Abstract
In the unicellular cyanobacterium Synechocystis sp. strain PCC 6803, the pentameric bidirectional Ni-Fe hydrogenase (HoxEFUYH) is the sole enzyme involved in hydrogen metabolism. Recent investigations implicated the transcription factor LexA in the regulation of the hox genes in this cyanobacterium, suggesting the factor to work as an activator. In this work, we show evidence that LexA cannot account exclusively for the regulation of the hox genes in this cyanobacterium. Therefore, we investigated which additional transcription factors interact in and may regulate the expression of the hox genes in Synechocystis sp. strain PCC 6803. By using DNA affinity assays, a transcription factor with similarity to the transition state regulator AbrB from Bacillus subtilis was isolated. Electrophoretic mobility shift assays showed that the AbrB-like protein specifically interacts with the promoter region of the hox genes as well as with its own promoter region. In addition, results obtained with two genetically modified strains of Synechocystis sp. strain PCC 6803, one with a not fully segregated inactivation mutation of the abrB-like gene and the other overexpressing the same abrB-like gene, suggest that this transcription factor functions as a regulator of hox gene expression.
Two different Ni-Fe hydrogenases have been characterized physiologically, biochemically, and on the molecular level in cyanobacteria: one is named uptake hydrogenase and catalyzes the consumption of H2 produced by the nitrogenase during N2 fixation, and the other is termed bidirectional hydrogenase and its function is still under debate (32, 37, 38).
Over the years, it has continuously been shown by reverse transcription-PCR and Northern blot analysis (2, 3, 4, 15, 23, 27, 33, 38) that the transcription of cyanobacterial hydrogenases varies under different environmental conditions. Nevertheless, an understanding of which key regulators are involved is just emerging and the inherent signal transduction pathways certainly deserve better attention. Therefore, it is imperative to study the mechanisms controlling the expression of both cyanobacterial hydrogenases more closely in order to acquire an improved comprehension of their functions and also to develop novel tools for enhancing a sustainable and reliable production of H2 from cyanobacteria (38).
Recently, the first reports on the transcription factors involved in the regulation of cyanobacterial hydrogenases became available. NtcA, the nitrogen control regulator in cyanobacteria, has been suggested to mediate the transcription of the uptake hydrogenase in different cyanobacterial strains, Nostoc punctiforme PCC 73102 (24), Gloeothece sp. strain ATCC 27152 (27), and Lyngbya majuscula CCAP 1446/4 (23), as well as that of the hydrogenase maturation proteins in Lyngbya majuscula CCAP 1446/4 (15). A LexA-related protein, which has been proposed to not be involved in the classical regulation of DNA repair genes (11), was shown to interact in two different regions of the promoter of the hox genes in Synechocystis sp. strain PCC 6803 (17, 28) and was further suggested to activate the transcription of the hox operon (17). Moreover, the LexA-related protein from Anabaena sp. strain PCC 7120 has also been shown to interact with the promoter regions of the two hox operons (33).
In the present work, we addressed the question of which transcription factors, in addition to LexA, might be involved in the regulation of the expression of the hox genes, encoding the single hydrogenase in the freshwater cyanobacterium Synechocystis sp. strain PCC 6803. We found that the protein product of sll0359, a gene annotated as a hypothetical protein, interacts specifically with the hox promoter region. Moreover, our data suggest that Sll0359 works as an activator of hox gene expression. The implications of these findings are further discussed.
MATERIALS AND METHODS
Organisms and growth conditions.
Synechocystis sp. strain PCC 6803 wild-type cells were grown in BG11 medium (35), supplemented with 10 mM HEPES, pH 7.5, and sparged with air at 25°C and with a continuous irradiance of 40 μmol of photons m−2 s−1. The cells were subjected to a combined nitrogen-depleted regimen for 24 h and grown as described previously (2). The Synechocystis sp. strain PCC 6803 mutant cells, SFM02 (the Δsll0359::Kmr/sll0359+ heteroploid mutant) and SFoe01 (harboring the self-replicating plasmid pFMoe01) (see below), were grown in the same conditions as the wild type was, except that the medium was supplemented with kanamycin to a final concentration of 100 μg/ml and with chloramphenicol to a final concentration of 15 μg/ml, respectively. The three different strains were inoculated to an optical density at 730 nm of 0.08 and grown for 2 days as described above. After this period, the medium was supplemented with additional CuSO4 to a final concentration of 1 μM (12) and the cells were left to grow for two more days before being harvested, washed once with BG110 (BG11 without added nitrate), and resuspended in fresh BG11 medium supplemented with 1 μM CuSO4 and the respective antibiotics. Finally, cells were harvested 24 h later for RNA extractions and measurements of the bidirectional hydrogenase activity. Escherichia coli strains were grown in LB liquid medium or LB agar plates supplemented with appropriate antibiotics at 37°C.
Nucleic acid isolation and analysis.
Genomic DNA and total RNA were isolated from Synechocystis sp. strain PCC 6803 cells as described previously (3, 39). Plasmid DNA was isolated from E. coli by using the GenElute Plasmid Miniprep kit (Sigma-Aldrich). Northern blot analyses were performed as described previously (2, 33) by using probes that were obtained by PCR with specific oligonucleotides (Table 1) and further labeled with [α-32P]dCTP by using the Rediprime II random prime labeling system (GE Healthcare). The even loading of the total RNA aliquots was controlled by verification of equal abundances of the rRNA bands on the agarose gel and of the constitutive RNA component of the ribozyme RNase P (43). For determining the transcription start point (TSP) upstream of sll0359, the system of rapid amplification of cDNA ends (5′RACE, version 2.0; Invitrogen) was used. The resulting PCR product was cloned into the pCR2.1-TOPO vector (Invitrogen), according to the manufacturer's instructions, before being sequenced at Macrogen, Inc. Sequence homology searches were performed with the BLAST program (1), while computer-assisted sequence analyses were performed using CLUSTAL W (40).
TABLE 1.
Oligonucleotides used in this studya
| Primer | Sequence 5′→3′ | Purpose of primer | Reference |
|---|---|---|---|
| ShoxEF | GGGAACGGCTTGCTACGTTAAA | Probe for Northern blotting | 28 |
| ShoxER | GCCAATACCGCTTCGTCATTCT | Probe for Northern blotting | 28 |
| SlexAF | GGATCCGAACCTCTCACCCGAGCCCAAAAAG | Probe for Northern blotting | 28 |
| SlexAR | AAGCTTCTAAACTCCCTGGAAATTGCGC | Probe for Northern blotting | 28 |
| SrnpBR | TTCTGTTCCAGGATGCGAGGCA | Probe for Northern blotting | 2 |
| SrnpBF | GAGAGTTAGGGAGGGAGTTG | Probe for Northern blotting | 2 |
| sll0359F | GAGCTCTTATATTTTATCTTATGTAGAC | PCR analysis | This work |
| sll0359sbF | TTTGACCGACTTCTATGACG | Probe for Southern blotting | This work |
| sll0359sbR | AAGCTTTTATACTTCCTCTTCGTCATCG | Probe for Southern blotting and PCR analysis | This work |
| ShoxprF | GCAATTGGGGTTGCGACTAT | DNA affinity assay and EMSA | 28 |
| ShoxprR | CCTCCACAATCTTGCCCACAATAA | DNA affinity assay and EMSA | 28 |
| sll0359RACER | TCAGATGAATATGCT | 5′ RACE (RT) | This work |
| sll0359RACER2 | TTAGTGCGGTGGAGGCGTT | 5′ RACE (PCR) | This work |
| Sll0359mutF | AGGCGAACTGGGTGAGAACCAT | Creation of knockout mutant | This work |
| Sll0359mutR | AAACTCCATCAAATTTTCCAT | Creation of knockout mutant | This work |
| pUC4KF | ACGCGTTGAGGTCTGCCTCGTGAAGAA | Creation of knockout mutant | This work |
| pUC4KR | ACGCGTAAAGCCACGTTGTGTCTCAAA | Creation of knockout mutant | This work |
| sll0359oeF | GAGCTCCCAAACGCCTCCACCGCACTAA | Introduced into self-replicating plasmid | This work |
| sll0359oeR | AAGCTTTTATACTTCCTCTTCGTCATCG | Introduced into self-replicating plasmid | This work |
| sll0359EMSAF | AGGCGAACTGGGTGAGAACCAT | EMSA | This work |
| sll0359EMSAR | TTAGTGCGGTGGAGGCGTT | EMSA | This work |
| vectorF | GTAAAACGACGGCCAGTGAA | EMSA | This work |
| vectorR | CAGGAAACAGCTATGACCAT | EMSA | This work |
| CmF | GCGAAGCTTATGCCCTTTCGTCTTCGAAT | Test presence of self-replicating plasmid | This work |
| CmR | GCGAAGCTTATGGGTCGAATTTGCTTTCG | Test presence of self-replicating plasmid | This work |
Underlined nucleotides correspond to restriction sites. RT, reverse transcription.
DNA affinity and EMSAs.
To address the question of possible DNA binding proteins/transcription factors interacting in the regulatory promoter region of the Synechocystis sp. strain PCC 6803 hox operon, DNA affinity assays were carried out as described previously (28) by using streptavidin-coated magnetic beads (Dynabeads M-280; Dynal Biotech-Invitrogen). A previously used DNA fragment of 462 bp (positions −415 to +47 relative to the transcription start point), referred to as ShoxPr (28), was used. Proteins were excised from the gels, cleaved with trypsin by in-gel digestion, and analyzed by electrospray ionization mass spectrometry according to the method of Wilm et al. (47) on a quadrupole time-of-flight mass spectrometer (Waters Ltd.) using MassLynx software. The sequence homology search was performed with the BLAST program (1). For the electrophoretic mobility shift assays (EMSAs), the different DNA fragments used were obtained by PCR and end labeled with [γ-32P]ATP as described previously (27). For each 20-μl reaction mixture, 20 fmol of each labeled DNA fragment was incubated with various amounts of Sll0359 (see below) (in a buffer described previously [26]), supplemented with 1 μg of salmon sperm DNA. After incubation at 30°C for 30 min, the reaction mixtures were separated by electrophoresis on a 6% (wt/vol) polyacrylamide nondenaturing gel and the relative positions of the isotope were visualized by using a BAS-2000II bioimaging analyzer (Fuji Film).
Cloning of sll0359 and purification of the gene product.
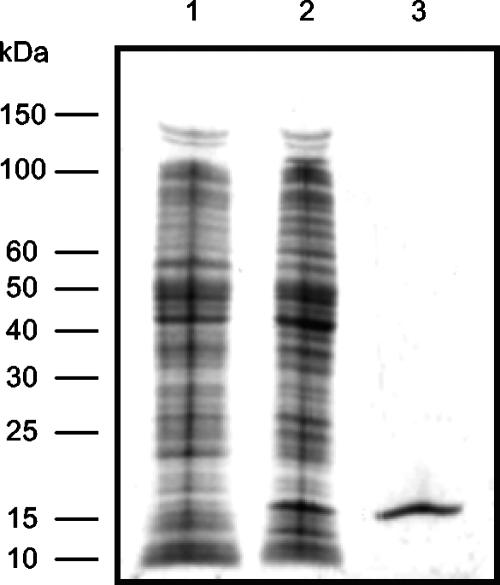
The sll0359 gene was amplified from genomic DNA by using the oligonucleotides sll0359oeF and sll0359oeR (Table 1). The obtained PCR fragment was cloned into the pCR2.1-TOPO (Invitrogen) vector, and its identity was confirmed by sequencing at Macrogen, Inc. The sll0359 gene was further subcloned into pQE-30 (Qiagen) and introduced into M15 (pREP4) cells (Qiagen). After confirming (by sequencing) that no mutations had been introduced, we overexpressed and purified the protein product of sll0359 by using Ni-nitrilotriacetic acid Superflow resin (Qiagen), according to the manufacturer's instructions. The obtained Sll0359 His-tag protein was more than 95% pure, as determined by Coomassie blue staining by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (see Fig. 4).
FIG. 4.
Coomassie-stained SDS-PAGE showing the overexpression and purification of the His-tagged AbrB-like protein Sll0359 in E. coli. Lanes: 1, noninduced crude extract; 2, induced crude extract; 3, purified His-tagged Sll0359. The molecular masses of the Fermentas protein marker are indicated on the left.
Construction of the sll0359 insertion mutant.
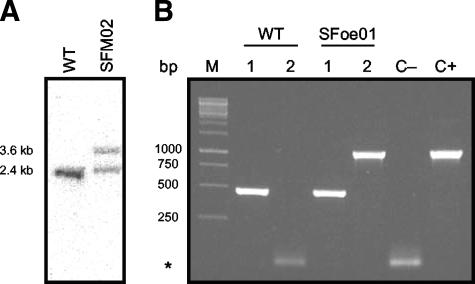
Using specific oligonucleotides (sll0359mutF and sll0359mutR [Table 1]), we amplified the sll0359 gene by PCR, along with the two 0.4-kb-long segments of flanking sequences providing the homologous platforms for recombination, mediating targeted gene replacement (22). The obtained PCR fragment was cloned in the plasmid pBluescript SK+ (Stratagene). Then, sll0359 was inactivated as follows: the kanamycin resistance cassette from the plasmid pUC4K (GE Healthcare) was amplified with the oligonucleotides pUC4KF and pUC4KR (both harboring MluI restriction sites), and the PCR product was cloned into the EcoRV site of the vector pBluescript SK+ (Stratagene). The Kmr cassette was further inserted at the MluI restriction site of sll0359 (i.e., 129 bp downstream of the ATG start codon), creating the vector pFM02, and its sequence was verified at Macrogen, Inc. Transformation of Synechocystis sp. strain PCC 6803 cells with the vector pFM02 was performed as described previously (21). The selection of mutants was carried out in plates initially supplemented with 25 μg/μl kanamycin. To analyze the extent of chromosome segregation, after we transformed Synechocystis sp. strain PCC 6803 with the vector pFM02, Southern blot hybridizations were performed as described previously (39).
Construction of the SFoe01 mutant strain.
The oligonucleotides sll0359oeSF and sll0359oeSR (Table 1) were used to amplify the sll0359 gene from genomic DNA of Synechocystis sp. strain PCC 6803. The PCR product was cloned into the pCR2.1-TOPO (Invitrogen) vector, and its identity was confirmed by sequencing. Then, sll0359 was further subcloned into the self-replicating plasmid pAWG1.1 (kindly provided by Matthias Rögner, Ruhr Universität, Bochum, Germany), which is a derivative of RSF1010 (12, 18), resulting in the vector pFMoe01. In this construct, the sll0359 gene was cloned downstream of the petE promoter region, which is inducible by copper (12, 18, 25). The obtained construct was subsequently sequenced to confirm that no mutations had been introduced. After the confirmation, pFMoe01 was transferred to Synechocystis sp. strain PCC 6803 by conjugation as described previously (13). The selection of mutants was carried out in plates supplemented with 20 μg/μl chloramphenicol. Individual colonies were transferred to liquid BG11 medium supplemented with 15 μg/μl chloramphenicol. In order to confirm that the cells were transformed with the vector pFMoe01, DNA extractions, followed by PCR analysis, were carried out (see Fig. 7B) by using oligonucleotides designed to amplify the chloramphenicol resistance cassette (Table 1). The obtained mutant strain used to express Sll0359 was named SFoe01.
FIG. 7.
Genetically modified strains of Synechocystis sp. strain PCC 6803 produced in this work. (A) Southern Blot analysis showing the extent of segregation of wild-type chromosomes. A total of 7.5 μg of genomic DNA was digested with HincII. Lanes: WT, wild type; SFM02, Δsll0359::Km/sll0359+ mutant. (B) PCR analysis showing the presence or absence of the self-replicating plasmid pFMoe01 in Synechocystis sp. strain PCC 6803. PCR amplifications were carried out by using genomic DNA extracted from WT or Synechocystis sp. strain PCC 6803 cells harboring the vector pFMoe01 (SFoe01) as the template and the respective primer pairs. Lanes 1, sll0359F and sll0359sbR (Table 1); lanes 2, CmF and CmR (Table 1). The PCR controls (negative control [C−], and positive control [C+]), were carried out with the primer pair CmF and CmR by using water or the plasmid pFMoe02 as the template, respectively. *, PCR artifacts (primer dimers) (4).
In vivo bidirectional hydrogenase activity measurement.
We used a Clarus 500 gas chromatograph with a Molecular Sieve 5A 60/80 mesh column (PerkinElmer) and Ar as the carrier gas to assay the activity of the bidirectional hydrogenase by determining the evolution of H2 from methyl viologen reduced by sodium dithionite, as described previously (39).
RESULTS
lexA and hox transcription do not follow the same pattern of expression.
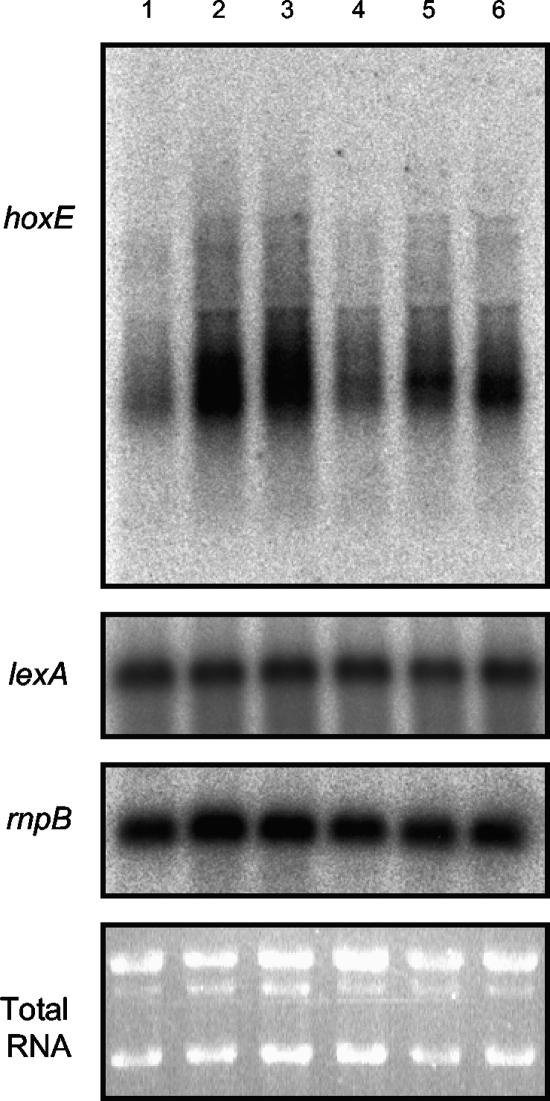
The relative expression of the hox genes in Synechocystis sp. strain PCC 6803 has previously been shown to change under combined nitrogen-depleted conditions (2). In this work, we examined whether lexA transcripts followed the same pattern of expression as the hox genes did, since LexA has previously been suggested to work as an activator of hox transcription (17). Interestingly, lexA transcription patterns did not follow the same trend as did hox expression under the conditions tested (Fig. 1), which may indicate that additional factors might be involved in the regulation of the hox gene transcription in Synechocystis sp. strain PCC 6803.
FIG. 1.
Northern blot analysis of the relative amount of hoxE, lexA, and rnpB transcripts of Synechocystis sp. strain PCC 6803 under different growth conditions. A culture of Synechocystis sp. strain PCC 6803 was grown in BG11, under light and aerobic conditions, before being split in six different conditions. Lanes: 1, BG11, no condition shift imposed; 2, BG110; 3, BG11 containing 1.76 mM of NaNO3; 4, BG11, fresh medium, containing 17.6 mM NaNO3; 5, BG110 supplemented with 2 mM of NH4Cl; 6, BG110 supplemented with 15 mM NH4Cl. Total RNA was extracted 24 h after the shift to the new growth conditions.
DNA affinity assays identified DNA binding proteins interacting with the hox promoter region.
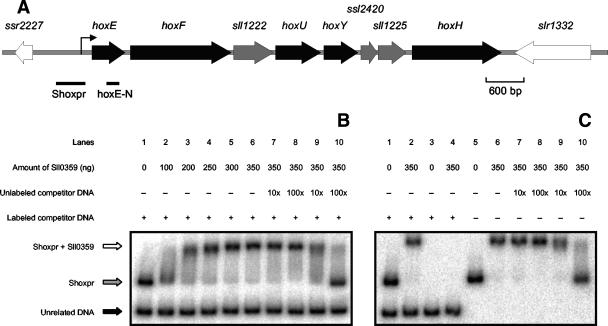
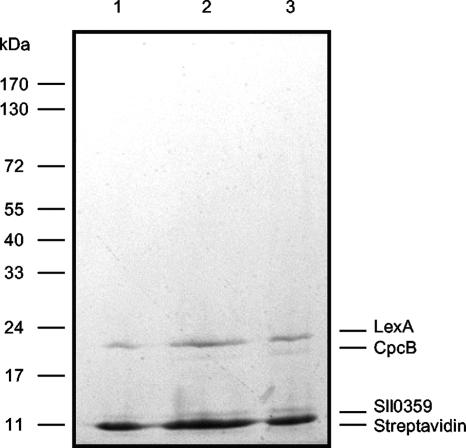
In recent studies, a LexA-related protein has been shown to interact with the promoter region of the hox genes in Synechocystis sp. strain PCC 6803 (17, 28). DNA affinity assays were carried out in order to disclose whether additional DNA binding proteins interact with the promoter region of the hox genes in Synechocystis sp. strain PCC 6803. For this purpose, a fragment of DNA (Shoxpr [see Fig. 5A]) partially covering the promoter region of the genes in study was incubated with Synechocystis sp. strain PCC 6803 cell extracts. The DNA-interacting proteins were visualized by SDS-PAGE (Fig. 2), excised from the gel and further analyzed by mass spectrometry. Two of the identified peptides have previously been picked up at our laboratory (28) and correspond to streptavidin and LexA (Fig. 2). The newly identified peptides correspond to phycocyanin β subunit CpcB and the hypothetical protein Sll0359 (Fig. 2).
FIG. 5.
EMSAs with purified AbrB-like protein from Synechocystis sp. strain PCC 6803 and the hox operon regulatory region. (A) Schematic representation of the hox locus in the genome of Synechocystis sp. strain PCC 6803. The arrow upstream of hoxE corresponds to the TSP of the hox operon, as identified previously (28). The positions of different probes used in this study are represented as lines and identified by their names: Shoxpr and hoxE-N were used as EMSA and Northern blot probes, respectively. (B) Analysis of the electrophoretic mobility of Shoxpr in the presence of increasing concentrations of purified AbrB-like protein. EMSAs were carried out, with the target fragment Shoxpr incubated with an unrelated DNA fragment, obtained from pBluescript, without protein (lane 1) or together with increasing amounts of AbrB-like protein (lanes 2 to 6). The unrelated DNA is indicated with a black arrow, and the Shoxpr fragment and its retardation are indicated with gray and white arrows, respectively. To further demonstrate the specific binding between the AbrB-like protein and the fragment Shoxpr, competition experiments were carried out by using a severalfold molar excess of unlabeled and unspecific (lanes 7 and 8) or Shoxpr DNA fragments (lanes 9 and 10). (C) Additional EMSAs were carried out to demonstrate that the presence of two labeled DNA fragments, working as possible targets in the same reaction, do not produce unexpected artifacts. Hence, the unrelated DNA fragment (lanes 3 and 4) and Shoxpr (lanes 5 and 6) were incubated alone with AbrB-like protein, in opposition to an assay where both DNA fragments were present (lanes 1 and 2). Lanes 7 to 10 represent supplementary competition experiments. The amount of AbrB-like protein used in each assay is indicated in nanograms in the figure. A plus indicates that the fragment was included in the assay, and a minus indicates that the fragment was excluded from the assay.
FIG. 2.
Coomassie-stained SDS-PAGE depicting the peptides obtained with DNA affinity assays. The experiments were carried out with 250 μg magnetic beads and incubated with 150 μg (lane 1), 300 μg (lane 2), and 600 μg (lane 3) of Synechocystis sp. strain PCC 6803 cell extract. The identities of the observed and analyzed peptides are shown on the right side. The molecular masses of the Fermentas protein marker are represented as lines on the left.
Sll0359 is an AbrB-like protein.
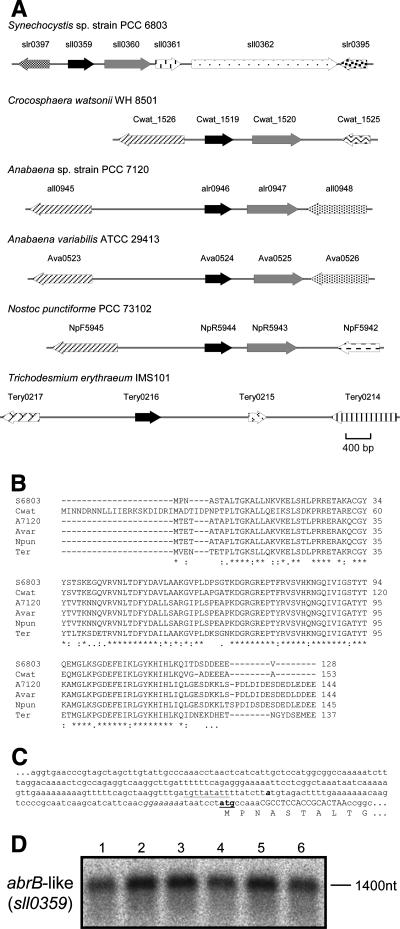
The open reading frame for sll0359 is annotated in the Synechocystis sp. strain PCC 6803 genome as a hypothetical protein (20). It is a member of the protein families TIGR01439 and IPR006339, which consist of proteins that contain DNA-binding domains similar to the one found in proteins like AbrB, a transition state regulator in Bacillus subtilis. Therefore, from here on, we will address Sll0359 as an AbrB-like protein. Interestingly, when performing a BLAST search using the sequence of the AbrB-like protein as the query, we observed that many of the cyanobacterial genomes sequenced so far possess homologues of this protein (Fig. 3), sharing a high degree of similarity (50 to 81%). Furthermore, the loci harboring the genes encoding the AbrB-like proteins in different cyanobacterial genomes also show a remarkable similarity between different species (Fig. 3).
FIG. 3.
The AbrB-like protein Sll0359 and homologues in different cyanobacteria. (A) Physical maps of the AbrB-like coding gene (black arrow), the putative protease (gray arrow), and additional neighboring open reading frames in several cyanobacterial strains. Identical representation patterns between different maps correspond to homologous genes. Locus tags are given based on the genome annotations at CyanoBase and the Joint Genome Institute, respectively. (B) CLUSTAL W alignment of the deduced AbrB-like protein sequences from the cyanobacterial strains illustrated in panel A: Synechocystis sp. strain PCC 6803 (S6803), Crocosphaera watsonii WH8501 (Cwat), Anabaena sp. strain PCC 7120 (A7120), Anabaena variabilis ATCC 29413 (Avar), Nostoc punctiforme PCC 73102 (Npun), and Trichodesmium erythraeum IMS101 (Ter). (C) Nucleotide sequence of the promoter region upstream of sll0359 (the Synechocystis sp. strain PCC 6803 AbrB-like coding gene). The sll0359 starting codon is indicated in boldface and underlined, and the deduced N-terminal amino acid sequence is given below. The primer used in 5′RACE to identify the TSP is shown in uppercase letters, while the putative ribosomal binding site is presented in italic. The TSP is depicted in boldface and italic, and the putative −10 extended box is underlined. (D) Northern blot analysis of the relative amount of sll0359 transcript under different growth conditions. Lanes 1 to 6 are as described in the legend for Fig. 1. The estimated size of the transcript is indicated in nucleotides.
Little is known about this AbrB-like protein in Synechocystis sp. strain PCC 6803. However, polypeptide sequences from Sll0359 have been detected in different proteomic studies (16, 31) and its transcript has also been identified in microarray experiments (36), demonstrating that this open reading frame is indeed transcribed and encodes a bona fide protein. For Synechocystis sp. strain PCC 6803, it was possible to confirm, by Northern blot analysis, that the AbrB-like coding gene and the open reading frame encoding the putative protease (sll0360) (Fig. 3A) are transcribed together (Fig. 3D). Moreover, when the transcription profile of sll0359 was analyzed by Northern blot analysis using RNA extracted from Synechocystis sp. strain PCC 6803 cells under combined nitrogen-depleted conditions, as described above (Fig. 1), we observed that in general, it follows the same trend as that of the hox operon (Fig. 1 and 3D).
Interestingly, although sll0359 is annotated in the CyanoBase genome database to be 387 bp long and, therefore, encoding a protein of 128 amino acids, other databases, namely ExPASy and GenBank, have annotated this open reading frame to be 468 bp long. This result has to do with the fact that 81 bp upstream of the CyanoBase-annotated translational start point of sll0359, it is possible to find an additional ATG triplet, which is in the same frame as sll0359, therefore suggesting the existence of a longer protein. Nevertheless, 5′RACE experiments carried out in this work showed that the TSP is localized 64 bp upstream of the CyanoBase-annotated sll0359 start codon, suggesting the existence of the shorter protein. Furthermore, based on N-terminal sequencing of proteins separated by two-dimensional gel electrophoresis, Sazuka et al. (31) could also show that Sll0359 is the shorter version. Therefore, we suggest that the databases should be updated. Moreover, bioinformatically it was possible to identify an extended −10 box in the form of TGNTAN3T (5, 10, 41) in the promoter region of sll0359, although no other characterized recognition motifs could be found.
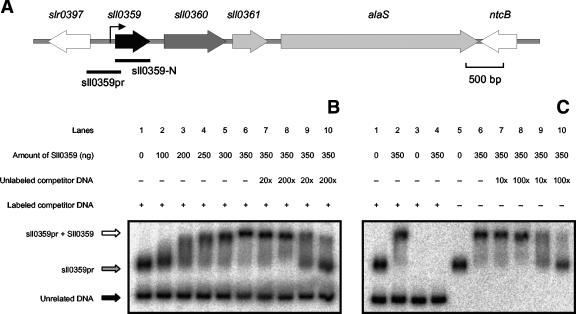
The AbrB-like protein interacts with both the hox and the sll0359 promoter regions.
The sll0359 gene was cloned and overexpressed in E. coli. After purification of the His-tagged AbrB-like protein (Fig. 4), its identity was further confirmed by mass spectrometry. Supporting the data obtained by DNA affinity assays, EMSAs showed a specific interaction between the purified AbrB-like protein and the DNA fragment Shoxpr, covering the hox genes promoter region (Fig. 5B). In these experiments, a mixture of two labeled DNA fragments was used: Shoxpr (462 bp) and an unrelated DNA fragment (227 bp) amplified by PCR from the vector pBluescript. When with AbrB-like protein, Shoxpr is clearly preferred, showing an obvious shift in its mobility, over the unrelated DNA fragment, which suffered no shift. Furthermore, when we used an excess of unlabeled DNA, either Shoxpr or the unrelated DNA fragment, it was possible to strengthen the previous observations that the AbrB-like protein shows high specificity toward Shoxpr (Fig. 5B and C). The purified AbrB-like protein was further used in similar EMSAs by using sll0359pr (457 bp), harboring the sll0359 promoter region, and the unrelated DNA fragment mentioned above. As shown in Fig. 6B and C, the AbrB-like protein interacts with significant specificity with its own promoter region, even in the presence of an excess of unrelated competitor DNA.
FIG. 6.
EMSAs with purified AbrB-like protein from Synechocystis sp. strain PCC 6803 and the sll0359 regulatory region. (A) Schematic representation of the sll0359 locus in the genome of Synechocystis sp. strain PCC 6803. The arrow upstream of sll0359 corresponds to the TSP identified by 5′RACE. The positions of different probes used in this study are represented as lines and identified by their names: sll0359pr and sll0359-N were used as EMSA and Northern blot probes, respectively. (B) Analysis of the electrophoretic mobility of sll0359pr in the presence of increasing concentrations of purified AbrB-like protein. EMSAs were carried out with the target fragment sll0359pr incubated with the unrelated DNA fragment, obtained from pBluescript, without protein (lane 1) or together with increasing amounts of AbrB-like protein (lanes 2 to 6). The unrelated DNA is indicated with a black arrow, and the sll0359pr fragment and its retardation are indicated with gray and white arrows, respectively. To further demonstrate the specific binding between the AbrB-like protein and the fragment sll0359pr, competition experiments were carried out by using a severalfold molar excess of unlabeled unspecific (lanes 7 and 8) or sll0359pr DNA fragments (lanes 9 and 10). (C) Additional EMSAs were carried out to demonstrate that the presence of two labeled DNA fragments, working as possible targets in the same reaction, do not produce unexpected artifacts. Hence, the unrelated DNA fragment (lanes 3 and 4) and sll0359pr (lanes 5 and 6) were incubated alone with AbrB-like protein, in opposition to an assay where both DNA fragments were present (lanes 1 and 2). Lanes 7 to 10 represent supplementary competition experiments. The amount of AbrB-like protein used in each assay is indicated in nanograms in the figure. A plus indicates that the fragment was included in the assay, and a minus indicates that the fragment was excluded from the assay.
The AbrB-like protein is essential for Synechocystis sp. strain PCC 6803 and works as an activator of hox gene expression.
As usual for Synechocystis sp. strain PCC 6803, the degree of chromosome segregation depends strongly on the general role of the studied gene. An attempt to create an sll0359 knockout mutant in Synechocystis sp. strain PCC 6803 was made by inserting the kanamycin resistance cassette within the coding sequence of this gene (see Material and Methods). However, it was not possible to get a fully segregated mutant, since cells harboring the sll0359::Kmr cassette invariably retained wild-type chromosome copies (Fig. 7A), irrespective of an increase of the selective pressure (kanamycin concentration) and duration of subcultivation. Therefore, our results suggest that sll0359 is a gene crucial to the viability of Synechocystis sp. strain PCC 6803. On the other hand, the transformation of Synechocystis sp. strain PCC 6803 with the vector pFMoe01 proved to be efficient (Fig. 7B), creating a strain (SFoe01) that expresses sll0359 under the regulation of the petE promoter region (12, 18).
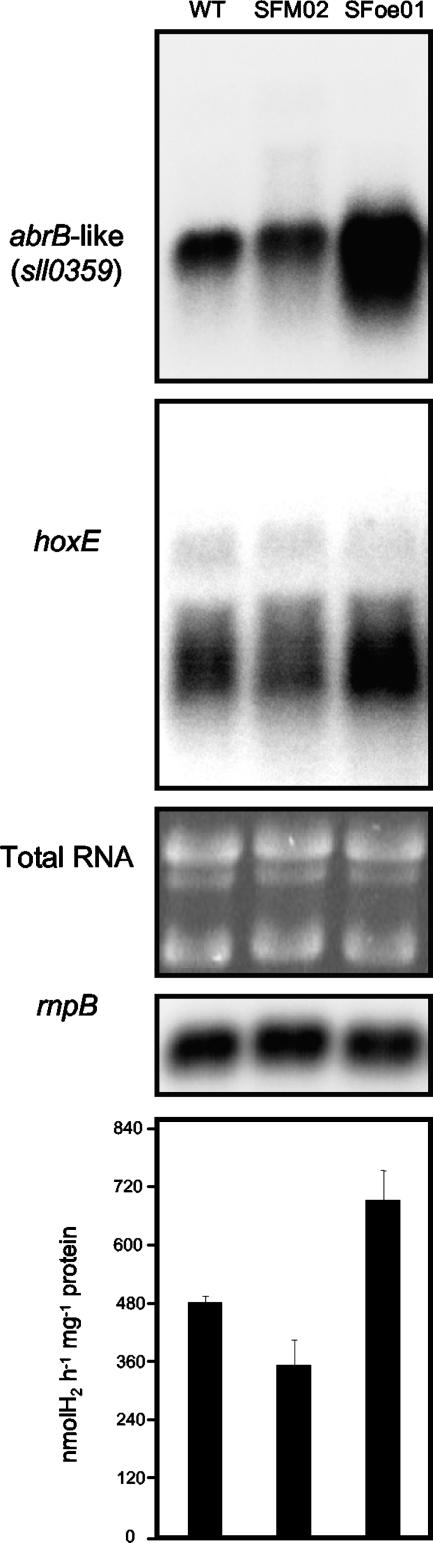
Wild-type and SFM02 and SFoe01 mutant cells of Synechocystis sp. strain PCC 6803 were grown as described previously, followed by RNA extractions and measurements of the bidirectional hydrogenase activity. Under the conditions tested, the sll0359 transcription levels were as expected, considering the genetic modifications introduced (Fig. 8). Interestingly, the expression of the hox genes was higher in SFoe01 (overexpressing sll0359) and lower in SFM02 (Δsll0359::Kmr/sll0359+ heteroploid mutant cells) than in the wild type (Fig. 8). In agreement, the bidirectional hydrogenase activity measurements showed a similar pattern (Fig. 8). All together, these results suggest that the AbrB-like protein works as a transcription activator of the hox gene expression.
FIG. 8.
Northern blot analysis of the relative amount of sll0359, hoxE, and rnpB of Synechocystis sp. strain PCC 6803 wild-type (WT) and SFM02 and SFoe01 mutant cells. The abundance of the rRNA bands stained with ethidium bromide on the agarose gel is shown to further demonstrate the even loading of the total RNA aliquots. Methyl viologen-dependent bidirectional hydrogenase activities from the three different strains in the study are shown in the graph. Error bars correspond to standard deviations from three independent experiments.
DISCUSSION
The genes encoding the bidirectional hydrogenase (hoxEFUYH) in Synechocystis sp. strain PCC 6803 are clustered in the genome and have been shown to be transcribed as one operon (2, 17, 28). The transcription regulator LexA has been suggested to work as an activator of the hox genes in this cyanobacterium (17). In addition, Patterson-Fortin et al. (30) recently suggested that LexA works as a repressor of the RNA helicase CrhR in Synechocystis sp. strain PCC 6803. In that work, the authors showed, by Northern blot analyses, that lexA and crhR are divergently expressed even when the cells are submitted to different conditions. This indicates that at least for crhR, it is possible to correlate the Synechocystis sp. strain PCC 6803 lexA and crhR transcript levels.
However, our results indicate that the expression of lexA and hox genes in Synechocystis sp. strain PCC 6803 does not follow the same pattern when the cells are facing combined nitrogen-limited conditions (Fig. 1). This result might indicate that LexA is not exclusively accounting for the regulation of the hox gene transcription. Alternatively, one cannot exclude the possibility of posttranslational modifications operated on, or cofactors binding to, LexA, which may affect its DNA binding capability and therefore have an effect on the downstream targets of its regulation. Recently, Espinosa et al. (14) reported that PipX interacts with both PII and NtcA, providing a mechanistic link between these two factors. Moreover, the authors showed that PipX is required for NtcA-dependent transcriptional activation, thus implying that it may function as a prokaryotic transcriptional coactivator. Interestingly, LexA has been identified in independent studies (34, 45) to be associated with the thylakoid membranes of Synechocystis sp. strain PCC 6803, which could indeed suggest that it is subject to modification. This hypothesis definitely deserves further attention.
In the present study, we focused on the possibility of additional DNA-binding proteins interacting and regulating the expression of the hox genes in Synechocystis sp. strain PCC 6803. By using streptavidin-coated magnetic beads in DNA affinity assays, it was possible for us to isolate and identify four proteins (Fig. 2), two of which have been described previously, streptavidin and LexA (28). The two novel proteins were identified as phycocyanin β-subunit CpcB and Sll0359.
The blue-green color typical of many cyanobacteria is due to the presence of pigments called phycobilins. These pigments are associated with proteins arranged in the phycobilisome in a distinctive order. The three major phycobiliproteins are allophycocyanin, phycocyanin, and phycoerythrin. In some cyanobacteria, the proportions of these pigments can be altered to increase the absorption of light of specific wavelengths (29). In general, phycocyanins represent approximately 40% of all the phycobiliproteins (46), which in turn can constitute up to 60% of the soluble protein content of cyanobacteria (8, 9, 44). As a result, the polypeptide isolated from the DNA affinity assay and further identified by mass spectrometry as the phycocyanin β-subunit most likely corresponds to an unspecific interaction with the DNA fragment used.
The second novel peptide identified in this study as interacting with the promoter region of the hox genes was Sll0359, an AbrB-like protein possessing homologues in numerous cyanobacterial genomes. Furthermore, when we analyzed the genome locus of different cyanobacterial strains in detail, it was clear that the arrangement of the neighboring genes is strikingly similar (Fig. 3). In fact, a consistent finding was the AbrB-like coding gene, followed by an open reading frame (Fig. 3A) predicted to encode a protein with several transmembrane segments, which belongs to the protein family PF02517. Members of this family are probably proteases, predicted to remove the AAX tripeptide from the C-terminal CAAX motif of a protein, after a prenyl group is attached to the Cys residue. Even though the AbrB-like protein does not have a CAAX motif at its C terminus, the possible interaction between the putative protease and the AbrB-like protein and its consequent modification remain to be evaluated.
When using the purified His-tagged AbrB-like protein (Fig. 4) in gel shift assays (Fig. 5 and 6), we observed a specific interaction with both the hox and the sll0359 promoter regions, supporting the earlier results obtained with the DNA affinity assays. Furthermore, when we used a severalfold molar excess of a number of different unspecific DNA fragments of the approximate sizes of Shoxpr and sll0359pr in additional EMSAs, those fragments failed to compete with the latter probes (data not shown), discarding the possibility that, e.g., the AbrB-like protein binds only to DNA fragments of a minimum size. During in silico analyzation of the DNA sequence of the two promoter regions, it was not possible to find putative recognition motifs, since a search for highly similar DNA stretches failed to give possible hits. Interestingly, AbrB from B. subtilis is well known for its significant promiscuity in DNA recognition (6, 7, 42) and the comparison between the described target regions reveals no apparent base sequence that can be defined as an AbrB consensus binding site (48).
After a strong interaction between the AbrB-like protein and the hox promoter was demonstrated, it became imperative for us to understand the regulatory action that this transcription factor has on the hox genes. For that purpose, the inactivation of sll0359 was tried, although it produced a not fully segregated strain, SFM02 (Fig. 7A). In addition, a mutant overexpressing the abrB-like gene was produced by transforming Synechocystis sp. strain PCC 6803 with the vector pFMoe01 and named SFoe01 (see Material and Methods and Fig. 7B), which is inducible by copper (18). These different strains (wild-type, SFM02 and SFoe01) were subsequently grown, and the transcript levels of the abrB-like gene and hoxE were analyzed by Northern blotting.
Compared to the wild type, the Δsll0359::Kmr/sll0359+ heteroploid mutant strain (SFM02) showed less abrB-like transcript and an additional band/smear when the exposure time was prolonged (data not shown), which might correspond to a readthrough from the antibiotic resistance cassette. On the other hand, SFoe01 showed higher levels of abrB-like transcript than the wild-type did, since the gene was under the regulation of the petE promoter. These observations are in agreement with the genetic modifications introduced in Synechocystis sp. strain PCC 6803. Interestingly, we show that the AbrB-like protein binds to its own promoter region, but the question of whether it activates or represses its transcription remains to be answered. Based on our results, it is premature to put forward any concrete suggestions.
The examinations of the hox transcript levels and the bidirectional hydrogenase activity (Fig. 8), linked with the levels of abrB-like transcript, suggested that the AbrB-like protein works as a transcription activator of the Synechocystis sp. strain PCC 6803 hox genes. However, double-knockout mutants, in addition to overexpressing strains, are needed to understand how LexA and the AbrB-like protein operate, alone and in combination, to control the activity of the bidirectional hydrogenase in this cyanobacterium.
In conclusion, to the best of our knowledge, this is the first time that Sll0359 has been connected to the regulation of the hox operon in the cyanobacterium Synechocystis sp. strain PCC 6803, by interacting with its promoter region. Consequently, this study opens up the possibility for further investigations: what is the overall function of this transcription factor? What other target genes are under its regulation? Which transduction pathways is this AbrB-like protein involved in? It is important to clarify that although the sll0359 and hox operon expressions follow the same trend under combined nitrogen-depleted conditions (Fig. 1 and 3D), there is no experimental evidence thus far that this AbrB-like protein is the direct regulator intermediating this environmental signal and the hox operon transcription response. Interestingly, the AbrB-like protein was recently connected to the expression of other genes in Synechocystis sp. strain PCC 6803 (A. Kaplan, The Hebrew University of Jerusalem, Israel, personal communication). In particular, Ishii and Hihara (19) reported on the possible involvement between this AbrB-like protein and both photosynthesis and pigment biosynthesis. Future investigations are needed to shed light onto the function of this novel transcription regulator in Synechocystis sp. strain PCC 6803 and in other cyanobacteria.
Acknowledgments
We thank Håkan Larsson (Department of Plant Biology and Forest Genetics, Swedish Agricultural University, Sweden) for sequencing the proteins, Matthias Rögner (Ruhr Universität, Bochum, Germany) for his generous gift of the plasmid pAWG1.1 and Fernando Lopes Pinto (Uppsala University) for designing the primers for 5′RACE.
This work was financially supported by the Swedish Energy Agency, the Knut and Alice Wallenberg Foundation, the Nordic Energy Research (project BioH2), the EU/NEST projects SOLAR-H (contract no. 516510) and BioModularH2 (contract no. 043340), and the EU/Energy project SOLAR-H2 (contract 212508).
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antal, T. K., P. Oliveira, and P. Lindblad. 2006. The bidirectional hydrogenase in the cyanobacterium Synechocystis sp. strain PCC 6803. Int. J. Hydrogen Energy 311439-1444. [Google Scholar]
- 3.Axelsson, R., and P. Lindblad. 2002. Transcriptional regulation of Nostoc hydrogenases: effects of oxygen, hydrogen, and nickel. Appl. Environ. Microbiol. 68444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axelsson, R., F. Oxelfelt, and P. Lindblad. 1999. Transcriptional regulation of Nostoc uptake hydrogenase. FEMS Microbiol. Lett. 17077-81. [DOI] [PubMed] [Google Scholar]
- 5.Barne, K. A., J. A. Bown, S. J. W. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the ‘extended-10’ motif at promoters. EMBO J. 164034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobay, B. G., L. Benson, S. Naylor, B. Feeney, A. C. Clark, M. B. Goshe, M. A. Strauch, R. Thompson, and J. Cavanagh. 2004. Evaluation of the DNA binding tendencies of the transition state regulator AbrB. Biochemistry 4316106-16118. [DOI] [PubMed] [Google Scholar]
- 7.Bobay, B. G., G. A. Mueller, R. J. Thompson, A. G. Murzin, R. A. Venters, M. A. Strauch, and J. Cavanagh. 2006. NMR structure of AbhN and comparison with AbrBN: first insights into the DNA binding promiscuity and specificity of AbrB-like transition state regulator proteins. J. Biol. Chem. 28121399-21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogorad, L. 1975. Phycobiliproteins and complementary chromatic adaptation. Annu. Rev. Plant Physiol. 26369-401. [Google Scholar]
- 9.Colyer, C. L., C. S. Kinkade, P. J. Viskari, and J. P. Landers. 2005. Analysis of cyanobacterial pigments and proteins by electrophoretic and chromatographic methods. Anal. Bioanal. Chem. 382559-569. [DOI] [PubMed] [Google Scholar]
- 10.deHaseth, P. L., M. L. Zupancic, and M. T. Record, Jr. 1998. RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 1803019-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domain, F., L. Houot, F. Chauvat, and C. Cassier-Chauvat. 2004. Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 5365-80. [DOI] [PubMed] [Google Scholar]
- 12.Dyczmons, N. G. 2006. Expression and regulation of membrane proteins: special focus on cytochrome bd-oxidase from Synechocystis sp. PCC 6803. Ph.D. thesis. Ruhr-Universität Bochum, Bochum, Germany.
- 13.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria, p. 747-754. In L. Packer and A. N. Glazer (ed.), Methods in enzymology, vol. 167. Academic Press, Inc., San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 14.Espinosa, J., K. Forchhammer, S. Burillo, and A. Contreras. 2006. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 61457-469. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira, D., E. Leitão, J. Sjöholm, P. Oliveira, P. Lindblad, P. Moradas-Ferreira, and P. Tamagnini. 2007. Transcription and regulation of the hydrogenase(s) accessory genes, hypFCDEAB, in the cyanobacterium Lyngbya majuscula CCAP 1446/4. Arch. Microbiol. 188609-617. [DOI] [PubMed] [Google Scholar]
- 16.Gan, C. S., K. F. Reardon, and P. C. Wright. 2005. Comparison of protein and peptide prefractionation methods for the shotgun proteomic analysis of Synechocystis sp. PCC 6803. Proteomics 52468-2478. [DOI] [PubMed] [Google Scholar]
- 17.Gutekunst, K., S. Phunpruch, C. Schwarz, S. Schuchardt, R. Schulz-Friedrich, and J. Appel. 2005. LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 58810-823. [DOI] [PubMed] [Google Scholar]
- 18.Hörnemann, A. 2001. Heterologe Überexpression in cyanobakterien. M.S. thesis. Ruhr-Universität Bochum, Bochum, Germany.
- 19.Ishii, A., and Y. Hihara. 2006. Characterization of putative transcriptional regulators having an AbrB-type DNA binding domain in Synechocystis sp. PCC 6803. Plant Cell Physiol. 47S87. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement). DNA Res. 3185-209. [DOI] [PubMed] [Google Scholar]
- 21.Kufryk, G. I., M. Sachet, G. Schmetterer, and W. F. J. Vermaas. 2002. Transformation of the cyanobacterium Synechocystis sp. PCC 6803 as a tool for genetic mapping: optimization of efficiency. FEMS Microbiol. Lett. 206215-219. [DOI] [PubMed] [Google Scholar]
- 22.Labarre, J., F. Chauvat, and P. Thuriaux. 1989. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC 6803. J. Bacteriol. 1713449-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leitão, E., F. Oxelfelt, P. Oliveira, P. Moradas-Ferreira, and P. Tamagnini. 2005. Analysis of the hupSL operon of the nonheterocystous cyanobacterium Lyngbya majuscula CCAP 1446/4: regulation of transcription and expression under a light-dark regimen. Appl. Environ. Microbiol. 714567-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindberg, P. 2003. Cyanobacterial hydrogen metabolism—uptake hydrogenase and hydrogen production by nitrogenase in filamentous cyanobacteria. Ph.D. thesis. Uppsala University, Uppsala, Sweden.
- 25.Malakhov, M. P., O. A. Malakhova, and N. Murata. 1999. Balanced regulation of expression of the gene for cytochrome cM and that of genes for plastocyanin and cytochrome c6 in Synechocystis. FEBS Lett. 444281-284. [DOI] [PubMed] [Google Scholar]
- 26.Montesinos, M. L., A. M. Muro-Pastor, A. Herrero, and E. Flores. 1998. Ammonium/methylammonium permeases of a cyanobacterium: identification and analysis of three nitrogen-regulated amt genes in Synechocystis sp. PCC 6803. J. Biol. Chem. 27331463-31470. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira, P., E. Leitão, P. Tamagnini, P. Moradas-Ferreira, and F. Oxelfelt. 2004. Characterization and transcriptional analysis of hupSLW in Gloeothece sp. ATCC 27152: an uptake hydrogenase from a unicellular cyanobacterium. Microbiology 1503647-3655. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira, P., and P. Lindblad. 2005. LexA, a transcription regulator binding in the promoter region of the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 25159-66. [DOI] [PubMed] [Google Scholar]
- 29.Oliver, R. L., and G. G. Ganf. 2000. Freshwater blooms, p. 149-194. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 30.Patterson-Fortin, L. M., K. R. Colvin, and G. W. Owttrim. 2006. A LexA-related protein regulates redox-sensitive expression of the cyanobacterial RNA helicase, crhR. Nucleic Acids Res. 343446-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sazuka, T., M. Yamaguchi, and O. Ohara. 1999. Cyano2Dbase updated: linkage of 234 protein spots to corresponding genes through N-terminal microsequencing. Electrophoresis 202160-2171. [DOI] [PubMed] [Google Scholar]
- 32.Schütz, K., T. Happe, O. Troshina, P. Lindblad, E. Leitão, P. Oliveira, and P. Tamagnini. 2004. Cyanobacterial H2 production—a comparative analysis. Planta 218350-359. [DOI] [PubMed] [Google Scholar]
- 33.Sjöholm, J., P. Oliveira, and P. Lindblad. 2007. Transcription and regulation of the bidirectional hydrogenase in the cyanobacterium Nostoc sp. strain PCC 7120. Appl. Environ. Microbiol. 735435-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srivastava, R., T. Pisareva, and B. Norling. 2005. Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 54905-4916. [DOI] [PubMed] [Google Scholar]
- 35.Stanier, R. Y., R. Kunisawa, M. Mandel, and G. Cohen-Bazire. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Microbiol. Mol. Biol. Rev. 35171-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, I., Y. Kanesaki, H. Hayashi, J. J. Hall, W. J. Simon, A. R. Slabas, and N. Murata. 2005. The histidine kinase Hik34 is involved in thermotolerance by regulating the expression of heat shock genes in Synechocystis. Plant Physiol. 1381409-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamagnini, P., R. Axelsson, P. Lindberg, F. Oxelfelt, R. Wünschiers, and P. Lindblad. 2002. Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol. Mol. Biol. Rev. 661-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamagnini, P., E. Leitão, P. Oliveira, D. Ferreira, F. Pinto, D. J. Harris, T. Heidorn, and P. Lindblad. 2007. Cyanobacterial hydrogenases: diversity, regulation and applications. FEMS Microbiol. Rev. 31692-720. [DOI] [PubMed] [Google Scholar]
- 39.Tamagnini, P., O. Troshina, F. Oxelfelt, R. Salema, and P. Lindblad. 1997. Hydrogenases in Nostoc sp. strain PCC 73102, a strain lacking a bidirectional enzyme. Appl. Environ. Microbiol. 631801-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valladares, A., A. M. Muro-Pastor, A. Herrero, and E. Flores. 2004. The NtcA-dependent P1 promoter is utilized for glnA expression in N2-fixing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 1867337-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaughn, J. L., V. Feher, S. Naylor, M. A. Strauch, and J. Cavanagh. 2000. Novel DNA binding domain and genetic regulation model of Bacillus subtilis transition state regulator AbrB. Nat. Struct. Mol. Biol. 71139-1146. [DOI] [PubMed] [Google Scholar]
- 43.Vioque, A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 206331-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viskari, P. J., C. S. Kinkade, and C. L. Colyer. 2001. Determination of phycobiliproteins by capillary electrophoresis with laser-induced fluorescence detection. Electrophoresis 222327-2335. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Y., J. Sun, and P. R. Chitnis. 2000. Proteomic study of the peripheral proteins from thylakoid membranes of the cyanobacterium Synechocystis sp. PCC 6803. Electrophoresis 211746-1754. [DOI] [PubMed] [Google Scholar]
- 46.Williams, R. C., J. C. Gingrich, and A. N. Glazer. 1980. Cyanobacterial phycobilisomes. Particles from Synechocystis 6701 and two pigment mutants. J. Cell Biol. 85558-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379466-469. [DOI] [PubMed] [Google Scholar]
- 48.Xu, K., and M. A. Strauch. 2001. DNA-binding activity of amino-terminal domains of the Bacillus subtilis AbrB Protein. J. Bacteriol. 1834094-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]