Abstract
A spermidine excretion protein in Escherichia coli was looked for among 33 putative drug exporters thus far identified. Cell toxicity and inhibition of growth due to overaccumulation of spermidine were examined in an E. coli strain deficient in spermidine acetyltransferase, an enzyme that metabolizes spermidine. Toxicity and inhibition of cell growth by spermidine were recovered in cells transformed with pUCmdtJI or pMWmdtJI, encoding MdtJ and MdtI, which belong to the small multidrug resistance family of drug exporters. Both mdtJ and mdtI are necessary for recovery from the toxicity of overaccumulated spermidine. It was also found that the level of mdtJI mRNA was increased by spermidine. The spermidine content in cells cultured in the presence of 2 mM spermidine was decreased, and excretion of spermidine from cells was enhanced by MdtJI, indicating that the MdtJI complex can catalyze excretion of spermidine from cells. It was found that Tyr4, Trp5, Glu15, Tyr45, Tyr61, and Glu82 in MdtJ and Glu5, Glu19, Asp60, Trp68, and Trp81 in MdtI are involved in the excretion activity of MdtJI.
Polyamines (putrescine, spermidine, and spermine) are essential for normal cell growth (3, 11, 12), and their content in cells is regulated by biosynthesis, degradation, uptake, and excretion (5, 9, 10, 26). With regard to transport, the properties of three polyamine transport systems were characterized in Escherichia coli (15, 16, 40). They include spermidine-preferential and putrescine-specific uptake systems, which belong to the family of ATP binding cassette transporters, and a protein, PotE, involved in the excretion of putrescine by a putrescine-ornithine antiporter activity. Furthermore, it has been reported that cadaverine and aminopropylcadaverine function as compensatory polyamines for cell growth (13), and CadB, a cadaverine-lysine antiporter, is strongly involved in cell growth at acidic pH, like PotE (23, 33, 34, 41). Analogous to the speF-potE operon (18), cadB is one component of the cadBA operon, in which cadA encodes lysine decarboxylase (22, 41) and is induced by acidic pH and lysine (23). The cadBA and speF-potE operons contribute to an increase in the pH of the extracellular medium through excretion of cadaverine and putrescine, the consumption of a proton, and a supply of carbon dioxide during the decarboxylation reaction (33, 38), so the expression of these two operons is important for cell growth at acidic pH.
Although PotE and CadB excrete putrescine and cadaverine at acidic pH, they function as uptake proteins for putrescine and cadaverine at neutral pH (16, 33). Thus, no polyamine excretion proteins that function at neutral pH have been identified to date. Overaccumulated spermidine is either metabolized by acetylation of spermidine, in a reaction catalyzed by spermidine acetyltransferase (6), or neutralized by the increase in l-glycerol 3-phosphate (27). In this study, we looked for spermidine excretion proteins among putative drug exporters comprising five families (the major facilitator family, the small multidrug resistance [SMR] family, the resistance-nodulation-cell division family, the ATP binding cassette family, and the multidrug and toxic compound extrusion family) (25) and found that the MdtJI complex in the SMR family can catalyze excretion of spermidine at neutral pH.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli CAG2242 (speG putE44 hsdR thi thr leu lacY1 tonA21), a spermidine acetyltransferase-deficient mutant (2), was kindly supplied by E. W. Gerner (University of Arizona Health Science Center). The cells were grown in Luria-Bertani (LB) medium (10 g of tryptone per liter, 5 g of yeast extract per liter, and 10 g of NaCl per liter). Where indicated, 2 or 12 mM spermidine was added at the onset of cell growth. Cell growth was monitored by measuring the A540. Cell viability was determined by counting colonies on 1.5% agar plates containing LB medium at 37°C. Thus, the definition of viable cells was cells that were able to grow on the agar plate. Ampicillin (100 μg/ml), tetracycline (15 μg/ml), and/or chloramphenicol (30 μg/ml) was added to the medium, if necessary. E. coli CAG2242 mdtJ::Kmr or mdtI::Kmr was constructed by means of P1 transduction, in which E. coli W3110 mdtJ::Kmr or mdtI::Kmr was used as a donor. E. coli W3110 strains were kindly supplied by H. Mori, Nara Institute of Science and Technology (1).
Plasmids and site-directed mutagenesis of mdtJI.
Plasmids encoding 33 putative drug transporters in pUC119 (Takara Shuzo) were prepared as described previously (25). These plasmids included the original promoters for the drug transporter genes. Since the functions of some proteins among the 33 putative drug transporters were identified, new genetic names (Geno Base [http://ecoli.naist.jp/GB6/search.jsp]) were also attached (Fig. 1). The plasmids pMWcusA, pMWmdtABC, pMWacrD, pMWmdtJI (ydgFE), pMWsugE, pMWmdtG (yceE), pMWydiM, pMWyieO, pMWybjYZ, pMWyddA, and pMWyojIH were prepared by inserting the corresponding genes in pUC119 into pMW119 (Nippon Gene), using the same restriction enzymes. For preparation of pMWacrAB, pMWemrE, and pMWmdfA, PCRs were performed using pUCacrAB, pUCemrE, and pUCmdfA as templates, with the following primers: acrAB (SphI), 5′-ATTTTGCATGCGTATGTACCATAGCACGACG-3′; acrAB (BamHI), 5′-ATTAGGATCCACTCCTTAATGTTCGTAGGT-3′; emrE (EcoRI), 5′-CAGAGAATTCCGATGAAACGGGTATTGAGG-3′; emrE (SphI), 5′-TATTGCATGCTTCTTACGCCATAATCTTGA-3′; mdfA (EcoRI), 5′-GTAAGAATCGCTTAACCGTGGTTTCAGCT-3′; and mdfA (HindIII), 5′-GAGAAAGCTTGATCGCACAAAGCAGTCAGG-3′. The PCR products thus obtained were digested with SphI and BamHI for acrAB, EcoRI and SphI for emrE, and EcoRI and HindIII for mdfA, and the fragments were inserted into the same restriction sites of pMW119.
FIG. 1.
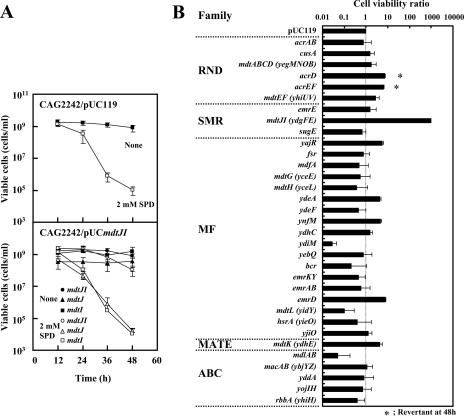
Effect of spermidine on cell viability of E. coli CAG2242 transformed with pUC119 encoding putative drug transporters. Cells were cultured in the presence and absence of 2 mM spermidine, and viable cells were counted at the designated times as described in Materials and Methods. (A) Effects of 2 mM spermidine, mdtJI, mdtJ, and mdtI on cell viability. Symbols shown in the figure indicate the transformed genes. Numbers of colonies (viable cells) are shown in the figure. SPD, spermidine. (B) Effects of 2 mM spermidine and 33 putative drug transporters on cell viability at 48 h. The number of colonies obtained from cells transformed with pUC119 was defined as 1, and cell viability ratios are shown in the figure, in which genes are classified into five families of putative drug exporters. *, a revertant appeared at 48 h. Values are the means ± standard deviations (SD) for three samples.
For preparation of pUCmdtJ (ydgF) and pUCmdtI (ydgE), PCRs were performed using pUCmdtJI (ydgFE) as the template, with the following primers: P-J1, 5′-GGTTTCGCTGGATCCAGCGAAAATTAA-3′; P-J2, 5′-CAAAAAGACGTTAGCAACGAATTCCAGCAC-3′; P-I1, 5′-CAGGTACCCGGATCCCGCGTAAACCTGAAC-3′; and P-I2, 5′-AAAATAGAATTCAAACGCTGCCCGACAGCG-3′. The PCR products thus obtained were digested with BamHI and EcoRI, and the fragments were inserted into the same restriction site of pUC119. Plasmids pUCmdtJ (ydgF) and pUCmdtI (ydgE) were under the control of the lacUV5 promoter instead of the original promoter.
Plasmid YEp-HA3 with 170 bp of the 3′-untranslated region (3′-UTR) of the UGA4 gene (YEp-HA3-3′-UTR·UGA4) (39) was used for preparation of YEp-mdtJ-HA3. The BamHI site of the HA3 region of YEp-HA3-3′-UTR·UGA4 was deleted by site-directed mutagenesis, and then the mdtJ gene, containing the promoter region and the open reading frame lacking the termination codon, was inserted into the SalI and BamHI sites of the plasmid to fuse to the HA3 tag. pUCmdtJ-HA3 was prepared by inserting the 0.7-kb SalI-EcoRI fragment of YEp-MdtJ-HA3 into the same restriction site of pUC119. pCA24N-mdtI with an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter, pT5/lac, and a His tag in the NH2 terminus of MdtI was kindly supplied by H. Mori, National BioResource Project (NIG, Japan) (20). Site-directed mutagenesis for the construction of mutated mdtJ (ydgF) and mdtI (ydgE) genes was performed using pUCmdtJI, encoding both MdtJ and MdtI in an operon, by overlap extension using PCR (8) or with a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The nucleotide sequences of the plasmids were confirmed with a CEQ8000 DNA genetic analysis system (Beckman Coulter).
Dot blot analysis.
Total RNA was prepared from E. coli CAG2242 transformed with pUCmdtJI (ydgFE) or pMWmdtJI (ydgFE) according to the method of Emory and Belasco (4). Dot blot analysis was performed using various amounts of total RNA and a 32P-labeled probe consisting of 400 bp of mdtI, which was labeled with [α-32P]dCTP by use of a BcaBEST labeling kit (Takara Shuzo) (30).
Western blot analysis.
Western blotting was performed by the method of Nielsen et al. (24), using 40 μg of cell lysate and ECL Western blotting reagents (GE Healthcare). Mouse monoclonal anti-hemagglutinin (anti-HA) antibody (clone HA-7) and rabbit anti-six-His antibody were purchased from Sigma and Bethyl Laboratories, respectively.
Measurement of polyamines.
Polyamine levels in E. coli were determined by high-performance liquid chromatography, as described previously (13), after homogenization and extraction of the polyamines with 5% trichloroacetic acid and centrifugation at 27,000 × g for 15 min at 4°C. The retention times for putrescine, spermidine, and spermine were 6.7, 13, and 25 min, respectively. Protein content in the precipitate was determined by the method of Lowry et al. (21).
Assay for spermidine excretion from cells.
E. coli CAG2242 cells transformed with either pUC119 or pUCmdtJI were cultured as described above and harvested at an A540 of 0.5. After cells were washed with buffer 1, containing 0.4% glucose, 62 mM potassium phosphate, pH 7.0, 1.7 mM sodium citrate, 7.6 mM (NH4)2SO4, and 0.41 mM MgSO4, [14C]spermidine was preloaded by the incubation of cells (0.2 mg protein/ml buffer 1) with 1 mM [14C]spermidine (37 MBq/mmol) for 90 min. After cells were washed with buffer 1, spermidine excretion from cells was measured by incubating cells (0.2 mg/ml buffer 1) at 37°C for the designated times. The cells were then removed by centrifugation at 17,000 × g for 5 min at 4°C, and the radioactivity of the supernatant (0.5 ml) was counted by a liquid scintillation counter. When excreted polyamines were analyzed by high-performance liquid chromatography, nonlabeled spermidine was used instead of [14C]spermidine.
RESULTS
Identification of spermidine excretion protein.
We previously reported that spermidine toxicity is increased due to overaccumulation of spermidine when spermidine acetyltransferase is deficient (6). To look for a spermidine excretion protein(s), cell viability was estimated using E. coli CAG2242, which is deficient in spermidine acetyltransferase (2), after transformation with a candidate gene potentially encoding a spermidine excretion protein. First, recovery of cell viability, i.e., spermidine excretion activity, was examined using cells transformed with 33 kinds of drug transporters (25). As shown in Fig. 1A, the viability of E. coli CAG2242 cultured in the presence of 2 mM spermidine was reduced to <0.1% compared with that of cells cultured without spermidine. When the mdtJI gene was transformed into E. coli CAG2242, cell viability during culture with 2 mM spermidine increased >1,000-fold (Fig. 1A and B). Essentially the same results were obtained when E. coli CAG2242 mdtJ::Kmr or mdtI::Kmr was used instead of E. coli CAG2242 (data not shown). When genes for the other 32 drug transporters were transformed, the viability of E. coli CAG2242 did not increase significantly (Fig. 1B). It has been reported that mdtJ and mdtI are coexpressed (25). When either mdtJ or mdtI was transformed alone, the cell viability of E. coli CAG2242 did not increase significantly (Fig. 1A), indicating that both the MdtJ and MdtI proteins are required to rescue cell viability during culture with 2 mM spermidine.
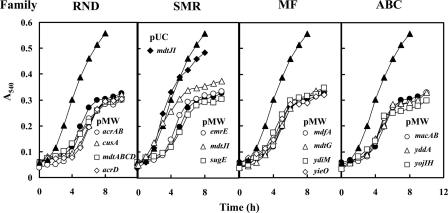
Among 33 genes for drug transporters, the expression of 13 of these genes by the multicopy vector pUC119 significantly inhibited cell growth in the absence of spermidine. Thus, the effect of expression of these genes on cell viability was examined using the low-copy-number vector pMW119. However, there was no significant effect of these genes on viability of E. coli CAG2242 cultured with 2 mM spermidine (data not shown). We then tested whether growth of E. coli CAG2242 cultured with a higher concentration of spermidine (12 mM) was influenced by the expression of these genes inserted in the low-copy-number pMW119 vector. As shown in Fig. 2, growth of E. coli CAG2242 was significantly inhibited by 12 mM spermidine. Expression of the same 13 genes as those studied previously did not influence the growth of E. coli CAG2242 cultured with 12 mM spermidine, whereas cell growth was rescued by the expression of mdtJI. The degree of the rescue of cell growth was greater with pUCmdtJI than with pMWmdtJI. The results strongly suggest that mdtJI can enhance cell viability and growth through excretion of spermidine when spermidine overaccumulates in cells.
FIG. 2.
Effect of spermidine on cell growth of E. coli CAG2242 transformed with pMW119 encoding putative transporters. Cells were cultured in the presence and absence of 12 mM spermidine, and cell growth was monitored by measuring the A540. ▴, cell growth of E. coli CAG2242/pMW119 without spermidine; •, cell growth of E. coli CAG2242/pMW119 with 12 mM spermidine. Data were shown by dividing the transporters into families. Symbols shown in the figure indicate the genes transformed with pMW119. As a control, cell growth of E. coli CAG2242/pUCmdtJI (⧫) was monitored. Values are the averages of duplicate determinations.
Excretion of spermidine by MdtJI.
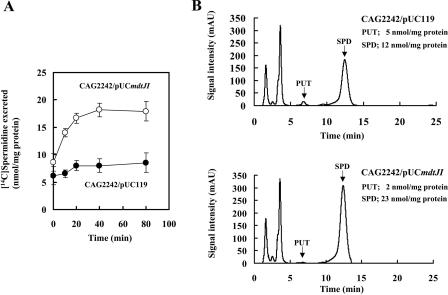
It was then determined whether spermidine can be excreted from E. coli CAG2242 by MdtJI. When the spermidine and putrescine contents were measured in E. coli CAG2242 cultured with or without 2 mM spermidine, overaccumulation of spermidine, but not putrescine, was observed in cells cultured with spermidine (Table 1). When mdtJI was transformed into E. coli CAG2242, accumulation of spermidine in E. coli CAG2242 cultured with 2 mM spermidine was greatly diminished in parallel with the recovery of cell viability (Table 1 and Fig. 1). Next, excretion of [14C]spermidine by MdtJI was examined. As shown in Fig. 3A, excretion of accumulated [14C]spermidine was observed in cells transformed with pUCmdtJI but not in cells carrying a vector. Excretion of spermidine from cells was confirmed by measuring the level of polyamines in the reaction mixture after removal of cells by centrifugation. As shown in Fig. 3B, the level of spermidine in the reaction mixture at 40 min increased significantly when pUCmdtJI was transformed into cells. These results indicate that MdtJI can catalyze the excretion of spermidine.
TABLE 1.
Effect of spermidine addition to the medium on the level of polyamines in cellsa
| Strain | Time of incubation (h) | Addition of 2 mM spermidine | Polyamine concn (mM)
|
|
|---|---|---|---|---|
| Putrescine | Spermidine | |||
| CAG2242/pUC119 | 12 | − | 4.86 ± 1.12 | 5.27 ± 0.88 |
| 12 | + | 0.19 ± 0.14 | 26.2 ± 2.34 | |
| 24 | − | 3.03 ± 1.14 | 5.73 ± 1.04 | |
| 24 | + | 0.17 ± 0.10 | 30.9 ± 2.28 | |
| 36 | − | 1.66 ± 0.16 | 4.58 ± 0.58 | |
| 36 | + | 0.07 ± 0.04 | 32.3 ± 3.06 | |
| CAG2242/pUCmdtJI | 12 | − | 0.96 ± 0.52 | 5.95 ± 1.42 |
| 12 | + | 0.15 ± 0.14 | 11.8 ± 0.66 | |
| 24 | − | 3.40 ± 1.26 | 4.71 ± 1.06 | |
| 24 | + | 0.24 ± 0.10 | 12.1 ± 2.06 | |
| 36 | − | 3.92 ± 0.46 | 2.83 ± 1.00 | |
| 36 | + | 0.33 ± 0.26 | 15.5 ± 1.34 | |
E. coli CAG2242/pUC119 and CAG2242/pUCmdtJI cells were cultured in the presence and absence of 2 mM spermidine for the indicated times, and the polyamine content in cells was measured as described in Materials and Methods. The concentrations of polyamines were calculated based on the data for 5 μl of cell volume per mg protein (19, 29). Values are means ± SD for three samples.
FIG. 3.
Excretion of spermidine from cells by MdtJI. (A) Assays for excretion of [14C]spermidine were performed as described in Materials and Methods. The amount of [14C]spermidine accumulated in E. coli CAG2242 cells transformed with either pUC119 or pUCmdtJI during a 90-min preincubation was 70.1 ± 0.5 or 65.2 ± 7.5 nmol/mg protein, respectively. (B) Assays of excretion of spermidine from cells were performed with nonlabeled spermidine, and excreted spermidine at 40 min was measured by high-performance liquid chromatography after the removal of cells by centrifugation. The retention times for putrescine (PUT) and spermidine (SPD) were 6.7 and 13 min, respectively. Levels of putrescine and spermidine in the supernatant per mg protein of cells in the precipitate are shown in the figure. Experiments were repeated twice, and the results were reproducible. AU, absorbance units.
Increase in the level of mdtJI mRNA by spermidine.
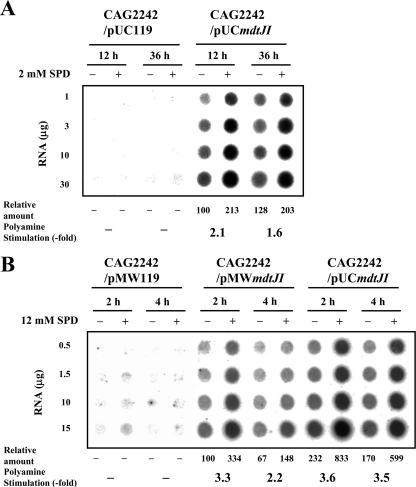
We examined whether the level of mdtJI mRNA was increased by spermidine. The level of mdtJI mRNA was measured by dot blot analysis. The level of mdtJI mRNA expressed from host genes was very low (Fig. 4). Expression of mdtJI mRNA was clearly observed only when pMWmdtJI or pUCmdtJI was transformed into cells. As shown in Fig. 4A, the level of mdtJI mRNA was increased about 1.5- to 2.0-fold by 2 mM spermidine during culture from 12 h to 36 h when pUCmdtJI was transformed into cells. When cells were cultured in the presence of 12 mM spermidine, the level of mdtJI mRNA was greatly increased in cells transformed with either pMWmdtJI or pUCmdtJI 2 h after the onset of cell growth (Fig. 4B). However, when pMWmdtJI was transformed into cells, the level of mdtJI mRNA decreased 4 h after the onset of cell growth (Fig. 4B), in parallel with the slowdown of cell growth (Fig. 2). The results indicate that the increase in the level of mdtJI mRNA by spermidine is important for the decrease in spermidine toxicity.
FIG. 4.
Dot blot analysis of mdtJI mRNA in E. coli CAG2242 cells cultured with or without spermidine (SPD). Dot blot analysis was performed as described in Materials and Methods. The radioactivity of each spot was quantified using a Fujix Bas 2000II imaging analyzer, and the averages for four dots are shown as relative amounts. Results were reproducible in two separate experiments. (A) Levels of mdtJI mRNA in E. coli CAG2242 cells transformed with pUCmdtJI or pUC119 and cultured for 12 or 36 h in the presence and absence of 2 mM spermidine. (B) Levels of mdtJI mRNA in E. coli CAG2242 cells transformed with pMWmdtJI, pMW119, or pUCmdtJI and cultured for 2 or 4 h in the presence and absence of 12 mM spermidine. Cell growth was started at an A540 of 0.05.
Identification of functional amino acids in MdtJI.
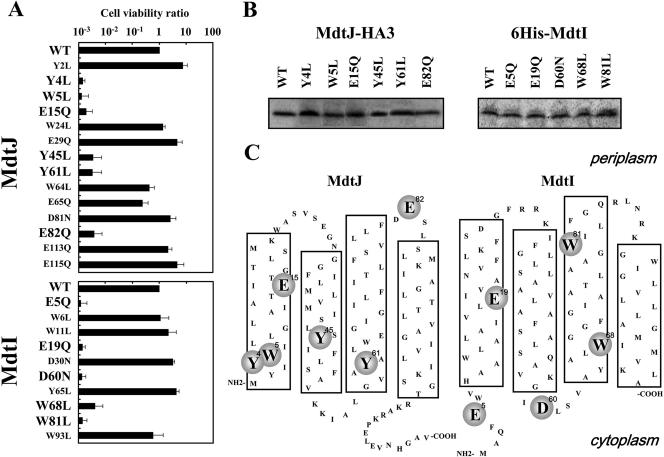
We previously identified the functional amino acids which recognize spermidine on the substrate binding protein PotD in the spermidine-preferential uptake system (17, 36). They were acidic amino acids (Glu and Asp) and aromatic amino acids (Trp and Tyr). To determine the functional amino acids in MdtJ and MdtI, Asp, Glu, Trp, and Tyr were replaced by Asn, Gln, Leu, and Leu, respectively, and the viability of cells carrying mutated pUCmdtJI cultured with 2 mM spermidine was measured 48 h after the onset of cell growth. As shown in Fig. 5A, mutation of Tyr4, Trp5, Glu15, Tyr45, Tyr61, and Glu82 in MdtJ and Glu5, Glu19, Asp60, Trp68, and Trp81 in MdtI decreased cell viability >100-fold compared to that of the wild-type MdtJI complex. The activity of spermidine excretion also decreased in these mutants, judging from the increased level of spermidine in cells (Table 2). The levels of the mutated proteins were nearly equal to those of wild-type MdtJ and MdtI (Fig. 5B). The results indicate that these amino acid residues are important for the activity of spermidine excretion by MdtJI. The results are in accordance with the idea that NH2 and NH groups of spermidine are recognized by Asp and Glu and that propyl and butyl groups of spermidine are recognized by Trp and Tyr in MdtJI proteins (17, 36).
FIG. 5.
Cell viability of various mutants of MdtJ and MdtI. (A) Cell viability of various mutants of MdtJ and MdtI was measured as described in Materials and Methods. Values are means ± SD for three samples. (B) E. coli CAG2242/pUCmdtJ-HA3 and its mutants and E. coli CAG2242/pCA24N-mdtI and its mutants were cultured for 24 h. E. coli CAG2242/pCA24N-mdtI and its mutants were cultured further for 2 h in the presence of 1 mM IPTG. Experiments were repeated twice, and the results were reproducible. The levels of mutated MdtJ and MdtI were evaluated as described in Materials and Methods, using antibodies against the HA tag and the His tag, respectively. (C) Amino acid residues of MdtJ and MdtI involved in relief from spermidine toxicity. Models of secondary structures of proteins were constructed according to the average hydropathy profiles obtained with a hydrophilicity/hydrophobicity plot (Genentyx-Mac, version 10). Putative transmembrane segments are shown in large boxes. Amino acid residues involved in relief from spermidine toxicity are shown in circles.
TABLE 2.
Levels of polyamines in CAG2242/pUCmdtJI and its mutants cultured with 2 mM spermidinea
| Strain | Polyamine concn (mM)
|
|
|---|---|---|
| Putrescine | Spermidine | |
| CAG2242/pUCmdtJI | 0.49 ± 0.12 | 11.7 ± 0.42 |
| mdtJ(Y4L) | 0.14 ± 0.05 | 36.0 ± 1.39 |
| mdtJ(W5L) | 0.23 ± 0.07 | 24.1 ± 0.99 |
| mdtJ(E15Q) | 0.07 ± 0.03 | 24.7 ± 1.83 |
| mdtJ(Y45L) | 0.33 ± 0.14 | 31.1 ± 0.64 |
| mdtJ(Y61L) | 0.36 ± 0.09 | 33.0 ± 1.56 |
| mdtJ(E82Q) | 0.19 ± 0.11 | 31.9 ± 2.01 |
| mdtI(E5Q) | 0.36 ± 0.18 | 30.9 ± 2.30 |
| mdtI(E19Q) | 0.60 ± 0.24 | 23.2 ± 1.35 |
| mdtI(D60N) | 0.65 ± 0.15 | 20.9 ± 0.49 |
| mdtI(W68L) | 0.24 ± 0.10 | 29.4 ± 2.14 |
| mdtI(W81L) | 0.18 ± 0.13 | 20.7 ± 0.88 |
E. coli CAG2242/pUCmdtJI and its mutants were cultured in the presence of 2 mM spermidine for 24 h, and the polyamine content in cells was measured as described in Materials and Methods. Values are means ± SD for three samples.
The MdtJI proteins belong to the SMR family. Proteins in the SMR family (EmrE, MdtJI, and SugE) are thought to have four transmembrane segments. It has also been reported that the structure of EmrE is a parallel dimer (35). If MdtJ and MdtI have parallel topology, most of the functional amino acid residues would be located in the cytoplasmic side (Fig. 5C), in a situation similar to that of other putrescine and cadaverine excretion proteins, such as PotE and CadB (16, 34).
DISCUSSION
It is known that overaccumulation of spermidine and/or spermine inhibits growth in both E. coli (6) and mammalian cells (7). Thus, the enzymes that metabolize spermidine and/or spermine—spermidine acetyltransferase in E. coli (6) and spermidine/spermine N1-acetyltransferase in mammalian cells (7)—are induced when spermidine and/or spermine overaccumulates (6, 7). Once spermidine and/or spermine is acetylated, it cannot interact with RNA, and inhibition of protein synthesis due to overaccumulation of spermidine and/or spermine is relieved (14). The second mechanism to decrease polyamine toxicity is induction of l-glycerol 3-phosphate, which makes a complex with spermidine (27). Accordingly, inhibition of protein synthesis due to overaccumulation of spermidine is relieved. This was observed when AcrD and AcrEF were overproduced (Fig. 1).
The third mechanism to decrease polyamine toxicity is excretion of polyamines from cells. Since spermidine acetyltransferase does not exist in Saccharomyces cerevisiae, there are five excretion proteins (TPO1 to TPO5) in yeast (37). There were no previous reports identifying a spermidine excretion protein in E. coli. To find the spermidine excretion protein(s), a spermidine acetyltransferase-deficient mutant, CAG2242, was used. In this study, we have shown that the MdtJI protein complex can excrete spermidine. It is likely that the MdtJI protein complex is the major spermidine excretor in E. coli and contributes to relief from spermidine toxicity. In connection with this, it is noted that the level of mdtJI mRNA is increased in the presence of spermidine. This is most likely due to the enhancement of transcription of mdtJI mRNA by spermidine rather than to stabilization of its mRNA. Such an increase in mdtJI mRNA by spermidine strongly contributes to the relief of toxicity by overaccumulation of spermidine.
It has been reported that expression of MdtJI causes resistance to sodium dodecyl sulfate and deoxycholate (25). Therefore, this is the first report that the MdtJI complex excretes positively charged substances such as spermidine. In this respect, it is noted that EmrE and SugE, which also belong to the SMR family, can recognize positively charged substances such as tetraphenylphosphonium (TPP+) (28, 31, 32). The total number of functional amino acid residues (Asp, Glu, Trp, and Tyr) in MdtJI was similar to that in the EmrE homodimer (28, 31). However, as for acidic amino acid residues, five molecules were necessary for excretion of spermidine by MdtJI, although only two molecules were necessary for excretion of TPP+ by the EmrE homodimer. This may be due to a difference in the numbers of positive charges in spermidine and TPP+.
Acknowledgments
We thank A. J. Michael for his kind help in preparing the manuscript. Thanks are also due to E. W. Gerner and H. Mori for kindly supplying E. coli CAG2242, E. coli W3110 mdtJ::Kmr or mdt::Kmr, and pCA24N-mdtI plasmid.
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture, Japan.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 22006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carper, S. W., D. G. Willis, K. A. Manning, and E. W. Gerner. 1991. Spermidine acetylation in response to a variety of stress in Escherichia coli. J. Biol. Chem. 26612439-12441. [PubMed] [Google Scholar]
- 3.Cohen, S. S. 1998. A guide to the polyamines, p. 1-543. Oxford University, Oxford, United Kingdom.
- 4.Emory, S. A., and J. G. Belasco. 1990. The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J. Bacteriol. 1724472-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fogel-Petrovic, M., S. Vujcic, P. J. Brown, M. K. Haddox, and C. W. Porter. 1996. Effect of polyamines, polyamine analogs, and inhibitors of protein synthesis on spermidine-spermine N1-acetyltransferase gene expression. Biochemistry 3514436-14444. [DOI] [PubMed] [Google Scholar]
- 6.Fukuchi, J., K. Kashiwagi, M. Yamagishi, A. Ishihama, and K. Igarashi. 1995. Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli. J. Biol. Chem. 27018831-18835. [DOI] [PubMed] [Google Scholar]
- 7.He, Y., K. Kashiwagi, J. Fukuchi, K. Terao, A. Shirahata, and K. Igarashi. 1993. Correlation between the inhibition of cell growth by accumulated polyamines and the decrease of magnesium and ATP. Eur. J. Biochem. 21789-96. [DOI] [PubMed] [Google Scholar]
- 8.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 9.Igarashi, K., K. Ito, and K. Kashiwagi. 2001. Polyamine uptake systems in Escherichia coli. Res. Microbiol. 152271-278. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi, K., and K. Kashiwagi. 1999. Polyamine transport in bacteria and yeast. Biochem. J. 344633-642. [PMC free article] [PubMed] [Google Scholar]
- 11.Igarashi, K., and K. Kashiwagi. 2000. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 271559-564. [DOI] [PubMed] [Google Scholar]
- 12.Igarashi, K., and K. Kashiwagi. 2006. Polyamine modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J. Biochem. (Tokyo) 13911-16. [DOI] [PubMed] [Google Scholar]
- 13.Igarashi, K., K. Kashiwagi, H. Hamasaki, A. Miura, T. Kakegawa, S. Hirose, and S. Matsuzaki. 1986. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J. Bacteriol. 166128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kakegawa, T., Y. Guo, Y. Chiba, T. Miyazaki, M. Nakamura, S. Hirose, Z. N. Canellakis, and K. Igarashi. 1991. Effect of acetylpolyamines on in vitro protein synthesis and on the growth of a polyamine-requiring mutant of Escherichia coli. J. Biochem. (Tokyo) 109627-631. [DOI] [PubMed] [Google Scholar]
- 15.Kashiwagi, K., A. Innami, R. Zenda, H. Tomitori, and K. Igarashi. 2002. The ATPase activity and the functional domain of PotA, a component of the spermidine-preferential uptake system in Escherichia coli. J. Biol. Chem. 27724212-24219. [DOI] [PubMed] [Google Scholar]
- 16.Kashiwagi, K., A. Kuraishi, H. Tomitori, A. Igarashi, K. Nishimura, A. Shirahata, and K. Igarashi. 2000. Identification of the putrescine recognition site on polyamine transport protein PotE. J. Biol. Chem. 27536007-36012. [DOI] [PubMed] [Google Scholar]
- 17.Kashiwagi, K., R. Pistocchi, S. Shibuya, S. Sugiyama, K. Morikawa, and K. Igarashi. 1996. Spermidine-preferential uptake system in Escherichia coli. Identification of amino acids involved in polyamine binding in PotD protein. J. Biol. Chem. 27112205-12208. [DOI] [PubMed] [Google Scholar]
- 18.Kashiwagi, K., T. Suzuki, F. Suzuki, T. Furuchi, H. Kobayashi, and K. Igarashi. 1991. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J. Biol. Chem. 26620922-20927. [PubMed] [Google Scholar]
- 19.Kellenberger, E. 1996. Structure and function at the subcellular level, p. 17-28. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 20.Kitagawa, M., T. Ara, M. Arifuzzaman, T. Ioka-Nakamichi, E. Inamoto, H. Toyonaga, and H. Mori. 2005. Complete set of ORF clones in Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12291-299. [DOI] [PubMed] [Google Scholar]
- 21.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 22.Meng, S. Y., and G. N. Bennett. 1992. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J. Bacteriol. 1742659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neely, M. N., and E. R. Olson. 1996. Kinetics of expression of the Escherichia coli cad operon as a function of pH and lysine. J. Bacteriol. 1785522-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, P. J., K. L. Manchester, H. Towbin, J. Gordon, and G. Thomas. 1982. The phosphorylation of ribosomal protein S6 in rat tissues following cycloheximide injection, in diabetes, and after denervation of diaphragm. A simple immunological determination of the extent of S6 phosphorylation on protein blots. J. Biol. Chem. 25712316-12321. [PubMed] [Google Scholar]
- 25.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 1835803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pegg, A. E. 1988. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 48759-774. [PubMed] [Google Scholar]
- 27.Raj, V. S., H. Tomitori, M. Yoshida, A. Apirakaramwong, K. Kashiwagi, K. Takio, A. Ishihama, and K. Igarashi. 2001. Properties of a revertant of Escherichia coli viable in the presence of spermidine accumulation: increase in l-glycerol 3-phosphate. J. Bacteriol. 1834493-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rotem, D., S. Steiner-Mordoch, and S. Schuldiner. 2006. Identification of tyrosine residues critical for the function of an ion-coupled multidrug transporter. J. Biol. Chem. 28118715-18722. [DOI] [PubMed] [Google Scholar]
- 29.Rottenberg, H. 1979. The measurement of membrane potential and ΔpH in cells, organelles, and vesicles. Methods Enzymol. 55547-569. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Dot and slot hybridization of purified RNA, p. 7.46-7.50. In J. Sambrook and S. W. Russell (ed.), Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- 31.Sharoni, M., S. Steiner-Mordoch, and S. Schuldiner. 2005. Exploring the binding domain of EmrE, the smallest multidrug transporter. J. Biol. Chem. 28032849-32855. [DOI] [PubMed] [Google Scholar]
- 32.Sikora, C. W., and R. J. Turner. 2005. SMR proteins SugE and EmrE bind ligand with similar affinity and stoichiometry. Biochem. Biophys. Res. Commun. 335105-111. [DOI] [PubMed] [Google Scholar]
- 33.Soksawatmaekhin, W., A. Kuraishi, K. Sakata, K. Kashiwagi, and K. Igarashi. 2004. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol. Microbiol. 511401-1412. [DOI] [PubMed] [Google Scholar]
- 34.Soksawatmaekhin, W., T. Uemura, N. Fukiwake, K. Kashiwagi, and K. Igarashi. 2006. Identification of cadaverine recognition site on the cadaverine-lysine antiporter CadB. J. Biol. Chem. 28129213-29220. [DOI] [PubMed] [Google Scholar]
- 35.Soskine, M., S. Mark, N. Tayer, R. Mizrachi, and S. Schuldiner. 2006. On parallel and antiparallel topology of an homomeric multidrug transporter. J. Biol. Chem. 28136205-36212. [DOI] [PubMed] [Google Scholar]
- 36.Sugiyama, S., D. G. Vassylyev, M. Matsushima, K. Kashiwagi, K. Igarashi, and K. Morikawa. 1996. Crystal structure of PotD, the primary receptor of the polyamine transport system in Escherichia coli. J. Biol. Chem. 2719519-9525. [DOI] [PubMed] [Google Scholar]
- 37.Tachihara, K., T. Uemura, K. Kashiwagi, and K. Igarashi. 2005. Excretion of putrescine and spermidine by the protein encoded by YKL174c (TPO5) in Saccharomyces cerevisiae. J. Biol. Chem. 28012637-12642. [DOI] [PubMed] [Google Scholar]
- 38.Takayama, M., T. Ohyama, K. Igarashi, and H. Kobayashi. 1994. Escherichia coli cad operon functions as a supplier of carbon dioxide. Mol. Microbiol. 11913-918. [DOI] [PubMed] [Google Scholar]
- 39.Uemura, T., K. Tachihara, H. Tomitori, K. Kashiwagi, and K. Igarashi. 2005. Characteristics of the polyamine transporter TPO1 and regulation of its activity and cellular localization by phosphorylation. J. Biol. Chem. 2809646-9652. [DOI] [PubMed] [Google Scholar]
- 40.Vassylyev, D. G., H. Tomitori, K. Kashiwagi, K. Morikawa, and K. Igarashi. 1998. Crystal structure and mutational analysis of the Escherichia coli putrescine receptor. Structural basis for substrate specificity. J. Biol. Chem. 27317604-17609. [DOI] [PubMed] [Google Scholar]
- 41.Watson, N., D. S. Dunyak, E. L. Rosey, J. L. Slonczewski, and E. R. Olson. 1992. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J. Bacteriol. 174530-540. [DOI] [PMC free article] [PubMed] [Google Scholar]