 | 39 ± 12 | 83 ± 19 | 56 ± 9.2 |
Data are expressed as averages ± standard deviations of maximum absorbance values obtained in duplicate runs. Values in boldface are significantly higher than values for the wt by ANOVA (P < 0.05). Values in italics are significantly lower than values for the wt by ANOVA (P < 0.05).
Strongly utilized substrates (>100 U).
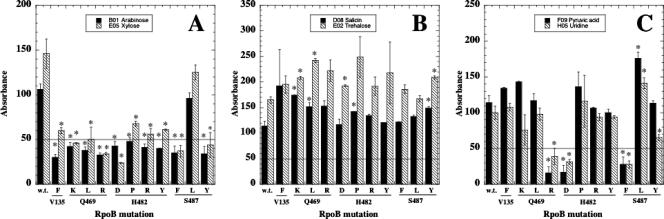
The combination of 11 Rifr mutants and 13 strongly utilized substrates resulted in 143 mutant/substrate combinations (Table 2). Out of this total, 15 examples of significant increase in substrate utilization by the mutants were observed, compared to 41 examples of significant decrease and 87 examples of no statistical difference between the wt and the Rifr mutant (Table 2). In other words, among examples in which significant differences were noted, in ∼73% of the cases (41/56) rpoB mutations caused reduced utilization of substrates normally used strongly by the wt, and in ∼27% of the cases (15/56) the rpoB mutations led to enhanced utilization. Mutation to Rifr had no significant effect on utilization of the sugars fructose, mannose, or sucrose; glucose utilization was lowered slightly in the H482Y mutant but not affected significantly in any other Rifr mutant (Table 2). Some patterns of substrate utilization effects by RpoB mutants could be discerned from the data. For example, utilization of arabinose and xylose was severely reduced to near- or below-background levels in all Rifr mutants except the S487L mutant (Table 2; Fig. 1A). In contrast, utilization of salicin and trehalose was not reduced in any Rifr mutants but was increased significantly by the Q469K, Q469L, H482D, H482P, and S487Y mutations (Table 2; Fig. 1B). Substrates such as pyruvic acid and uridine exhibited a more complex pattern; mutations at Q469R, H482D, and S487F/Y significantly reduced utilization, the S487L mutation significantly increased utilization, and the rest had no significant effect (Table 2; Fig. 1C).
FIG. 1.
Substrate utilization patterns in wt and Rifr mutant B. subtilis strains for substrates utilized strongly (>100 U) by the wt. The horizontal line at 50 U denotes the background level (see text for details). Asterisks denote significant differences (P < 0.05) from the wt. (A) Arabinose and xylose. (B) Salicin and trehalose. (C) Pyruvic acid and uridine.
Utilization of ribose.
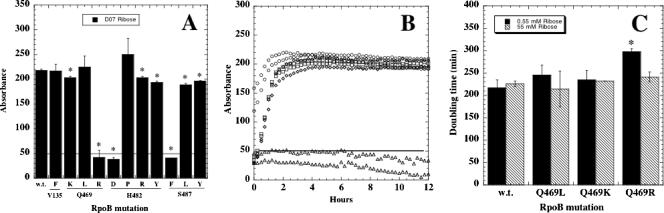
Utilization of ribose presented an interesting situation. While some rpoB mutations exerted either no significant effect (V135F, Q469L, and H482P) or only a slight reduction (Q469K, H482R/Y, and S487L/Y), other mutations at the same positions (Q469R, H482D, and S487F) caused dramatic reductions in ribose utilization to below background level (Fig. 2A). In particular it was intriguing that replacement of S487 by the similar aromatic residues F versus Y or of Q469 by the similar basic amino acids K versus R should result in such disparate effects (Fig. 2A). Examination of the MicroPlate kinetic data for utilization of ribose by the wt and the Q469 mutants confirmed that the Q469R mutant was indeed unable to utilize ribose at all time points and in fact was at or below the 50 U “background” level (Fig. 2B). We wondered if this low level of ribose utilization in the Q469R mutant was severe enough to manifest itself as a growth defect on ribose as a sole carbon source, i.e., whether this particular rpoB mutation would lead to ribose auxotrophy. We tested this notion by measuring the growth rates of the wt strain and all Q469 mutants in SMM supplemented with d-ribose at various concentrations (55, 5.5, 0.55, and 0.055 mM) (d-ribose is supplied in GP2 MicroPlates at 22 mM [Jeffrey Carlson, Biolog, personal communication]). We noted that the Q469R mutant grew at the same rate as the wt and the Q469K and Q469L mutants at 55 mM (Fig. 2C) and 5.5 mM (data not shown) ribose. Only at 0.55 mM ribose did the Q469R mutant grow significantly more slowly than the wt and the other Q469 mutants (Fig. 2C). At 0.055 mM ribose, none of the strains were able to grow (data not shown). Interestingly, it appeared that GP2 MicroPlates were able to detect a defect in ribose utilization by the Q469R rpoB mutant that did not manifest itself as a slower growth rate phenotype until cells were stressed by growth at a 40-fold-lower ribose concentration than that used in the GP2 plate. Therefore, in some cases phenotypic profiling may actually be more sensitive at uncovering certain subtle defects than traditional auxotrophic growth assays.
FIG. 2.
Ribose utilization in wt and Rifr mutant B. subtilis strains. (A) Ribose utilization patterns. The horizontal line at 50 U denotes the background level. Asterisks denote significant differences from the wt (P < 0.05). (B) Kinetics of ribose utilization in the wt (circles) and RpoB Q469K (squares), Q469L (diamonds), and Q469R (triangles) mutants. Only the first 12 h are shown for clarity. (C) Growth rates (doubling times) of wt and Q469 mutant strains in SMM containing d-ribose at 55 mM or 0.55 mM. Data are averages and standard deviations of triplicate determinations. The asterisk denotes significant difference (P < 0.05).
Substrates weakly utilized (50 to 100 U) by the wt.
The combination of 11 Rifr mutants and 19 substrates utilized weakly by the wt resulted in 209 mutant/substrate combinations (Table 3). Out of this total, 31 examples of significant increase in substrate utilization by the mutants was observed, compared to 18 examples of significant decrease and 160 examples of no statistical difference between the wt and the Rifr mutant (Table 3). Opposite to what was observed with strongly utilized substrates, in ∼63% of the cases (31/49) rpoB mutations caused increased utilization of substrates normally used weakly by the wt, and in ∼37% of the cases (18/49) the rpoB mutations led to further-reduced utilization (Table 3). Mutation to Rifr had no significant effect on utilization of amygdalin, palatinose, malic acid, or glycerol (Table 3). Utilization of glycogen, 3-methyl-d-glucose, thymidine, and d-l-α-glycerol phosphate was either unaffected or further reduced significantly in at least one of the Rifr mutants (Table 3).
Utilization of β-glucosides.
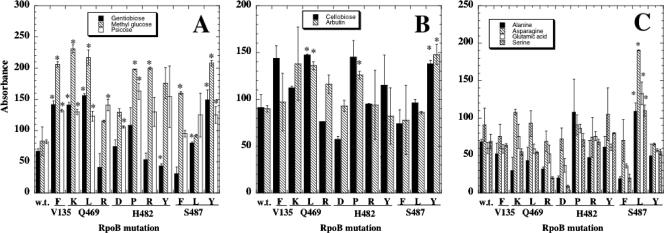
It was noted that the utilization of several substrates was significantly increased by Rifr mutants, in particular, β-glucosides such as gentiobiose (6-O-β-d-glucopyranosyl-d-glucose), cellobiose (4-O-β-d-glucopyranosyl-d-glucose), arbutin (4-hydroxyphenyl-β-d-glucopyranoside), and β-methyl-d-glucoside (Table 3; Fig. 3A and B). This apparent general up-regulation of β-glucoside utilization was also consistent with the observation of increased utilization of salicin [2-(hydroxymethyl)phenyl-β-d-glucopyranoside] (Table 2; Fig. 1B). Interestingly, increased utilization of these five β-glucosides was associated with the same subset of Rifr mutants (V135F, Q469K, Q469L, H482P, S487F, and S487Y mutants), and it is particularly notable that the Q469L and S487Y mutants significantly increased utilization of all five β-glucosides (Tables 2 and 3). The transport and utilization of β-glucosides in B. subtilis result from a complex network of genes which to date has been incompletely explored (15, 30, 32, 36). Up-regulation of utilization for multiple β-linked carbohydrates by Rifr mutants indicates that there may exist a common mechanism of transcriptional regulation of these genes. How might Rifr mutants play into this? Activation of the licRBCAH operon is mediated by the transcriptional activator LicR, and the bglPH and licT-bglS operons are regulated through the LicT antiterminator protein. The activities of both LicR and LicT are in turn modulated by PTS phosphorylation (14-17). It is well documented that Rifr mutations in the β subunit perturb interactions among prokaryotic RNA polymerases, regulatory proteins, and initiation/termination complexes (10, 13; reviewed in reference 37). It is thus reasonable to presume that changes in substrate utilization by Rifr mutants may result from altered interactions between RNA polymerase and small ligands, regulatory proteins, and/or regulatory RNA secondary structures.
FIG. 3.
Substrate utilization patterns in wt and Rifr mutant B. subtilis strains for substrates utilized weakly (50 to 100 U) by the wt. Asterisks denote significant differences (P < 0.05) from the wt. (A) Gentiobiose, β-d-methyl glucoside, and psicose. (B) Cellobiose and arbutin. (C) Amino acids l-alanine, l-asparagine, l-glutamic acid, and l-serine.
Gentiobiose and β-methyl-d-glucoside.
We found that the β-glucosides gentiobiose and β-methyl-d-glucoside were utilized weakly by the wt and that utilization was increased significantly in several Rifr mutants (V135F, Q469K/L, H482P/R/Y, and S487Y mutants) (Table 3; Fig. 3A). To our knowledge prior to this study these two sugars were considered nonutilizable growth substrates by B. subtilis. However, previous studies showed gentiobiose and β-methyl-d-glucoside to be powerful attractants for B. subtilis chemotaxis (24, 25), strongly suggesting that B. subtilis may indeed utilize these sugars. At present it is unknown whether gentiobiose and β-methyl-d-glucoside are transported and/or utilized by the bgl or lic gene products or if there exist specific genes coding for their transport and utilization. It should also be noted that a number of currently uncharacterized genes are present in the B. subtilis genome database encoding additional putative β-glucan and β-glucoside transport and utilization proteins which may have also been activated in Rifr mutants.
d-Psicose.
In 7 of the 11 Rifr mutants (V135F, Q469K/L/R, H482D/P, and S487Y mutants), we observed increased utilization of the sugar d-psicose (d-ribo-2-hexulose), a C-3 epimer of d-fructose (Table 3; Fig. 3A). d-Psicose is considered an ultralow-energy “rare sugar,” found in nature only in small quantities in agricultural products and commercially prepared carbohydrate complexes (11). Prior to this communication, B. subtilis had not been reported to utilize d-psicose. However, d-psicose utilization has recently been described to occur in a newly discovered Bacillus species, B. decisifrondis (38). This discovery of utilization of d-psicose in Rifr mutants of B. subtilis will certainly be of value in elucidating the genes involved in its uptake and utilization.
Amino acids.
Included as substrates in GP2 MicroPlates are 10 amino acids, amino acid analogs, and dipeptides. None of these compounds was utilized strongly by wt or Rifr mutant strains (Table 2). Furthermore, all strains were negative (<50 U) for utilization of d-alanine; the amino acid analogs N-acetyl glutamic acid, l-alaninamide, and l-pyroglutamic acid; and the dipeptides l-alanyl-glycine and glycyl-l-glutamic acid (Table 1). The amino acids l-alanine, l-asparagine, l-glutamic acid, and l-serine were utilized weakly by the wt (Table 3). Interestingly, only the S487L mutant was significantly increased in utilization of all four of these amino acids (Table 3; Fig. 3C). Which target genes for amino acid utilization might be affected by the S487L mutation? In B. subtilis, utilization of l-glutamate, l-asparagine, and l-alanine are controlled at the transcriptional level by the products of the rocR, ansR (1, 2, 34, 35), and alaR (communicated to the Subtilist database by A. L. Sonenshein) genes, respectively. Although none of the Rifr mutants led to amino acid auxotrophy on SMM plates (data not shown), the altered phenotypes conferred by the S487L mutant can be explored further to better understand regulation of amino acid utilization in B. subtilis.
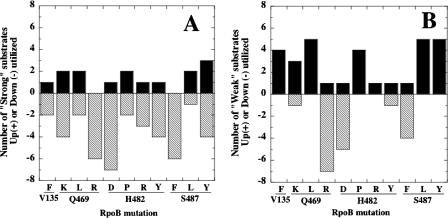
Patterns of substrate utilization by Rifr mutants.
We examined substrate utilization among the 11 Rifr mutants in an attempt to discern underlying patterns (Fig. 4). In general, compared to the wt, Rifr mutations led to a decrease in strongly utilized substrates (41/56) (Fig. 4A) and an increase in weakly utilized substrates (31/49) (Fig. 4B). Specific rpoB mutations exhibited pronounced stimulatory or inhibitory effects. For example, the Q469R, H482D, and S487F mutants affected utilization of 14, 14, and 11 substrates, respectively, and in the vast majority of cases (35/39) decreased substrate utilization, regardless of whether these substrates were used strongly or weakly by the wt (Fig. 4). In contrast, the Q469L, H482P, S487L, and S487Y mutants affected utilization of 9, 8, 8, and 12 substrates, respectively, and in the majority of cases (28/37) increased substrate utilization (Fig. 4). No consistent patterns could be discerned among allelic substitutions at specific residues. For example, mutation of Q469 to a basic R residue caused decreased utilization of most substrates (12/13), while mutation to the closely related basic K residue was as likely to lead to an increase (5/10) as a decrease (5/10) in substrate utilization (Fig. 4). Similarly, closely related aromatic amino acid substitutions at S487 exerted very distinct effects; the S487F mutant was decreased in 9 out of 10 substrates, while the S487Y mutant was increased in 8 out of 12 substrates (Fig. 4).
FIG. 4.
Summary of substrate utilization by Rifr mutants. (A) Substrates utilized strongly (>100 U) by the wt. (B) Substrates utilized weakly (50 to 100 U) by the wt. Numbers of substrates whose utilization was significantly increased (black bars) or decreased (hatched bars) relative to the wt (P < 0.05 by ANOVA) are shown above the corresponding mutation.
Results from earlier experiments suggested that in the absence of Rif selection, an uncharacterized set of mutations to Rifr in rpoB exerted a net reduction in B. subtilis fitness, as measured using competition experiments performed with rich brain heart infusion medium (9). Indeed, compared to the growth rate of the wt, we have also noted lowered growth rates of certain Rifr rpoB alleles (e.g., in the S487L mutant) but not others (e.g., in the Q469R and H482R/Y mutants) in rich liquid LB medium (19). However, in its soil environment B. subtilis would not likely be bathed in such rich sources of nutrients and would more likely need to rely on the ability to utilize ephemeral, rare, or less readily metabolized substrates. In particular, decomposing plant matter in soil is rich in β-glucosides released through breakdown of cellulose and hemicelluloses (38), and it was precisely these substrates which were utilized more effectively by Rifr mutants in our phenotypic profiles. Thus, it is entirely possible that under more realistic environmental conditions B. subtilis rpoB mutants may indeed exhibit greater fitness than the wt strain. Competition experiments to test this notion are under way.
In addition to new and testable insights gained into B. subtilis ecology and evolution, global phenotype profiling of Rifr B. subtilis mutants can be a useful technique for a number of postgenomic applications. First, as has been shown here with gentiobiose, β-methyl-d-glucose, and psicose, hitherto-unknown metabolic capabilities can be uncovered, leading to discovery of new genes encoding novel functions. In this exploratory study we used GP2 MicroPlates, which assay 95 substrates. However, the Omnilog instrument is designed to be used with a collection of 20 Biolog phenotype microarrary plates to test a total of 1,900 different phenotypes (6). Thus, testing our collection of Rifr mutants on the entire battery of phenotype microarray plates might be expected to uncover a large number of additional novel phenotypes, many of them likely to be encoded by genes of currently unknown or putative function. Second, Rifr mutants were shown in this study to be altered in the expression of a large number of already-known phenotypes. Elucidating the molecular details of how Rifr mutations in β lead to altered regulation of known target genes will certainly enhance our understanding of transcription regulation in general and could lead to the discovery of novel global regulatory circuitries. Both of the above-mentioned aims will necessarily require identification of the specific gene targets involved. Because Rifr mutations alter RNA polymerase function, comparison of global transcription patterns in wt versus specific Rifr mutants by use of transcriptional microarrays is the ideal next step to identify novel genes whose transcription are up- or down-regulated in particular Rifr mutants, for further molecular and metabolic analyses. These experiments are ongoing.
Acknowledgments
We thank Nobuo Munakata and Marion Hulett for the generous donation of strains, Mike Roberts and Jeffrey Carlson for kind assistance with the Omnilog instrument, and Rick Gourse and Tina Henkin for helpful discussions.
This work was supported by grants from NASA (NNA06CB58G) and the USDA-Florida Agriculture Experiment Station (FLA-MCS-04602) to W.L.N.
Footnotes
Published ahead of print on 20 July 2007.
REFERENCES
- 1.Belitsky, B. R., and A. L. Sonenshein. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 1806298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky, B. R., and A. L. Sonenshein. 1999. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 9610290-10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochner, B. R. 1989. Sleuthing out bacterial identities. Nature 339157-158. [DOI] [PubMed] [Google Scholar]
- 4.Bochner, B. R. 1989. “Breathprints” at the microbial level. ASM News 55536-539. [Google Scholar]
- 5.Bochner, B. R. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4309-314. [DOI] [PubMed] [Google Scholar]
- 6.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 111246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boor, K. J., M. L. Duncan, and C. W. Price. 1995. Genetic and transcriptional organization of the region encoding the beta subunit of Bacillus subtilis RNA polymerase. J. Biol. Chem. 27020329-20336. [DOI] [PubMed] [Google Scholar]
- 8.Boylan, R. J., N. Mendelson, D. Brooks, and F. E. Young. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J. Bacteriol. 110281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohan, F. M., E. C. King, and P. Zawadzki. 1994. Amelioration of the deleterious pleiotropic effects of an adaptive mutation in Bacillus subtilis. Evolution 4881-95. [DOI] [PubMed] [Google Scholar]
- 9a.Inaoka, T., K. Takahashi, H. Yada, M. Yoshida, and K. Ochi. 2004. RNA polymerase mutation activates the production of a dormant antibiotic 3,3′-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 2793885-3892. [DOI] [PubMed] [Google Scholar]
- 10.Ingham, C. J., and P. A. Furneaux. 2000. Mutations in the β subunit of the Bacillus subtilis RNA polymerase that confer both rifampicin resistance and hypersensitivity to NusG. Microbiology 1463041-3049. [DOI] [PubMed] [Google Scholar]
- 11.Izumori, K. 2002. Bioproduction strategy for rare hexose sugars. Naturwissenschaften 89120-124. [DOI] [PubMed] [Google Scholar]
- 12.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 20245-58. [DOI] [PubMed] [Google Scholar]
- 13.Jin, D. J., and C. A. Gross. 1991. RpoB8, a rifampicin-resistant termination-proficient RNA polymerase, has an increased Km for purine nucleotides during transcription elongation. J. Biol. Chem. 26614478-14485. [PubMed] [Google Scholar]
- 14.Krüger, S., and M. Hecker. 1995. Regulation of the putative bglPH operon for aryl-beta-glucoside utilization in Bacillus subtilis. J. Bacteriol. 1775590-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Coq, D., C. Lindner, S. Krüger, M. Steinmetz, and J. Stülke. 1995. New beta-glucoside (bgl) genes in Bacillus subtilis: the bglP gene product has both transport and regulatory functions similar to those of BglF, its Escherichia coli homolog. J. Bacteriol. 1771527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindner, C., A. Galinier, M. Hecker, and J. Deutscher. 1999. Regulation of the activity of the Bacillus subtilis antiterminator LicT by multiple PEP-dependent, enzyme I- and HPr-catalysed phosphorylation. Mol. Microbiol. 31995-1006. [DOI] [PubMed] [Google Scholar]
- 17.Lindner, C., M. Hecker, D. Le Coq, and J. Deutscher. 2002. Bacillus subtilis mutant LicT antiterminators exhibiting enzyme I- and HPr-independent antitermination affect catabolite repression of the bglPH operon. J. Bacteriol. 1844819-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maughan, H., V. Callicotte, A. Hancock, C. W. Birky, Jr., W. L. Nicholson, and J. Masel. 2006. The population genetics of trait deterioration in experimental populations of Bacillus subtilis. Evolution 60686-695. [PubMed] [Google Scholar]
- 19.Maughan, H., B. Galeano, and W. L. Nicholson. 2004. Novel rpoB mutations conferring rifampin resistance on Bacillus subtilis: global effects on growth, competence, sporulation, and germination. J. Bacteriol. 1862481-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. 1972. Experiments in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Munakata, N., T. Natsume, K. Takahashi, K. Hieda, C. Panitz, and G. Horneck. 2004. Mutagenesis of Bacillus subtilis spores exposed to simulated space environment, abstr. 04-A-00898. Abstr. 35th Committee Space Res. Sci. Assembly. Committee on Space Research, Paris, France.
- 22.Nicholson, W. L., and H. Maughan. 2002. The spectrum of spontaneous rifampin resistance mutations in the rpoB gene of Bacillus subtilis 168 spores differs from that of vegetative cells and resembles that of Mycobacterium tuberculosis. J. Bacteriol. 1844936-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Sussex, England.
- 24.Ordal, G. W., L. Márqez-Magaña, and M. J. Chamberlin. 1993. Motility and chemotaxis, p. 765-784. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology and molecular genetics. ASM Press, Washington, DC.
- 25.Ordal, G. W., D. P. Villani, and M. S. Rosendahl. 1979. Chemotaxis towards sugars by Bacillus subtilis. J. Gen. Microbiol. 118471-478. [DOI] [PubMed] [Google Scholar]
- 26.Paidhungat, M., and P. Setlow. 2002. Spore germination and outgrowth, p. 537-548. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 27.Perego, M., and J. A. Hoch. 2002. Two-component systems, phosphrelays, and regulation of their activities by phosphatases, p. 473-481. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 28.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 29.Qi, Y., and F. M. Hulett. 1998. PhoP∼P and RNA polymerase sigma A holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP∼P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 281187-1198. [DOI] [PubMed] [Google Scholar]
- 30.Reizer, J., S. Bachem, A. Reizer, M. Arnaud, M. H. Saier, and J. Stulke. 1999. Novel phosphotransferase system genes revealed by genome analysis—the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology 1453419-3429. [DOI] [PubMed] [Google Scholar]
- 31.Rothstein, D. M., C. L. Keeler, and A. L. Sonenshein. 1976. Bacillus subtilis RNA polymerase mutants temperature-sensitive for sporulation, p. 601-616. In R. L. Losick and M. Chamberlin (ed.), RNA polymerase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Setlow, B., R. M. Cabrera-Martinez, and P. Setlow. 2004. Mechanism of the hydrolysis of 4-methylumbelliferyl-beta-d-glucoside by germinating and outgrowing spores of Bacillus species. J. Appl. Microbiol. 961245-1255. [DOI] [PubMed] [Google Scholar]
- 33.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 441072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, D., and P. Setlow. 1991. Cloning, nucleotide sequence, and expression of the Bacillus subtilis ans operon, which codes for l-asparaginase and l-aspartase. J. Bacteriol. 1733831-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun, D., and P. Setlow. 1993. Cloning and nucleotide sequence of the Bacillus subtilis ansR gene, which encodes a repressor of the ans operon coding for l-asparaginase and l-aspartase. J. Bacteriol. 1752501-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tobisch, S., J. Stulke, and M. Hecker. 1999. Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the LicR regulator protein by site-directed mutagenesis. J. Bacteriol. 1814995-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trinh, V., M.-F. Langelier, J. Archambault, and B. Coulombe. 2006. Structural perspective on mutations affecting the function of multisubunit RNA polymerases. Microbiol. Mol. Biol. Rev. 7012-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, L., Z. Xu, and B. K. Patel. 2007. Bacillus decisifrondis sp. nov., isolated from soil underlying decaying leaf foliage. Int. J. Syst. Evol. Microbiol. 57974-978. [DOI] [PubMed] [Google Scholar]