Abstract
Corynebacterium glutamicum can grow on l-lactate as a sole carbon and energy source. The NCgl2816-lldD operon encoding a putative transporter (NCgl2816) and a quinone-dependent l-lactate dehydrogenase (LldD) is required for l-lactate utilization. DNA affinity chromatography revealed that the FadR-type regulator LldR (encoded by NCgl2814) binds to the upstream region of NCgl2816-lldD. Overexpression of lldR resulted in strongly reduced NCgl2816-lldD mRNA levels and strongly reduced LldD activity, and as a consequence, a severe growth defect was observed in cells grown on l-lactate as the sole carbon and energy source, but not in cells grown on glucose, ribose, or acetate. Deletion of lldR had no effect on growth on these carbon sources but resulted in high NCgl2816-lldD mRNA levels and high LldD activity in the presence and absence of l-lactate. Purified His-tagged LldR bound to a 54-bp fragment of the NCgl2816-lldD promoter, which overlaps with the transcriptional start site determined by random amplification of cDNA ends-PCR and contains a putative operator motif typical of FadR-type regulators, which is −1TNGTNNNACNA10. Mutational analysis revealed that this motif with hyphenated dyad symmetry is essential for binding of LldD to the NCgl2816-lldD promoter. l-Lactate, but not d-lactate, interfered with binding of LldRHis to the NCgl2816-lldD promoter. Thus, during growth on media lacking l-lactate, LldR represses expression of NCgl2816-lldD. In the presence of l-lactate in the growth medium or under conditions leading to intracellular l-lactate accumulation, the l-lactate utilization operon is induced.
Lactate is a major product of anaerobic metabolism, but it also serves as a carbon and energy source for anaerobic and aerobic microorganisms. Lactate can be fermented to acetate, propionate, or butyrate by, e.g., sulfate-reducing bacteria, propionibacteria, or Eubacterium hallii (11). When oxygen becomes available but glucose is limiting, Lactobacillus plantarum converts its fermentation product, lactate, to acetate (18). Aerobic growth with l-lactate as the sole carbon and energy source has been studied in Escherichia coli in some detail. l-Lactate is taken up into the E. coli cell either by the l-lactate permease LldP or by the glycolate permease GlcA (39). l-Lactate is oxidized to the central metabolite pyruvate by quinone-dependent l-lactate dehydrogenase (LldD; EC 1.1.2.3) (10). For growth on l-lactate, E. coli requires lldD, which forms an operon with lldP and the putative lactate regulator gene lldR (10). Transcription of lldDRP is repressed by ArcAB under anaerobic reducing conditions (24) and is maximal in the presence of l-lactate (10); however, regulation of lldDRP by the putative regulator LldR encoded in this operon has not been analyzed yet in detail.
Recently, we identified the l-lactate utilization operon in Corynebacterium glutamicum, a nonpathogenic gram-positive soil bacterium that is widely used for biotechnological production of amino acids such as l-glutamate and l-lysine. C. glutamicum can grow aerobically on a variety of sugars, sugar alcohols, and organic acids, including l-lactate, as sole carbon and energy sources (9, 17, 27, 31, 36, 59). C. glutamicum forms l-lactate with the soluble NAD+-dependent l-lactate dehydrogenase (EC 1.1.1.27) encoded by ldhA (3, 22) under oxygen deprivation (22) and as a by-product during glutamate and lysine production (27, 28, 53). For l-lactate utilization, on the other hand, C. glutamicum requires the quinone-dependent l-lactate dehydrogenase LldD (EC 1.1.2.3) (53), which is a peripheral membrane protein (51) catalyzing oxidation of l-lactate to pyruvate (3, 53).
The C. glutamicum l-lactate utilization operon comprises the quinone-dependent l-lactate dehydrogenase gene lldD and a gene encoding a putative permease (NCgl2816), and its expression is maximal in the presence of l-lactate (53). C. glutamicum reutilizes l-lactate formed during glutamate production in the presence of glucose (53) and coutilizes l-lactate with glucose when it is grown on glucose-l-lactate mixtures. Coutilization of glucose with acetate (57), propionate (5), protocatechuate and vanillate (35), serine (37), and fructose (8) has also been observed, while C. glutamicum utilizes glucose before it utilizes glutamate and ethanol (2, 31). During coutilization of glucose and l-lactate, the specific activity of the quinone-dependent l-lactate dehydrogenase LldD was almost as high as it was with l-lactate alone, while it was about sevenfold lower with glucose as a sole carbon source (53). The apparent absence of glucose repression and the approximately 17-fold-higher levels of mRNA of NCgl2816-lldD during growth on l-lactate than during growth on pyruvate as a sole carbon and energy source as determined by transcriptome analyses (53) suggest that the NCgl2816-lldD operon is subject to l-lactate-specific regulation. However, a putative regulatory gene is not present in the C. glutamicum NCgl2816-lldD lactate utilization operon. Here, we identified a previously unknown FadR-type regulator of the NCgl2816-lldD operon, which we designated LldR, and characterized its role in l-lactate-dependent regulation of NCgl2816-lldD expression.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All strains and plasmids used in this work are listed in Table 1. C. glutamicum type strain ATCC 13032 (30) was used as the wild type (WT). For growth experiments, determination of the LldD activity, and DNA microarray experiments, the first preculture, in 70 ml Luria-Bertani (LB) medium (48), was inoculated from a fresh LB agar plate and incubated in a 500-ml baffled shake flask at 30°C and 120 rpm. For DNA microarray experiments, cells were washed in culture medium without any carbon source, and the second preculture and the main culture were inoculated to obtain optical densities at 600 nm of 0.1 and 0.5, respectively, in 60 ml CgXII minimal medium (26), which contained 0.03 g/liter protocatechuic acid and 0.2 mg/liter biotin. For growth experiments and determination of the LldD activity, the main culture was inoculated to obtain an optical density of 1. The following compounds were used as carbon and energy sources: 200 mM glucose, 200 mM potassium acetate, 200 mM sodium pyruvate, 200 mM sodium l-lactate, 100 mM ribose, 50 mM fructose plus 100 mM sodium l-lactate, and 50 mM glucose plus 100 mM sodium l-lactate. Media contained 50 μg/ml kanamycin, 50 μg/ml ampicillin, or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), when appropriate. For all cloning experiments, E. coli DH5α (20) was used as the host and was cultivated aerobically at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−thi-1 endA1 hsdr17(rK− mK−) supE44 ΔlacU169 (Φ80lacZΔM15) recA1 gyrA96 relA1 | Bethesda Research Laboratories (20) |
| BL21(DE3) | ompT hsdSB(rB− mB−) gal dcm (DE3) | 54 |
| C. glutamicum strains | ||
| WT | Wild-type strain ATCC 13032, auxotrophic for biotin | 1 |
| WTΔlldR | In-frame deletion of lldR gene | This study |
| Plasmids | ||
| pK19mobsacB | Kanr, E. coli/C. glutamicum shuttle vector for construction of insertion and deletion mutants in C. glutamicum (pK18 oriVE.c.sacB lacZα) | 49 |
| pK19mobsacB-ΔlldR | Kanr, pK19mobsacB with deletion construct of the lldR gene | This study |
| pVWEx1 | Kanr, PtaclacIq | 43 |
| pVWEx1-lldR | Kanr, pVWEx1 with a 761-bp fragment of the lldR gene and an artificial ribosome-binding site | This study |
| pET16b | Ampr, overproduction of proteins with an N-terminal decahistidine tag in E. coli (pBR322 oriVE.c.PT7lacI) | Novagen |
| pET16b-lldRHis | Ampr, pET16b with a 707-bp fragment of the lldR gene | This study |
Recombinant DNA experiments.
The enzymes used for recombinant DNA work were obtained from Roche Diagnostics (Mannheim, Germany). The oligonucleotides were obtained from Operon (Cologne, Germany) or MWG (Ebersberg, Germany). Standard methods, including PCR, restriction, or ligation, were carried out as described by Sambrook and Russell (48). Plasmids were isolated from E. coli with a QIAprep spin miniprep kit (Qiagen, Hilden, Germany). E. coli was transformed by the RbCl method (19), and C. glutamicum was transformed by electroporation (55) using the following conditions: 25 μF, 600 Ω, and 2.5 kV/cm (Bio-Rad Gene Pulser Xcell; Bio-Rad Laboratories, Hercules, CA). After electroporation, 4 ml LB medium was immediately added to the sample. After a heat shock at 46°C for 6 min, the cells were incubated at 30°C for 50 min to regenerate before they were plated. DNA sequencing was performed by Agowa GmbH (Berlin, Germany).
Construction of an lldR deletion mutant.
An in-frame lldR (NCgl2814) deletion mutant of C. glutamicum was constructed by a two-step homologous recombination procedure as described previously (38). The lldR up- and downstream regions (∼450 bp each) were amplified using the oligonucleotide pairs lldR-A/lldR-B and lldR-C/lldR-D. The PCR products served as templates for crossover PCR performed with oligonucleotides lldR-A and lldR-D. The resulting ∼0.9-kb PCR product was restricted with SphI and XbaI and cloned into SphI/XbaI-restricted plasmid pK19mobsacB. After DNA sequence analysis of the resulting plasmid, pk19mobsacB-ΔlldR, confirmed that the cloned PCR product did not contain mutations, the plasmid was transferred into C. glutamicum by electroporation. Screening for the first and second recombination events was performed as described previously (38). Kanamycin-sensitive and sucrose-resistant clones were tested by PCR analysis of chromosomal DNA with the primer pair lldR-0/lldR-1. Clones that had the desired in-frame deletion of the lldR gene, in which all of the nucleotides except the first 6 codons and the last 12 codons were replaced by a 21-bp tag, had an 0.93-kb PCR fragment instead of the 1.6-kb PCR fragment obtained with wild-type DNA.
Overproduction and purification of LldRHis.
E. coli BL21(DE3) carrying plasmid pET16b-lldR was grown at 37°C in 500 ml of LB medium with 50 μg/ml ampicillin to an optical density at 600 nm of 0.6 before 1 mM IPTG was added. After cultivation for another 4 h at room temperature, cells were harvested by centrifugation (10 min, 11,325 × g, 4°C), washed in 20 ml TNI5 buffer (20 mM Tris-HCl [pH 7.9], 300 mM NaCl, 5% [vol/vol] glycerol, 5 mM imidazole), and stored at −20°C. For preparation of cell extracts, thawed cells were resuspended in 10 ml of TNI5 buffer containing 1 mM diisopropylfluorophosphate and 1 mM phenylmethylsulfonyl fluoride. The cell suspension was passed five times through a French pressure cell (SLM Aminco, Spectronic Instruments, Rochester, NY) at 1,800 lb/in2. Cell debris and intact cells were removed by centrifugation (20 min, 5,292 × g, 4°C). The cell extract was then subjected to ultracentrifugation (1 to 1.5 h, 150,000 × g, 4°C). After ultracentrifugation, the supernatant was purified by nickel affinity chromatography using nickel-activated nitrilotriacetic acid-agarose (Novagen, San Diego, CA). The column was washed with TNI175 buffer (which contained 175 mM imidazole). Then the LldRHis protein was eluted with TNI400 buffer (which contained 400 mM imidazole). Dominant protein-containing fractions were pooled, and the elution buffer was exchanged against BS buffer (100 mM Tris-HCl, 20% [vol/vol] glycerol, 100 mM KCl, 20 mM MgCl2, 1 mM EDTA; pH 7.5) using PD10 columns.
Quinone-dependent l-lactate dehydrogenase assay.
For determination of enzyme activities, exponentially growing cells were harvested by centrifugation (4,500 × g, 5 min, 4°C), and crude extracts were prepared as described previously by Stansen et al. (53). The quinone-dependent l-lactate dehydrogenase activity was measured using a spectrophotometric assay mixture containing 100 mM KH2PO4 (pH 7.5), 0.05 mM 2,6-dichloroindophenol, and an appropriate amount of crude extract. The assay was started by addition of 20 mM l-lactate at 30°C, and the decrease in absorbance of 2,6-dichloroindophenol (ɛ600 = 20 mM−1 cm−1) was determined.
Determination of the transcriptional start site.
The transcriptional start site of the NCgl2816-lldD operon was determined by random amplification of cDNA ends (RACE)-PCR using a 5′/3′ second-generation RACE kit (Roche, Mannheim, Germany) as recommended by the manufacturer. The primers used were 2816-RT for reverse transcription, 2816-PCR1 for the first PCR, and 2816-PCR2 for the nested PCR.
Gel shift assays.
Gel shift assays with LldRHis were performed as described previously (58). Briefly, purified LldRHis (at concentrations ranging from 0 to 2.4 μM) was mixed with the full-length promoter of NCgl2816-lldD fragment F0 or promoter subfragments F1 to F5 in a 20-μl (total volume) mixture that contained 50 mM Tris-HCl, 10% (vol/vol) glycerol, 50 mM KCl, 10 mM MgCl2, and 0.5 mM EDTA (pH 7.5). Then a nontarget promoter fragment was added at a concentration of 40 to 46 nM as a negative control. The full-length promoter of NCgl2816-lldD covering the region from position −252 to position 79 relative to the translational start was obtained by performing PCR with the primers listed in Table 2. After incubation for 30 min at room temperature, the samples were separated on a 10% native polyacrylamide gel at room temperature and 170 V (constant voltage) using 1× Tris-borate-EDTA (89 mM Tris base, 89 mM boric acid, 2 mM EDTA; pH 8.3) as the electrophoresis buffer. The gels were subsequently stained with SYBR green I according to the instructions of the supplier (Sigma, Rödermark, Germany) and photographed. To test for possible effectors, the protein was incubated with glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate, phosphoenolpyruvate, pyruvate, l-lactate, d-lactate, and acetyl-coenzyme (acetyl-CoA) (20 mM each) in the binding buffer for 15 min before promoter DNA fragment F0 was added and the mixture was incubated for an additional 30 min. All PCR products used in the gel shift assays were purified with a PCR purification kit (Qiagen, Hilden, Germany) and eluted in 10 mM Tris-HCl (pH 8.5).
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′→3′)a | Use |
|---|---|---|
| Overexpression | ||
| 310-Kom-for | GCGTCGACAAGGAGATATAGATATGAGTGTGAAAGCACATGAATC (SalI) | Amplification of NCgl2814 (forward) |
| 310-Kom-rev | GCGTCGACCGTGTAGATCTGAAACCGC (SalI) | Amplification of NCgl2814 (reverse) |
| 310-His-for | GCCATATGAGTGTGAAAGCACATGAATCTGTC (NdeI) | Amplification of NCgl2814 for purification (forward) |
| 310-His-rev | GCGTCGACTTAGGCCTCGGCGGCAG (SalI) | Amplification of NCgl2814 for purification (reverse) |
| Deletion of NCgl2814 | ||
| 310-A | GCGCATGCAGGTTCCGCGGATAAACAG (SphI) | Amplification of the left internal fragment (forward) |
| 310-B | CCCATCCACTAAACTTAAACATTCATGTGC TTTCACACTCATTAG | Amplification of the left internal fragment (reverse) |
| 310-C | TGTTTAAGTTTAGTGGATGGGGGCTACTAC GAAGAAACCG | Amplification of the right internal fragment (forward) |
| 310-D | GCTCTAGATTGTTTCGCGGTGAGGC (XbaI) | Amplification of the right internal fragment (reverse) |
| 310-0 | TCAAAGCTTTCAACGTGCC | Proof of deletion (forward) |
| 310-1 | GCTAGTTCGTCGTCTAGC | Proof of deletion (reverse) |
| Gel shift | ||
| 312-F1 | GCCACGTGGAGGATCCTTTGGG (BamHI) | Amplification of fragments F0 and F1 (forward) |
| 312-R3 | CTGCCACTCGAGCTCCCCAGC (SacI) | Amplification of fragments F0 and F5 (reverse) |
| 312-R1 | CCCATTTAAGCAACAGAGTTAGTTAATC | Amplification of fragment F1 (reverse) |
| 312-F12 | GCGTCCGTGGCCGTTTCC | Amplification of fragment F2 (forward) |
| 312-R21 | GGCGTGTCACCTTTAATTGTCAATGG | Amplification of fragments F2 and F12 (reverse) |
| 312-F2 | GGTAATTGGATTCGACTGTTTTCC | Amplification of fragment F3 (forward) |
| 312-R2 | TAAACGGGCTGAAACCGATTGG | Amplification of fragment F3 (reverse) |
| 312-F22 | TTACATTCTTGTGGTCTGACCATG | Amplification of fragment F4 (forward) |
| 312-R31 | TTTTGATCTACTGCGGTTGTCATG | Amplification of fragments F4, F4WT, F4M1, F4M2, F4M3, F4M4, F4M5, F4M6, and F4M12 (reverse) |
| 312-F3 | TCCCGCCGTCCGTTTCAGAGAAGAGG | Amplification of fragment F5 (forward) |
| 0430-for | GAAAGCTCAGAAGAAGGTCCAGAG | Amplification of control fragment 0430 (forward) |
| 0430-rev | GCTGGATGGGATAACGGAGGTC | Amplification of control fragment 0430 (reverse) |
| 312-F4-WT | TTACATTCTTGTGGTCTGACCATGAGGTTGGG | Amplification of fragment F4WT (forward) |
| 312-F4-Mut1 | TTACATTCTTGGGTGCTGACCATGAGGTTGGG | Amplification of fragment F4M1 (forward) |
| 312-F4-Mut2 | TTACATTCTTGTGGTCTGCACGTGAGGTTGGG | Amplification of fragment F4M2 (forward) |
| 312-F4-Mut12 | TTACATTCTTGGGTGCTGCACGTGAGGTTGGG | Amplification of fragment F4M12 (forward) |
| 312-F4-Mut3 | TGCAATTCTTGGGTGCTGCACGTGAGGTTGGG | Amplification of fragment F4M3 (forward) |
| 312-F4-Mut4 | TTACCAACTTGGGTGCTGCACGTGAGGTTGGG | Amplification of fragment F4M4 (forward) |
| 312-F4-Mut5 | TTACCAACTTGGGTGCTGCACGTGAGTGGGGGCCAATCG | Amplification of fragment F4M5 (forward) |
| 312-F4-Mut6 | TTACATTCTTGGGTGCTGCACGTGAGGTTTTTCCAATCGGTTC | Amplification of fragment F4M6 (forward) |
| RACE-PCR | ||
| 2816-RT | CGCCACCCATGAGCATCAAGG | Primer for reverse transcription of NCgl2816 |
| 2816-PCR1 | GATGTGGGCGAAGATGAC | PCR primer 1 for NCgl2816 |
| 2816-PCR2 | CAGGGAGAAGGATGCGTACG | Nested PCR primer for NCgl2816 |
Restriction sites are underlined, and the restriction enzymes are indicated in parentheses. The overlapping complementary sequences for crossover PCR and start codons are indicated by bold type.
DNA microarray analysis.
Generation of C. glutamicum whole-genome DNA microarrays (56), synthesis of fluorescently labeled cDNA from total RNA, microarray hybridization, washing, and gene expression analysis were performed as described previously (23, 32, 33, 44). Genes that exhibited significantly changed mRNA levels (changed by at least a factor of two; P ≤ 0.05, as determined by Student's t test) were determined in two different DNA microarray experiments performed with RNA isolated from two independent cultures in CgXII minimal medium.
Affinity chromatography.
Enrichment of DNA-binding proteins interacting with the upstream regions of NCgl2816-lldD was performed as described previously (13). A 331-bp NCgl2816 promoter fragment was amplified by PCR using genomic DNA from C. glutamicum and oligonucleotides 312-F1 and 312-R3, one of which (312-F1) was tagged with biotin via a TEG linker (Operon, Cologne, Germany). Proteins that bound nonspecifically were washed off with TGED buffer (50 mM Tris-HCl [pH 7.6], 1 mM dithiothreitol, 10 mM MgCl2, 1 mM EDTA, 10% [vol/vol] glycerol, 10 μM phenylmethylsulfonyl fluoride) containing 400 μg chromosomal DNA, and specifically bound proteins were subsequently eluted with TGED buffer containing 2 M NaCl. The proteins present in the high-salt eluate were separated on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels and subsequently analyzed by matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectrometry (see below).
MALDI-TOF mass spectrometry.
For peptide mass fingerprinting, a protein band of interest was excised from colloidal Coomassie blue-stained gels and subjected to in-gel digestion with trypsin essentially as described previously (50). Data acquisition and analysis were performed using a Voyager DE-STR mass spectrometer (Applied Biosystems, Weiterstadt, Germany), Voyager Control Panel software (version 5.0), and Voyager Data Explorer software (version 3.5) as described previously (50).
RESULTS
Identification of LldR as a protein binding to the upstream region of the NCgl2816-lldD operon.
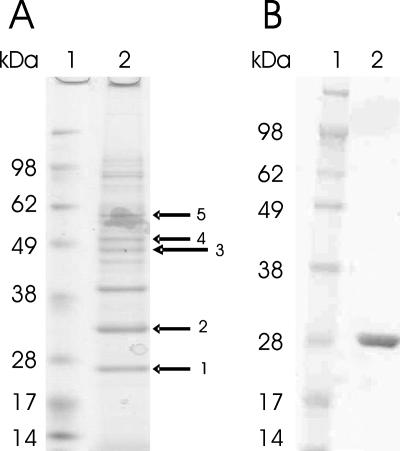
To identify a transcriptional regulator(s) of the l-lactate utilization operon NCgl2816-lldD, which appears to be regulated by l-lactate (53), proteins specifically binding to its upstream region were enriched by DNA affinity chromatography. A 331-bp biotinylated promoter DNA probe (positions −251 to 80 relative to the NCgl2816 start codon) was linked to streptavidin-coated magnetic beads and incubated with crude extracts from C. glutamicum grown on minimal medium containing l-lactate as the sole carbon source. After washing with TGED buffer containing 400 μg of genomic DNA from C. glutamicum as a competitor, specifically bound proteins were eluted with buffer containing 2 M NaCl and identified by peptide mass fingerprint analysis as described previously (4, 13, 32). Among these proteins was a putative transcriptional regulator (Fig. 1A), which was designated LldR and was subsequently shown to regulate the NCgl2816-lldD operon. The lldR gene (corresponding to NCgl2814) is located close to the NCgl2816-lldD operon and is separated only by NCgl2815 encoding a hypothetical protein. As deduced from the gene sequence, LldR consists of 213 amino acids and has a predicted molecular mass of 25.1 kDa, which corresponds well with the apparent mass observed by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 1). Its N terminus contains a predicted helix-turn-helix domain for DNA binding, and over its entire length LldR shares similarities with FadR-type regulators, a subfamily of GntR-type regulators (47) that belongs to the cluster of orthologous genes COG2186.
FIG. 1.
(A) SDS-PAGE of C. glutamicum proteins eluted in a DNA affinity chromatography experiment with TGED buffer containing 2 M NaCl using the NCgl2816-lldD promoter as a probe and cell extracts from l-lactate-grown cells of C. glutamicum WT (lane 2). MALDI-TOF mass spectrometry of tryptic peptides from the protein bands revealed LldR (arrow 1), RamA (NCgl2472) (arrow 2), a subunit of DNA polymerase I (NCgl1299) (arrow 3), a subunit of DNA polymerase III (NCgl2035) (arrow 4), and a subunit of a putative restriction nuclease (NCgl1705) (arrow 5). The prominent band between arrows 2 and 3 could not be identified. Lane 1 contained protein standards. (B) SDS-PAGE of purified LldR. Lane 2 contained purified LldR with a His tag, and lane 1 contained protein standards.
Binding of purified LldRHis to the NCgl2816-lldD promoter.
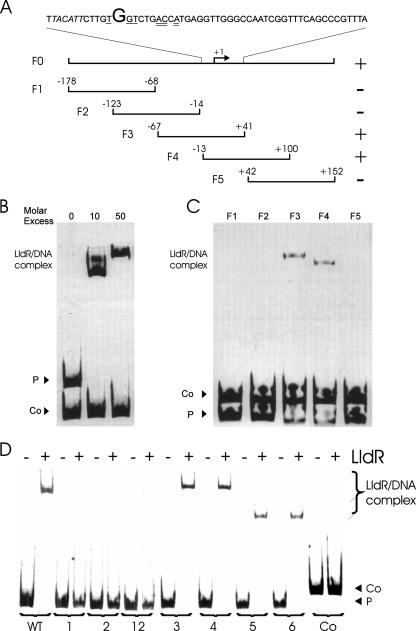
First, the transcriptional start site of the NCgl2816-lldD operon was identified by RACE-PCR. Transcription of the NCgl2816-lldD operon starts with a G which is located 73 bp upstream of the ATG start codon. The promoter contains a TACATT motif (from position −11 to position −6) (Fig. 2A), which closely resembles the consensus −10 hexamer TA(C/T)AAT of C. glutamicum promoters (41), as well as the hexamer TTGACA (from position −36 to position −31), which is similar to the consensus −35 region (TTGCCA).
FIG. 2.
Binding of LldR to the NCgl2816-lldD promoter. (A) DNA fragments used to analyze LldRHis binding to the NCgl2816-lldD promoter region. The numbers indicate the ends of the fragments relative to the NCgl2816 transcriptional start site (+1). Binding of LldR to the fragments is indicated by a plus sign, and a lack of binding is indicated by a minus sign. Oligonucleotides used for amplification of the six fragments are listed in Table 2. The sequence above the fragments shows the region between position −13 and position 41 relative to the transcriptional start site. The transcriptional start is indicated by a large letter, the −10 region is italicized, and the half-sites of the consensus operator sequence for FadR-type regulators are underlined (single and double underlining). (B) Purified His-tagged LldR protein was used in 0-, 10- and 50-fold molar excess over DNA fragment F0 (P) before separation by native PAGE and SYBR green I staining. A 175-bp fragment of the NCgl0430 (43 nM) promoter served as a negative control DNA fragment (Co). (C) Subfragments F1, F2, F3, F4, and F5 of NCgl2816-lldD promoter fragment F0 were incubated with an 11-fold molar excess of purified LldR, separated by PAGE, and stained with SYBR green I. A 190-bp fragment of the NCgl2027 promoter (40 nM) served as a control fragment (Co). (D) Subfragment F4 and derived fragments with different mutations in or near the consensus sequence for FadR-type regulators M1, M2, M12, M3, M4, M5, and M6 (P) were incubated with an 11-fold molar excess of LldRHis. Lanes WT, wild type; lanes 1, the nucleotides in panel A underlined with one line were changed by PCR from TGT to GTG (fragment M1); lanes 2, the nucleotides in panel A underlined with two lines were changed by PCR from ACA to TAG (fragment M2); lanes 12, all the underlined nucleotides in panel A were changed by PCR (fragment 12). Changes outside the consensus sequence were introduced into fragments M3 (7 bp upstream; TCA → GCA) (lanes 3), M4 (4 bp upstream; ATT → CAA) (lanes 4), M5 (3 bp downstream; GTT → TGG) (lanes 5), and M6 (7 bp downstream; GGG → TTT) (lanes 6). A PCR product from position −178 to position −14 relative to the NCgl2816-lldD transcriptional start site (46 nM) served as a negative control (lanes Co). Oligonucleotides used for amplification of the fragments are listed in Table 2.
To characterize the binding of LldR to the upstream region of NCgl2816-lldD, the LldR protein containing an amino-terminal His tag was overproduced in E. coli and purified to near homogeneity by nickel chelate chromatography (see Materials and Methods) (Fig. 1B). For gel shift assays, DNA fragments (24 nM) were incubated with various concentrations of the LldRHis protein (0 to 2.4 μM) and then separated on 10% polyacrylamide gels. LldRHis bound to the upstream region of the NCgl2816-lldD operon with high affinity as a 10-fold molar excess of LldRHis protein resulted in a complete gel shift (Fig. 2B). Two LldR-DNA complexes having different gel mobilities were observed. In contrast, LldRHis did not bind to a negative control DNA, the promoter fragment of NCgl0430 (encoding an uncharacterized putative transcriptional regulator).
Gel shift assays with different and partially overlapping subfragments (72 nM) of the NCgl2816-lldD upstream region allowed confinement of the LldR binding site(s) to a region from position −13 to position 41 relative to the transcriptional start site of NCgl2816 (Fig. 2C). Subfragments F1, F2, and F5 were not bound by LldR (11-fold molar excess), whereas an LldR-DNA complex was formed after incubation of LldR with subfragments F3 and F4. These data indicate that the overlapping region of subfragments F3 and F4 (positions −13 to 41) contains the binding site of LldR. By comparing known or putative operator sites of several FadR-type regulators, Rigali et al. (47) postulated that TNGTNNNACNA is the consensus operator motif for FadR-type regulators. This motif with hyphenated dyad symmetry is present in the LldR-binding region at positions −1 to 10 (Fig. 2A). To test whether the putative consensus operator motif plays a role in binding of LldR to the NCgl2816-lldD promoter, we performed gel shift assays with subfragment F4 and three derived variants containing mutations in the left and/or right putative operator half-sites. In mutant M1, the left three conserved nucleotides of the inverted repeat (Fig. 2A) were changed from TNGT to GNTG, while in mutant M2 the right three conserved nucleotides of the inverted repeat (Fig. 2A) were changed from ACNA to CANG. Mutant M12 had these changes in both half-sites (Fig. 2D). In gel shift assays, wild-type subfragment F4 was completely shifted by LldR at a 10-fold molar excess, whereas the mutations in both half-sites of the putative consensus motif for FadR-type regulators described above abolished the formation of an LldR-DNA complex (Fig. 2D). Mutations outside this motif (mutants M3 to M6) did not affect binding of LldR (Fig. 2D, lanes 3 to 6). The data reveal that LldR binds to the motif TGGTCTGACCA in the promoter region of the NCgl2816-lldD operon and that both the TNGT nucleotides at positions −1, 2, and 3 and the ACNA nucleotides at positions 7, 8, and 10 are essential for this interaction.
l-Lactate prevents binding of LldRHis to the NCgl2816-lldD promoter region.
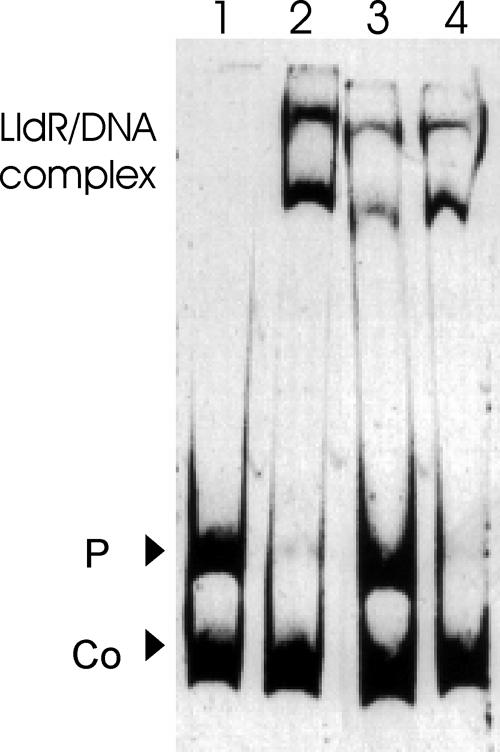
As expression of the NCgl2816-lldD operon is maximal when l-lactate is present in the medium and as the binding affinity of FadR-type regulators can be modulated by an effector molecule, whether binding of LldR to the NCgl2816-lldD promoter region was affected by intermediates of the central carbon metabolism was tested. To do this, the purified LldRHis protein was incubated with the putative effectors at a concentration of 20 mM for 15 min before addition of NCgl2816-lldD promoter fragment F0 (24 nM), and after further incubation for 30 min free DNA and protein-DNA complexes were separated on 10% nondenaturing polyacrylamide gels. The presence of 20 mM phosphoenolpyruvate, glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate, pyruvate, or acetyl-CoA had no effect on the affinity of binding of LldRHis to promoter fragment F0 (data not shown). However, 40 mM l-lactate (and to a lesser extent 20 mM l-lactate [data not shown]) prevented binding of LldRHis to the DNA region upstream of NCgl2816-lldD (Fig. 3, lane 3), while 40 mM d-lactate did not prevent this binding (lane 4). Thus, l-lactate could be identified as an inducer of LldR.
FIG. 3.
Binding of LldR in the presence of d-lactate or l-lactate. The 331-bp F0 fragment (24 nM) of the NCgl2816-lldD promoter was incubated without protein (lane 1) or with a 20-fold molar excess of purified LldR in the absence of an effector (lane 2), in the presence of 40 mM l-lactate (lane 3), or in the presence of 40 mM d-lactate (line 4). A 175-bp promoter fragment of NCgl0430 (43 nM) served as a negative control (Co).
Effects of inactivation and overexpression of lldR on growth and LldD activity.
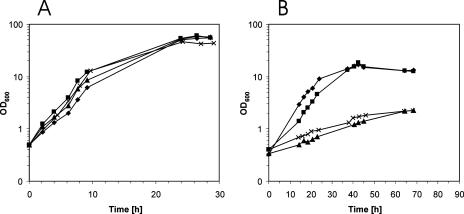
For functional analysis of the lldR gene, an in-frame deletion mutant was constructed by two-step homologous recombination. In the resulting mutant, WTΔlldR, the whole lldR coding region except the 6 5′-terminal codons and 12 3′-terminal codons was replaced by a 21-bp tag (see Materials and Methods). For IPTG-inducible overexpression of the lldR gene, the gene was cloned into the E. coli/C. glutamicum shuttle vector pVWEx1. There were no significant differences in growth rate and biomass formation between C. glutamicum strains WT(pVWEx1), WTΔlldR(pVWEx1), and WTΔlldR(pVWEx1-lldR) in minimal medium containing glucose, pyruvate, acetate, or ribose as the sole carbon source (Fig. 4 and data not shown). However, when 200 mM l-lactate was the sole carbon source, the growth of C. glutamicum WTΔlldR(pVWEx1-lldR) and WT(pVWEx1-lldR) was perturbed as the growth rates (0.04 and 0.03 h−1, respectively) and biomass formation were reduced compared to those of C. glutamicum WT(pVWEx1) and WTΔlldR(pVWEx1) (growth rates, 0.10 and 0.12 h−1, respectively) (Fig. 4). A lag phase in lactate medium was observed for WT(pVWEx1) but not for WTΔlldR(pVWEx1), which is consistent with the view that in the wild type some time is required for induction of the NCgl2816-lldD operon, while the operon is always derepressed in the lldR deletion mutant.
FIG. 4.
Growth of C. glutamicum strains WT(pVWEx1) (▪), WTΔlldR(pVWEx1) (⧫), WT(pVWEx1-lldR) (×), and WTΔlldR(pVWEx1-lldR) (▴) on minimal medium with 200 mM glucose (A) or 200 mM sodium l-lactate (B). IPTG (1 mM) was added immediately after inoculation.
The specific activity of the quinone-dependent l-lactate dehydrogenase LldD was determined using crude extracts of C. glutamicum WT(pVWEx1), WTΔlldR(pVWEx1), and WTΔlldR(pVWEx1-lldR) grown on minimal medium with l-lactate, l-lactate plus glucose, glucose, pyruvate, acetate, or ribose. On media lacking l-lactate, the specific activities of LldD were low (0.01 to 0.02 μmol min−1 mg [dry weight]−1) in C. glutamicum WT(pVWEx1), while they were 6- to 15-fold higher during growth on 200 mM l-lactate and 50 mM glucose plus 100 mM l-lactate (0.13 and 0.15 μmol min−1 mg−1, respectively) (Table 3). In the strain lacking lldR, the specific activities of LldD were high on all media tested [7- to 16-fold higher than the specific activity in C. glutamicum WT(pVWEx1) on media lacking l-lactate] (Table 3). The finding that the specific activities of LldD were slightly higher in C. glutamicum WT(pVWEx1) grown on l-lactate and on glucose plus l-lactate than in the strain lacking lldR might indicate that an additional regulator(s) is involved (Table 3). Overexpression of lldR led to very low specific activities of LldD on all carbon sources tested even in the presence of l-lactate (Table 3).
TABLE 3.
Specific activities of the quinone-dependent l-lactate dehydrogenase in C. glutamicum strains WT(pVWEx1), WTΔlldR(pVWEx1), and WTΔlldR(pVWEx1-lldR)
| Carbon source(s) | Sp act of quinone-dependent l-lactate dehydrogenase LldD (μmol min−1 mg−1)a
|
||
|---|---|---|---|
| WT(pVWEx1) | WTΔlldR(pVWEx1) | WTΔlldR(pVWEx1-lldR) | |
| l-Lactate | 0.14 | 0.08 | <0.01 |
| Glucose + l-lactate | 0.15 | 0.12 | 0.01 |
| Glucose | 0.01 | 0.13 | 0.01 |
| Pyruvate | 0.01 | 0.10 | 0.01 |
| Acetate | 0.01 | 0.16 | <0.01 |
| Ribose | 0.02 | 0.14 | <0.01 |
All data are mean values of at least two determinations for at least two independent cultures with errors of <14%. The cultures contained 1 mM IPTG.
DNA microarray analysis of the transcriptomes of C. glutamicum WT(pVWEx1), WTΔlldR(pVWEx1), and WTΔlldR(pVWEx1-lldR).
In order to determine the effects of LldR on global gene expression, whole-genome DNA microarrays of C. glutamicum (56) were used to compare the mRNA levels of strains WT(pVWEx1), WTΔlldR(pVWEx1), and WTΔlldR(pVWEx1-lldR). In the absence of LldR, only the genes of the l-lactate-utilizing NCgl2816-lldD operon showed significantly increased mRNA levels (NCgl2816, 8.8-fold increased; and lldD, 6.8-fold increased). On the other hand, overexpression of lldR led to twofold decreases in the mRNA levels of NCgl2715 and ldhA, as well as to strongly decreased mRNA levels of lldD and NCgl2816 (25- and 11-fold decreased levels, respectively, compared to the control). However, as only lldD and NCgl2816 showed increased mRNA levels in the absence of LldR and decreased mRNA levels when lldR was overexpressed, LldR likely regulates only the NCgl2816-lldD operon for l-lactate utilization.
DISCUSSION
In this study we showed that the C. glutamicum protein LldR, which belongs to the FadR subfamily of GntR family regulators, represses expression of the NCgl2816-lldD operon. Homologs of LldR from C. glutamicum are encoded in the genomes of other Corynebacterineae, like C. glutamicum R (97% sequence identity; cgR_2816), Corynebacterium efficiens (76% sequence identity; CE2757), Corynebacterium diphtheriae (38% sequence identity; DIP0011), Rhodococcus sp. (42% sequence identity; RHA1_ro03478), and Mycobacterium smegmatis (42% sequence identity; MSMEG_0895), while other mycobacterial genomes apparently lack homologous genes. In addition to Corynebacterineae, LldR homologs also occur in species of other suborders of the Actinomycetales, like Arthrobacter aurescens (33% sequence identity; AAur_3797), Saccharopolyspora erythraea (37% sequence identity; SACE_3508), and Nocardioides sp. (37% sequence identity; Noca_2132). There is also considerable sequence identity between LldR from C. glutamicum and proteins from distantly related species, including clostridia like Clostridium perfringens (33% sequence identity; CPR_0301) or Caulobacter crescentus (32% sequence identity; CC_2813). LldR from C. glutamicum shares only 19 and 22% sequence identity with the proteins for which the regulator family and subfamily were named, GntR, the gluconate-responsive repressor of the gluconate operon of Bacillus subtilis (15), and FadR, the acyl-CoA-responsive regulator of fatty acid degradation and biosynthesis of E. coli (7, 40), respectively.
LldR (formerly LctR) from E. coli, a putative regulator of the l-lactate utilization operon of this bacterium (10), and LldR from C. glutamicum share 25% identical amino acids over the entire length, 42% identical amino acids in the N-terminal helix-turn-helix DNA-binding domain, and 26% identical amino acids in the first half of the C-terminal domain (amino acids 97 to 164 in C. glutamicum and amino acids 100 to 167 in E. coli), which typically is important for ligand binding in FadR-type regulators. A regulatory role for LldR from E. coli has been inferred only indirectly as anaerobic expression of an lldD-lacZ fusion was elevated when multiple copies of the region upstream of lldP were present (34). Binding of LldR from E. coli to the promoter region of the lldPRD operon has not been demonstrated experimentally, but it was postulated to involve a sequence similar to the binding site of PdhR from E. coli (46) and similar to the consensus sequence for FadR-type regulators, TNGTNNNACNA (47). Alternatively, PdhR, rather than LldR, could bind to this sequence and regulate lldPRD in response to pyruvate availability (34). The binding site of LldR of C. glutamicum could be identified experimentally by gel shift assays and mutational analysis. When binding of LldR from C. glutamicum to the promoter of NCgl2816-lldD was assayed, two LldR-DNA complexes were observed (Fig. 2). The LldR-DNA complex with higher gel mobility was dominant at lower molar excess of LldR. This might have been due either to a second binding event (although a sequence similar to the identified binding site could not be found) or to binding of a higher-order multimer of LldR (e.g., LldR tetramer rather than LldR dimer). The different gel mobilities of subfragments F3 and F4 (Fig. 2C) might be due to a small difference in length (subfragment F4 is 5 bp longer) and/or to the position of the LldR binding site within the fragments (more to the center in subfragment F3). The sequence motif upstream of NCgl2816, −1TGGTCTGACCA10, shows hyphenated dyad symmetry containing the two half-sites, TNGT and ACNA, of the consensus sequence for FadR-type regulators. Mutational analysis revealed that both half-sites are essential for binding of LldR to the NCgl2816-lldD promoter, while mutations outside this motif did not affect LldR binding. The motif overlaps the transcriptional start site of the NCgl2816-lldD operon, which is consistent with a repression mechanism involving interference with the RNA polymerase-promoter interaction.
The inducer of the C. glutamicum NCgl2816-lldD operon could be identified as l-lactate as this compound prevents binding of LldR to the NCgl2816-lldD promoter in vitro at a concentration of 40 mM, while, for comparison, 1 mM pyruvate abolished binding of the FadR-like regulator PdhR from E. coli to the promoter of the pdhR-aceEF-lpd operon (46). However, detection of intracellular l-lactate concentrations of 32 to 39 mM in glucose-grown C. glutamicum ATCC 17965 cells (42) indicates that l-lactate affects LldR function at physiologically relevant concentrations. Besides l-lactate antagonizing repression by LldR, effectors for only two other transcriptional regulators of carbon metabolism are known in C. glutamicum: fructose-6-phosphate inhibits repression of the PTS genes ptsG, ptsS, and ptsF by SugR (14), and cyclic AMP is required for repression of the malate synthase gene aceB by GlxR (29). Regulation of the NCgl2816-lldD operon by a mechanism other than l-lactate via LldR (e.g., by oxygen availability or pH) has not been studied yet, but it was observed that in long-term lactic acid-adapted C. glutamicum cells grown in continuous culture at pH 5.7, the mRNA levels of NCgl2816 and lldD were not changed compared to the levels in continuous cultures at pH 7.5 (25). The finding that RamA binds to the promoter of the NCgl2816-lldD operon (Fig. 1) suggests that RamA represses or activates this operon. Whether RamA is indeed involved in regulation of NCgl2816-lldD remains to be studied.
In C. glutamicum, l-lactate may accumulate in the medium at concentrations up to >200 mM under oxygen deprivation conditions (22). Transient accumulation of l-lactate in the medium can be observed during growth on glucose (45) and to a greater extent during growth on fructose (9, 45) even under fully aerobic conditions. l-Lactate occurs as a by-product during l-lysine production on glucose, fructose, and sucrose (27, 28), as well as during glutamate production (53). Cells grown on l-lactate showed altered mRNA levels for other genes (e.g., the isocitrate lyase gene aceA and the phosphotransacetylase gene pta) in addition to the NCgl2816-lldD operon compared with pyruvate-grown cells (53). l-Lactate was also shown to stimulate S-layer formation of C. glutamicum strain ATCC 14067 (52). These expression differences are likely due to control by the regulators of carbon metabolism RamA (6) and RamB (16), as shown previously for aceA, pta, and the S-layer protein gene cspB (21). As only NCgl2816 and lldD showed >6-fold-higher mRNA levels when lldR was deleted, as well as >10-fold-lower mRNA levels when lldR was overexpressed (Table 4), LldR appears to be a specific regulator of the NCgl2816-lldD operon. According to the current model for regulation of the l-lactate utilization operon NCgl2816-lldD in C. glutamicum, LldR binds to its operator sequence, −1TGGTCTGACCA10, upstream of NCgl2816 and represses transcription of NCgl2816-lldD. In the presence of l-lactate, l-lactate binds to LldR, preventing repression of NCgl2816-lldD by LldR. Thus, transcription of the NCgl2816-lldD operon is controlled by l-lactate availability.
TABLE 4.
Genes showing at least twofold-altered mRNA levels in transcriptome comparisons of C. glutamicum WT(pVWEx1) with WTΔlldR(pVWEx1) and WTΔlldR(pVWEx1-lldR)
| Gene (identifier) | Annotation | mRNA level relative to WT(pVWEx1)
|
|
|---|---|---|---|
| WTΔlldR(pVWEx1)a | WTΔlldR(pVWEx1-lldR)b | ||
| NCgl2715 | Sulfate adenyltransferase subunit | 1.1 | 0.47 |
| NCgl2737 | Putative membrane protease subunit | 0.28 | 0.25 |
| NCgl2810 (ldhA) | LdhA, NAD+-dependent l-lactate dehydrogenase | 1.0 | 0.45 |
| NCgl2814 (lldR) | LldR, repressor of the NCgl2816-lldD operon | 0.15 | 24.9 |
| NCgl2816 | Transporter | 8.8 | 0.04 |
| NCgl2817 (lldD) | LldD, quinone-dependent l-lactate dehydrogenase | 6.8 | 0.09 |
The relative mRNA levels of strains WTΔlldR(pVWEx1) and WT(pVWEx1) were compared during exponential growth on minimal medium containing 100 mM ribose.
The relative mRNA levels of strains WTΔlldR(pVWEx1-lldR) and WT(pVWEx1) were compared during exponential growth on minimal medium containing 50 mM fructose plus 100 mM l-lactate.
Acknowledgments
We thank Hermann Sahm (Jülich, Germany) for support, Michael Bott (Jülich, Germany) for access to and help with the mass spectrometry facility, and Ursula Degner for technical assistance.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Abe, S., K.-I. Takayama, and S. Kinoshita. 1967. Taxonomical studies on glutamic acid-producing bacteria. J. Gen. Appl. Microbiol. 13279-301. [Google Scholar]
- 2.Arndt, A., M. Auchter, T. Ishige, V. F. Wendisch, and B. J. Eikmanns. Ethanol catabolism in Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 3.Bott, M., and A. Niebisch. 2003. The respiratory chain of Corynebacterium glutamicum. J. Biotechnol. 104129-153. [DOI] [PubMed] [Google Scholar]
- 4.Brocker, M., and M. Bott. 2006. Evidence for activator and repressor functions of the response regulator MtrA from Corynebacterium glutamicum. FEMS Microbiol. Lett. 264205-212. [DOI] [PubMed] [Google Scholar]
- 5.Claes, W. A., A. Puhler, and J. Kalinowski. 2002. Identification of two prpDBC gene clusters in Corynebacterium glutamicum and their involvement in propionate degradation via the 2-methylcitrate cycle. J. Bacteriol. 1842728-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cramer, A., R. Gerstmeir, S. Schaffer, M. Bott, and B. J. Eikmanns. 2006. Identification of RamA, a novel LuxR-type transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 1882554-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronan, J. E., Jr., and S. Subrahmanyam. 1998. FadR, transcriptional co-ordination of metabolic expediency. Mol. Microbiol. 29937-943. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez, H., M. Cocaign-Bousquet, and N. D. Lindley. 1997. Simultaneous consumption of glucose and fructose from sugar mixtures during batch growth of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 47600-603. [Google Scholar]
- 9.Dominguez, H., C. Rollin, A. Guyonvarch, J. L. Guerquin-Kern, M. Cocaign-Bousquet, and N. D. Lindley. 1998. Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur. J. Biochem. 25496-102. [DOI] [PubMed] [Google Scholar]
- 10.Dong, J. M., J. S. Taylor, D. J. Latour, S. Iuchi, and E. C. Lin. 1993. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J. Bacteriol. 1756671-6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan, S. H., P. Louis, and H. J. Flint. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ Microbiol. 705810-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Engels, S., J. E. Schweitzer, C. Ludwig, M. Bott, and S. Schaffer. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor σH. Mol. Microbiol. 52285-302. [DOI] [PubMed] [Google Scholar]
- 14.Engels, V., and V. F. Wendisch. 2007. The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J. Bacteriol. 1892955-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, Y., and T. Fujita. 1987. The gluconate operon gnt of Bacillus subtilis encodes its own transcriptional negative regulator. Proc. Natl. Acad. Sci. USA 844524-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerstmeir, R., A. Cramer, P. Dangel, S. Schaffer, and B. J. Eikmanns. 2004. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 1862798-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerstmeir, R., V. F. Wendisch, S. Schnicke, H. Ruan, M. Farwick, D. Reinscheid, and B. J. Eikmanns. 2003. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 10499-122. [DOI] [PubMed] [Google Scholar]
- 18.Goffin, P., F. Lorquet, M. Kleerebezem, and P. Hols. 2004. Major role of NAD-dependent lactate dehydrogenases in aerobic lactate utilization in Lactobacillus plantarum during early stationary phase. J. Bacteriol. 1866661-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning, vol. 1. IRL Press, Washington, DC. [Google Scholar]
- 20.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 21.Hansmeier, N., A. Albersmeier, A. Tauch, T. Damberg, R. Ros, D. Anselmetti, A. Puhler, and J. Kalinowski. 2006. The surface (S)-layer gene cspB of Corynebacterium glutamicum is transcriptionally activated by a LuxR-type regulator and located on a 6 kb genomic island absent from the type strain ATCC 13032. Microbiology 152923-935. [DOI] [PubMed] [Google Scholar]
- 22.Inui, M., S. Murakami, S. Okino, H. Kawaguchi, A. A. Vertes, and H. Yukawa. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate production under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 7182-196. [DOI] [PubMed] [Google Scholar]
- 23.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 1854519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iuchi, S., and E. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 851888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakob, K., P. Satorhelyi, C. Lange, V. F. Wendisch, B. Silakowski, S. Scherer, and K. Neuhaus. 2007. Gene expression analysis of Corynebacterium glutamicum subjected to long-term lactic acid adaptation. J. Bacteriol. 1895582-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 1755595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiefer, P., E. Heinzle, and C. Wittmann. 2002. Influence of glucose, fructose and sucrose as carbon sources on kinetics and stoichiometry of lysine production by Corynebacterium glutamicum. J. Industrial Microbiol. Biotechnol. 28338-343. [DOI] [PubMed] [Google Scholar]
- 28.Kiefer, P., E. Heinzle, O. Zelder, and C. Wittmann. 2004. Comparative metabolic flux analysis of lysine-producing Corynebacterium glutamicum cultured on glucose or fructose. Appl. Environ Microbiol. 70229-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, H. J., T. H. Kim, Y. Kim, and H. S. Lee. 2004. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J. Bacteriol. 1863453-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinoshita, S., S. Udaka, and M. Shimono. 1957. Studies on the amino acid fermentation. I. Production of l-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3193-205. [PubMed] [Google Scholar]
- 31.Krämer, R., C. Lambert, C. Hoischen, and H. Ebbighausen. 1990. Uptake of glutamate in Corynebacterium glutamicum. 1. Kinetic properties and regulation by internal pH and potassium. Eur. J. Biochem. 194929-935. [DOI] [PubMed] [Google Scholar]
- 32.Krug, A., V. F. Wendisch, and M. Bott. 2005. Identification of AcnR, a TetR-type repressor of the aconitase gene acn in Corynebacterium glutamicum. J. Biol. Chem. 280585-595. [DOI] [PubMed] [Google Scholar]
- 33.Lange, C., D. Rittmann, V. F. Wendisch, M. Bott, and H. Sahm. 2003. Global expression profiling and physiological characterization of Corynebacterium glutamicum grown in the presence of l-valine. Appl. Environ. Microbiol. 692521-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch, A. S., and E. C. C. Lin. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding to target promoters. J. Bacteriol. 1786238-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkens, H., G. Beckers, A. Wirtz, and A. Burkovski. 2005. Vanillate metabolism in Corynebacterium glutamicum. Curr. Microbiol. 5159-65. [DOI] [PubMed] [Google Scholar]
- 36.Moon, M. W., H. J. Kim, T. K. Oh, C. S. Shin, J. S. Lee, S. J. Kim, and J. K. Lee. 2005. Analyses of enzyme II gene mutants for sugar transport and heterologous expression of fructokinase gene in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol. Lett. 244259-266. [DOI] [PubMed] [Google Scholar]
- 37.Netzer, R., P. Peters-Wendisch, L. Eggeling, and H. Sahm. 2004. Cometabolism of a nongrowth substrate: l-serine utilization by Corynebacterium glutamicum. Appl. Environ. Microbiol. 707148-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niebisch, A., and M. Bott. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175282-294. [DOI] [PubMed] [Google Scholar]
- 39.Nunez, M. F., O. Kwon, T. H. Wilson, J. Aguilar, L. Baldoma, and E. C. Lin. 2002. Transport of l-lactate, d-lactate, and glycolate by the LldP and GlcA membrane carriers of Escherichia coli. Biochem. Biophys. Res. Commun. 290824-829. [DOI] [PubMed] [Google Scholar]
- 40.Nunn, W. D., and R. W. Simons. 1978. Transport of long-chain fatty acids by Escherichia coli: mapping and characterization of mutants in the fadL gene. Proc. Natl. Acad. Sci. USA 753377-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patek, M., J. Nesvera, A. Guyonvarch, O. Reyes, and G. Leblon. 2003. Promoters of Corynebacterium glutamicum. J. Biotechnol. 104311-323. [DOI] [PubMed] [Google Scholar]
- 42.Pequignot, C., C. G. Dussap, A. Pons, and J. B. Gros. 1997. Intra- and extracellular concentrations of glutamate, lactate and acetate during growth of Corynebacterium glutamicum on different media. J. Ind. Microbiol. Biotechnol. 18312-318. [Google Scholar]
- 43.Peters-Wendisch, P. G., B. Schiel, V. F. Wendisch, E. Katsoulidis, B. Mockel, H. Sahm, and B. J. Eikmanns. 2001. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3295-300. [PubMed] [Google Scholar]
- 44.Polen, T., and V. F. Wendisch. 2004. Genomewide expression analysis in amino acid-producing bacteria using DNA microarrays. Appl. Biochem. Biotechnol. 118215-232. [DOI] [PubMed] [Google Scholar]
- 45.Pons, A., C. G. Dussap, C. Pequinot, and J. B. Gros. 1996. Metabolic flux distribution in Corynebacterium melassecola ATCC 17965 for various carbon sources. Biotechnol. Bioeng. 51177-189. [DOI] [PubMed] [Google Scholar]
- 46.Quail, M. A., and J. R. Guest. 1995. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol. Microbiol. 15519-529. [DOI] [PubMed] [Google Scholar]
- 47.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 27712507-12515. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 50.Schaffer, S., B. Weil, V. D. Nguyen, G. Dongmann, K. Gunther, M. Nickolaus, T. Hermann, and M. Bott. 2001. A high-resolution reference map for cytoplasmic and membrane-associated proteins of Corynebacterium glutamicum. Electrophoresis 224404-4422. [DOI] [PubMed] [Google Scholar]
- 51.Schluesener, D., F. Fischer, J. Kruip, M. Rogner, and A. Poetsch. 2005. Mapping the membrane proteome of Corynebacterium glutamicum. Proteomics 51317-1330. [DOI] [PubMed] [Google Scholar]
- 52.Soual-Hoebeke, E., C. de Sousa-D'Auria, M. Chami, M. F. Baucher, A. Guyonvarch, N. Bayan, K. Salim, and G. Leblon. 1999. S-layer protein production by Corynebacterium strains is dependent on the carbon source. Microbiology 1453399-3408. [DOI] [PubMed] [Google Scholar]
- 53.Stansen, C., D. Uy, S. Delaunay, L. Eggeling, J. L. Goergen, and V. F. Wendisch. 2005. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 715920-5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 55.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52541-545. [DOI] [PubMed] [Google Scholar]
- 56.Wendisch, V. F. 2003. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J. Biotechnol. 104273-285. [DOI] [PubMed] [Google Scholar]
- 57.Wendisch, V. F., A. A. de Graaf, H. Sahm, and B. J. Eikmanns. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 1823088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wennerhold, J., A. Krug, and M. Bott. 2005. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J. Biol. Chem. 28040500-40508. [DOI] [PubMed] [Google Scholar]
- 59.Yokota, A., and N. D. Lindley. 2005. Central metabolism: sugar uptake and conversion, p. 215-240. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum, vol. 10. CRC Press, Boca Raton, FL. [Google Scholar]