Abstract
PhyR represents a novel alphaproteobacterial family of response regulators having a structure consisting of two domains; a predicted amino-terminal extracytoplasmic function (ECF) sigma factor-like domain and a carboxy-terminal receiver domain. PhyR was first described in Methylobacterium extorquens AM1, in which it has been shown to be essential for plant colonization, probably due to its suggested involvement in the regulation of a number of stress proteins. Here we investigated the PhyR regulon using microarray technology. We found that the PhyR regulon is rather large and that most of the 246 targets are under positive control. Mapping of transcriptional start sites revealed candidate promoters for PhyR-mediated regulation. One of these promoters, an ECF-type promoter, was identified upstream of one-third of the target genes by in silico analysis. Among the PhyR targets are genes predicted to be involved in multiple stress responses, including katE, osmC, htrA, dnaK, gloA, dps, and uvrA. The induction of these genes is consistent with our phenotypic analyses which revealed that PhyR is involved in resistance to heat shock and desiccation, as well as oxidative, UV, ethanol, and osmotic stresses, in M. extorquens AM1. The finding that PhyR is involved in the general stress response was further substantiated by the finding that carbon starvation induces protection against heat shock and that this protection is at least in part dependent on PhyR.
Aerial parts of plants are exposed to UV radiation, rapid changes in temperature, and low water and carbon availability. As a consequence, they have been described as a harsh environment (26). Members of the Alphaproteobacteria belonging to the genus Methylobacterium are ubiquitous and abundant on plant leaf surfaces (8, 18, 43). A previous study using Methylobacterium extorquens AM1 as a model organism led to identification of candidate proteins potentially involved in bacterial phyllosphere colonization (13). PhyR (for phyllosphere-induced regulator) is one of these proteins. Using a phyR deletion mutant strain, we demonstrated that PhyR is essential for plant surface colonization (13).
PhyR is composed of an amino-terminal effector domain that exhibits sequence identity with the family of extracytoplasmic function (ECF) sigma factors and a carboxy-terminal response regulator receiver domain. Proteome analysis revealed that different proteins related to stress responses were more abundant in a phyR-overexpressing strain than in a phyR deletion strain, suggesting that PhyR acts as a positive regulator. Among the proteins induced in the phyR-overexpressing strain were proteins involved in stress response, such as a heat shock protein (Hsp20), catalase (KatA), the DNA protection protein (Dps), and lactoylglutathione lyase (GloA) (13). These results, in addition to the prediction that PhyR orthologues are present in essentially all free-living Alphaproteobacteria (12, 13), suggest that PhyR not only is essential for epiphytic growth but also has a more global role in stress response physiology in Alphaproteobacteria.
In response to various stress or starvation stimuli, bacteria change their physiology to become more resistant to the present conditions and to future stress that they may encounter in their natural habitat. This so-called general stress response has been extensively studied in Escherichia coli and Bacillus subtilis, in which σS (RpoS) and σB (SigB), respectively, were identified as master regulators controlling the response (4, 11, 15, 24, 28). The signal transduction pathways of these key regulators have been intensively investigated, and their complexity has been demonstrated (for reviews, see references 14, 22, and 34). Pioneering conclusions that emerged from the study of σS and σB were successfully applied to a number of other bacteria that contain homologues of these two regulatory proteins, including the human pathogens Salmonella, Listeria, and Staphylococcus (25, 51, 52). Some bacterial taxa, such as the alpha subdivision of the Proteobacteria, however, lack orthologues of σS and σB genes (37). The general stress response has been described for some Alphaproteobacteria. It was notably shown that Rhizobium leguminosarum bv. phaseoli and Caulobacter crescentus acquire cross-resistance to various stresses upon starvation (36, 44); however, little is known about the underlying molecular basis.
In the present study, we investigated the PhyR regulon at the transcriptional level. The results led us to test and demonstrate the importance of PhyR in M. extorquens AM1 during various stress conditions and further supported our previous hypothesis that PhyR is involved in the general stress response of this member of the Alphaproteobacteria. The results are discussed with respect to the occurrence of phyR in other Alphaproteobacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. extorquens wild-type strain AM1 and strains ΔphyR::kanR and ΔphyR::kanR/pBG11 (13) were used in this study. The strains were grown at 28°C on minimal medium (MM) (10) containing 18.5 mM sodium succinate and 120 mM methanol.
RNA sample preparation.
Overnight cultures of the wild-type, ΔphyR::kanR, and ΔphyR::kanR/pBG11 strains were diluted 1/10 in fresh MM supplemented with methanol and succinate and grown to mid-exponential phase (optical density at 600 nm [OD600] = 1). For heat shock, 10-ml mid-exponential-phase cultures were incubated at 42 or 28°C (control) for 5 min prior to harvest. Then 0.1 volume of an ice-cold stop solution (5% phenol in ethanol) was added, and the cultures were harvested by centrifugation at 4,000 rpm for 10 min at 4°C. Total RNA was extracted using an RNeasy kit (Qiagen). Lysis was performed for 5 min by bead beating with a TissueLyser (Qiagen), using 0.1-mm zirconia beads (Biospect Products Inc.). After elution, SuperAsin was added at a final concentration of 1 U/μl. Total RNA was treated twice with DNase I (Ambion) using 0.5 U of DNase per μg of RNA for 1 h at 37°C. DNase-treated RNA was cleaned up with the RNeasy kit after each DNase treatment. The absence of contaminating DNA was checked by PCR. RNA concentrations were determined using a NanoDrop ND1000 spectrophotometer, and RNA integrity was determined using an Agilent 2100 bioanalyzer according to the manufacturer's instructions.
Labeling of hybridization probes, microarray hybridization, and analyses.
Sixty-mer oligonucleotide microarrays of M. extorquens AM1 were generated by Agilent Technologies (Agilent Technologies, Santa Clara, CA). They were based on the gapped sequence M. extorquens AM1 genome (6.5× coverage) (http://www.integratedgenomics.com/genomereleases.html#list6) and were designed and tested as described previously (30). Hybridization probe preparation, microarray hybridization, and scanning were performed by MOgene, LC, United States, as were basic data analyses. A SAM analysis (one class; default settings; false discovery route (FDR), 4.415904%) was performed with normalized data from three independent replicates. From this list, genes with an average change of at least twofold and an initial P value of <0.05 for each replicate were considered to be significantly differentially expressed. Differentially expressed genes were manually annotated.
RACE PCR.
5′ Rapid amplification of cDNA ends (RACE) PCR was performed using a 5′/3′ RACE kit (second generation; Roche) according to the manufacturer's instructions. Briefly, 2 μg of RNA from strains ΔphyR and ΔphyR/pBG11 for the RMQ09549, RMQ01575, RMQ02894, RMQ06760, RMQ05258, and RMQ12010 genes or from the wild type with and without heat shock treatment for the RMQ12794, RMQ08198, and RMQ08196 genes was used for cDNA synthesis. After poly(A) tailing, cDNA was used in a first PCR, and a nested PCR was performed when necessary (for the primers used, see Table S1 in the supplemental material). PCR products were either gel purified and sequenced or cloned into the TOPO cloning vector (Invitrogen) and transformed into TOP10 cells (Invitrogen). Selected plasmids were then sequenced.
Disk diffusion assays.
Bacteria were grown in MM to an OD600 of 1. Bacterial suspensions (5 ml) were then mixed with 100 ml of 42°C prewarmed MM soft agar (0.75% agar), and 4-ml portions of this mixture were poured on solid MM. Disks were placed at the center of the plates, and aliquots (5 μl) of 5.55 M methylglyoxal, 10 M hydrogen hydroperoxide, or 12 M formaldehyde were deposited on the filter disks. The diameters of growth inhibition areas were measured after incubation at 28°C for 3 days.
Heat shock resistance assays.
Bacteria were grown in 20 ml of MM in 100-ml flasks to an OD600 of 1. The flasks were then transferred to 55°C, and 100-μl aliquots were collected before and after 20, 30, and 40 min of heat exposure. The suspensions were diluted and plated, and colonies were counted after 3 days of incubation at 28°C. For starvation experiments, cultures that reached an OD of 1 were centrifuged to collect the cells. Cells were washed once with 20 ml of carbon-free MM and resuspended in the same solution. Bacteria were then incubated in flasks overnight at 28°C with shaking before transfer to 55°C.
Desiccation resistance assays.
Dilution series of exponentially growing cells that reached an OD600 of 1 were spotted on the surface of cellulose filters (type HA; 0.45 μm; Millipore, United States). Filters were then briefly dried for 1 min (control) or for 3 days under a sterile airflow in a hood before they were placed on MM plates.
UV resistance assays.
Dilution series of exponentially growing cell cultures that reached an OD600 of 1 were plated. The plates were exposed to 254-nm UV light (400 μW per cm2) using an LF-206.LS lamp (UVITEC, United Kingdom) for 30 s (the control was not exposed to UV light). The CFU on plates were counted after 3 days of incubation at 28°C.
Microarray data accession number.
The microarray data have been deposited in the GEO database.
RESULTS
Identification and analysis of the PhyR regulon.
In our previous study, we identified a set of genes that are regulated by PhyR via proteome analysis using two-dimensional gel electrophoresis (13). Since this analysis was not exhaustive and the structure prediction suggested that PhyR may act as a transcriptional regulator, we investigated the influence of PhyR on the transcriptome of M. extorquens AM1 using microarray technology (30). To do this, we compared the transcriptional profile of a phyR overexpression strain with that of a phyR deletion strain as described previously, assuming that PhyR overproduction mimics PhyR activation and in order to avoid any basal activity (13). In total, we found 246 genes whose levels of expression were significantly different in the two strains (see Table S2 in the supplemental material). The majority of these genes (229 genes) were upregulated in the phyR-overexpressing strain, reinforcing the hypothesis that PhyR acts as a transcriptional activator. Genes differentially expressed at the transcriptional level encoded 19 of the 42 gene products previously identified at the protein level. The level of a 20th gene, encoding a putative superoxide dismutase (RMQ02531), was just below the threshold level used in this study. This overlap is in agreement with the results obtained in comparable studies (40, 49).
Of the 17 downregulated genes in the phyR-overexpressing strain, 11 code for hypothetical proteins and 3 code for putative sensor or regulatory proteins (see Table S2 in the supplemental material). As mentioned above, the majority of PhyR-regulated genes were induced. About one-half of these genes encode hypothetical proteins; about 60 encode proteins involved in metabolism, and 11 encode putative proteins involved in sensing and regulation. Interestingly, we found several genes that are typically associated with stress responses (Table 1). The katE gene is one of these genes. katE encodes a protein that is homologous (42% identity) to the RpoS-regulated E. coli catalase KatE (38) that detoxifies hydrogen peroxide, a reactive oxygen species produced during aerobic metabolism. The htrA, ibpA, grpE, dnaK, and cbpA genes are induced in the phyR-overexpressing strain. These genes encode chaperone/cochaperone proteins or proteases which have been found to be important for resistance to stress, such as heat shock and oxidative stress, in other bacteria (21, 27, 42). An rpoH gene is present in the phyR regulon; in E. coli, this gene encodes an alternative sigma factor necessary for growth at elevated temperatures (53). Other genes involved in resistance to oxidative stress (dps [29] and osmC [7]), UV radiation (uvrA [45] and dps [29]), and osmotic stress (mscL [2]) belong to the regulon, as do two gloA homologues which encode lactoylgluthatione lyase. This enzyme is involved in methylglyoxal conversion; it detoxifies this electrophilic compound that accumulates during metabolism imbalance and damages DNA and proteins (3). Other genes that have unknown functions but typically are induced by various stresses, such as csbD and yciF (1, 17), are also among the PhyR-induced genes (Table 1).
TABLE 1.
Subset of genes upregulated in phyR-overexpressing strain
| Gene | Gene homolog | Function | Change (fold)a | Promoterb |
|---|---|---|---|---|
| Stress responses | ||||
| RMQ09549 | katE | Catalase | 10.2 | TGCAGAAACGAATGGCTCCAAGCTCGGCCTCACGCGTTGGGGCGGCAACG-N64-ATG |
| RMQ05258 | dps | DNA protection during starvation | 12.1 | GCCATGCGGAGCGGAACAAAGCTTCCCGCTTCCTCATTCTGCCGATCGAG-N59-ATG |
| RMQ06760 | osmC | Osmotically inducible, OsmC/Ohr family | 11.3 | GGCTGCCCCTCGACATGACCGTCCACCGCTCCACGTCGAAGGCCACGCGG-N34-ATG |
| RMQ05835 | csbD | Unknown | 6.3 | CATCATAGGAGTGGAACCAAGCGCTTGGGGCTCCCGTTCAGTTTGCCGAG-N42-ATG |
| RMQ06983 | grpE | Hsp70 cofactor | 3.2 | TCAGCACCGCCGGGAACGGCGGCACATATTCGCGAATTATTGACGCGCCT-N120-ATGc |
| RMQ06982 | dnaK | Chaperone Hsp70 | 2.5 | —c |
| RMQ00525 | cbpA | Curved DNA-binding protein, DnaJ family | 2.3 | —c |
| RMQ05310 | ibpA | Small heat shock protein | 2.8 | TTCAAAAAATTGGGAACCGGACTCC-TTTGGCCGCGTTACGCGCGTGCGA-N85-ATG |
| RMQ09060 | htrA | Serine protease | 3.5 | ND |
| RMQ12030 | uvrA | DNA excision repair enzyme subunit | 2.1 | ND |
| RMQ09593 | mscL | Mechanosensitive channel | 5.1 | ND |
| RMQ01575 | gloA | Lactoylglutathione lyase | 4.9 | GTCGCCCCGTTGACGGGTTTGCGTGCCGGGCCAGATAGCCGGACATCCCT-N37-ATG |
| RMQ02894 | gloA | Lactoylglutathione lyase | 6.1 | CAAAAACCCTTTCCGCCCGGCCGGACTGCGCCAGATGGGCGCTCATCCAC-N29-ATG |
| RMQ01469 | yciF | Unknown | 3.2 | CTAACTCTGTCAGGAACGTCGGCAGA-AGTTGGAGGTTGGCCGGAGGAGC-N246-ATG |
| RMQ10257 | yciF | Unknown | 4.0 | ND |
| RMQ10020 | yciF | Unknown | 6.5 | GGCTGGCATATCGGAACCGCCGCAGCGCCGTCGGGATTCCCCTGTGATGG-N43-ATG |
| Carotenoid biosynthesis | ||||
| RMQ04997 | ctrQ | Glycosyl transferase involved in synthesis of the carotenoid staphyloxanthin | 2.2 | ND |
| RMQ04999 | crtI | Phytoen desaturase | 2.2 | ND |
| Metabolism | ||||
| RMQ06018 | fdh | NAD(P)-dependent formaldehyde dehydrogenase | 8.4 | ND |
| Regulation | ||||
| RMQ12010 | rpoH | RNA polymerase sigma 32 factor | 9.0 | ACGCGCCCGTAGGGAACAAGCGTCG-CACGGATGTGTTGTGCCTCTGCCA-N190-TTG |
| RMQ09061 | rpoE | RNA polymerase ECF-type sigma factor | 2.7 | ND |
| RMQ08139 | Regulatory protein, Crp family | 24.6 | GACACCCTTGCGGGAACATCGGTCC-GCAGGGGCGGTTTCTCGCAAAACT-N47-ATG | |
| phyR region | ||||
| RMQ12794 | Conserved hypothetical putative membrane protein | 22.3 | GGAAACGCAAAAGGAACCTGCGGCGAAAACTTGGCGTTATTGCTTCCAAG-N45-ATG | |
| RMQ08196 | Hypothetical protein, putative exported protein | 4.5 | CGGGACAGTGAAGGAACCGAAGATCTGAAAGCAGCGTTGTGATGACAACC-N28-ATG | |
| RMQ08197 | Hypothetical protein | 10.5 | AGAGCCAATTTCGGAACCTGGCCAAT-GGTCATTCGTTAGAGAATCACCCd | |
| Homologs of RMQ12794 | ||||
| RMQ05048 | Conserved hypothetical putative membrane protein | 10.1 | CGGGCGCTTCCCGGAACGCTTCGCTCCCGGCAACTGTTTTCAGCGCCATC-N96-ATG; AGCGCCATCGACGGAACGATTCCGGCAGCTTG GCAATTTCTCAGCCAGGA-N55-ATGe | |
| RMQ01992 | Conserved hypothetical putative membrane protein | 6.1 | GCAGGCCGCGACGGAACGCCGCCGGCTCGTTGGCCGTTACTGCTCCGCTT-N20-ATG |
Average changes in three experiments (Q value, <0.05).
Putative promoters, with their distances to the start codon. Bold type indicates a transcription start site that was mapped by 5′RACE PCR. Putative ECF −35 and −10 elements are double underlined, and putative σ32 −35 and −10 elements are underlined. ND, not detected.
RMQ06983, RMQ06982, and RMQ00525 are probably part of the same operon.
For RMQ08197, the consensus was found in the CDS.
Two putative ECF promoters were found for RMQ05048.
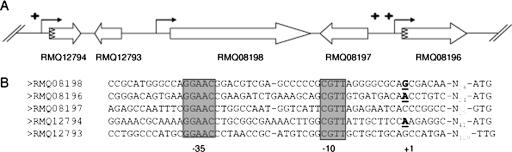
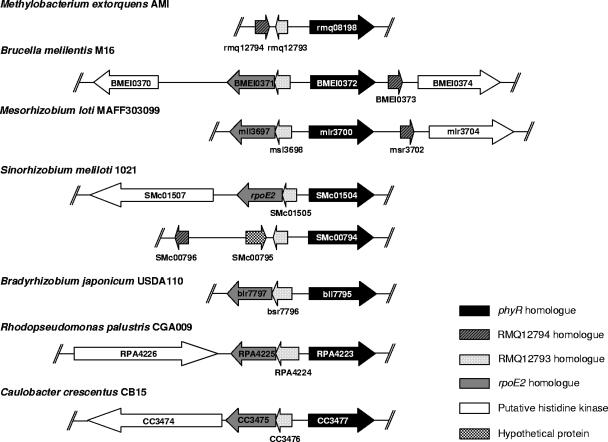
The following three genes located in the phyR region on the M. extorquens AM1 chromosome are upregulated in the overexpressing strain: RMQ12794, RMQ08197, and RMQ08196, which encode hypothetical proteins (Table 1 and Fig. 1). RMQ12794 is highly conserved in Alphaproteobacteria and colocalizes with phyR in some bacteria of this group (see below). The M. extorquens AM1 genome contains two gene paralogues of RMQ12794, RMQ05048 and RMQ01992. Interestingly, all three paralogues were found to be induced in a PhyR-dependent way (Table 1).
FIG. 1.
(A) Organization of the M. extorquens AM1 phyR region. Genes are represented by large open arrows. Transcript start sites of phyR (RMQ08198), RMQ08196, and RMQ12794 were identified by RACE and are indicated by bent arrows. Putative signal peptides are represented by a zigzag symbol. Genes induced in the phyR-overexpressing strain are labeled with a plus sign at the 5 prime end. (B) Alignment of promoter regions. Conserved −35 and −10 elements are indicated by shading. Transcription start sites identified experimentally are in bold type and underlined. Distances to the start codon are indicated.
In order to identify common features in the promoter region of the genes of the PhyR regulon, we mapped the transcriptional start sites of nine PhyR-activated genes. These genes were selected based on their typical stress-related functions or their colocalization with phyR on the chromosome. We used the same RNA sample preparation that was used for microarray experiments for all genes except phyR itself and genes located in the same region, for which we used RNA extracted from the wild-type strain due to the eventual disturbance of the region caused by the phyR deletion. RACE was successfully used to determine the transcriptional start sites of these genes, and the results are shown in Table 1 and Fig. 1. One of these genes (RMQ09549) did not exhibit any recognizable promoter, and three of them (RMQ02894, RMQ01575, and RMQ06760) had a −35 element exhibiting some similarities to σ32- or σ70-dependent promoters (see Fig. S1 in the supplemental material). Five genes (RMQ05258, RMQ12010, RMQ12794, RMQ08196, and RMQ08198 [phyR]) have a typical ECF-type promoter with the consensus sequence GGAAC-N16,17-CGTT (23, 39) (Table 1 and Fig. 1). Two of these genes, RMQ12794 and RMQ08196, are present in the phyR region. We then searched for this ECF-type promoter sequence upstream of all PhyR-induced genes. We localized this strikingly conserved element upstream of a total of 102 genes (81 transcriptional units) (see Fig. S2 in the supplemental material).
PhyR is involved in resistance to various stresses.
Microarrays and proteomic analyses revealed that PhyR upregulates a number of genes that have been shown to be involved in stress resistance in different organisms. This prompted us to evaluate the importance of PhyR for resistance to various stress conditions. The involvement of phyR in resistance to hydrogen peroxide, methylglyoxal, heat shock, ethanol, osmotic stress, UV radiation exposure, and desiccation was evaluated and is described below.
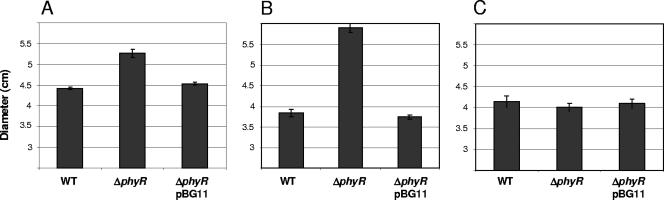
The induction of a predicted catalase and lactoylglutathione lyase in the phyR-overexpressing strain at the protein level as well as the transcriptional level (13) (Table 1) led us investigate the eventual importance of PhyR in resistance to hydrogen peroxide and methylglyoxal, respectively. Resistance to formaldehyde was also tested, as a putative glutathione-dependent formaldehyde dehydrogenase was identified in the PhyR regulon (13) (Table 1). Disk diffusion assays were used to evaluate the relevance of PhyR. The phyR deletion strain was found to be more sensitive to hydrogen peroxide and methylglyoxal than the wild type and the complemented strain. However, no significant difference was observed for formaldehyde (Fig. 2). This is not surprising, as conversion via the tetrahydromethanopterin-dependent pathway is the principal route used for detoxification of this toxic metabolite (5, 6, 48).
FIG. 2.
Hydrogen peroxide, methylglyoxal, and formaldehyde resistance of the M. extorquens wild-type AM1, ΔphyR, and complemented ΔphyR/pBG11 strains as determined by disk diffusion assays. Exponential-phase cells were mixed with MM soft agar and overlaid onto MM agar plates. Disks (diameter, 5 mm) were placed on each plate, 5 μl of 10 M H2O2 (A), 40% methylglyoxal (B), or 12 M formaldehyde (C) was added to each disk, and the plates were incubated at 28°C for 3 days. The data indicate the diameters of the inhibition halos and are the means for three plates for each strain from three independent experiments; the error bars indicate the standard errors of the means. WT, wild type.
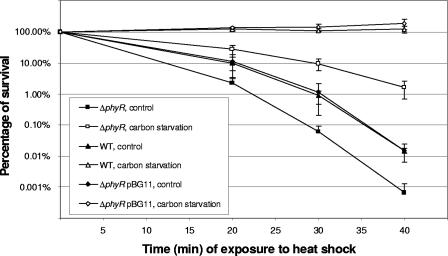
The presence of homologues of rpoH, htrA, and dnaK in the phyR regulon led us evaluate the importance of phyR during heat shock, as these genes are known to be involved in resistance to this stress in other bacteria (27, 42, 53). The levels of survival of the three test strains were compared after different periods of incubation at 55°C. The ΔphyR strain lost viability more rapidly over time than the wild type and the complemented strain. For example, the proportion of surviving cells of the phyR mutant strain was 10-fold lower than the proportion of surviving cells of the wild type or the complemented strain after 40 min of exposure to elevated temperatures (Fig. 3) (see below), clearly demonstrating the importance of phyR for heat shock resistance.
FIG. 3.
Thermal resistance of the M. extorquens wild-type AM1, ΔphyR derivative, and complemented ΔphyR/pBG11 strains. Cultures were grown at 28°C in MM containing methanol and succinate to an OD600 of 1. They were then directly challenged at 55°C or subjected to overnight carbon starvation before heat shock. Viability was determined by plating on solid minimal medium. WT, wild type.
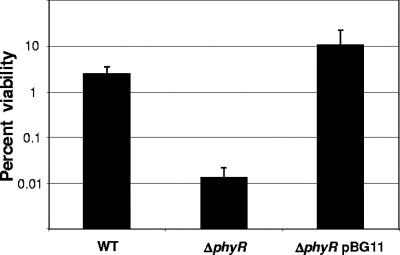
The data for a few genes present in the PhyR regulon, including homologues of dps and uvrA, suggest that these genes could be involved in UV light resistance,. These two genes encode a DNA binding protein induced during starvation and a subunit of the UvrABC complex that is responsible for DNA lesion recognition and repair, respectively (29, 45). Although no differences between the three strains were observed on the unexposed control plate, the proportion of the ΔphyR mutant cells able to form colonies after UV exposure was strongly affected and was found to be 100 times lower than the proportions of the wild-type and complemented strain cells (Fig. 4). This also shows the importance of PhyR for protection against UV light. Intriguingly, we noted that the phyR mutant strain is less pigmented than the wild type, as judged by the less intense pink color (Fig. 5). Diminished pigmentation of the mutant compared to the wild type might be explained by the presence of RMQ04997 and RMQ04999 in the phyR regulon (Table 1). These two genes encode a putative homologue of Staphylococcus aureus CtrQ, which is a glycosyl transferase involved in the synthesis of the carotenoid staphyloxanthin (32), and a phytoen desaturase known to be essential for pigmentation in M. extorquens AM1 (46), respectively. Carotenoids are known to be major scavenger molecules protecting against radiation and oxidative stress (41). Thus, their production is linked directly to stress resistance. It is therefore interesting to find that pigmentation seems to be at least partially controlled by PhyR in this phyllosphere colonizer.
FIG. 4.
UV light tolerance of the M. extorquens wild-type AM1, ΔphyR, and complemented ΔphyR pBG11/strains. Cell viability was tested by spotting dilution series of cells in exponential growth phase and exposing them to UV light (254 nm) for 30 s. WT, wild type.
FIG. 5.
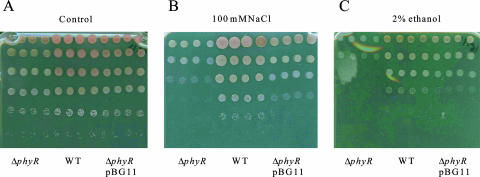
Salt and ethanol tolerance of the M. extorquens wild-type AM1, ΔphyR, and complemented ΔphyR/pBG11 strains. Dilution series of cells in exponential growth phase were plated on MM (control) (A) or MM containing 100 mM NaCl (B) or 2% ethanol (C). WT, wild type.
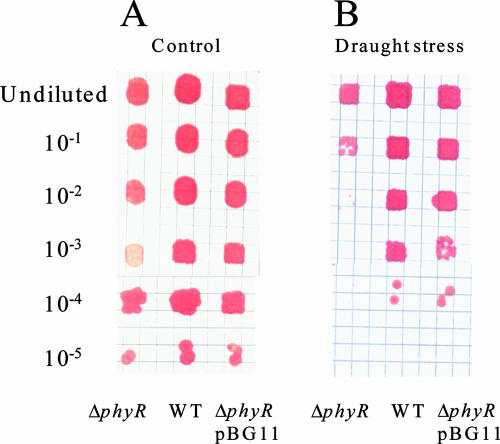
Besides radiation, desiccation is a stress that is encountered in the phyllosphere. We therefore determined whether the phyR mutant has a defect in drought tolerance. We compared the resistance levels of the phyR deletion strain and the wild-type and complemented strains under drought conditions. To do this, we determined the number of bacteria able to form colonies after exposure on filters to a sterile airflow for 3 days. In a control experiment, filters were placed directly on agar plates and incubated. No difference between strains was observed on untreated filters. However, the desiccation experiment revealed that the phyR mutant was more sensitive than the wild type and the complemented strain, showing a >100-fold reduction in viability (Fig. 6).
FIG. 6.
Desiccation test with the M. extorquens wild-type AM1, ΔphyR, and complemented ΔphyR/pBG11 strains. Cell viability was tested by spotting dilution series of cells in exponential growth phase on filter paper. Filters were laid on solid minimal medium immediately after a short drying period (control) (A) or after 3 days under a sterile airflow (B). WT, wild type.
In order to determine whether PhyR is involved in salt and ethanol tolerance, we compared the abilities of all three strains to grow on agar plates in the presence of 100 mM NaCl or 2% ethanol. No growth differences were observed on control medium. However, growth of the ΔphyR strain was delayed compared to growth of the wild type in the presence of NaCl or ethanol; after 3 days of incubation the ΔphyR strain was hardly visible (Fig. 5).
In Gammaproteobacteria, σS, the master gene regulator involved in the general stress response, is responsible for resistance to multiple stresses. It is also responsible for the capacity of cells to become more resistant to stress after a sublethal stress treatment (24, 28). As phyR plays a role in resistance to different stresses and no general stress response has been described so far in Methylobacterium, we decided to test if PhyR is involved in preadaptation to stress. We found that overnight carbon starvation completely protected the wild type against an elevated temperature, whereas nonpretreated cells gradually lost viability over time during heat shock. After 40 min, only 0.02% of the cells remained viable (Fig. 3). As described above, the phyR deletion mutant was less viable than the wild type during heat shock. After starvation pretreatment, a protective effect could also be observed for the phyR mutant strain, although it was incomplete (Fig. 3), which was indicative of partial involvement of PhyR in this cross-protection test.
DISCUSSION
In the present study, we investigated the PhyR regulon of M. extorquens AM1 using microarray technology. We found that PhyR positively regulates 229 genes, including a conspicuous number of genes encoding predicted stress proteins. This finding is consistent with our phenotypic analysis that revealed that PhyR is involved in resistance to heat shock, desiccation, UV radiation, oxidative, and osmotic stresses. The multitude of stress resistance responses associated with PhyR and our demonstration that PhyR is involved in cross-protection provide striking parallels with the insights that have been obtained for σS and σB and have resulted in the concept of master regulators of the general stress response in Gammaproteobacteria (in particular, E. coli) and gram-positive bacteria (in particular, B. subtilis), respectively. Similar phenotypic observations regarding PhyR and these global regulators of the general stress response were made. Both σS and σB have been shown to be involved in heat shock, ethanol, oxidative, and osmotic stress (11, 15, 24, 28, 47). Also, the regulons of these master regulators have been described to be large and to share common key genes involved in the stress response, such as katE, dps, and osmC, which are also part of the PhyR regulon (31, 33, 35, 50). The significance of these master regulators of general stress in the natural environment of bacteria has been widely discussed (14, 16), and it is interesting to note that we demonstrated the importance of PhyR for the phyllosphere lifestyle of M. extorquens AM1 in our previous study (13). Considering all the data, we concluded that PhyR is involved in the general stress response of this member of the Alphaproteobacteria.
The conclusion that PhyR is involved in the general stress response, however, is of interest not only for M. extorquens. The master regulators σS and σB do not have homologues in the alpha subdivision of the Proteobacteria (37). Since PhyR orthologues are present in essentially all free-living Alphaproteobacteria and PhyR is restricted to this taxon, we may speculate that PhyR is important for stress resistance in other Alphaproteobacteria as well (13). The fact that phyR homologues are induced in response to various stressful conditions in other Alphaproteobacteria (9, 19, 20, 39) is in agreement with this assumption.
Although we have strong indications that PhyR is involved in the general stress response of M. extorquens AM1, so far it is not known how PhyR itself is regulated. Furthermore, it is currently not known how PhyR activates its targets. Intriguingly, PhyR contains a predicted ECF sigma factor-like domain, and whether PhyR acts as a bona fide sigma factor is a question that remains to be answered (13). We identified a conserved promoter motif for ECF sigma factors upstream of about one-third of the 248 genes regulated by PhyR, which could be in agreement with the hypothesis that PhyR recognizes these sequences. However, the possibility that PhyR may not function as a sigma factor or a DNA-binding protein and the possibility that the protein is indirectly involved in gene regulation should also be considered. In this context, it is worth noting that we identified an ECF sigma factor as part of the PhyR regulon. This might be an alternative candidate involved in promoter recognition of at least some of the PhyR-regulated genes. Additional studies have to be performed to discriminate between these hypotheses. Interestingly, an ECF sigma factor encoded by rpoE2, located next to SMc01504, a phyR-like gene in Sinorhizobium meliloti (Fig. 7), has recently been found to act as a general stress response regulator since many typical stress genes are under rpoE2 control (39). No stress resistance defect in an rpoE2 mutant strain could be demonstrated (39). Therefore, questions regarding general stress response regulation remain open and require more in-depth analysis. It is, however, striking that the same ECF-type promoter was described for genes in both regulons. Taken together, phyR-like genes and rpoE2 homologues are prime candidates for further investigation of the general stress response in Alphaproteobacteria. In addition, our study and that of Sauviac et al. (39) provide circumstantial evidence that additional genes in the phyR genome region might be important in the same signaling pathway. Interestingly, there is a microsyntheny at the phyR locus of Alphaproteobacteria; a conserved small open reading frame followed by an rpoE2 homologue is present in the opposite direction and upstream of phyR-like genes (Fig. 7). Also, one or two histidine kinases are associated with phyR-like genes in various Alphaproteobacteria (12) (Fig. 7). With respect to the genomic context, the genus Methylobacterium appears to be an exception, since no ECF sigma factor and no histidine kinase-encoding genes are located close to phyR. Elucidating the PhyR signaling pathway that enables the cell to cope with multiple stresses and determining whether its regulation is conserved in other Alphaproteobacteria remain major objectives.
FIG. 7.
Genetic organization of the phyR locus in different Alphaproteobacteria. The gene tags and scale are based on data deposited in the Uniref public database, with the exception of the two CDS located upstream of BMEI0372 and SMc0794, which show 38 and 31% amino acid identity with RMQ12793, respectively, and are not annotated as genes in the Uniref database.
Supplementary Material
Acknowledgments
This work was supported by ETH Zurich and by a fellowship from the French Ministère de l'Enseignement Supérieur et de la Recherche to B.G.
We thank Elizabeth Skovran, University of Washington, for valuable advice on RNA preparation and Claude Bruand, Laboratoire des Interactions Plantes-Microorganismes, for critical reading of the manuscript.
Footnotes
Published ahead of print on 16 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akbar, S., S. Y. Lee, S. A. Boylan, and C. W. Price. 1999. Two genes from Bacillus subtilis under the sole control of the general stress transcription factor σB. Microbiology 1451069-1078. [DOI] [PubMed] [Google Scholar]
- 2.Booth, I. R., M. D. Edwards, S. Black, U. Schumann, and S. Miller. 2007. Mechanosensitive channels in bacteria: signs of closure? Nat. Rev. Microbiol. 5431-440. [DOI] [PubMed] [Google Scholar]
- 3.Booth, I. R., G. P. Ferguson, S. Miller, C. Li, B. Gunasekera, and S. Kinghorn. 2003. Bacterial production of methylglyoxal: a survival strategy or death by misadventure? Biochem. Soc. Trans. 311406-1408. [DOI] [PubMed] [Google Scholar]
- 4.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J. Bacteriol. 1757931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chistoserdova, L., S.-W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 1852980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science 28199-102. [DOI] [PubMed] [Google Scholar]
- 7.Conter, A., C. Gangneux, M. Suzanne, and C. Gutierrez. 2001. Survival of Escherichia coli during long-term starvation: effects of aeration, NaCl, and the rpoS and osmC gene products. Res. Microbiol. 15217-26. [DOI] [PubMed] [Google Scholar]
- 8.Corpe, W. A., and S. Rheem. 1989. Ecology of methylotrophic bacteria on living leaf surfaces. FEMS Microbiol. Ecol. 62243-250. [Google Scholar]
- 9.Dominguez-Ferreras, A., R. Perez-Arnedo, A. Becker, J. Olivares, M. J. Soto, and J. Sanjuan. 2006. Transcriptome profiling reveals the importance of plasmid pSymB for osmoadaptation of Sinorhizobium meliloti. J. Bacteriol. 1887617-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fulton, G. L., D. N. Nunn, and M. E. Lidstrom. 1984. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J. Bacteriol. 160718-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaidenko, T. A., and C. W. Price. 1998. General stress transcription factor σB and sporulation transcription factor σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 1803730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galperin, M. Y. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 1884169-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gourion, B., M. Rossignol, and J. A. Vorholt. 2006. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc. Natl. Acad. Sci. USA 10313186-13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecker, M., J. Pané-Farré, and U. Völker. 2007. SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria. Annu. Rev. Microbiol. 61215-236. [DOI] [PubMed] [Google Scholar]
- 15.Hecker, M., and U. Völker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the σB regulon. Mol. Microbiol. 291129-1136. [DOI] [PubMed] [Google Scholar]
- 16.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 17.Hindupur, A., D. Liu, Y. Zhao, H. D. Bellamy, M. A. White, and R. O. Fox. 2006. The crystal structure of the E. coli stress protein YciF. Protein Sci. 152605-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano, S. S., and C. D. Upper. 1991. Bacterial community dynamics, p. 271-294. In J. H. Andrews and S. S. Hirano (ed.), Microbial ecology of leaves. Springer, New York, NY.
- 19.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 1861448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, P., E. L. Brodie, Y. Suzuki, H. H. McAdams, and G. L. Andersen. 2005. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J. Bacteriol. 1878437-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagawa, M., Y. Matsumura, and T. Tsuchido. 2000. Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol. Lett. 184165-171. [DOI] [PubMed] [Google Scholar]
- 22.Klauck, E., A. Typas, and R. Hengge. 2007. The σS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci. Prog. 90103-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane, W. J., and S. A. Darst. 2006. The structural basis for promoter −35 element recognition by the group IV sigma factors. PLoS Biol. 4e269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 549-59. [DOI] [PubMed] [Google Scholar]
- 25.Lee, I. S., J. Lin, H. K. Hall, B. Bearson, and J. W. Foster. 1995. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17155-167. [DOI] [PubMed] [Google Scholar]
- 26.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 691875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 4493-140. [DOI] [PubMed] [Google Scholar]
- 28.McCann, M. P., J. P. Kidwell, and A. Matin. 1991. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J. Bacteriol. 1734188-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair, S., and S. E. Finkel. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 1864192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okubo, Y., E. Skovran, X. Guo, D. Sivam, and M. E. Lidstrom. 2007. Implementation of microarrays for Methylobacterium extorquens AM1. Omics 11325-340. [DOI] [PubMed] [Google Scholar]
- 31.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272580-591. [DOI] [PubMed] [Google Scholar]
- 32.Pelz, A., K. P. Wieland, K. Putzbach, P. Hentschel, K. Albert, and F. Götz. 2005. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 28032493-32498. [DOI] [PubMed] [Google Scholar]
- 33.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 1835617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria, p. 179-198. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress response. ASM Press, Washington, DC.
- 35.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41757-774. [DOI] [PubMed] [Google Scholar]
- 36.Rava, P. S., L. Somma, and H. M. Steinman. 1999. Identification of a regulator that controls stationary-phase expression of catalase-peroxidase in Caulobacter crescentus. J. Bacteriol. 1816152-6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roop, R. M., II, J. M. Gee, G. T. Robertson, J. M. Richardson, W. L. Ng, and M. E. Winkler. 2003. Brucella stationary-phase gene expression and virulence. Annu. Rev. Microbiol. 5757-76. [DOI] [PubMed] [Google Scholar]
- 38.Sak, B. D., A. Eisenstark, and D. Touati. 1989. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc. Natl. Acad. Sci. USA 863271-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauviac, L., H. Philippe, K. Phok, and C. Bruand. 2007. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J. Bacteriol. 1894204-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu, T., K. Shima, K. Yoshino, K. Yonezawa, T. Shimizu, and H. Hayashi. 2002. Proteome and transcriptome analysis of the virulence genes regulated by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 1842587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sies, H., and W. Stahl. 2004. Carotenoids and UV protection. Photochem. Photobiol. Sci. 3749-752. [DOI] [PubMed] [Google Scholar]
- 42.Skorko-Glonek, J., D. Zurawa, E. Kuczwara, M. Wozniak, Z. Wypych, and B. Lipinska. 1999. The Escherichia coli heat shock protease HtrA participates in defense against oxidative stress. Mol. Gen. Genet. 262342-350. [DOI] [PubMed] [Google Scholar]
- 43.Sy, A., A. C. Timmers, C. Knief, and J. A. Vorholt. 2005. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ. Microbiol. 717245-7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thorne, S. H., and H. D. Williams. 1997. Adaptation to nutrient starvation in Rhizobium leguminosarum bv. phaseoli: analysis of survival, stress resistance, and changes in macromolecular synthesis during entry to and exit from stationary phase. J. Bacteriol. 1796894-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truglio, J. J., D. L. Croteau, B. Van Houten, and C. Kisker. 2006. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 106233-252. [DOI] [PubMed] [Google Scholar]
- 46.Van Dien, S. J., C. J. Marx, B. N. O'Brien, and M. E. Lidstrom. 2003. Genetic characterization of the carotenoid biosynthetic pathway in Methylobacterium extorquens AM1 and isolation of a colorless mutant. Appl. Environ. Microbiol. 697563-7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 1813942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vorholt, J. A. 2002. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch. Microbiol. 178239-249. [DOI] [PubMed] [Google Scholar]
- 49.Wan, X. F., N. C. Verberkmoes, L. A. McCue, D. Stanek, H. Connelly, L. J. Hauser, L. Wu, X. Liu, T. Yan, A. Leaphart, R. L. Hettich, J. Zhou, and D. K. Thompson. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J. Bacteriol. 1868385-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 1871591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 1803650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 1786036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, Y. N., N. Kusukawa, J. W. Erickson, C. A. Gross, and T. Yura. 1988. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor sigma 32. J. Bacteriol. 1703640-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.