Abstract
Under anoxic conditions, the Escherichia coli oxygen sensor FNR (fumarate nitrate reductase regulator) is in the active state and contains a [4Fe-4S] cluster. Oxygen converts [4Fe-4S]FNR to inactive [2Fe-2S]FNR. After prolonged exposure to air in vitro, apoFNR lacking a Fe-S cluster is formed. ApoFNR can be differentiated from Fe-S-containing forms by the accessibility of the five Cys thiol residues, four of which serve as ligands for the Fe-S cluster. The presence of apoFNR in aerobically and anaerobically grown E. coli was analyzed in situ using thiol reagents. In anaerobically and aerobically grown cells, the membrane-permeable monobromobimane labeled one to two and four Cys residues, respectively; the same labeling pattern was found with impermeable thiol reagents after cell permeabilization. Alkylation of FNR in aerobic bacteria and counting the labeled residues by mass spectrometry showed a form of FNR with five accessible Cys residues, corresponding to apoFNR with all Cys residues in the thiol state. Therefore, aerobically growing cells contain apoFNR, whereas a significant amount of Fe-S-containing FNR was not detected under these conditions. Exposure of anaerobic bacteria to oxygen caused conversion of Fe-S-containing FNR to apoFNR within 6 min. ApoFNR from aerobic bacteria contained no disulfide, in contrast to apoFNR formed in vitro by air inactivation, and all Cys residues were in the thiol form.
The transcriptional regulator FNR (fumarate nitrate reductase regulator) of Escherichia coli functions as an O2 sensor (3, 7, 11, 15, 16, 19, 39). Under anoxic conditions, the protein is in the active state and is predominately found as a homodimer with one [4Fe-4S] cluster per monomer. In the presence of O2, [4Fe-4S]FNR is converted to monomeric [2Fe-2S]FNR, which is no longer active in gene regulation (6, 7, 12, 18, 24). The Fe-S clusters are bound by four cysteine residues (Cys20, Cys23, Cys29, and Cys122) of the five Cys residues of FNR. The only other Cys residue (Cys16) is not essential and not involved in binding of the FeS cluster (13, 25, 29). The Fe-S clusters in FNR can be detected by Mössbauer spectroscopy, which has been essential for characterizing the Fe-S-containing forms in vitro and in vivo (18, 27, 32).
The properties of [4Fe-4S]FNR and [2Fe-2S]FNR, their presence in aerobically and anaerobically growing E. coli in vivo and in vitro, and their significance in O2 sensing are well documented. In vivo [2Fe-2S]FNR degrades further upon continued exposure of the bacteria to O2 (18, 32). In vitro, apoFNR is formed within a few minutes after exposure of [4Fe-4S]FNR to O2, which points toward a physiological relevance of this form of FNR (1). ApoFNR, like [2Fe-2S]FNR, is inactive in DNA binding and gene regulation (14). The conditions for apoFNR formation from [4Fe-4S]FNR have been characterized in vitro (1). In vivo, only indirect evidence for the formation of apoFNR has been provided: the [4Fe-4S]2+ and [2Fe-2S]2+ clusters of FNR are lacking in aerobically grown cells under conditions in which the FNR protein is still present (31). From this, it has been concluded that the remaining FNR is apoFNR, but its identity has not been directly demonstrated. It is not known whether apoFNR in vivo is of the same type as in vitro, where apoFNR contains disulfides Cys16/20 and Cys23/29 which are of unknown physiological significance (1, 2).
Here we studied the formation and presence of apoFNR in aerobically and anaerobically growing E. coli in situ by differentiating apoFNR from [2Fe-2S]FNR and [4Fe-4S]FNR using a method for measuring the accessibility of the Cys residues to thiol reagents (1). Up to five Cys residues are accessible to thiol reagents in apoFNR, whereas only Cys16 is accessible in [4Fe-4S]FNR and [2Fe-2S]FNR; the other four Cys residues are ligands of the Fe-S clusters in [4Fe-4S]FNR and [2Fe-2S]FNR and are thereby protected from labeling. The amount of accessible Cys residues was determined by the use of Cys-specific reagents which introduce a fluorescence label into FNR or change the apparent Mr of FNR in sodium dodecyl sulfate (SDS)-polyacrylamide gels. For labeling, membrane permeable and impermeable thiol reagents were used. Membrane permeable labels are supposed to allow labeling of cytoplasmic proteins in situ without cell disruption and with minimal perturbation of cellular metabolism and functional state of FNR. The results demonstrate that the major form of FNR in aerobically growing E. coli is apoFNR lacking disulfides and that the kinetics of apoFNR formation in vivo and in vitro is similar to that of [4Fe-4S]FNR/[2Fe-2S]FNR conversion.
MATERIALS AND METHODS
Growth of E. coli and in vivo labeling of proteins with thiol reagents.
E. coli CAG627 [lacZ(Am) trp(Am) pho(Am) supC(Ts) mal rpsL lon] (34) carrying plasmid pMW32 (pTrc99A with the 750-bp NcoI/BamHI fnr fragment of pGS199 [30]) overproduces FNR from the plasmid-encoded fnr gene. The strain was grown under oxic or anoxic conditions (36) in 2×YT medium (28) with glucose (100 mM), and the oxic and anoxic conditions were strictly maintained throughout the labeling procedure. At an optical density at 578 nm (OD578) of 0.6 to 0.8, the culture was diluted 1:10 in the same medium, and fnr transcription was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM). After 100 min of incubation (OD578 of 0.7; mid-exponential phase), 1 or 2 mM monobromobimane (mBBr) (Invitrogen) (20-22) from a freshly prepared stock solution (10 mM) was added, and incubation continued for 5 min in the dark. The aerobic sample (1 ml in a 15-ml plastic tube) was shaken at 150 rpm under air. The anaerobic sample (1 ml in a 2-ml plastic tube) was handled and incubated in a glove box under N2, and anoxic solutions were used throughout. Labeling reached maximal levels after 1 min or longer (not shown). The labeling was stopped after 5 min by adding 2 mM dithiothreitol (DTT). The cells were collected by centrifugation and resuspended and lysed in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 5% SDS (23, 28).
For measuring the kinetics of the appearance of accessible Cys residues, E. coli CAG627(pMW32) was grown anaerobically in the presence of IPTG to an OD578 of 1.0. Aliquots (1 ml) were transferred to petri dishes (diameter, 8.5 cm) and shaken under air (130 rpm). At different time points, 1 mM mBBr was added, and the suspensions were incubated for 1 min. The reaction was stopped by the addition of 1 mM DTT, and the samples were prepared for SDS-PAGE as described above. Proteins were labeled with monobromotrimethylammoniobimane (qBBr) (Calbiochem) (21) following the same method as for mBBr, except that cells were permeabilized with chloroform (50 μl) before adding qBBr (2 mM) from a freshly prepared qBBr stock solution (10 mM in methanol).
Proteins were labeled with 4-acetamido-4′-maleimidylstilbene-2′2-disulfonic acid (AMS) (8 mM) in 1 ml of a cell suspension (OD578 of 0.8) containing 50 μl chloroform for 30 min (17) following a method similar to that described for qBBr.
Alkylation of FNR in vivo by NEM or iodoacetate for MS.
FNR was produced as a GST′-′FNR (GST is glutathione S-transferase) fusion in E. coli CAG627(pMW68) growing under aerobic conditions as described previously (37) by induction with IPTG. Two hours after the start of induction, N-ethylmaleimide (NEM) (10 mM in water) or iodoacetate (10 mM in water, pH 8.0) was added for 15 min (1, 4). The bacteria were harvested, and GST-FNR was prepared by chromatography on glutathione-Sepharose 4B (1, 37). The GST tag was removed by incubation with thrombin for 2 h, and the isolated FNR (600 μg/ml) was analyzed by mass spectrometry (MS). All analyses were carried out on a Q-TOF II system (Waters Corp., Manchester, United Kingdom). Prior to mass measurements, the samples were separated by reversed-phase high-performance liquid chromatography (HPLC) (Waters Alliance 2795; Waters Corp., Manchester, United Kingdom) on a Vydac C18 column (250 by 2.1 mm) connected online to the MS instrument. The HPLC instrument was operated using a binary gradient system at a flow rate of 300 μl/min from 80% to 20% solvent A in 10 min and isocratic for 20 min (solvent A is 0.025% trifluoroacetic acid in water, and solvent B is 0.023% trifluoroacetic acid in acetonitrile). The sample concentration was 0.5 mg/ml. Ten microliters of each sample was injected.
Labeling of isolated FNR.
FNR was isolated as apoFNR and reconstituted to [4Fe-4S]FNR as described previously (1). mBBr binding was quantitatively calibrated using FNR with one, two, three, and five Cys residues in the thiol state. Isolated FNR contains three Cys thiols (1). To obtain FNR with one or two free Cys thiol groups, isolated FNR was incubated with 1 and 2 mol NEM per mol FNR, respectively. FNR with five Cys thiols was produced by reducing isolated FNR with an equimolar amount of DTT. Samples of the various forms of FNR in 50 mM Tris-HCl (pH 7.6) were incubated with 2 mM mBBr for 5 min and then subjected to SDS-PAGE, followed by Western blotting.
For labeling of purified FNR with AMS, reconstituted [4Fe-4S]FNR was incubated with AMS (8 mM) in a glove box under N2 for 20 min. For air inactivation, a solution (1 ml) of reconstituted FNR (1) was spread onto a petri dish and shaken under air for 20 min at 130 rpm.
Quantitative evaluation of mBBr and qBBr labeling of FNR.
The labeling by the various reagents was quantitatively evaluated using the Western blots of the labeled proteins (cell homogenates or isolated FNR). For each set of experiments, the samples were run and evaluated on the same gel and blot. The fluorescence of protein bands labeled with mBBr or qBBr was measured with a fluorescence imager (Kodak ImageStation CF440) from 400 to 500 nm. The fluorescence of individual bands was quantified using Kodak software by integrating the amount of fluorescence of a specific band after subtraction of background fluorescence (“net intensity”). The integrated fluorescence is given in pixel intensity (arbitrary units). From the same blot, the FNR content was determined using anti-FNR serum (17, 35) and a secondary antibody coupled to horseradish peroxidase. The stain intensity was recorded by the imager. For quantitative experiments, all samples from one blot were treated the same, and at least four independent labeling experiments were performed. The amount of label is given as the pixel intensity of the fluorescence and of the absorption of the anti-FNR stain. For quantitation, FNR with one or five accessible Cys residues was used as the reference (20% and 100% labeling, respectively). For determining the amount of labeled thiols of FNR in vivo, the cells were labeled as described above. Samples were run on a gel with isolated FNR labeled on five Cys residues as the reference (100% labeling). After SDS-PAGE and blotting, mBBr fluorescence and FNR immunostaining were measured, and the specific amount of labeling was calculated (pixel intensity of mBBr fluorescence/pixel intensity of FNR immunostain). For each sample, the ratio was determined and compared to the ratio of fivefold-labeled FNR as the reference.
RESULTS
Accessible and reactive Cys residues of FNR in aerobically and anaerobically grown E. coli.
ApoFNR can be differentiated from FNR containing Fe-S clusters by measuring the accessibility of the Cys residues (1). The accessible Cys residues of the cellular proteins of E. coli, including FNR, were labeled with the membrane-permeable thiol reagent mBBr. By penetrating the cell membrane without need of cell disruption or treatment with detergents or solvents (22), the reagent should allow labeling with minimal perturbation of the functional state of the cell and of FNR. mBBr becomes fluorescent only after reaction with thiol groups. E. coli CAG627(pMW32) overproduces FNR to levels which are sufficient for direct quantitation of FNR from cell extracts by immunoblotting, whereas wild-type FNR contents are too low for direct quantitation (35, 38, 39). FNR from bacteria with (moderately) overproduced FNR is supposed to show normal response to the presence of O2 (35).
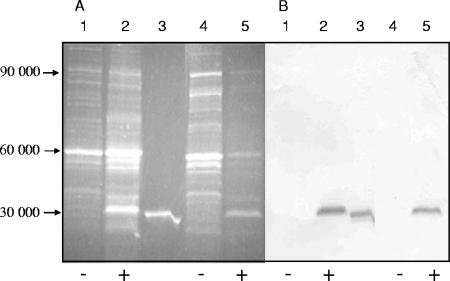
The bacteria were grown aerobically or anaerobically and then labeled with mBBr. The cellular proteins were separated by SDS-PAGE and blotted onto a nitrocellulose membrane. In the blots of noninduced aerobic or anaerobic E. coli CAG627(pMW32), a limited number of bands with strong fluorescence was found; the band were mainly in the Mr range of 50,000 to 60,000 and around 90,000 (Fig. 1A). In induced cells of aerobically or anaerobically grown E. coli, an additional major fluorescent band in the 30,000-Mr region was observed. A similar labeling pattern was obtained when the bacteria were permeabilized by chloroform or after cell disruption before labeling, demonstrating that mBBr gains sufficient access to the cytoplasm of E. coli, as suggested earlier for other types of cells (20, 22). Purified and mBBr-labeled apoFNR formed a fluorescent band with a mobility similar to that of the prominent 30,000-Mr band (Fig. 1A). The band corresponding to the fluorescent 30,000-Mr protein reacted specifically with anti-FNR serum (Fig. 1B). When the expression of plasmid-encoded fnr was not induced, neither the fluorescent band (Fig. 1A) nor the band responding to anti-FNR in the immunoblot was produced (Fig. 1B). Altogether, the findings identified the reactive 30,000-Mr protein as FNR and the corresponding fluorescence as FNR derived. The identities of the other labeled proteins are not known.
FIG. 1.
In vivo labeling with mBBr of proteins of aerobically and anaerobically grown E. coli CAG627(pMW32). Detection of bound mBBr by fluorescence (A) and FNR by immunostaining (B). The bacteria were grown aerobically (lanes 1 and 2) or anaerobically (lanes 4 and 5) to an OD578 of 0.7 under fnr-inducing (+) (1 mM IPTG; lanes 2 and 5) or noninducing (−) (lanes 1 and 4) conditions. Lane 3 contains 1.2 μg purified FNR. Cells were incubated for 5 min with 2 mM mBBr. Sedimented bacteria were resuspended and lysed in SDS-PAGE sample buffer (approximately 30 μg/lane); samples were subjected to SDS-PAGE and blotted onto nitrocellulose. (A) Fluorescence was measured using a Kodak ImageStation CF440 with emission at 400 to 500 nm. (B) The same blot was used subsequently for immunostaining with anti-FNR. Staining was recorded as the absorbance at 400 to 500 nm. The positions of proteins corresponding to Mr 30,000 to Mr 90,000 (calculated from Mr markers) are shown to the left of the blots.
When proteins were labeled with the impermeable thiol reagent monobromo-trimethylammoniobimane, a very similar pattern of fluorescence labeling was observed (not shown); proteins were labeled only after permeabilization of the cells. Again a labeled band with an Mr of approximately 30,000 was observed in induced cells.
Aerobically grown E. coli contains FNR with increased amounts of accessible Cys residues.
To calculate the specific content of labeled Cys residues in FNR, the amounts of bound mBBr and of FNR were determined by quantifying the fluorescence and immunostaining in Western blots, as in Fig. 1 (Table 1). The specific amount of fluorescence per FNR from aerobically grown E. coli showed some variation but was reproducibly twofold higher than that from anaerobically grown E. coli. The increased labeling was specific for FNR; the fluorescence of other labeled bands did not increase in the same way. qBBr-labeled proteins were evaluated in a similar way. Although the intensity of qBBr fluorescence was lower than that of mBBr fluorescence, aerobically grown cells again showed a twofold increase in labeling of FNR compared to anaerobically grown bacteria (Table 1). Since [4Fe-4S]FNR and [2Fe-2S]FNR require the same number of Cys residues (four) for liganding the FeS cluster, the difference should indicate increased amounts of apoFNR.
TABLE 1.
In vivo labeling with mBBr and qBBr of Cys residues in FNR in aerobically and anaerobically growing E. coli CAG627(pMW32)a
| Thiol reagent | Aerobic
|
Anaerobic
|
||||
|---|---|---|---|---|---|---|
| Fluorescence label (PIfluor) | Anti-FNR stain (PIimmuno) | Specific labeling FNR (PIfluor/PIimmuno) | Fluorescence label (PIfluor) | Anti-FNR stain (PIimmuno) | Specific labeling FNR (PIfluor/PIimmuno) | |
| mBBr | 67,860 ± 7,000 | 113,230 ± 11,000 | 0.6 ± 0.06 | 30,530 ± 3,000 | 117,800 ± 12,000 | 0.3 ± 0.03 |
| qBBr | 2,520 ± 250 | 20,740 ± 2,100 | 0.12 ± 0.01 | 1,385 ± 0,140 | 24,890 ± 2,500 | 0.056 ± 0.006 |
The amount of label was determined from blots (compare Fig. 1 and 2) by measuring the fluorescence of mBBr and qBBr by fluorescence imaging (integrated pixel intensity of fluorescence [PIfluor] of the complete bands). The amount of FNR was determined from immunostaining with anti-FNR by absorption imaging with a Kodak ImageStation CF440 (integrated pixel intensity of immunostaining [PIimmuno] of the complete bands). The specific labeling of FNR was calculated from the two values (PIfluor/PIimmuno). All values are means ± standard deviations from at least four replicate and independent samples.
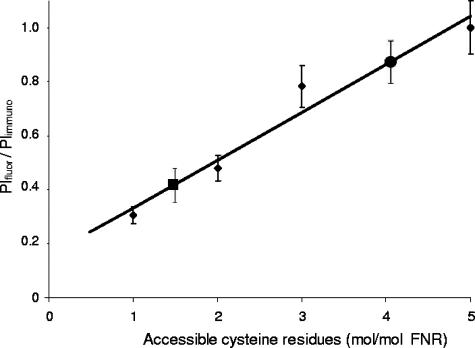
The specific amount of label incorporated into FNR was determined using purified apoFNR as a reference. In purified apoFNR, defined numbers of Cys residues were blocked by reaction with defined amounts of NEM (1). The amount of reactive Cys residues was quantified by reaction with 5,5′-dithiobis-nitrobenzoate (1). In this way, FNR with one, two, three, or five Cys residues in the thiol state was available, which then reacted with mBBr. The fluorescence of the proteins was determined in Western blots as in Fig. 1 and normalized for the amount of FNR protein in immunoblots. The specific amount of fluorescence per FNR protein showed a linear relationship to the number of accessible Cys residues (Fig. 2). The specific labeling varied by a factor of approximately 5 when FNR with one and five accessible Cys residues was labeled, confirming that by this method up to five Cys residues per FNR become labeled.
FIG. 2.
Specific fluorescence of mBBr-labeled FNR in aerobically and anaerobically grown E. coli CAG627(pMW32). The specific fluorescence was calibrated using purified FNR with one, two, three, and five Cys thiols/FNR (small solid diamonds) labeled with mBBr. After separation by SDS-PAGE and blotting onto nitrocellulose, mBBr fluorescence (integrated pixel intensity of fluorescence [PIfluor]) and FNR immunostaining (integrated pixel intensity of immunostaining [PIimmuno]) were measured by fluorescence and absorption imaging as described in the legend to Fig. 1. The specific labeling of the different FNR species (PIfluor/PIimmuno) was determined as described in footnote a of Table 1, and the specific labeling of FNR with five accessible Cys residues was taken as 100%. Following the same method, the PIfluor/PIimmuno of protein from aerobically grown cells (one solid circle) and anaerobically grown cells (one solid square) was determined. The mean experimental values are derived from four or more independent experiments, and the standard deviations (error bars) are given.
The calibration curve was used to determine the specific content of mBBr-labeled FNR of E. coli cells. The label which was found in FNR from aerobically and anaerobically growing cells amounted to approximately 80% and 40%, respectively, of FNR labeled at five Cys residues (Fig. 2). Therefore, in aerobically and anaerobically growing E. coli, on average four and two Cys residues are accessible to modification. In aerobic bacteria, labeling of five Cys residues is expected. The lower degree of labeling might be due to incomplete application of aerobic conditions or to incomplete chemical labeling in vivo.
Kinetics of [4Fe-4S]FNR conversion to apoFNR upon exposure to air.
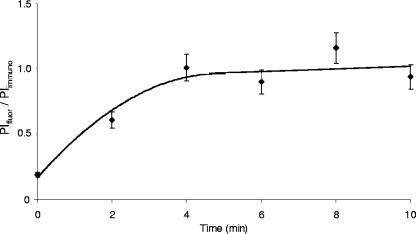
Labeling of the Cys residues of FNR by mBBr was a rapid process, and after 1 min of labeling, no further increase was observed (not shown). Due to the rapid reaction, the method can be used to determine the kinetics of the appearance of accessible Cys residues after shifting anaerobically growing bacteria to oxic conditions. At distinct time points after the shift, samples were withdrawn, and the cells were incubated with mBBr (Fig. 3). Before the shift, about 20% of the Cys residues (corresponding to about one labeled Cys residue/FNR) were accessible to mBBr. With increasing time of O2 exposure, the amount of label increased, and maximal labeling was achieved 5 to 6 min after the shift. The degree of labeling was similar to that of FNR from E. coli grown aerobically only.
FIG. 3.
Kinetics of [4Fe-4S]FNR conversion to apoFNR in vivo, as determined by labeling of FNR with mBBr. E. coli CAG627(pMW32) was grown anaerobically to an OD578 of 1.0 in the presence of IPTG. Aliquots were placed in petri dishes and shaken under air; at specific time points, the samples were labeled with mBBr. After the reaction was stopped by the addition of DTT, the cellular proteins were subjected to SDS-PAGE and Western blotting. FNR fluorescence (integrated pixel intensity of fluorescence [PIfluor]) and immunostaining (integrated pixel intensity of immunostaining [PIimmuno]) were quantitated using isolated FNR with five accessible Cys residues as a reference as described in the legend to Fig. 3 and in footnote a of Table 1. Values are shown as means ± standard deviations (error bars) from three independent experiments.
Distinguishing the Fe-S-containing forms of FNR from apoFNR in vivo by AMS.
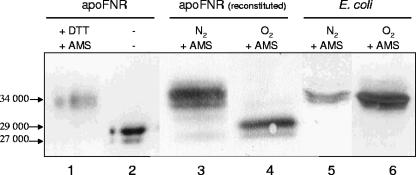
AMS has been widely used for the labeling of Cys residues in the hydrophilic space of proteins (33). When aerobically purified apoFNR (Mr of 29,000) was reduced by DTT and incubated with AMS, the apparent Mr increased from 29,000 to 34,000 according to SDS-PAGE (Fig. 4). It can be assumed that this form of FNR is labeled at five Cys residues, indicating an approximate increase in Mr of 1,000 per molecule of bound AMS. The increase corresponds to the mass of AMS (536.44 Da), but the negative charge of AMS might contribute to the changed mobility as well. The labeling was compared to the labeling of reconstituted FNR in vitro. Purified ApoFNR was reconstituted in vitro using Fe(II), cysteine, and cysteine desulfurase NifSAV as described previously (1). When [Fe-4S]FNR reconstituted in this way was reacted with AMS, FNR with an apparent Mr of 34,000 and small amounts of FNR with an Mr of 32,000 were found. Thus, apoFNR, which was reduced by DTT, and reconstituted [4Fe-4S]FNR showed the same Mr of 34,000 after labeling by AMS, suggesting that AMS replaces the FeS cluster and labels FNR at up to five Cys residues. FNR with an Mr of 32,000 is presumably labeled at three (or two) Cys residues by AMS.
FIG. 4.
AMS labeling of Cys thiols of purified apoFNR and FNR from aerobically or anaerobically grown E. coli CAG627(pMW32). Purified apoFNR was reduced with excess DTT (+ DTT) and subsequently labeled with AMS (8 mM) (+ AMS) (lane 1). Lane 2 contains purified unlabeled apoFNR for comparison. Reconstituted [4Fe-4S]FNR was labeled with AMS either directly (lane 3) or after incubation under air for 20 min (lane 4). For labeling in vivo, E. coli CAG627(pMW32) was grown under inducing conditions under anoxic (N2) (lane 5) or oxic (O2) (lane 6) conditions to an OD578 of 0.8, permeabilized with chloroform, and incubated with 10 mM AMS for 5 min. After the reaction was stopped with the SDS sample buffer, the isolated proteins (2 to 5 μg of purified FNR) and the cellular proteins (30 μg) were separated by SDS-PAGE, blotted onto nitrocellulose, and used for immunoblotting. The positions of proteins corresponding to Mr 27,000, 29,000, and 34,000 are shown.
After exposure of reconstituted FNR to air (20 min) and subsequent labeling with AMS, the 29,000-Mr form of FNR and a small amount of a 27,000-Mr form of FNR were left. ApoFNR with an Mr of 27,000 represents an oxidized form of FNR, which can be converted to FNR with an Mr of 29,000 by reduction with DTT (1). The experiments indicate that AMS displaces [Fe-S] clusters from FNR and labels the newly accessible Cys ligands. In contrast, air-exposed apoFNR contains large amounts of Cys disulfides (1), which are not labeled by AMS (and other thiol reagents). Treatment of the air-oxidized apoFNR with DTT results in AMS-labeled apoFNR similar to that of lane 1 in Fig. 4 (not shown).
Labeling of FNR with the impermeable reagent AMS was performed also in vivo after permeabilizing the cells with chloroform. The proteins were analyzed after SDS-PAGE by immunoblotting with anti-FNR. The immunoblot revealed a form of FNR with an apparent Mr of 34,000 in both anaerobically and aerobically growing cells. FNR proteins with other Mr values were not found (Fig. 4, lanes 5 and 6). The 34,000-Mr form of FNR can be explained by assuming labeling of all Cys residues of FNR. The 34,000-Mr form of FNR (Fig. 4) from the aerobically and anaerobically grown bacteria is supposed to originate from [4Fe-4S]FNR in the anaerobically grown bacteria and from fully reduced apoFNR in the aerobically grown bacteria.
Lack of Cys disulfides in apoFNR of aerobically growing E. coli.
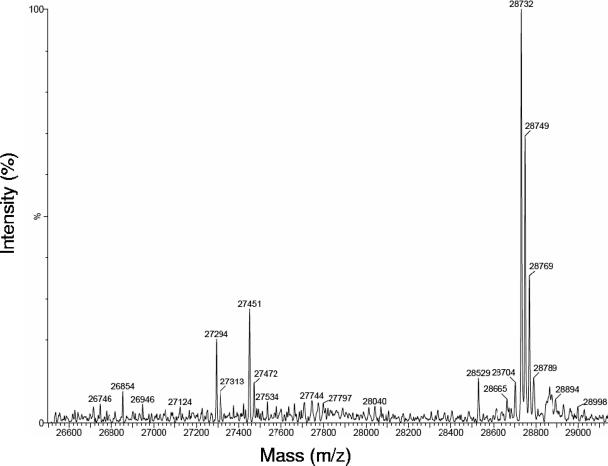
The labeling pattern of apoFNR from aerobically grown E. coli suggested that all Cys residues are accessible to labeling by AMS (Fig. 4), indicating that no intramolecular disulfides are formed. For a more quantitative analysis of the redox state of the cysteine residues, apoFNR from aerobically growing E. coli was labeled in vivo in nonpermeabilized bacteria with NEM. Isolated FNR was then subjected to MS (Fig. 5). The major signal with a mass of 28,732 Da corresponds to FNR (28,111 Da) alkylated at five Cys residues by NEM, each label causing an increment of 124 Da. No significant masses corresponding to smaller FNR species were detectable. In particular, there were no signals equivalent to FNR labeled at only one or three Cys residues which would be characteristic for the presence of two or one intramolecular disulfide bonds. Instead, minor bands of higher masses were detected; these bands may represent other proteins or products of alternative thrombin cleavage during FNR preparation. A similar result was obtained when FNR was labeled in aerobically growing bacteria by iodoacetate (not shown). Here, MS analysis revealed a major signal at a mass of 28,398 Da corresponding to apoFNR alkylated at five Cys residues by acetate (increase in molecular mass by 58 Da/acetate) which confirms the presence of apoFNR with five reduced thiol residues. In addition, a minor signal at a much lower intensity was detected corresponding to fourfold alkylated apoFNR (28,339 Da). ApoFNR with one nonreactive Cys residue could be due to temporary binding of a monothiolate or a metal ion. Again, there were no significant signals with masses corresponding to that of unlabeled FNR or to the masses of forms of FNR with only one, two, or three alkylated Cys residues. The labeling pattern obtained with NEM and iodoacetate therefore verifies that no significant amount of disulfide is present in apoFNR from aerobically grown E. coli.
FIG. 5.
MS of FNR from aerobically grown bacteria after alkylation by NEM in vivo. E. coli CAG627(pMW68) with induced GST-FNR production (OD578 of 1.0) was grown under inducing conditions and labeled by adding NEM (10 mM) to the medium for 15 min without permeabilization of the bacteria. FNR was isolated and separated from GST by thrombin. FNR (600 μg/ml) was subjected to reversed-phase HPLC on a C18 column connected online to the MS instrument. Unmodified FNR has a mass of 28,111 Da (cleavage product of FNR-GST); modified forms with one, two, three, four, and five NEM residues have predicted masses of 28,235, 28,359, 28,483, 28,607, and 28,781 Da, respectively.
DISCUSSION
Direct demonstration of apoFNR in vivo: apoFNR as the main form of FNR in aerobically growing E. coli.
FNR of aerobically growing bacteria is predominately found as apoFNR. Labeling reagents like mBBr which selectively label accessible Cys residues without removing the labile FeS cluster allowed identification of apoFNR and differentiation from FeS-containing FNR in vivo. FNR with nonaccessible Cys residues (i.e., FeS-containing FNR) was found under the same conditions as in earlier studies by Mössbauer spectroscopy (27, 31, 32). The labeling methods used here allowed direct demonstration of apoFNR, and in addition, of the redox state of the Cys residues. Apparently, [2Fe-2S]- and [4Fe-4S]-containing FNR showed similar sensitivities to the displacement of the FeS cluster, and both forms were not differentiated in the labeling studies. Earlier approaches to identify accessible Cys residues of FNR in vivo (10) were performed when the presence of FeS clusters in FNR was not known. The experiments therefore were not designed to differentiate between FeS-containing FNR and apoFNR as in the present study.
ApoFNR in the aerobically grown bacteria is either newly synthesized with not yet incorporated FeS cluster or the conversion product of [4Fe-4S]FNR. The relative contribution of both processes to apoFNR formation is not known. The anaerobic/aerobic shift experiments in bacteria (9, 10, 27, 31, 32) and in vitro (1) demonstrate that conversion takes place efficiently.
The reagent mBBr allowed quantitative in situ studies on apoFNR. Up to five Cys residues per FNR become labeled by mBBr, and the number of reactive residues varies between one and two and about four Cys residues in vivo per FNR depending on the presence of O2. Principally, for [4Fe-4S]FNR/[2Fe-2S]FNR and apoFNR. one and five accessible and labeled Cys residues are expected. The presence of one or two accessible residues in anaerobic cells suggests that the bacteria contain some apoFNR. Mössbauer spectroscopy of anaerobic bacteria also showed significant amounts apoFNR without incorporated FeS cluster (27). The suboptimal labeling in aerobic bacteria (labeling of four compared to five Cys labels) could be due either to incomplete labeling and accessibility or to the presence of some residual FNR with bound FeS cluster. Labeling of FNR in vivo in aerobic bacteria with NEM showed indeed the theoretical value of five accessible residues predicted for apoFNR.
The quantitative conversion of [4Fe-4S]FNR to apoFNR in vivo within a few minutes demonstrates the physiological relevance of the reaction. In vitro, air inactivation of [4Fe-4S]FNR yields apoFNR with 1.5 Cys-disulfide residues (1). The present experiments show that apoFNR in bacterial cells contains no significant amounts of disulfides. Obviously the disulfides are formed in vitro only in the absence of the highly reduced thiol/glutathione redox buffer of E. coli. In the bacterial cells, more than 90% of the free thiols are in the thiol state (8) and provide strongly reducing conditions.
Before the presence and function of the [4Fe-4S] cluster in FNR was known, the Cys residues of aerobically growing E. coli were shown to be more readily accessible to alkylation than those from anaerobically growing E. coli (35). The change in the labeling behavior was then attributed to a thiol/disulfide change and to metal ion binding by FNR in response to O2 availability. After identification of the O2-sensitive [4Fe-4S] cluster and the [4Fe-4S]/[2Fe-2S] switch of FNR, this model had to be dismissed. It now becomes apparent that the increase in the number of accessible Cys residues in FNR of aerobically growing E. coli is functionally relevant, but it has to be attributed to the release of the FeS clusters and the formation of apoFNR.
Two-step inactivation of [4Fe-4S]FNR by oxygen: roles for [2Fe-2S]FNR and apoFNR.
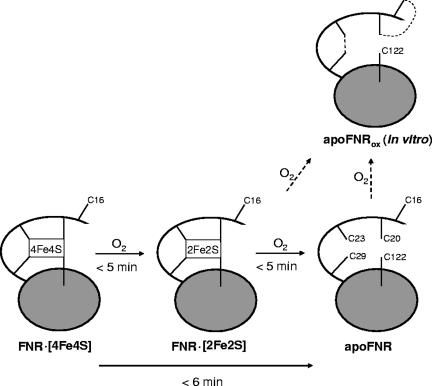
Overall, the earlier experiments and the present experiments identify three different functional forms of FNR in vivo: [4Fe-4S]FNR, [2Fe-2S]FNR, and apoFNR (Fig. 6). The rapid formation of the two inactive forms, [2Fe-2S]FNR and apoFNR, suggests that both play a role in the bacterial cells. [2Fe-2S]FNR is formed from [4Fe-4S]FNR and then degraded within a few minutes (31, 32). In the absence of further sufficient oxygen (or superoxide) [2Fe-2S]FNR is rather stable (32). Here a similar kinetics in the minute range for the formation of apoFNR from [4Fe-4S]FNR in aerobically growing cells was found. Thus, under oxic conditions, [2Fe-2S]FNR appears to be an intermediate in the formation of apoFNR, and apoFNR represents the inactive form of FNR in aerobically growing E. coli. Switch experiments suggest that the active form of FNR and apoFNR can be interconverted in both directions (9, 10). ApoFNR, however, exhibits a decreased half-life (26).
FIG. 6.
Scheme depicting the various forms of FNR and the accessibility and redox state of the five Cys residues. The state of the Cys residues in the N terminus (C16, C20, C23, and C29) and the central part (C122) of [4Fe-4S]FNR, [2Fe-2S]FNR, and apoFNR are shown. ApoFNRox indicates the form of apoFNR obtained after isolation of the protein under oxic conditions or after air inactivation of reconstituted [4F-4S]FNR in vitro; apoFNR is the form found in aerobically grown bacteria or of isolated apoFNR after reduction by DTT. The broken lines in oxidized apoFNR (apoFNRox) represent Cys disulfides. For the dimeric [4Fe-4S]FNR, only one monomer is shown. See text for details. ApoFNR could be derived from [2Fe-2S]FNR or de novo synthesized. The times for the interconversion of the different forms refer to air-saturated conditions and are approximate times from in vivo and in vitro experiments (1, 7, 24, 27, 31, 32).
When O2 is supplied only briefly or at low concentrations, inactivation of [4Fe-4S]FNR might stop at the level of [2Fe-2S]FNR. The presence of [2Fe-2S]FNR instead of apoFNR could possibly allow a rapid return to the [4Fe-4S]FNR state upon reversal to anoxic conditions and thereby rapid restoration of anaerobic growth. Such a situation could be important for bacteria growing in microaerobic or other biotopes where oxygen supply is not permanent or close to the regulatory pO0.5 value of FNR of 1 to 5 μM O2 (5). For long-term growth under oxic conditions, on the other hand, a stable repression of genes encoding enzymes of anaerobic metabolism is appropriate; the presence of apoFNR instead of [2Fe-2S]FNR might suppress frequent shifts to anaerobic metabolism during short-term lack of oxygen. Such a two-step inactivation of FNR might provide a more stable regulation, which on the one hand is sensitive to oxygen, and on the other hand absorbs or buffers small changes in oxygen supply without permanent aerobic/anaerobic shifting.
Acknowledgments
This work was supported by a grant from Deutsche Forschungsgemeinschaft.
We are grateful to R. E. Streeck (Universität Mainz) for enabling access to the fluorescence imager.
Footnotes
Published ahead of print on 30 November 2007.
REFERENCES
- 1.Achebach, S., T. Selmer, and G. Unden. 2005. Properties and significance of apoFNR as a second form of air-inactivated [4Fe-4S]·FNR of Escherichia coli. FEBS J. 2724260-4269. [DOI] [PubMed] [Google Scholar]
- 2.Achebach, S., Q. H. Tran, A. Vlamis-Gardikas, M. Müllner, A. Holmgren, and G. Unden. 2004. Stimulation of Fe-S cluster insertion into apoFNR by Escherichia coli glutaredoxins 1, 2, and 3 in vitro. FEBS Lett. 565203-206. [DOI] [PubMed] [Google Scholar]
- 3.Achebach, S., Y. Zeuner, and G. Unden. 2003. Control of the O2-sensor/regulator FNR by O2 and reducing agents in vitro and in vivo, p. 93-99. In P. Durre and B. Friedrich (ed.), Regulatory networks in prokaryotes. Horizon Scientific Press, Norfolk, United Kingdom.
- 4.Baty, J. W., M. B. Hampton, and C. C. Winterbourn. 2002. Detection of oxidant sensitive thiol proteins by fluorescence labelling and two-dimensional electrophoresis. Proteomics 91261-1266. [DOI] [PubMed] [Google Scholar]
- 5.Becker, S., G. Holighaus, T. Gabrielczyk, and G. Unden. 1996. O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J. Bacteriol. 1784515-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crack, J., J. Green, M. R. Cheeseman, N. E. Le Brun, and A. J. Thomson. 2007. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc. Natl. Acad. Sci. USA 1042092-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crack, J., J. Green, and A. J. Thompson. 2004. Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR). J. Biol. Chem. 2799278-9286. [DOI] [PubMed] [Google Scholar]
- 8.De Crouy-Chanel, A., and G. Richarme. 2001. Amount and redox state of cytoplasmic, membrane and periplasmic proteins in Escherichia coli redox mutants. Res. Microbiol. 152663-669. [DOI] [PubMed] [Google Scholar]
- 9.Dibden, D. P., and J. Green. 2005. In vivo cycling of the Escherichia coli transcription factor FNR between active and inactive states. Microbiology 1514063-4070. [DOI] [PubMed] [Google Scholar]
- 10.Engel, P., M. Trageser, and G. Unden. 1991. Reversible interconversion of the functional state of the gene regulator FNR from Escherichia coli in vivo by O2 and iron availability. Arch. Microbiol. 156463-470. [DOI] [PubMed] [Google Scholar]
- 11.Green, J., and M. S. Paget. 2004. Bacterial redox sensors. Nat. Rev. Microbiol. 2954-966. [DOI] [PubMed] [Google Scholar]
- 12.Green, J., B. Bennet, P. Jordan, E. T. Ralph, A. J. Thomson, and J. R. Guest. 1996. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, J., A. D. Sharrocks, B. Green, M. Geisow, and J. R. Guest. 1993. Properties of FNR proteins substituted at each of the five cysteine residues. Mol. Microbiol. 861-68. [DOI] [PubMed] [Google Scholar]
- 14.Green, J., M. Trageser, S. Six, G. Unden, and J. R. Guest. 1991. Characterization of the FNR protein of Escherichia coli, an iron-binding transcriptional regulator. Proc. Biol. Sci. 244137-144. [DOI] [PubMed] [Google Scholar]
- 15.Guest, J. R., J. Green, A. Irvine, and S. Spiro. 1996. The FNR modulon and FNR-regulated gene expression. Chapman & Hall, New York, NY.
- 16.Gunsalus, R. P. 1992. Control of electron flow in Escherichia coli: coordinated transcription of respiratory pathway genes. J. Bacteriol. 1747069-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joly, J. C., and J. R. Schwartz. 1997. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry 3610067-10072. [DOI] [PubMed] [Google Scholar]
- 18.Khoroshilova, N., C. Popescu, E. Münck, H. Beinert, and P. J. Kiley. 1997. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. USA 946087-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22341-352. [DOI] [PubMed] [Google Scholar]
- 20.Kosower, E. M., and N. S. Kosower. 1995. Bromobimane probes for thiols. Methods Enzymol. 251133-148. [DOI] [PubMed] [Google Scholar]
- 21.Kosower, N. S., and E. M. Kosower. 1987. Thiol labelling with bromobimanes. Methods Enzymol. 14376-84. [DOI] [PubMed] [Google Scholar]
- 22.Kosower, N. S., E. M. Kosower, G. L. Newton, and M. Ranney. 1979. Bimane fluorescence labels: labelling of normal human red cells under physiological conditions. Proc. Natl. Acad. Sci. USA 763382-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lazazzera, B. A., H. Beinert, N. Khoroshilova, M. C. Kennedy, and P. J. Kiley. 1996. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 2712762-2768. [DOI] [PubMed] [Google Scholar]
- 25.Melville, S. B., and R. P. Gunsalus. 1990. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J. Biol. Chem. 26518733-18736. [PubMed] [Google Scholar]
- 26.Mettert, E. L., and P. J. Kiley. 2005. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J. Mol. Biol. 354220-232. [DOI] [PubMed] [Google Scholar]
- 27.Popescu, C. V., D. M. Bates, H. Beinert, E. Münck, and P. J. Kiley. 1998. Mössbauer spectroscopy as a tool of activation/inactivation of the transcriptional regulator FNR in whole cells of Escherichia coli. Proc. Natl. Acad. Sci. USA 9513431-13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Sharrocks, A. D., J. Green, and J. R. Guest. 1990. In vivo and in vitro mutants of FNR, the anaerobic transcriptional regulator of E. coli. FEBS Lett. 270119-122. [DOI] [PubMed] [Google Scholar]
- 30.Spiro, S., and J. R. Guest. 1987. Regulation and overexpression of the fnr gene of Escherichia coli. J. Gen. Microbiol. 1333279-3288. [DOI] [PubMed] [Google Scholar]
- 31.Sutton, V. R., E. L. Mettert, H. Beinert, and P. J. Kiley. 2004. Kinetic analysis of oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J. Bacteriol. 1868018-8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutton, V. R., A. Stubna, T. Patschkowski, E. Münck, H. Beinert, and P. J. Kiley. 2004. Superoxide destroys the [2Fe-2S]2+ cluster of FNR from Escherichia coli. Biochemistry 43791-798. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, B. L., K. J. Watts, and M. S. Johnson. 2007. Oxygen and redox sensing by two-component systems that regulate behavioral responses: behavioral assays and structural studies of aer using in vivo disulfide cross-linking. Methods Enzymol. 422190-232. [DOI] [PubMed] [Google Scholar]
- 34.Trageser, M., and G. Unden. 1990. Isolation of intact FNR protein (Mr 30,000) of Escherichia coli. Mol. Microbiol. 421-27. [DOI] [PubMed] [Google Scholar]
- 35.Trageser, M., and G. Unden. 1989. Role of the cysteine residues and of metal ions in the regulatory functioning of FNR, the transcriptional regulator of amaerobic respiration in Escherichia coli. Mol. Microbiol. 3593-599. [DOI] [PubMed] [Google Scholar]
- 36.Tran, Q. H., and G. Unden. 1998. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur. J. Biochem. 251538-543. [DOI] [PubMed] [Google Scholar]
- 37.Tran, Q. H., T. Arras, S. Becker, G. Holighaus, G. Ohlberger, and G. Unden. 2000. Role of glutathione in the formation of the active form of the oxygen sensor FNR ([4Fe-4S]FNR) and in the control of FNR function. Eur. J. Biochem. 2674817-4824. [DOI] [PubMed] [Google Scholar]
- 38.Unden, G., and A. Duchene. 1987. On the role of cyclic AMP and the FNR protein in Escherichia coli growing anaerobically. Arch. Microbiol. 147195-200. [DOI] [PubMed] [Google Scholar]
- 39.Unden, G., and J. Schirawski. 1997. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Mol. Microbiol. 25205-210. [DOI] [PubMed] [Google Scholar]