Abstract
The plasticity region of the Helicobacter pylori genome comprises strain-specific gene loci. We performed genotyping and functional biology analysis of one such locus (jhp940) that was previously found to be functionally unknown but present in gastric cancer-associated strains from many different countries. We found its geographic prevalence to be independent of cagA presence and disease status. Cloning, expression, and purification of JHP940 revealed a novel, ∼36-kDa protein in a biologically active form which elicited strong and significant levels of tumor necrosis factor alpha and interleukin-8 in human macrophages. Also, JHP940 was able to induce enhanced translocation of the transcription factor NF-κB complex in cultured macrophages. The induction of the proinflammatory cytokines by JHP940, therefore, points to its putative role in chronic gastric inflammation and, possibly, the various other outcomes of H. pylori infection, including gastric cancer.
The gastric pathogen Helicobacter pylori is one of the most successful pestilences of mankind and infects almost half of the world's population. However, a small fraction of infected individuals experience H. pylori-associated diseases, such as gastritis, peptic ulcers, and, more rarely, the gastric adenocarcinomas (3). It is believed that proinflammatory cytokines released in response to H. pylori infection might increase the risk of severe pathological outcomes such as cancer (1, 22, 23). The postgenomic biology of H. pylori has been quite successful, wherein the mechanisms of transmission, invasion, persistence, survival, adaptation, and pathogenesis have been unraveled. However, its complex pathology, especially in the case of gastric cancer, has not been fully elucidated. Some studies reported interleukin-8 (IL-8) and tumor necrosis factor alpha (TNF-α) to be potent chemoattractants and the cardinal agents involved in the pathogenesis of H. pylori (6, 18). The stimulation of these proinflammatory cytokines is mediated by NF-κB via the recognition of Toll-like receptors on the cell surface (16). Mononuclear cells and macrophages are the main sources of IL-8 and TNF-α, and hence, they play a critical role in the progression of the lesions. The molecular mechanism behind the secretion of these cytokines is mediated through NF-κB (p50/RelA complex), a nuclear transcription factor (sequestered in the cytoplasm by IκB complexes) which upon experiencing various stimuli is translocated to the nucleus and regulates the expression of various chemokines (5, 15).
Like many other microbial genomes, the H. pylori genome harbors hundreds of genes with no known function; many of them are unexplored as yet. Some loci within the plasticity region have been thought previously to serve as markers of virulence (gastritis or cancer) (9, 11, 13), and hence, the assumption has been made that these could be strain-specific genes gained or lost at liberty during adaptation to a new host. The identification of biologically active proteins from the plasticity region, especially those with a role in virulence, might potentiate the thinking that a gene pool encoding different virulence factors than the classical ones is indeed maintained within the nonessential compartment of the genome.
We describe the geographical conservation and functional characterization of an unknown protein encoded by the open reading frame (ORF) jhp940 of the H. pylori plasticity region. This was found to be possibly interacting with the host immune system, and it augmented the release of proinflammatory cytokines IL-8 and TNF-α from cultured macrophages.
Genomic PCR amplification was carried out to geographically analyze the distribution of jhp940 (Fig. 1) in a set of 120 H. pylori genomic DNA samples (irrespective of disease status), with 20 samples from each geographic region (India, France, Spain, Peru, Japan, South Africa, and Costa Rica). Also, the stability of this locus was analyzed through assessment of the same in serial isolates obtained a decade apart (12) from different niches (corpus and antrum) of the stomach of a single patient. PCR amplification of jhp940 was carried out using ∼100 ng genomic DNA, 10 pmol of each primer, 200 μmol of each deoxynucleoside triphosphate, and 1 unit of AccuTaq DNA polymerase (Fermentas, Hanover, MD) in a standard PCR buffer supplied by the manufacturer. Amplification was performed in a MasterCycler (Eppendorff, Westbury, NY) under the following conditions: an initial denaturation at 94°C for 5 min was followed by 35 cycles of 94°C for 1 min, 50°C for 1 min, 72°C for 5 min, and a final extension of 10 min at 72°C. Amplicons were separated in a 1.5% agarose gel and visualized under UV light to ascertain the proper amplification. The PCR amplicons (amplified with primers having HindIII and XhoI sites included as follows: forward, ATGCCAACCATTGATTTTACTTTT, and reverse, TTATCGTCTACGCTTAGGTGTG) were cloned after double digestion followed by overnight ligation to the pRSET-A vector (Invitrogen, Carlsbad, CA). The clone was confirmed by releasing the insert following restriction endonuclease digestion and by sequencing using T7 primer. Later, the construct was used to transform Escherichia coli BL21(DE3) cells, and the recombinant colonies were picked up against ampicillin selection. Isopropyl-β-d-thiogalactopyranoside (IPTG) (0.1 M) (Sigma, United States)-induced recombinant E. coli cells grown to an optical density at 600 nm of 0.4 to 0.6 were centrifuged at 6,000 rpm. The cell pellet was lysed in 20 mM Tris-HCl and 200 mM NaCl, pH 8.0 (lysis buffer), by sonication, the resultant lysate was centrifuged at 12,000 rpm for 45 min at 4°C, and its supernatant was loaded onto a Ni-nitrilotriacetic acid column (Qiagen, Hilden, Germany) to purify the His-tagged recombinant protein. The column was washed extensively with washing buffer (lysis buffer and 20 mM imidazole, pH 8.0), and the overexpressed His-tagged protein was eluted using elution buffer (lysis buffer and 250 mM imidazole). The homogeneity of the protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%) analysis, and the amount of protein was estimated with Bradford's method (4). The protein was treated with 20 μg/ml of polymyxin B (Sigma, United States) to circumvent the effects of any possible endotoxin contamination.
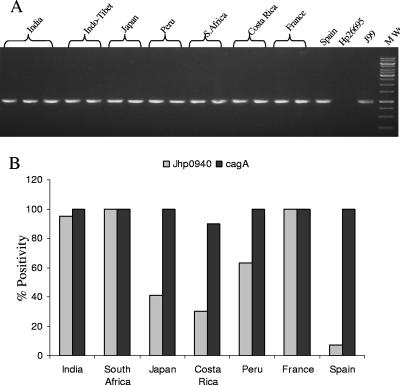
FIG. 1.
(A) Geographic distribution of ORF jhp940 as analyzed by PCR amplification (forward primer, ATGCCAACCATTGATTTTACTTTT, and reverse primer, TTATCGTCTACGCTTAGGTGTG). Lanes are marked with respect to geographic region of origin of the isolates; MW denotes the molecular weight marker. (B) Distribution of jhp940 as juxtaposed to the preponderance of the cagA gene analyzed in representative isolates. There were 20 isolates each from India, France, Spain, Peru, Japan, South Africa, and Costa Rica. On the y axis of the bar diagram is the percentage of strains carrying intact jhp940 and/or cagA, and on the x axis are the categories of isolates based on geographical descent.
Thp1 cells (ATCC, United States) were grown in RPMI 1640 medium (Invitrogen, United States) supplemented with 10% fetal bovine serum (FBS) (vol/vol) and 1% antibacterial and antimycotic solution. These cultures were maintained in an incubator at 37°C in a humidified atmosphere of 5% CO2. To differentiate them into the adherent macrophage-like cell state, Thp1 cells were treated with 5 ng/ml of phorbol 12-myristate 13-acetate (PMA; Sigma, United States). The cells were seeded in six-well tissue culture plates at a density of approximately 1 million/ml. Prior to stimulation with recombinant protein, the PMA-containing medium was aspirated and differentiated cells were washed with RPMI medium. These cells were later treated with the following concentrations of recombinant JHP940 (rJHP940): 0.1 μg, 0.25 μg, 0.5 μg, 0.75 μg, and 1.0 μg per ml. Untreated cells (differentiated macrophages without any protein treatment) and cells treated with proteinase K-lysed rJHP940 were used as negative controls. Lipopolysaccharide (LPS) (1 μg/ml) (LPS-E.coli; Sigma) was used as a positive control. After treatment, cells were kept in a humidified atmosphere containing 5% CO2 at 37°C. Culture supernatants were collected at 24 h and 48 h and stored at −80°C until analyzed further.
Peripheral blood mononuclear cells were separated from the blood of the voluntary donor(s) by using Ficoll-Hypaque density gradient centrifugation (14). The level of cell viability was checked by using the Trypan blue exclusion method and was found to be 90%. Approximately 0.5 million cells/well were seeded into 12-well plates in RPMI 1640 medium supplemented with 10% FBS and 2 mM glutamine and incubated at 37°C in a humidified atmosphere containing 5% CO2. The cells were allowed to adhere for 3 to 4 h; nonadherent cells were washed off before treatment. Following treatment with recombinant JHP940, culture supernatants were collected and assayed for cytokine induction.
The induction of proinflammatory cytokines IL-8 and TNF-α was assessed by using a commercially available enzyme-linked immunosorbent assay (ELISA) kit, OptEIA (BD Biosciences, San Jose, CA). Briefly, ELISA plates (Corning, United States) were coated with capture antibody and incubated overnight at 4°C. Nonspecific binding was blocked by treatment with 10% heat-inactivated FBS (in phosphate-buffered saline). After the plates were washed thoroughly with PBST (phosphate-buffered saline with 0.05% Tween 20), 100 μl of culture supernatants was added to the plates and they were incubated at 37°C for 2 h. This was followed by the addition of the appropriate biotinylated polyclonal antibodies at concentrations recommended by the manufacturer. The color development was carried out with streptavidin-horseradish peroxidase, and the plates were read in an ELISA reader at 490 nm. The sensitivities of the ELISA for IL-8 and TNF-α were 3.1 pg/ml and 7.8 pg/ml, respectively (BD Biosciences, San Jose, CA). Student's t test was used to analyze the data, and the values are represented as the standard errors of the means (SEM); P values less than 0.05 were considered statistically significant.
The DNA-protein binding reaction for the NF-κB complex was carried out by the electrophoretic mobility shift assay (EMSA) method as described earlier (20). Immunoblotting for the NF-κB complex was performed using antibodies against p65 and p50 (polyclonal anti-rabbit p65, anti-rabbit p50, and anti-rabbit α-actin; Santa Cruz) as described previously (21). Immunoreactive protein was detected by using an enhanced chemiluminescence kit according to the instructions of the manufacturer (Amersham, Inc.). β-Actin was used as an equal-loading control.
The jhp940 ORF was found to be fairly stable and widely prevalent geographically in the majority of strains we looked at (Fig. 1). Also, the locus was observed to be conserved across a decade in serial isolates derived from different niches of the stomach of a single patient. The preponderance of this locus was found to be independent of that of the cagA gene and irrespective of the disease condition (Fig. 1). The lowest prevalence of jhp940 was seen in the Spanish and Costa Rican isolates, followed by the Japanese and Peruvian isolates. The translated protein sequence of JHP940, when analyzed with the Protean software in the DNAStar (DNAStar, Inc., United States) package, revealed regions with a high hydrophilicity and antigenic index.
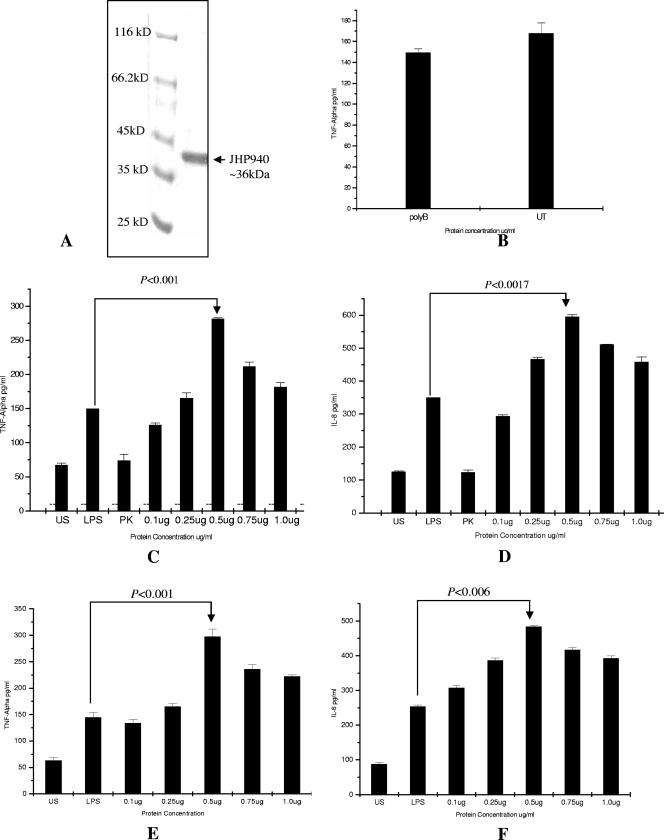
The human macrophage cells, after treatment with recombinant protein, revealed significant induction of IL-8 and TNF-α at 48 h postinduction. We attempted induction at various concentrations of the protein, beginning with 0.1 μg/ml and increasing to 1.0 μg/ml. Sustained and significant induction was, however, achieved at a 0.5-μg/ml concentration in both cases (Fig. 2C and D). We compared these titers with those in LPS-induced cells as a positive control. Our recombinant protein showed IL-8 and TNF-α titers that were nearly double those observed with LPS. The P values calculated for both the cytokine titers in comparing the effects of JHP940 and LPS were found to be highly significant. The cytokine induction was found to be dose dependent up to a certain extent, but the levels declined (although not significantly) at the highest concentration of recombinant protein (1.0 μg/ml). Similar results were obtained in freshly isolated human peripheral blood mononuclear cells (Fig. 2). It appears that macrophages excited by proteins such as JHP940 could be a significant source of proinflammatory cytokines in the gastric mucosa.
FIG. 2.
(A) Protein purification using a Ni-nitrilotriacetic acid column. Protein homogeneity was checked on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel stained with Coomassie blue. Molecular masses are shown. (B) Estimation of levels of TNF-α in culture supernatants after stimulation of the cells with rJHP940 protein pretreated with polymyxin B (labeled on the x axis as polyB) and with recombinant protein without polymyxin B treatment (labeled on the x axis as UT). The values are expressed as the means ± SEMs of the results. (C) Rate of induction of TNF-α estimated in culture supernatants collected after induction with rJHP940 protein. Data represent the means ± SEMs of the results of three independent experiments. (D) Rate of induction of IL-8 estimated in culture supernatant after induction with rJHP940. Data represent the means ± SEMs of the results of three independent experiments. US, cytokine response from uninduced cells; LPS, cytokine response from LPS-treated cells; PK, cytokine response from cells treated with proteinase K-lysed fractions of JHP940. Bars showing responses from cells treated with different concentrations of JHP940 are labeled on the x axis with the concentration in micrograms. (E) Rate of induction of TNF-α estimated in culture supernatants after stimulation of human peripheral blood mononuclear cells with rJHP940 at different concentrations (μg/ml). (F) Rate of induction of IL-8 estimated in culture supernatants after stimulation of human peripheral blood mononuclear cells with rJHP940.
We do not believe that the proinflammatory responses induced by JHP940 were chiefly due to the effects of contaminating endotoxin and LPS; this is evident from the consistent results that we observed with polymyxin B-treated fractions of the recombinant protein. Also, proteinase K-treated fractions did not produce any significant induction of the two cytokines (Fig. 2). We also used isocitrate dehydrogenase of H. pylori as an unrelated negative control for the induction of proinflammatory cytokines; this protein was cloned, expressed, and purified under conditions similar to those used for JHP940. The isocitrate dehydrogenase protein did not significantly induce any IL-8 or TNF-α responses (data not shown).
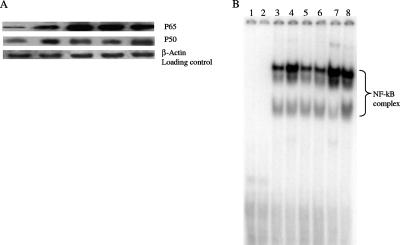
To further determine whether the increased expression of IL-8 was mediated through NF-κB, a DNA binding assay and immunoblotting were performed as described above. Compared to the results for uninduced cells, the NF-κB complex was significantly translocated in the cells induced with recombinant protein. The specificity of the complex was checked by competition analysis using unlabeled probe (Fig. 3). Furthermore, the kinetic analysis of NF-κB complex activation at various time points showed induction of the complex at 6 h and the levels increased significantly up to 24 h. Immunoblot analysis using nuclear extract prepared from cells induced with increasing protein concentrations revealed recombinant JHP940 to be a potent stimulator of the NF-κB complex (Fig. 3). LPS, a known inducer of the NF-κB complex, was used as a positive control.
FIG. 3.
(A) Western blot analysis to demonstrate translocation of NF-κB complex following induction with rJHP940 protein. Immunoblot shows significant differences in the induction of p65 and p50 complex compared to their levels in uninduced control as detected by specific antibodies (see the text). A significant increase in RelA (p65) and p50 complex was observed with increasing protein concentrations. First lane, nuclear extract from uninduced cells; second lane, nuclear extract of LPS-induced cells; third to fifth lanes, nuclear extracts from cells induced with concentrations of 0.1 μg/ml, 0.5 μg/ml, and 1.0 μg/ml of rJHP940, respectively. (B) EMSA with the nuclear extracts obtained from different control macrophage cells and those induced with rJHP940. Lane 1, free probe; lane 2, cold competition; lane 3, nuclear extract from uninduced macrophages; lane 4, nuclear extract from LPS-treated (1 μg/ml) cells; lane 5, nuclear extracts obtained from macrophages treated with proteinase K-digested rJHP940; lanes 6 to 8, nuclear extracts obtained from macrophages induced with rJHP940 for durations of 6 h, 12 h, and 24 h (at the rate of 0.5 μg/ml), respectively.
Although H. pylori has been widely argued to be a natural inhabitant of the human gut (1), evidence exists for its being a coevolved bacterium (8). Recommendations against eradication and in favor of eradication are confusing the scenario, despite the fact that H. pylori poses an obvious threat of gastric adenocarcinoma in susceptible populations (1). TNF-α is a cytokine implicated in the pathology of H. pylori, and IL-8 has been the cardinal effector molecule involved in H. pylori-induced gastroduodenal pathology owing to its being a neutrophil chemotactic agent (10, 18, 22). cagPAI, the cag pathogenicity island that carries cagA, has been considered to be the anciently acquired virulence determinant, and the strains lacking its acquisition are generally considered benign (2, 17). However, the presence and functional activity of several other virulence factors (irrespective of the presence of cagPAI), such as vacA, urease, porins, flagellins, oipA, and several outer membrane proteins, weaken this assumption (7, 19, 22).
We found it interesting to observe that an unknown protein having no known sequence homology in existing databases induced high levels of cytokines; this might suggest acquisition from an unknown organism, as lateral gene transfer is extremely common in H. pylori. More interesting is the observation that the presence of the jhp940 locus was independent of the cagA status and disease category, conveying the possibility that it could be a stand-alone virulence factor that might be present in a majority of H. pylori strains. We found this locus to be broadly conserved in strains from different regions; this contrasts with the finding of Santos and colleagues (13), who recorded a much lower prevalence (1.5%) in Brazilian isolates. A characteristic absence of this ORF from H. pylori 26695 could be a rare exception to its presence in all other European isolates (genogroup, HPEurope), something similar to the fact that some strain 26695-specific loci, such as hp0986, are absent only from strain J99 and not from other J99-like isolates (13; A. Alvi, unpublished data) of genogroup HPAfrica. Given that H. pylori is a highly recombining organism, revealing the jhp940 locus to be conserved across a decade and within different gastric niches, conveys the possibility that its product might be an essential protein which is not rearranged and is evolutionarily stable.
We selected locus jhp940 primarily based on the results of a DNA profiling study (11) using different clinical strains. Similarly, another gene from the plasticity region cluster (dupA) was shown to have roles in the promotion of duodenal ulcers (9). Also, the functional prediction of the ORF based on computational analyses (DNAStar, NCBI BLAST, Protean, etc.) revealed it to be a putative immunogen of potential merit. The high antigenicity of JHP940 led us to look at it as a potential antigen, capable of bringing about sustained and chronic inflammatory responses toward carcinogenic triggering in case of a long-term persistence. Our data on cytokine profiles do not rule out this possibility, and it is possible that JHP940 might act like a T-cell mitogen, similar to porins, LPS, and urease, which activate macrophages to bring about cytokine-induced changes in gastric physiology, mainly through the activities of IL-1, IL-8, IL-12, and TNF-α (18). Several cytokines are expressed in gastric epithelial cells in response to H. pylori infection, and it is known that cytokines IL-8 and TNF-α have a central role in the modification of the cellular microenvironment (3, 18, 23). IL-8, in particular, helps the recruitment of neutrophils across the proteoglycan scaffolding, and TNF-α induces fas-mediated apoptosis and disruption of the epithelial barrier to facilitate the translocation of bacterial antigens (18). Also, the cytokine-mediated damage could be a possible survival mechanism as H. pylori feeds on inflammatory exudates due to epithelial injury.
In the future, it will be interesting to further dissect the underlying signaling mechanisms, particularly aspects of the induction of IL-8, TNF-α, and NF-κB by JHP940, including the role of Toll-like receptors. Nonetheless, it is tempting to suggest that JHP940 possibly induces proinflammatory cytokines through a NF-κB-dependent pathway, and our data do not rule out this possibility. In view of the current findings, it may be possible to look at the JHP940 protein as a putative virulence factor, one of several such molecules that determine the outcome of H. pylori infection. It might be possible in the future to explore the biology of this protein in greater detail for further downstream effects of the observed cytokine responses.
Acknowledgments
We thank the Director of the Center for DNA Fingerprinting and Diagnostics for infrastructure and core support and Seyed E. Hasnain for helpful suggestions, discussion, and guidance.
A.A. was supported by the Department of Science and Technology of the Indian government through the Fast Track Young Scientist scheme. N.A. is the corresponding fellow of the European Helicobacter Study Group.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Ahmed, N., and L. A. Sechi. 2005. Helicobacter pylori and gastroduodenal pathology: new threats of the old friend. Ann. Clin. Microbiol. Antimicrob. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, M., A. A. Khan, S. K. Tiwari, N. Ahmed. L. V. Rao, and C. M. Habibullah. 2005. Association between cag-pathogenicity island in Helicobacter pylori isolates from peptic ulcer, gastric carcinoma, and non-ulcer dyspepsia subjects with histological changes. World J. Gastroenterol. 116815-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atherton, J. C. 2006. The pathogenesis of Helicobacter pylori induced gastro-duodenal diseases. Annu. Rev. Pathol. 163-96. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-252. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16225-260. [DOI] [PubMed] [Google Scholar]
- 6.Huang, J., P. W. O'Toole, P. Doig, and T. J. Trust. 1995. Stimulation of interleukin-8 production in epithelial cell lines by Helicobacter pylori. Infect. Immun. 631732-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilver, D., A. Arnqvist, J. Ogren. I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279373-377. [DOI] [PubMed] [Google Scholar]
- 8.Linz, B., F. Balloux, Y. Moodley, A. Manica, H. Liu, P. Roumagnac, D. Falush, C. Stamer, F. Prugnolle, S. W. van der Merwe, Y. Yamaoka, D. Y. Graham, E. Perez-Trallero, T. Wadstrom, S. Suerbaum, and M. Achtman. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu, H., P. I. Hsu, D. Y. Graham, and Y. Yamaoka. 2005. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology 128833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naumann, M., and J. E. Crabtree. 2004. Helicobacter pylori-induced epithelial cell signaling in gastric carcinogenesis. Trends Microbiol. 1229-36. [DOI] [PubMed] [Google Scholar]
- 11.Occhialini, A., A. Marais, R. Alm, F. Garcia, R. Sierra, and F. Mégraud. 2000. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect. Immun. 686240-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prouzet-Mauléon, V., M. A. Hussain, H. Lamouliatte, F. Kauser, F. Mégraud, and N. Ahmed. 2005. Pathogen evolution in vivo: genome dynamics of two isolates obtained 9 years apart from a duodenal ulcer patient infected with a single Helicobacter pylori strain. J. Clin. Microbiol. 434237-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos, A., D. M. Magalhães Queiroz, M. Ménard, A. Marais, G. A. Rocha, C. A. Oliveira, A. M. M. Ferreira Nogueira, M. Uzeda, and F. Mégraud. 2003. New pathogenicity marker found in the plasticity region of the Helicobacter pylori genome. J. Clin. Microbiol. 411651-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savill, J. S., A. H. Wyllie, J. E. Henson, M. J. Walport, P. M. Hensen, and C. Heslett. 1989. Macrophage phagocytosis of aging neutrophils in inflammation. Programmed cell death in the neutrophil leads to its recognition by macrophages. J. Clin. Investig. 83865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma, S. A., M. K. Tummuru, M. J. Blaser, and L. D. Kerr. 1998. Activation of IL-8 gene expression by Helicobacter pylori is regulated by transcription factor nuclear factor-kappaB in gastric epithelial cells. J. Immunol. 1602401-2407. [PubMed] [Google Scholar]
- 16.Smith, M. F., Jr., A. Mitchell, G. Li, S. Ding, A. M. Fitzmaurice, K. Ryan, S. Crowe, and J. B. Goldberg. 2003. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-kappa B activation and chemokine expression by epithelial cells. J. Biol. Chem. 27832552-32560. [DOI] [PubMed] [Google Scholar]
- 17.Stein, M., R. Rappuoli, and A. Covacci. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. USA 971263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 3471175-1186. [DOI] [PubMed] [Google Scholar]
- 19.Tanahashi, T., M. Kita, T. Kodama, Y. Yamaoka, N. Sawai, T. Ohno, S. Mitsufuji, Y.-P. Wei, K. Kashima, and J. Imanishi. 2000. Cytokine expression and production by purified Helicobacter pylori urease in human gastric epithelial cells. Infect. Immun. 68664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visvanathan, K., and S. Goodbourn. 1989. Double-stranded RNA activates binding of NF-κB, an inducible element in the human gamma interferon promoter. EMBO J. 81129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weih, F., D. Carrasco, and R. Bravo. 1994. Constitutive and inducible Rel/NF-kappa B activities in mouse thymus and spleen. Oncogene 93289-3297. [PubMed] [Google Scholar]
- 22.Yamaoka, Y., D. H. Kwan, and D. Y. Graham. 2000. A Mr 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 977533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaoka, Y., M. Kita, T. Kodama, N. Sawai, K. Kashima, and J. Imanishi. 1997. Induction of various cytokines and development of severe mucosal inflammation by cagA gene positive Helicobacter pylori strains. Gut 41442-451. [DOI] [PMC free article] [PubMed] [Google Scholar]