Abstract
Catheter-associated urinary tract infection (CAUTI) is the most common nosocomial infection in the United States. Uropathogenic Escherichia coli (UPEC), the most common cause of CAUTI, can form biofilms on indwelling catheters. Here, we identify and characterize novel factors that affect biofilm formation by UPEC strains that cause CAUTI. Sixty-five CAUTI UPEC isolates were characterized for phenotypic markers of urovirulence, including agglutination and biofilm formation. One isolate, E. coli MS2027, was uniquely proficient at biofilm growth despite the absence of adhesins known to promote this phenotype. Mini-Tn5 mutagenesis of E. coli MS2027 identified several mutants with altered biofilm growth. Mutants containing insertions in genes involved in O antigen synthesis (rmlC and manB) and capsule synthesis (kpsM) possessed enhanced biofilm phenotypes. Three independent mutants deficient in biofilm growth contained an insertion in a gene locus homologous to the type 3 chaperone-usher class fimbrial genes of Klebsiella pneumoniae. These type 3 fimbrial genes (mrkABCDF), which were located on a conjugative plasmid, were cloned from E. coli MS2027 and could complement the biofilm-deficient transconjugants when reintroduced on a plasmid. Primers targeting the mrkB chaperone-encoding gene revealed its presence in CAUTI strains of Citrobacter koseri, Citrobacter freundii, Klebsiella pneumoniae, and Klebsiella oxytoca. All of these mrkB-positive strains caused type 3 fimbria-specific agglutination of tannic acid-treated red blood cells. This is the first description of type 3 fimbriae in E. coli, C. koseri, and C. freundii. Our data suggest that type 3 fimbriae may contribute to biofilm formation by different gram-negative nosocomial pathogens.
Catheter-associated urinary tract infection (CAUTI) is the most common nosocomial infection in the United States, where it accounts for approximately 40% of all nosocomial infections (49). Although CAUTI is usually asymptomatic, it is a frequent cause of bacteremia and infected patients can also experience fever, acute pyelonephritis, and death (59). The risk of bacteriuria following urethral catheterization is estimated to be 5 to 10% per day (60). Most patients with indwelling urinary catheters for 30 days or longer develop bacteriuria (49).
Nosocomial CAUTI is caused by a range of gram-negative and gram-positive organisms, including Escherichia coli, Proteus mirabilis, Pseudomonas aeruginosa, Providencia stuartii, Staphylococcus epidermidis, and Enterococcus faecalis (60). These infections are often polymicrobial and can last from several days to months (29). E. coli is one of the most common organisms isolated from the urine of CAUTI patients. Like uropathogenic E. coli (UPEC) strains that cause cystitis and pyelonephritis, CAUTI E. coli strains possess a range of virulence factors, including adhesins (e.g., P and type 1 fimbriae) and toxins (e.g., hemolysin), and express certain O antigen and capsule (K) types (29). Adherence is important for the colonization of the urinary tract, and the best-characterized adhesins of UPEC are P and type 1 fimbriae from the chaperone-usher subclass. P fimbriae are associated most strongly with pyelonephritis and contribute to the establishment of bacteriuria by binding to the α-d-galactopyranosyl-(1-4)-β-d-galactopyranoside receptor epitope in the globoseries of glycolipids (22, 27). Type 1 fimbriae are produced by most E. coli strains and contribute to the colonization of the bladder by binding to α-d-mannosylated proteins, such as uroplakins (62). Both P and type 1 fimbriae recognize their receptor targets by virtue of organelle tip-located adhesins, namely PapG and FimH, respectively (25).
CAUTI results from the growth of bacterial biofilms on the inner surface of the urinary catheter. Biofilm formation promotes encrustation and protects bacteria from the hydrodynamic forces of urine flow, host defenses, and antibiotics (58). The removal and replacement of the infected catheter is often the only option for patients with symptomatic CAUTI. Treatment with antibiotics is thought to merely postpone the onset of bacteriuria and may result in the emergence of resistant bacteria in the patient and in the medical unit (58). Indeed, in intensive care units, CAUTI can be caused by bacteria that are resistant to all known antibiotics (34).
The mechanisms by which CAUTI E. coli strains adhere to and form biofilms on the surfaces of urinary catheters have not been well described. Several different factors have been associated with biofilm formation by E. coli, including type 1 and F9 fimbriae, flagella, curli, and antigen 43 (24, 29, 37, 38, 53). Here we examined in detail the biofilm-forming properties of the E. coli strain MS2027, which was isolated from a patient with nosocomial CAUTI. Genes associated with the formation of O antigen, capsule, and type 3 fimbriae were found to influence biofilm growth. This is the first report to describe the production and functional role of type 3 fimbriae in E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are described in Table 1. Clinical UTI isolates were obtained from urine samples of patients at the Princess Alexandra Hospital (Brisbane, Australia). The E. coli reference (ECOR) collection was obtained from the STEC Center, Michigan State University. E. coli MS427 (MG1655 flu) and E. coli MS528 (MG1655 fim flu) have previously been described (23, 37). E. coli MS661 is E. coli MS427 recA::tet. E. coli MS1355 is a spontaneous rifampin-resistant mutant of E. coli MS2027. Cells were routinely grown at 37°C on solid or in liquid Luria-Bertani (LB) medium supplemented with appropriate antibiotics unless otherwise stated. M9 minimal medium consisted of 42 mM Na2HPO4, 22 mM KH2O4, 9 mM NaCl, 18 mM NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, and 0.2% glucose (41) supplemented with appropriate antibiotics.
TABLE 1.
Bacterial strains and plasmids
| Strains/plasmids | Description | Reference |
|---|---|---|
| E. coli strains | ||
| MS427 | K-12 MG1655 flu | 39 |
| MS528 | K-12 MG1655 fim flu | 25 |
| MS661 | MS427recA::tet | 39 |
| MS1219 | E. coli S17-1 + pUT(mini-Tn5kan) | 30 |
| MS1355 | Rifampin resistant MS2027 | This study |
| MS1520 | E. coli DH5α containing pCO10 | This study |
| MS1486 | MS2027 transconjugant P4-6E (mrkD::Tn5kan) | This study |
| MS1488 | MS2027 transconjugant P20-5B (mrkB::Tn5kan) | This study |
| MS1489 | MS2027 transconjugant P20-11A (mrkA::Tn5kan) | This study |
| MS1502 | MS2027 transconjugant P17-9H (rmlC::Tn5kan) | This study |
| MS1505 | MS2027 transconjugant P20-11B (manB::Tn5kan) | This study |
| MS1506 | MS2027 transconjugant P22-3H (kpsM::Tn5kan) | This study |
| MS1998 | MS528 + pCO12 | This study |
| MS2000 | MS528 + pBR322 (no insert) | This study |
| MS2001 | MS1486 + pCO12 | This study |
| MS2003 | MS1486 + pBR322 (no insert) | This study |
| MS2004 | MS1488 + pCO12 | This study |
| MS2006 | MS1488 + pBR322 (no insert) | This study |
| MS2007 | MS1505 + pCO12 | This study |
| MS2009 | MS1505 + pBR322 (no insert) | This study |
| MS2027 | E. coli CAUTI isolate | This study |
| Plasmids | ||
| pCO10 | Plasmid from MS1486 (mrkD::Tn5kan) | This study |
| pCO12 | mrkABCDF amplified from E. coli MS2027 with primers 871 and 872 and ligated into EcoRV site of pBR322 | This study |
| pBR322 | Cloning vector | 63 |
DNA manipulations and genetic techniques.
Plasmid DNA was isolated by using the QIAprep Spin Miniprep kit (Qiagen, Australia). Restriction endonucleases were used according to the manufacturer's specifications (New England Biolabs). Chromosomal DNA was purified by using the GenomicPrep cell and tissue DNA isolation kit (Amersham Pharmacia Biotech, Inc., Australia). PCR was performed by using the Expand Long Template PCR system (for amplification of the mrk gene cluster) or Taq polymerase (for screening assays) according to the manufacturer's instructions (Roche, Australia). DNA sequencing was performed by the Australian Genome Research Facility. The mrk gene cluster was amplified from E. coli MS2027 by PCR using primers 871 (5′-CGCGCGATATCGCAGCATAACCGAACAAATG) and 872 (5′-CCGGGGATATCTAAATTTTCTGCGGCAAACC). The PCR product was digested with EcoRV and ligated to the EcoRV-digested plasmid pBR322 to construct plasmid pCO12.
Phenotypic assays.
Type 1 fimbria expression was assayed by the ability of bacterial cells to cause mannose-sensitive (MS) agglutination of yeast (Saccharomyces cerevisiae) cells on glass slides (46). Bacterial strains were grown overnight as shaking cultures in LB broth. Those strains with negative results by this assay were retested after three successive rounds of 48 h of static growth in LB broth. Mannose-resistant (MR) agglutination was assessed as described previously (15). Briefly, a 5% suspension (10 μl) of human type A red blood cells (RBC) washed in phosphate-buffered saline (PBS) was mixed with a 10-μl bacterial suspension on glass slides in the presence and in the absence of d-mannose. The bacterial suspension was prepared by transferring cells from a freshly grown LB agar colony into 50 μl PBS. Bacterial agglutination of tannic acid-treated human RBC (MR/K agglutination) was performed as described previously (10). Curli production was detected by the ability of colonies to stain with Congo red (63).
Transposon mutagenesis.
Transposon mutagenesis was performed via filter paper bacterial conjugation (7, 9). An overnight culture of the donor strain was concentrated 10-fold and left to stand at 37°C for 30 min to allow growth of the sex pili. The donor and recipient were then mixed in a ratio of 1:10 and left to incubate on filter paper for 3 to 4 h. The filter paper mixture was then resuspended in LB and plated out on selective antibiotic medium. Colonies confirmed as kanamycin resistant and ampicillin sensitive were tested for biofilm growth in the microtiter assay. Transposon insertion sites of transconjugants with altered biofilm abilities were identified by using inverse PCR as described previously (57). Primers 390 (5′-GGTTCTTTTTGTCAAGACCGACCTGT) and 391 (5′-CAGTCTAGCTATCGTCATGTAAGCCCACT) were used in combination with TaqαI digestion and religation; primers 395 (5′-AAGCTTGCTCAATCAATCACC) and 465 (5′-CGCCAACTATTGCGATAACA) were used in combination with HhaI digestion and religation.
Biofilm study.
Biofilm formation on polyvinyl chloride (PVC) surfaces was monitored by using 96-well microtiter plates (Falcon) essentially as described previously (45). Briefly, cells were grown for 24 h in M9 minimal medium (containing 0.2% glucose) at 37°C, washed to remove unbound cells and stained with crystal violet. The quantification of bound cells was performed by the addition of acetone-ethanol (20:80) and measurement of the dissolved crystal violet at an optical density of 595 nm (OD595). Flow chamber biofilm experiments were performed as described previously (23, 44), with the exception that cells were detected by using BacLight green fluorescent stain (Molecular Probes). Briefly, biofilms were allowed to form on glass surfaces in a multichannel flow system that permitted online monitoring of community structures. Flow cells were inoculated with OD600-standardized pregrown overnight cultures in M9 medium. BacLight green fluorescent stain was used at a concentration of 0.1 mM, according to the manufacturer's instructions. Biofilm development was monitored by confocal scanning laser microscopy at 20 to 24 h after inoculation. All experiments were performed in triplicate.
Scanning electron microscopy (SEM).
Cells were grown as described for the biofilm study on polystyrene surfaces, with the exception that the experiment was performed by using a 12-well microtiter plate (Greiner Bio-One) with a polystyrene disc placed at the bottom. The disc was fixed with 3% glutaraldehyde in 0.1 M cacodylate buffer and postfixed with 1% osmium tetroxide in 0.1 M cacodylate buffer. The sample was then infiltrated with glycerol and frozen in liquid nitrogen. The sample was freeze-substituted in 100% ethanol containing a molecular sieve and left at −80°C for 10 h, and then the temperature was increased from −80°C to −20°C over a 10-h period and critical point dried. The sample was then mounted on carbon tabs and sputter coated with platinum 15 mA for 120 s.
Phylogenetic analysis.
PCR products obtained from screening for the presence of mrkB were sequenced from 36 strains. Sequences were trimmed to obtain 130 nucleotides of high-quality sequence corresponding to the central region of mrkB (i.e., nucleotides 179 to 308 of mrkB in K. pneumoniae MGH78578). Phylogenetic analyses of 36 aligned mrkB sequences were carried out by using PHYLIP (8, 12, 16, 20). Consensus trees of bootstrap analyses were prepared by using the consensus network method (8, 12, 16, 20) as implemented by SplitsTree, version 4 (8, 12, 16, 20). Evidence for recombination was assessed by using the pairwise homoplasy index recombination test (8, 12, 16, 20).
Statistical analysis.
Differences in comparison of phenotypes from CAUTI E. coli and E. coli from other UTI syndromes were determined by using a chi-square test for the differences between two groups. Differences in biofilm phenotypes (mean optical density values) were compared by using a t test with a linear mixed model; each microtiter plate well was treated as a random effect, and each gene modification was treated as a fixed effect. All comparisons were against the values for E. coli MS2027. Both analyses were performed by using the statistical analysis program R (36a).
Nucleotide sequence accession numbers.
The mrkB sequence fragments from 34 strains were deposited in GenBank under accession numbers EU109428 to EU109460. The complete 9.3-kb mrk cluster (and adjacent regions) from E. coli MS2027 was deposited in GenBank under accession number EU105468.
RESULTS
Biofilm formation by CAUTI E. coli strains.
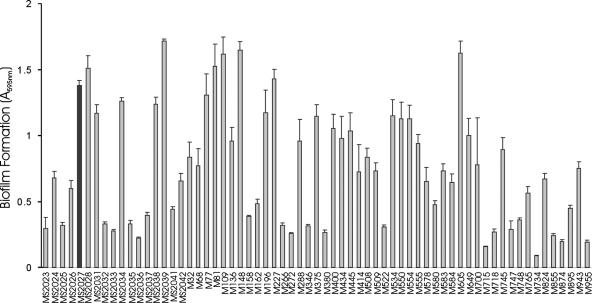
Sixty-five CAUTI E. coli strains from our UTI collection were examined for the ability to cause MS agglutination of yeast cells, MR agglutination of human RBC, and biofilm formation. These phenotypes were compared to those of strains collected from patients with asymptomatic bacteriuria (ABU), cystitis, and pyelonephritis from the same geographic location (Table 2). Sixty-two percent of the CAUTI E. coli strains were positive by the biofilm assay, while 42% of ABU (P = 0.05) and 15% of pyelonephritis strains were positive (P < 0.05). There was no correlation between biofilm formation and MR agglutination of human RBC. Biofilm analysis of the 65 CAUTI E. coli strains revealed a range of phenotypes (Fig. 1). To identify novel factors associated with biofilm formation by CAUTI E. coli, we studied strains that exhibited strong biofilm growth for the expression of type 1 fimbriae (by MS agglutination of yeast cells) and curli production (by Congo red staining). E. coli MS2027 was identified as a unique strong biofilm former that did not express type 1 fimbriae or curli under the growth conditions employed in this study. We therefore chose E. coli MS2027 for an in-depth molecular analysis.
TABLE 2.
Comparison of phenotypes from CAUTI E. coli and E. coli from other UTIs
| Source of isolates | Value for phenotypea
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MS agglutination
|
MR agglutination
|
Biofilm
|
|||||||
| No. of isolates (%) | PR | P | No. of isolates (%) | PR | P | No. of isolates (%) | PR | P | |
| CAUTI (n = 65) | 54 (83) | 10 (15%) | 40 (62%) | ||||||
| Pyelonephritis (n = 26) | 24 (92) | 1.11 | 0.42 | 13 (50%) | 3.33 | 0.00 | 4 (15%) | 0.24 | 0.00 |
| Cystitis (n = 19) | 19 (100) | 1.20 | 0.12 | 5 (26%) | 1.73 | 0.45 | 8 (42%) | 0.68 | 0.21 |
| ABU (n = 57) | 42 (74) | 0.89 | 0.30 | 14 (25%) | 1.67 | 0.30 | 24 (42%) | 0.68 | 0.05 |
Where the prevalence ratios (PR) and P values are relative to CAUTI isolates.
FIG. 1.
Biofilm formation by CAUTI E. coli strains. Strains were grown at 37°C in PVC microtiter plates containing M9 medium (supplemented with 0.2% glucose) for 16 h under shaking conditions, washed to remove unbound cells, and stained with 0.1% crystal violet. Biofilm formation was quantified by resuspending adhered cells in ethanol-acetate (80:20) and measuring the absorbance at 595 nm. The results are presented as the average of eight individual replicates (± standard deviation [error bars]). An arbitrary cutoff of OD595 at 0.5 was used, and strains were scored as either positive or negative for biofilm formation. The black bar highlights E. coli MS2027.
Generation of E. coli MS2027 mini-Tn5 mutants altered in biofilm formation.
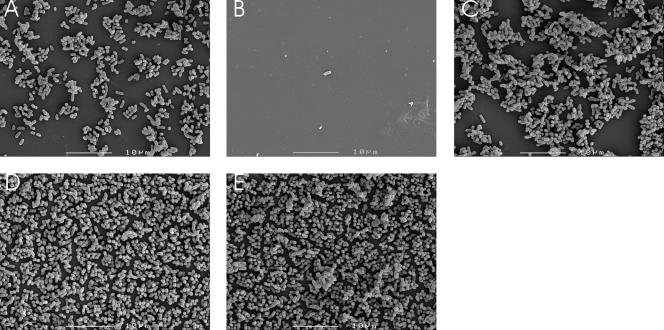
A collection of approximately 2,000 Kmr transposon insertion mutants were screened for their ability to form biofilms. Two types of biofilm mutants were obtained: (i) mutants displaying an enhanced biofilm phenotype and (ii) mutants displaying a decreased biofilm phenotype. SEM was performed to examine the structure of the biofilms produced by E. coli MS2027 and selected mutants representing each phenotype (Fig. 2). The enhanced biofilm growth of E. coli MS1502 and E. coli MS1506 correlated with a more densely packed arrangement of cells than the growth of E. coli MS2027. In contrast, cells from the biofilm-deficient mutant E. coli MS1486 were virtually undetectable. All of the mutants exhibited a growth rate indistinguishable from that of the wild-type strain (data not shown).
FIG. 2.
SEM micrographs depicting the biofilm structures of E. coli MS2027 and representative Tn5 mutants. (A) E. coli MS2027, (B) E. coli MS1486 (MS2027 mrkD::Tn5kan), (C) E. coli MS2001 (MS1486 complemented with pCO12), (D) MS1502 (MS2027 rmlC::Tn5kan), and (E) MS1506 (MS2027 kpsM::Tn5kan). Cells were grown in 12-well microtiter plates (Greiner Bio-One) containing a polystyrene disc placed at the bottom. Following overnight growth, the disc was removed, fixed, stained, and examined by SEM as described in Materials and Methods. Scale bar, 10 μm.
The insertion site of each mutant was determined by inverse PCR. The sequencing of the DNA flanking the transposon insertion site for the mutants displaying the enhanced biofilm phenotype (i.e., MS1502, MS1505, and MS1506) revealed that the transposon insertions disrupted genes associated with O antigen or capsule production (Table 3). In the case for the mutants displaying the decreased biofilm phenotype (i.e., MS1486, MS1488, and MS1489), all of the transposon insertions disrupted the same genetic locus, but at different positions. Blast analysis revealed that this genetic locus was homologous to the mrk (type 3 fimbriae) genetic locus of K. pneumoniae (Table 3). Unlike the E. coli MS2027 parent strain, all of these mutants were unable to exhibit MR/K agglutination, a standard assay for monitoring the expression of type 3 fimbriae in K. pneumoniae.
TABLE 3.
Transposon mutagenesis results for strain E. coli MS2027
| Strain | Biofilm type | Gene disrupted | Gene function |
|---|---|---|---|
| MS1486 | Reduced | mrkD | Adhesin of type 3 fimbriae |
| MS1488 | Reduced | mrkB | Chaperone of type 3 fimbriae |
| MS1489 | Reduced | mrkA | Major subunit of type 3 fimbriae |
| MS1502 | Increased | rmlC | dTDP-4-dehydrorhamnose 3,5- epimerase of OAg gene cluster |
| MS1505 | Increased | manB | Phosphomannomutase of OAg gene cluster |
| MS1506 | Increased | kpsM | Polysialic acid transport protein, membrane component |
The E. coli mrk gene locus is carried on a conjugative plasmid.
E. coli MS2027 contains several plasmids, and we suspected that the mrk genes were located on one of these. Thus, we attempted to transfer the mrk genes (containing Tn5-kanamycin) from the Tn5 mutants MS1486, MS1488, and MS1489 into a tetracycline-resistant E. coli MG1655 derivative strain (MS661) via conjugation. Indeed, we were able to obtain kanamycin-resistant E. coli MS661 transconjugants using all three Tn5 mutants by this method. PCR using primers specific for the mrk sequence identified previously by inverse PCR demonstrated that the mrk genes were located on this conjugative plasmid. The plasmid that originated from E. coli MS1486 was designated pCO10.
Characterization of the E. coli mrk gene locus.
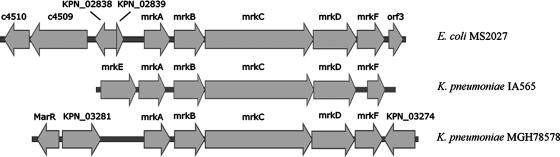
The mrk locus was sequenced directly from pCO10 on both DNA strands by primer walking. The sequence of the region disrupted by the Tn5 insertion was confirmed by PCR and sequencing from the intact plasmid in E. coli MS2027 (designated pMS2027). The mrk gene cluster contains six open reading frames (ORFs) arranged in the same transcriptional orientation, including genes that encode a putative major subunit protein (mrkA) as well as putative chaperone (mrkB)-, usher (mrkC)-, adhesin (mrkD)-, and anchor (mrkF)-encoding genes (Fig. 3). The E. coli mrk locus does not contain a putative regulator gene (mrkE) upstream of mrkA, unlike the K. pneumoniae mrk locus. Instead, this DNA sequence has a mosaic structure. The nucleotide sequence immediately upstream of mrkA is 98% identical (771/781 nucleotides) to the region spanning nucleotides 3116637 to 3117411 of the K. pneumoniae MGH78578 genome. This region contains genes encoding a putative IS1 transposase (KPN_02838) and an insertion element protein (KPN_02839). Further upstream we identified two putative ORFs (c4509 and c4510) that are highly similar to hypothetical genes found on the UPEC CFT073 chromosome (this region shares 96% [2010 of 2081] nucleotide sequence similarity with the corresponding region from CFT073). Downstream of mrkF, we identified an ORF (orf1) that encodes a putative cytoplasmic protein from Salmonella enterica SC-B67 (93% [296 of 315] nucleotide sequence similarity). The mrk gene cluster from E. coli MS2027 has an overall G+C content of 56.6%, which is closer to that found in K. pneumoniae MGH78578 (57.5%) than to that found in E. coli K-12 (50.8%). The nucleotide sequence similarity of each mrk gene (and the amino acid identity of its product) to the corresponding sequence from K. pneumoniae pIA565 (gb M55912) and K. pneumoniae MGH78578 (gb CP000647) is shown in Table 4.
FIG. 3.
Physical map of the mrk gene cluster and adjacent regions from E. coli MS2027, K. pneumoniae IA565, and K. pneumoniae MGH78578. The mrk genes are indicated and include mrkE (putative regulatory gene), mrkA (major subunit encoding gene), mrkB (chaperone encoding gene), mrkC (usher encoding gene), mrkD (adhesin encoding gene), and mrkF (encoding a putative anchor protein). The mrk genes from E. coli MS2027 and K. pneumoniae IA565 are plasmid located, while the mrk genes from K. pneumoniae MGH78578 are chromosomally located. ORFs adjacent to the mrk genes from both E. coli MS2027 and K. pneumoniae MGH78578 are also shown. E. coli MS2027-adjacent ORFs were KPN_02838 (encodes a putative IS1 transposase from K. pneumoniae MGH78578), KPN_02839 (encodes a putative insertion element protein from K. pneumoniae MGH78578), orf3 (encodes a putative cytoplasmic protein), and c4510 and c4509 (encode putative hypothetical proteins from E. coli UPEC CFT073). K. pneumoniae MGH78578-adjacent ORFs were marR (encodes a putative regulatory protein) and KPN_03281 and KPN_03274 (encode hypothetical proteins). Sequence information outside the mrk cluster is not known for K. pneumoniae IA565. Arrows indicate the direction of transcription for each gene.
TABLE 4.
Comparison of the E. coli MS2027 Mrk protein and nucleotide sequences with the corresponding sequences from K. pneumoniae IA565(pIA565) and K. pneumoniae MGH78578
| Mrk protein | Putative function | % Amino acid identity (nucleotide sequence conservation) of indicated protein in K. pneumoniae strain
|
|
|---|---|---|---|
| IA565 plasmid-encoded Mrk | MGH78578 chromosome-encoded Mrk | ||
| MrkA | Major subunit | 90 (86) | 94 (99) |
| MrkB | Chaperone | 91 (83) | 100 (99) |
| MrkC | Usher | 87 (82) | 99 (99) |
| MrkD | Adhesin | 54 (81) | 97 (98) |
| MrkF | Anchor | 78 (76) | 99 (99) |
The mrk locus promotes biofilm formation.
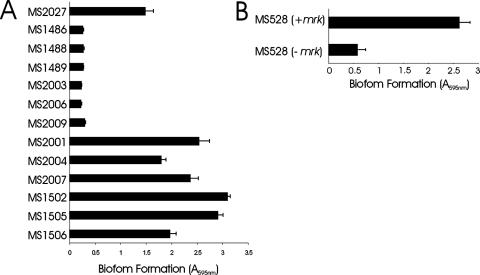
The mrk locus was amplified by PCR from E. coli MS2027 and cloned into the EcoRV site of pBR322 to generate plasmid pCO12. When this plasmid was introduced into E. coli MS528 (an MG1655 fim flu strain deficient in biofilm formation), we observed strong biofilm growth (Fig. 4). Biofilm formation was observed in static and dynamic growth conditions in M9 minimal medium (but not in LB broth). Plasmid pCO12 was also transformed into the three Tn5 mutants deficient in biofilm formation. The introduction of pCO12 into these strains restored their ability to form biofilms (Fig. 4), which was also confirmed by SEM (Fig. 2E).
FIG. 4.
(A) Biofilm formation by E. coli MS2027 and derivatives. Strains were grown at 37°C in PVC microtiter plates containing M9 medium (supplemented with 0.2% glucose) for 16 h under shaking conditions, washed to remove unbound cells, and stained with 0.1% crystal violet. Biofilm formation was quantified by resuspending adhered cells in ethanol-acetate (80:20) and measuring the absorbance at 595 nm. The results are presented as the average of eight individual replicates (± standard deviation). Shown are the results for MS1486 (MS2027 mrkD::Tn5kan), MS1488 (MS2027 mrkB::Tn5kan), MS1489 (MS2027 mrkA::Tn5kan), MS2003, MS2006, and MS2009, mrk mutants containing pBR322; MS2001, MS2004, and MS2007, mrk mutants containing pCO12 (mrk+); MS1502 (rmlC mutant); MS1505 (manB mutant); and MS1506 (kpsM mutant). (B) Biofilm formation by E. coli MS528 and E. coli MS528 containing pCO12. Cells were grown and analyzed for biofilm formation as described above. The introduction of plasmid pCO12 (containing the mrk gene cluster from MS2027) into MS528 promoted strong biofilm growth. −, absence of; +, presence of.
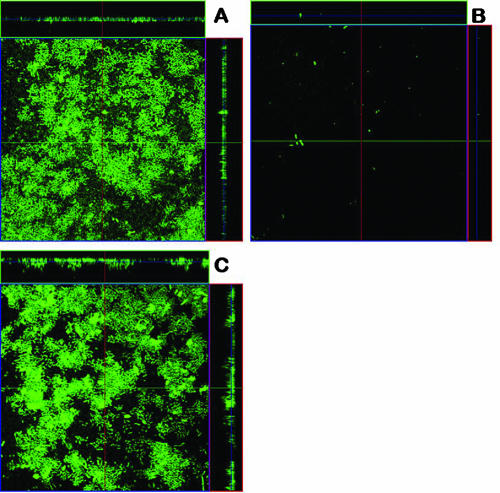
We next employed a continuous flow chamber system to examine the ability of type 3 fimbriae to promote biofilm formation by E. coli in dynamic conditions. Consistent with the results of our microtiter plate biofilm assays, E. coli MS2027 produced a strong biofilm (with a depth of approximately 10 μm), while E. coli MS2003 (mrkD::Tn5) was unable to form a biofilm (Fig. 5). The defect in biofilm growth by E. coli MS2003 could be complemented by the addition of plasmid pCO12 (which contains the mrk gene cluster from E. coli MS2027). Thus, type 3 fimbriae can promote strong biofilm growth by E. coli in two distinct in vitro model systems.
FIG. 5.
Flow chamber biofilm formation of E. coli MS2027 (A), E. coli MS2003 (B), and E. coli MS2001 (C). Biofilm development was monitored by confocal scanning laser microscopy 24 h after inoculation. Micrographs represent horizontal sections. Depicted to the right and below are vertical sections through the biofilm collected at the positions indicated by the lines.
Type 3 fimbrial expression is not associated with colonization of the mouse urinary tract.
E. coli MS2027 and the mrkD mutant MS1486 were tested for their ability to colonize and survive in the mouse urinary tract. We did not observe any significant difference in the ability of either strain to colonize the mouse bladder following transurethral infection. We also tested these two strains in a competition challenge experiment using a 50:50 mixture of each strain as the inoculum. However, no difference in colonization of the bladder was observed (data not shown). Neither strain was isolated from the mouse kidney at the time points assayed.
Distribution of mrk genes in UPEC and other UTI pathogens.
The finding that the mrk genes were located on a conjugative plasmid in E. coli prompted us to test for the prevalence of these genes in UPEC and other CAUTI pathogens. Primers were designed to PCR amplify an internal segment of the mrkB gene. First we tested for the presence of mrkB in the remaining 64 CAUTI E. coli strains from our collection. Two of these strains had positive PCR results and were confirmed by DNA sequencing. Next, we tested for the presence of mrkB in 70 CAUTI pathogens representing different gram-negative organisms isolated from UTI patients from the same location (Table 5). The mrk genes were detected from K. pneumoniae, K. oxytoca, C. koseri, and C. freundii CAUTI isolates (Table 5). The identity of each PCR product was confirmed by DNA sequencing. To determine whether the presence of the mrk genes was specific to CAUTI strains, we also tested for their prevalence in 45 E. coli strains isolated from cystitis and pyelonephritis patients. Among these strains, 2 of 45 contained the mrkB gene as determined by PCR amplification and DNA sequencing. Finally, we tested for the prevalence of mrkB in strains from the ECOR collection; three strains had positive PCR products for these genes, with results confirmed as correct by DNA sequencing. All of the organisms that contained the mrkB gene displayed a positive MR/K agglutination phenotype following growth in M9 minimal medium. We note that the C. koseri and C. freundii strains required growth in M9 minimal medium for 72 h under static conditions to induce MR/K agglutination.
TABLE 5.
Prevalence of mrkB gene and MR/K agglutination phenotype among UTI strains
| Bacterial species and strain type | Total no. of strains | Presencea of:
|
|
|---|---|---|---|
| mrkB | MR/K HA | ||
| Escherichia coli | |||
| CAUTI | 93 | 3 (3.2) | 3 (3.2) |
| ABU | 23 | 0 (0) | 0 (0) |
| Cystitis | 19 | 1 (5.3) | 1 (5.3) |
| Pyelonephritis | 26 | 1 (3.8) | 1 (3.8) |
| ECOR | 72 | 3 (4.2) | 3 (4.2) |
| Other species | |||
| C. freundii | 7 | 1 (14.3) | 1 (14.3) |
| C. koseri | 9 | 9 (100) | 9 (100) |
| K. oxytoca | 2 | 2 (100) | 2 (100) |
| K. pneumoniae | 15 | 13 (86.7) | 13 (86.7) |
Values are shown as no. (%).
Comparison of mrkB DNA sequences from different UTI pathogens.
An unrooted maximum likelihood DNA phylogram was constructed from 36 aligned mrkB gene fragments (130 nucleotides in length, comprising 35 polymorphic sites, 28 of which were informative). A consensus network tree of 1,000 bootstrap replicates revealed five distinct groups, ranging from 20 members (represented by K. pneumoniae MG78758) to one member (C. freundii) (Fig. 6). The most common primary allele is found in both E. coli and K. pneumoniae (e.g., E. coli MS2039 and Klebsiella pneumoniae MGH78578 mrkB sequences are identical). Furthermore, two E. coli mrkB sequences (M184 and ECOR28) are identical to the plasmid-borne Klebsiella pneumoniae pIA565 mrkB, which shares only 85% nucleotide identity with mrkB from strain MGH78578. These observations strongly suggest lateral gene transfer of mrkB in E. coli and K. pneumoniae. Comparisons of full-length mrkA and mrkC sequences from K. pneumoniae IA565 and MGH78578 with E. coli MS2027 show similar levels of divergence to that observed for mrkB (Table 4). This result suggests that the mrkB phylogenetic tree is representative of mrkA and mrkC and that en bloc transfer of the mrk cluster has occurred.
FIG. 6.
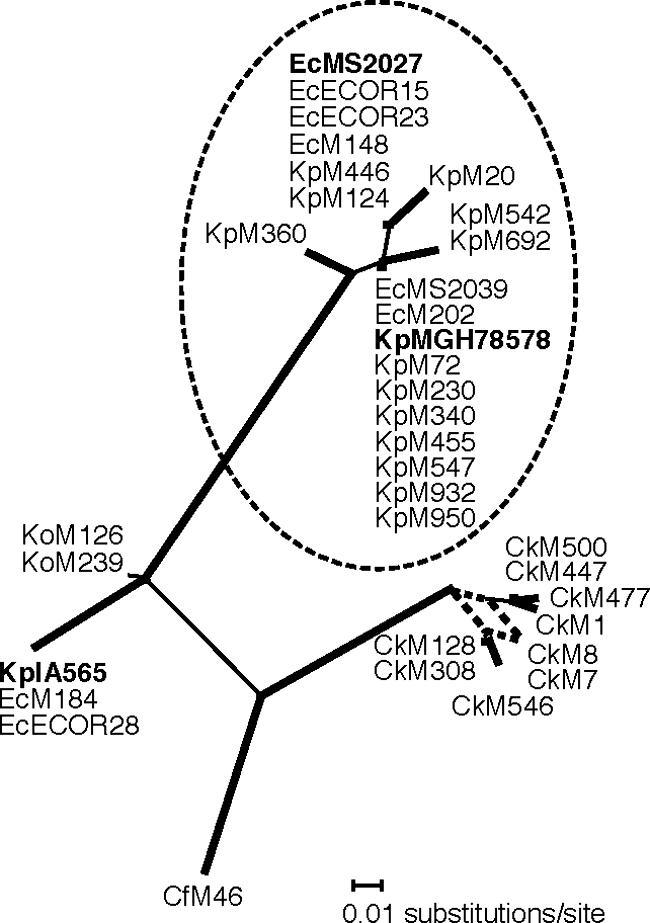
Unrooted phylogram of type 3 family chaperone genes (mrkB). Branch confidence levels are indicated by line thickness: thick, >90%; thin, >50%; dashed, <50%. Confidence levels were determined from 1,000 bootstrap replicates of DNA maximum likelihood trees prepared by using PHYLIP DNAML (8, 12, 16, 20). A network of all 1,000 trees was prepared by using the consensus network method (8, 12, 16, 20), as implemented by SplitsTree, version 4 (8, 12, 16, 20). Taxon IDs include species name abbreviations as suffixes (Ec, E. coli; Cf, C. freundii; Ck, C. koseri; Ko, K. oxytoca; and Kp, K. pneumoniae), followed by a unique strain identifier. The major group containing E. coli MS2027 and K. pneumoniae MGH78578 (shown in bold) is circled for greater clarity. K. pneumoniae IA565 is also shown in bold.
The long internal branches represent the divergence of primary mrkB alleles (Fig. 6), with an average interallelic nucleotide diversity of 9.8%. Secondary variant types are observed only within the KpMGH78578 and C. koseri groups (1% and 1.8% intra-allelic nucleotide diversity, respectively). No statistically significant evidence of recombination was detected between groups according to the pairwise homoplasy index test (8, 12, 16, 20). According to the consensus network (Fig. 6), there is some incompatibility in the divergence of C. koseri clones (as indicated by more than one branch leading to the CkM7/CkM8 tip); however, there are too few informative sites (n = 3) to accurately discriminate recombination from recurrent mutation among this group. Tree topologies very similar to those obtained by using DNA maximum likelihood were obtained by using parsimony- and distance-based phylogenetic methods (data not shown).
DISCUSSION
CAUTI is associated with biofilm formation on the inner surfaces of indwelling catheters. E. coli is a major cause of CAUTI, and several surface factors that contribute to biofilm growth by E. coli have been characterized. Type 1 fimbria expression is associated with persistence in the long-term-catheterized urinary tract (29), and minor modifications in the FimH adhesin can enhance biofilm formation (45). Flagella are motility organelles that play a role in the initial adhesion phase of biofilm formation (36). Curli are thin aggregative fibers involved in bacterial attachment and biofilm formation (38). In many E. coli strains, the expression of curli is temperature sensitive and occurs at 28°C but not at 37°C. Antigen 43 (Ag43) is a self-recognizing autotransporter adhesin that is associated with cell aggregation, biofilm formation, and urovirulence (6, 24, 54). Other factors that promote biofilm growth by E. coli include conjugative pili (14, 37) and several recently described cryptic autotransporter proteins (39). In this study, we have demonstrated that genes involved in capsule synthesis, O antigen production, and type 3 fimbriae influence biofilm formation by the CAUTI E. coli strain MS2027.
Three Tn5 mutants of E. coli MS2027 that possessed an increased biofilm formation phenotype were identified. The Tn5 insertions resulted in the disruption of genes associated with O antigen synthesis (rmlC and manB) and capsule synthesis (kpsM). Capsular polysaccharides are produced by many uropathogenic bacteria and provide protection from host cell phagocytosis. Capsular polysaccharides also contribute to UPEC biofilm formation in the bladder (6, 11). The enhanced biofilm growth of E. coli MS1506 might at first sight appear contradictory to these comments. However, our observations may reflect the limitations of the microtiter plate biofilm assay in comparison to in vivo biofilm growth. E. coli MS1506 contained a Tn5 insertion in the kpsM gene, which encodes an integral membrane protein involved in the translocation of the polysialic capsule. Previous studies have shown that an E. coli kpsM mutant is defective in capsule synthesis (35). While we have not shown that MS1506 is defective in capsule production, it is tempting to speculate that its enhanced biofilm growth is associated with the unmasking of other adhesins. In support of this hypothesis, a recent study reported that the expression of the nonfimbrial adhesin Ag43 in unencapsulated Klebsiella strains results in enhanced biofilm growth compared to capsulated strains that express Ag43, suggesting that the capsule might block the activity of other surface-located adhesins (43). The capsule shielding effect has also been demonstrated in adherence studies of other organisms, including E. coli (40, 43), Neisseria meningitidis (56), and Haemophilus influenzae (50). Furthermore, the function of type 1 fimbriae has been shown to be impeded by the presence of a capsule on the bacterial cell surface (42) and thus reduced capsule synthesis by E. coli MS1506 may enhance the contribution of fimbriae to biofilm formation. We note that soluble polysaccharide secreted by UPEC strains that produce a group II capsule was recently shown to inhibit biofilm growth by preventing adhesion (55). Therefore, we cannot rule out the possibility that the enhanced biofilm growth of MS1506 is due to reduced secretion of antiadhesive polysaccharide material. The enhanced biofilm growth by E. coli MS1508 and E. coli MS1509 might be associated with a similar mechanism, since many UPEC strains are known to produce large O antigen structures. We are currently attempting to elucidate the role of the capsule and O antigen in UPEC colonization and biofilm formation in an in vivo model of CAUTI.
The decreased biofilm growth by Tn5 mutants of E. coli MS2027 was due to the disruption of genes encoding type 3 fimbriae. Three biofilm-deficient mutants were identified, all of which contained a Tn5 insertion in the mrk operon. The role of type 3 fimbriae in biofilm formation was confirmed by the complementation of each of these mutants with a plasmid (pCO12) containing the mrk genes. Type 3 fimbriae are thin, filamentous structures (4 to 5 nm wide and 0.5 to 2 μm long) that extend from the surface of the cell (10) and are morphologically similar to K88 and K99 fimbriae (26). Type 3 fimbriae are characterized by their ability to mediate MR agglutination of tannic acid-treated RBC (which is referred to as MR Klebsiella-like or MR/K agglutination) (26). MR/K agglutination is conferred by the MrkD adhesin (5, 19). MrkD also mediates binding to the basolateral surface of renal tubular, tracheal, and bronchial cells via a high-affinity interaction with type V collagen (17, 51, 52). Type 3 fimbriae from K. pneumoniae have also been shown to mediate biofilm formation (21).
Type 3 fimbriae are most commonly associated with Klebsiella spp. (13). However, they are also produced by other members of the Enterobacteriaceae family, including Enterobacter, Morganella, Proteus, Providencia, Serratia, Salmonella, and Yersinia species (1-4, 13, 30-33, 48). Here we demonstrate that type 3 fimbriae are also produced by E. coli, C. koseri, and C. freundii. In E. coli MS2027, the mrk genes are located on a conjugative plasmid of approximately 45 kb (C.-L. Y. Ong, A. G. McEwan, and M. A. Schembri, unpublished data). Plasmid-carried mrk genes have previously been identified for K. pneumoniae and Y. enterocolitica (5, 13). K. pneumoniae IA565 possesses both chromosomal and plasmid-carried mrk genes; the plasmid pIA565 contains a functional copy of mrkA and mrkD, while only mrkA has been detected on the K. pneumoniae IA565 chromosome. The mrk genes from pIA565 have been well characterized and possess the same genetic arrangement as do the mrk genes on plasmid pMS2027. However, the sequence upstream of mrkA is different between the two gene clusters (the sequence downstream of mrkF on pIA565 has not been reported). On plasmid pIA565, a gene (mrkE) encoding a putative regulator protein is located immediately upstream of mrkA (5). This gene is not present on pMS2027. Instead, we identified a putative transposase-encoding gene upstream of mrkA and the entire cluster is flanked by two putative insertion sequence elements. Thus, it seems likely that the mrk cluster on plasmid pMS2027 is associated with a mobile genetic element. Importantly, the presence of mrk genes in E. coli was not unique to strains in our UTI collection, as three strains from the ECOR collection also contained mrkABC and caused MR/K hemagglutination. The location of the mrk genes from these strains remains to be determined.
We observed that the expression of type 3 fimbriae was dependent on the growth medium. E. coli MS2027 produced a strong biofilm and caused characteristic MR/K agglutination when grown in M9 minimal medium supplemented with glucose but not when grown in LB medium. This finding is consistent with the results of previous reports of type 3 fimbria expression in K. pneumoniae, where bacteria grown in minimal medium in the presence of glycerol or glucose resulted in a stronger MR/K hemagglutination reaction than did bacteria grown in complex medium (18, 47). This result suggests a similar method of regulation of the type 3 fimbrial genes of K. pneumoniae IA565 and E. coli MS2027, despite the absence of the putative mrkE regulator gene on pMS2027. It is interesting that all of the mrkABC-positive strains identified in this study caused MR/K agglutination, since previous studies have shown that not all Klebsiella spp. possess the adhesin-encoding mrkD gene (19).
We compared the mrk cluster from E. coli MS2027 with the mrk clusters from K. pneumoniae MGH78578 and pIA565 (Table 4). All five mrkABCDE genes showed remarkable sequence similarity to the chromosomally located MGH78578 sequences (98.8% ± 0.4%) compared to that shown by the respective pIA565 sequences (81.6% ± 3.6%). The presence of insertion sequences adjacent to the plasmid-borne E. coli MS2027 mrk cluster and the G+C content suggest that there was relatively recent lateral transfer from a K. pneumoniae strain, although we cannot rule out the possibility that both strains acquired the cluster independently from a third species. To assess the distribution of this cluster among UTI organisms, we amplified and sequenced a fragment of the chaperone gene (mrkB), which is typically the most highly conserved gene within chaperone/usher fimbrial clusters. Phylogenetic analyses indicated there were five primary alleles which, with the exception of the two K. oxytoca strains, are strongly supported by long internal branches (Fig. 6). The most common allele is that shared by K. pneumoniae MGH78578 and E. coli MS2027. This allele is also shared by E. coli CAUTI strains MS2039 and M148 and cystitis strain M202. Interestingly, mrkB from the pyelonephritis E. coli strain M184 is identical to that found in K. pneumoniae pIA565. The observation that two alleles (represented by strains MGH78578 and IA565) contain sequences that are identical in both E. coli and K. pneumoniae species, but substantially divergent from each other, is strong evidence of recurrent and recent lateral gene transfer of the mrk cluster among K. pneumoniae and E. coli UTI strains.
In conclusion, we have identified the capsule, O antigen, and type 3 fimbriae as factors that affect biofilm growth by CAUTI E. coli. Type 3 fimbriae are produced by many members of the Enterobacteriaceae family that are associated with opportunistic infections. Biofilm growth mediated by type 3 fimbriae may be important for the survival of these organisms on the surfaces of urinary catheters and within the hospital environment. We speculate that the mrk gene cluster in E. coli MS2027 may have originated from K. pneumoniae, and we are currently investigating this possibility.
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (grant no. 455914) and the Australian Research Council (grant no. DP0666852). C.-L.Y.O. was supported by an International Postgraduate Research Scholarship from the University of Queensland. S.A.B. was supported by an NHRMC Howard Florey Centenary fellowship.
The K. pneumoniae genome sequence data were produced by the Genome Sequencing Center at the Washington University School of Medicine in St. Louis, MO.
Footnotes
Published ahead of print on 30 November 2007.
REFERENCES
- 1.Adegbola, R. A., and D. C. Old. 1985. Fimbrial and non-fimbrial hemagglutinins in enterobacter-aerogenes. J. Med. Microbiol. 1935-43. [DOI] [PubMed] [Google Scholar]
- 2.Adegbola, R. A., and D. C. Old. 1983. Fimbrial hemagglutinins in enterobacter species. J. Gen. Microbiol. 1292175-2180. [DOI] [PubMed] [Google Scholar]
- 3.Adegbola, R. A., and D. C. Old. 1982. New fimbrial hemagglutinin in Serratia species. Infect. Immun. 38306-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adegbola, R. A., D. C. Old, and S. Aleksic. 1983. Rare Mr/K-like hemagglutinins (and type-3-like fimbriae) of salmonella strains. FEMS Microbiol. Lett. 19233-238. [Google Scholar]
- 5.Allen, B. L., G. F. Gerlach, and S. Clegg. 1991. Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J. Bacteriol. 173916-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301105-107. [DOI] [PubMed] [Google Scholar]
- 7.Blaesing, F., A. Muhlenweg, S. Vierling, G. Ziegelin, S. Pelzer, and E. Lanka. 2005. Introduction of DNA into Actinomycetes by bacterial conjugation from E-coli—an evaluation of various transfer systems. J. Biotechnol. 120146-161. [DOI] [PubMed] [Google Scholar]
- 8.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 1722665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 1726568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duguid, J. P. 1959. Fimbriae and adhesive properties in Klebsiella strains. J. Gen. Microbiol. 21271-286. [DOI] [PubMed] [Google Scholar]
- 11.Eto, D. S., J. L. Sundsbak, and M. A. Mulvey. 2006. Actin-gated intracellular growth and resurgence of uropathogenic Escherichia coli. Cell. Microbiol. 8704-717. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 2004. PHYLIP (phylogeny inference package), version 3.6. Department of Genome Sciences, Seattle, WA.
- 13.Gerlach, G. F., B. L. Allen, and S. Clegg. 1989. Type 3 fimbriae among enterobacteria and the ability of spermidine to inhibit Mr/K hemagglutination. Infect. Immun. 57219-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412442-445. [DOI] [PubMed] [Google Scholar]
- 15.Hagberg, L., U. Jodal, T. K. Korhonen, G. Lidin-Janson, U. Lindberg, and C. Svanborg Eden. 1981. Adhesion, hemagglutination, and virulence of Escherichia coli causing urinary tract infections. Infect. Immun. 31564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holland, B., and V. Moulton. 2003. Consensus networks: a method for visualizing incompatibilities in collections of trees, p. 165-176. In G. Benson and R. Page (ed.), Algorithms in bioinformatics, WABI 2003. Springer-Verlag, Berlin, Germany.
- 17.Hornick, D. B., B. L. Allen, M. A. Horn, and S. Clegg. 1992. Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect. Immun. 601577-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornick, D. B., B. L. Allen, M. A. Horn, and S. Clegg. 1991. Fimbrial types among respiratory isolates belonging to the family Enterobacteriaceae. J. Clin. Microbiol. 291795-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornick, D. B., J. Thommandru, W. Smits, and S. Clegg. 1995. Adherence properties of an mrkD-negative mutant of Klebsiella pneumoniae. Infect. Immun. 632026-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23254-267. [DOI] [PubMed] [Google Scholar]
- 21.Jagnow, J., and S. Clegg. 2003. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 1492397-2405. [DOI] [PubMed] [Google Scholar]
- 22.Kallenius, G., R. Mollby, S. B. Svenson, I. Helin, H. Hultberg, B. Cedergren, and J. Winberg. 1981. Occurrence of P-fimbriated Escherichia coli in urinary tract infections. Lancet ii1369-1372. [DOI] [PubMed] [Google Scholar]
- 23.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2695-702. [DOI] [PubMed] [Google Scholar]
- 24.Klemm, P., L. Hjerrild, M. Gjermansen, and M. A. Schembri. 2004. Structure-function analysis of the self-recognizing antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51283-296. [DOI] [PubMed] [Google Scholar]
- 25.Klemm, P., and M. A. Schembri. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 29027-35. [DOI] [PubMed] [Google Scholar]
- 26.Korhonen, T. K., E. Tarkka, H. Ranta, and K. Haahtela. 1983. Type 3 fimbriae of Klebsiella sp.: molecular characterization and role in bacterial adhesion to plant roots. J. Bacteriol. 155860-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leffler, H., and C. Svanborg-Eden. 1981. Glycolipid receptors for uropathogenic Escherichia coli on human erythrocytes and uroepithelial cells. Infect. Immun. 34920-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahapatra, N. R., S. Ghosh, P. K. Sarkar, and P. C. Banerjee. 2003. Generation of novel plasmids in Escherichia coli S17-1(pSUP106). Curr. Microbiol. 46318-323. [DOI] [PubMed] [Google Scholar]
- 29.Mobley, H. L., G. R. Chippendale, J. H. Tenney, R. A. Hull, and J. W. Warren. 1987. Expression of type 1 fimbriae may be required for persistence of Escherichia coli in the catheterized urinary tract. J. Clin. Microbiol. 252253-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Old, D. C., and R. A. Adegbola. 1985. Antigenic relationships among type-3 fimbriae of Enterobacteriaceae revealed by immunoelectronmicroscopy. J. Med. Microbiol. 20113-121. [DOI] [PubMed] [Google Scholar]
- 31.Old, D. C., and R. A. Adegbola. 1982. Hemagglutinins and fimbriae of Morganella, Proteus and Providencia. J. Med. Microbiol. 15551-564. [DOI] [PubMed] [Google Scholar]
- 32.Old, D. C., and R. A. Adegbola. 1984. Relationships among broad-spectrum and narrow-spectrum mannose-resistant fimbrial hemagglutinins in different Yersinia species. Microbiol. Immunol. 281303-1311. [DOI] [PubMed] [Google Scholar]
- 33.Old, D. C., and S. S. Scott. 1981. Hemagglutinins and fimbriae of Providencia spp. J. Bacteriol. 146404-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paterson, D. L., and J. Lipman. 2007. Returning to the pre-antibiotic era in the critically ill: the XDR problem. Crit. Care Med. 351789-1791. [DOI] [PubMed] [Google Scholar]
- 35.Pigeon, R. P., and R. P. Silver. 1997. Analysis of the G93E mutant allele of KpsM, the membrane component of an ABC transporter involved in polysialic acid translocation in Escherichia coli K1. FEMS Microbiol. Lett. 156217-222. [DOI] [PubMed] [Google Scholar]
- 36.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30285-293. [DOI] [PubMed] [Google Scholar]
- 36a.R Development Core Team. 2007. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 37.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48933-946. [DOI] [PubMed] [Google Scholar]
- 38.Römling, U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 621234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roux, A., C. Beloin, and J. M. Ghigo. 2005. Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J. Bacteriol. 1871001-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Runnels, P. L., and H. W. Moon. 1984. Capsule reduces adherence of enterotoxigenic Escherichia coli to isolated intestinal epithelial cells of pigs. Infect. Immun. 45737-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Schembri, M. A., J. Blom, K. A. Krogfelt, and P. Klemm. 2005. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 734626-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 1861249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48253-267. [DOI] [PubMed] [Google Scholar]
- 45.Schembri, M. A., and P. Klemm. 2001. Biofilm formation in a hydrodynamic environment by novel FimH variants and ramifications for virulence. Infect. Immun. 691322-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schembri, M. A., E. V. Sokurenko, and P. Klemm. 2000. Functional flexibility of the FimH adhesin: insights from a random mutant library. Infect. Immun. 682638-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schurtz, T. A., D. B. Hornick, T. K. Korhonen, and S. Clegg. 1994. The type 3 fimbrial adhesin gene (mrkD) of Klebsiella species is not conserved among all fimbriate strains. Infect. Immun. 624186-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sebghati, T. A. S., T. K. Korhonen, D. B. Hornick, and S. Clegg. 1998. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect. Immun. 662887-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamm, W. E. 1991. Catheter-associated urinary tract infections: epidemiology, pathogenesis, and prevention. Am. J. Med. 9165S-71S. [DOI] [PubMed] [Google Scholar]
- 50.St. Geme, J. W., III, and S. Falkow. 1991. Loss of capsule expression by Haemophilus influenzae type b results in enhanced adherence to and invasion of human cells. Infect. Immun. 591325-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarkkanen, A. M., B. L. Allen, B. Westerlund, H. Holthofer, P. Kuusela, L. Risteli, S. Clegg, and T. K. Korhonen. 1990. Type V collagen as the target for type-3 fimbriae, enterobacterial adherence organelles. Mol. Microbiol. 41353-1361. [DOI] [PubMed] [Google Scholar]
- 52.Tarkkanen, A. M., R. Virkola, S. Clegg, and T. K. Korhonen. 1997. Binding of the type 3 fimbriae of Klebsiella pneumoniae to human endothelial and urinary bladder cells. Infect. Immun. 651546-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ulett, G. C., A. N. Mabbett, K. C. Fung, R. I. Webb, and M. A. Schembri. 2007. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology 1532321-2331. [DOI] [PubMed] [Google Scholar]
- 54.Ulett, G. C., J. Valle, C. Beloin, O. Sherlock, J. M. Ghigo, and M. A. Schembri. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 753233-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valle, J., S. Da Re, N. Henry, T. Fontaine, D. Balestrino, P. Latour-Lambert, and J. M. Ghigo. 2006. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. USA 10312558-12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Virji, M., K. Makepeace, D. J. Ferguson, M. Achtman, and E. R. Moxon. 1993. Meningococcal Opa and Opc proteins: their role in colonization and invasion of human epithelial and endothelial cells. Mol. Microbiol. 10499-510. [DOI] [PubMed] [Google Scholar]
- 57.Vrionis, H. A., A. J. Daugulis, and A. M. Kropinski. 2002. Identification and characterization of the AgmR regulator of Pseudomonas putida: role in alcohol utilization. Appl. Microbiol. Biotechnol. 58469-475. [DOI] [PubMed] [Google Scholar]
- 58.Warren, J. W. 2001. Catheter-associated urinary tract infections. Int. J. Antimicrob. Agents 17299-303. [DOI] [PubMed] [Google Scholar]
- 59.Warren, J. W., D. Damron, J. H. Tenney, J. M. Hoopes, B. Deforge, and H. L. Muncie, Jr. 1987. Fever, bacteremia, and death as complications of bacteriuria in women with long-term urethral catheters. J. Infect. Dis. 1551151-1158. [DOI] [PubMed] [Google Scholar]
- 60.Warren, J. W., J. H. Tenney, J. M. Hoopes, H. L. Muncie, and W. C. Anthony. 1982. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J. Infect. Dis. 146719-723. [DOI] [PubMed] [Google Scholar]
- 61.Watson, N. 1988. A new revision of the sequence of plasmid pBR322. Gene 70399-403. [DOI] [PubMed] [Google Scholar]
- 62.Wu, X. R., T. T. Sun, and J. J. Medina. 1996. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc. Natl. Acad. Sci. USA 939630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 391452-1463. [DOI] [PubMed] [Google Scholar]