Abstract
The Mycobacterium avium complex is distributed ubiquitously in the environment. It is an important cause of pulmonary and extrapulmonary diseases in humans and animals. The species in this complex produce polar glycopeptidolipids (GPLs); of particular interest is their serotype-specific antigenicity. Several reports have described that GPL structure may play an important role in bacterial physiology and pathogenesis and in the host immune response. Recently, we determined the complete structure of the GPL derived from Mycobacterium intracellulare serotype 7 and characterized the serotype 7 GPL-specific gene cluster. The structure of serotype 7 GPL closely resembles that of serotype 12 GPL, except for O methylation. In the present study, we isolated and characterized the serotype 12-specific gene cluster involved in glycosylation of the GPL. Ten open reading frames (ORFs) and one pseudogene were observed in the cluster. The genetic organization of the serotype 12-specific gene cluster resembles that of the serotype 7-specific gene cluster, but two novel ORFs (orfA and orfB) encoding putative methyltransferases are present in the cluster. Functional analyses revealed that orfA and orfB encode methyltransferases that synthesize O-methyl groups at the C-4 position in the rhamnose residue next to the terminal hexose and at the C-3 position in the terminal hexose, respectively. Our results show that these two methyltransferase genes determine the structural difference of serotype 12-specific GPL from serotype 7-specific GPL.
The Mycobacterium avium complex (MAC) consists of two species, M. avium and Mycobacterium intracellulare, which are opportunistic pathogens of humans and animals. Human exposure to the MAC is common because organisms of this complex are ubiquitous in the environment: they have been isolated from water, soil, plants, house dust, and other sources. In fact, the MAC is the most common cause of disease attributable to nontuberculous mycobacteria in humans (9). The majority of MAC infections are acquired environmentally, and person-to-person transmission is considered to be rare. The treatment of MAC infection is difficult because the organisms are often resistant to standard antituberculosis drugs.
Many antigenic or immunoregulatory glycolipids with structural diversity are expressed on the mycobacterial cell wall. These molecules are considered to be involved in bacterial virulence through host immune responses (5, 14, 22, 23). It is necessary to elucidate the molecular structure, biochemical characteristics, and biological functions of the lipid components to better understand the mechanisms of pathogenesis and drug resistance of the MAC. The most prominent feature of the MAC is the presence of antigenic glycolipids, the glycopeptidolipids (GPLs), which are present on the cell surface (1). The standard method for differentiation of MAC strains is serologic typing based on the oligosaccharide (OSE) residue of the GPL. GPLs contain a tetrapeptide-amino alcohol core, d-phenylalanine-d-allo-threonine-d-alanine-l-alaninol (d-Phe-d-allo-Thr-d-Ala-l-alaninol), with an amido-linked 3-hydroxy or 3-methoxy C26-to-C34 fatty acid at the N terminus of d-Phe (4). The d-allo-Thr and terminal l-alaninol are further linked with 6-deoxy-talose (6-d-Tal) and 3,4-di-O-methyl-rhamnose (3,4-di-O-Me-Rha), respectively. This core GPL is present in all species of the MAC and shows a common antigenicity (1). In the serotype-specific GPLs, a haptenic OSE is linked with the 6-d-Tal residue. To date, 31 distinct serotype-specific polar GPLs have been identified biochemically; the complete structures of GPLs are partly defined for serotype 1 to 4, 7, 8, 9, 12, 14, 17, 19 to 21, 25, and 26 GPLs (7, 10). On the other hand, it has been reported that serotype-specific GPLs participate in pathogenesis and immunomodulation in the host (2, 13). Modification of the GPL structure might play an important role not only in antigenicity but also in host immune responses and bacterial physiology (18). Recently, chemical synthesis of various haptenic OSEs was demonstrated, and the genes encoding glycosylation pathway enzymes for the biosynthesis of GPLs were identified and characterized (8, 12, 19, 21). However, genes responsible for serotype-specific glycosylation have yet to be analyzed for most of the serotypes.
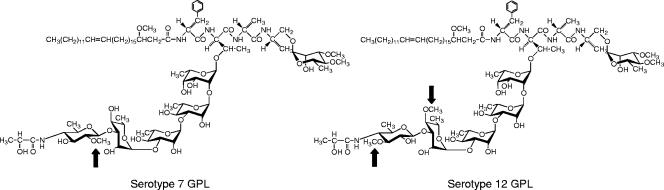
In a previous study, we determined the complete structure of the GPL derived from M. intracellulare serotype 7 and characterized the serotype 7-specific gene cluster for GPL synthesis (10). The structure of serotype 7 GPL closely resembles that of serotype 12 GPL, except for O methylation (Fig. 1). In the present study, we determined the nucleotide sequence of the serotype 12-specific gene cluster involved in the glycosylation of the GPL and characterized two novel open reading frames (ORFs) encoding O-methyltransferases that determine the difference of serotype 12 GPL from serotype 7 GPL.
FIG. 1.
Structures of serotype 7 and 12 GPLs. O-methyl groups specific to the serotypes are indicated by arrows.
MATERIALS AND METHODS
Bacterial strains and construction of M. intracellulare cosmid library.
M. intracellulare serotype 12 strain ATCC 35762 (NF 103), M. intracellulare serotype 7 strain ATCC 35847 (NF 027), and M. intracellulare serotype 7 strain NF 112 were used for this study. A cosmid library of M. intracellulare NF 103 was constructed as described previously (10). Briefly, genomic DNA of M. intracellulare NF 103 was prepared by mechanical disruption of bacterial cells in phosphate-buffered saline containing 50 mM EDTA, followed by phenol-chloroform extraction and precipitation with ethanol. Genomic DNA fragments randomly sheared to 30-kb to 50-kb fragments during the extraction process were fractionated and electroeluted from agarose gels. These DNA fragments were ligated to dephosphorylated arms of pYUB412 (XbaI-EcoRV and EcoRV-XbaI). After in vitro packaging using Gigapack III Gold extracts (Stratagene, La Jolla, CA), recombinant cosmids were introduced into Escherichia coli STBL2.
Isolation of cosmid clones carrying the GPL biosynthesis gene cluster and sequence analysis.
PCR was used to isolate cosmid clones carrying the rhamnosyltransferase gene (rtfA), using primers rtfA-F (5′-TTTTGGAGCGACGAGTTCATC-3′) and rtfA-R (5′-GTGTAGTTGACCACGCCGAC-3′). The insert of cosmid clone 161 was sequenced using a kit (BigDye Terminator cycle sequencing kit, version 3.1; Applied Biosystems, Foster City, CA) and a sequence analyzer (ABI Prism 310; Applied Biosystems). The putative function of each ORF was identified by similarity searches between the deduced amino acid sequences and those of known proteins, using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and FramePlot (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl) with the DNASIS computer program (Hitachi Software Engineering, Yokohama, Japan).
Transformation of M. intracellulare.
PCR was used to amplify and clone orfA and orfB into the plasmid vector pVV16. M. intracellulare NF 027 and NF 112 were transformed with the resultant plasmids by electroporation. Primers used to amplify orfA, orfB, and orfA-orfB were orfA-F (5′-GCGGATCCAGTGTGCAGACGAGCGGAACT-3′), orfA-R (5′-GCGAATTCTTATCGAGAAAAAATAAAAG-3′), orfB-F (5′-GCGGATCCACTGCTAGACTCCGCCACCAT-3′), and orfB-R (5′-GCGAATTCCTACACCTTCACGGCGAGTC-3′).
Preparation of GPLs and OSE moieties.
GPL 7 and GPL 12 were purified from M. intracellulare NF 027 and NF 103, respectively. The preparation of GPLs was performed as described previously (10, 15, 17). Briefly, each strain was grown in Middlebrook 7H9 broth (Difco Laboratories, Detroit, MI) with 0.5% glycerol and 10% Middlebrook oleic acid-albumin-dextrose-catalase enrichment (Difco Laboratories) at 37°C for 2 to 3 weeks. The heat-killed bacteria were sonicated and extracted using chloroform-methanol (2:1 [vol/vol]). The extractable lipids were hydrolyzed with 0.2 N sodium hydroxide in methanol at 37°C for 2 h. After neutralization using 6 N hydrochloride, chloroform-methanol (2:1 [vol/vol]) and water were added. The organic phase containing alkaline-stable lipids was recovered and evaporated, with subsequent addition of acetone to remove any acetone-insoluble components. The supernatant was dried up. It was then treated using a Sep-Pak silica cartridge (Waters Corp., Milford, MA) with washing (chloroform-methanol [95:5 {vol/vol}]) and elution (chloroform-methanol [1:1 {vol/vol}]) for partial purification. The GPL was then purified completely by preparative thin-layer chromatography (TLC) with silica gel G (Uniplate; 20 cm × 20 cm × 250 μm; Analtech, Inc., Newark, DE). The TLC was developed repeatedly, using chloroform-methanol-water (60:16:2 [vol/vol/vol]), until a single spot was obtained. To prepare the OSE moiety, purified GPL was processed using β-elimination with alkaline borohydride, and then the carbohydrate chain moiety elongated from d-allo-Thr was released as described previously (10, 15). Briefly, GPL was treated with 5 mg/ml sodium borohydride or borodeuteride in 0.5 N sodium hydroxide-ethanol (1:1 [vol/vol]) at 60°C for 16 h, with stirring. The reaction mixture was decationized with Dowex 50W X8 beads (The Dow Chemical Company, Midland, MI). The supernatant was collected and evaporated under nitrogen to remove boric acid. The dried residue was partitioned into two layers, using chloroform-methanol (2:1 [vol/vol]) and water. The upper aqueous phase was recovered and evaporated. In these processes, the OSE was purified as an oligoglycosyl alditol.
MALDI-TOF MS and MALDI-TOF/TOF MS analyses.
The molecular species of the intact GPLs were detected using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) with an Ultraflex II spectrophotometer (Bruker Daltonics, Billerica, MA). Each GPL was dissolved in chloroform-methanol (2:1 [vol/vol]) at a concentration of 1 mg/ml; 1 μl of a sample was then applied directly to the sample plate, followed by the addition of 1 μl of 10-mg/ml 2,5-dihydroxybenzoic acid in chloroform-methanol (1:1 [vol/vol]) as a matrix. The intact GPL was analyzed in the reflectron mode, with an accelerating voltage operating in positive mode at 20 kV (3). The OSE was analyzed by the fragment pattern with MALDI-TOF/TOF MS to determine the glycosyl composition. The OSE was dissolved with ethanol-water (3:7 [vol/vol]); the matrix was 10 mg/ml 2,5-dihydroxybenzoic acid in ethanol-water (3:7 [vol/vol]). The OSE and matrix were added to the sample plate by the same method as that for intact GPL. They were then analyzed in the lift-lift mode.
GC-MS analyses of alditol acetate derivatives.
Gas chromatography (GC) and GC-MS analyses of partially methylated alditol acetate derivatives were performed to determine glycosyl compositions and linkage positions. Perdeuteromethylation was conducted using a modified procedure of Hakomori, as described previously (10, 11). Briefly, the dried OSE was dissolved with a mixture of dimethyl sulfoxide and sodium hydroxide, and deuteromethyl iodide was added. The reaction mixture was stirred at room temperature for 15 min, followed by the addition of water and chloroform. After centrifugation at 2,400 × g for 15 min, the upper water layer was discarded. The chloroform layer was washed twice with water and evaporated completely. To prepare partially deuteromethylated alditol acetates, perdeuteromethylated OSE was hydrolyzed using 2 N trifluoroacetic acid at 120°C for 2 h, reduced with 10 mg/ml sodium borodeuteride at 25°C for 2 h, and acetylated with acetic anhydride at 100°C for 1 h (6, 10, 16). GC-MS was then performed using a benchtop ion-trap mass spectrometer (Trace DSQ GC/MS; Thermo Electron Corporation, Austin, TX) equipped with a fused capillary column (30 m; 0.25-mm internal diameter) (Equity-1 or SP-2380; Supelco, Bellefonte, PA). Helium was used as the carrier gas, and the flow rate was 1 ml/min. The SP-2380 column was used for the analysis of alditol acetate derivatives. The temperature program was started at 60°C, with an increase of 40°C/min to 260°C and a hold at 260°C for 25 min. The Equity-1 column was used for analysis of perdeuteromethylated alditol acetate derivatives. The temperature program was 80°C for 1 min, with an increase of 20°C/min to 180°C followed by an increase of 8°C/min to 280°C.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the NCBI GenBank database under accession number AB353739.
RESULTS
Cloning and sequence of the serotype 12 GPL biosynthesis cluster.
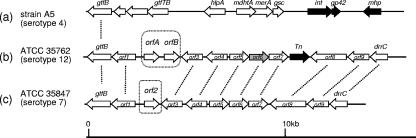
To isolate the serotype 12-specific GPL biosynthesis gene cluster, a genomic cosmid library of an M. intracellulare serotype 12 strain, NF 103, was constructed. DNA was extracted from each clone by boiling. Using colony PCR with rftA primers, the positive clone 161 was isolated from the E. coli transductants. Sequencing analysis revealed that cosmid clone 161 carried the DNA region from gtfB to drrC. Ten ORFs and one pseudogene other than gtfB and drrC were observed in the cluster (Table 1 and Fig. 2). The genetic organization between the gtfB and drrC genes (15.6 kb) of M. intracellulare NF 103 (serotype 12) closely resembled that of the same region of M. intracellulare NF 027 (serotype 7), except for three loci (Fig. 2). The first difference between them was an additional ORF encoding a transposase between orf7 and orf8 in NF 103 (Fig. 2). The second difference was that the orf6 homologous sequence in NF 103 had a frame shift, indicating that this locus does not encode a protein. The third difference is that two novel ORFs (orfA and orfB) instead of orf2 were found between orf1 and orf3 in NF 103.
TABLE 1.
Similarity of Orfs in M. intracellulare serotype 12 strain ATCC 35762 to known protein sequences
| Orf | Predicted molecular mass (Da) | Predicted pI | Similar protein | Identity (no. of matched amino acids/total no. of amino acids) | E value | GenBank accession no. |
|---|---|---|---|---|---|---|
| GtfB | 45,830 | 6.87 | Glycosyltransferase GtfB | 412/418 | 0.0 | BAF45360 |
| Orf1 | 45,203 | 6.10 | Putative glycosyltransferase | 414/417 | 0.0 | BAF45361 |
| OrfA | 28,904 | 7.42 | Putative methyltransferase | 182/224 | 5e−88 | NP_218045 |
| OrfB | 29,930 | 5.15 | Putative methyltransferase | 102/204 | 1e−19 | EAZ88812 |
| Orf3 | 32,151 | 10.41 | Putative glycosyltransferase | 196/223 | 1e−108 | BAF45363 |
| Orf4 | 40,742 | 5.41 | Putative aminotransferase | 338/374 | 0.0 | BAF45364 |
| Orf5 | 35,812 | 5.26 | Hypothetical protein | 303/329 | 4e−162 | BAF45365 |
| Orf7 | 27,693 | 5.99 | Putative metallophosphoesterase | 223/241 | 1e−122 | BAF45367 |
| Tn | 28,538 | 11.85 | Putative transposase | 213/255 | 6e−107 | AAL61662 |
| Orf8 | 80,044 | 9.16 | Putative acyltransferase | 689/747 | 0.0 | BAF45368 |
| Orf9 | 37,797 | 8.26 | Putative glycosyltransferase | 310/337 | 7e−169 | BAF45369 |
| DrrC | 28,549 | 12.01 | Daunorubicin resistance protein C | 261/263 | 3e−141 | BAF45370 |
FIG. 2.
Comparison of genetic organization of GPL biosynthesis clusters. (a) M. avium strain A5 organization, based on the annotated sequence obtained from GenBank (accession no. AY130970). (b) M. intracellulare ATCC 35762 (NF 103), sequenced in this study. (c) M. intracellulare ATCC 35847 (NF 027), sequenced in our previous study (GenBank accession no. AB274811). The orientation of each gene is shown by the arrow direction. The black arrows represent mobile elements, and the gray arrow represents a pseudogene. Mutually homologous ORFs and sequences are indicated with dotted lines.
Functional analysis of the two unique ORFs found in the serotype 12 GPL biosynthesis cluster.
Based on sequence homology, orfA and orfB were able to encode methyltransferases responsible for producing serotype 12 GPLs (Table 1). We constructed three plasmids carrying orfA and/or orfB downstream of the hsp-60 promoter to test this. These plasmids and a control vector plasmid were introduced individually into M. intracellulare serotype 7 (NF 027 and NF 112), and transformants were obtained. The GPLs produced from each transformant were analyzed.
The alkaline-stable lipids derived from six transformants of NF 027 and NF 112 in addition to the control strains (vector only) were developed by TLC, and the produced GPLs were compared to the spots of GPL 7 and GPL 12 (Fig. 3). The Rf values for GPLs synthesized in NF 027 transformed with orfA and NF 027 transformed with orfA and orfB (GPL 7-orfA and GPL 7-orfAB, respectively) were almost identical to that for GPL 12; the Rf value for the GPL synthesized in NF 027 transformed with orfB (GPL 7-orfB) was intermediate between those of GPL 7 and GPL 12, although the GPL synthesized in the control strain (GPL vector) was not changed from GPL 7. These results suggest that orfA, orfB, and orfA-orfB introduced into serotype 7 strain NF 027 were expressed and that they functioned for the modification of GPLs. We investigated the structural definition of these modified GPLs.
FIG. 3.
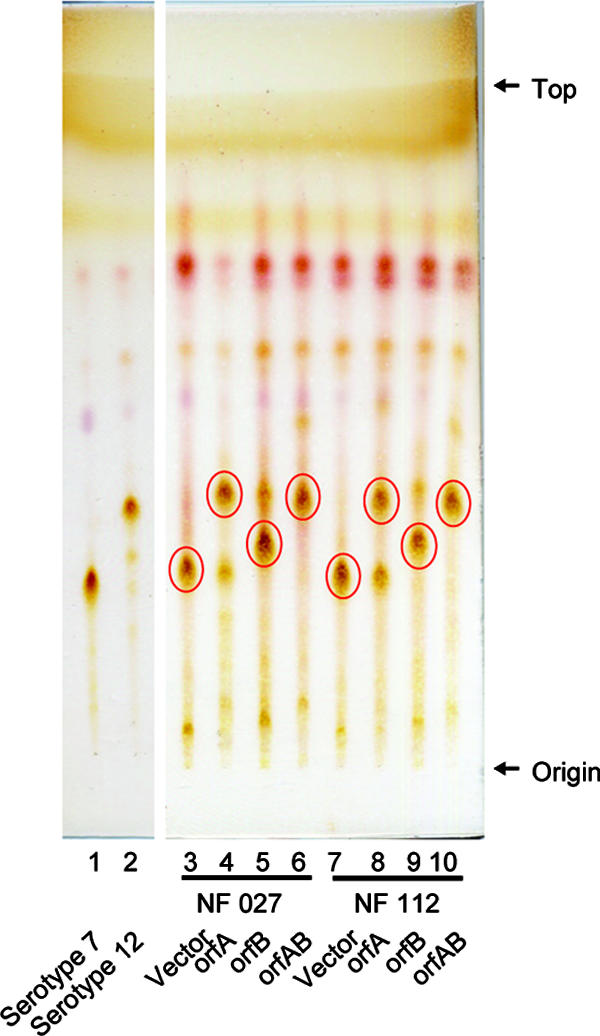
TLC patterns of alkaline-stable lipids derived from M. intracellulare serotype 7 transformants. GPL 7 and GPL 12 were purified from M. intracellulare serotype 7 strain ATCC 35847 (NF 027) and serotype 12 strain ATCC 35762 (NF103). TLC was developed with a solvent system of chloroform-methanol-water (65:25:4 [vol/vol/vol]). Circled spots indicate prominent GPLs.
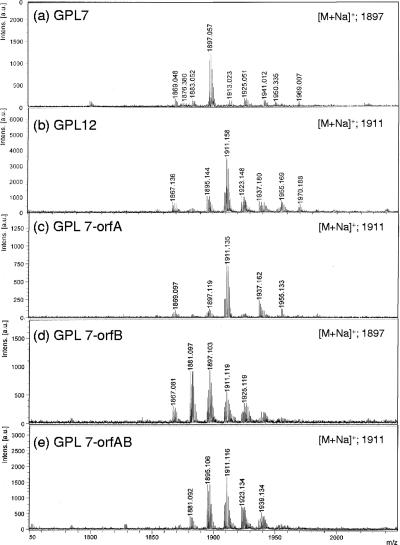
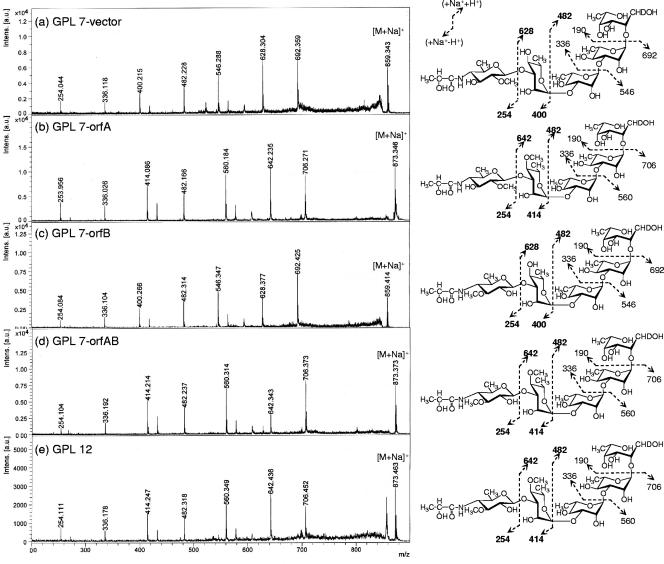
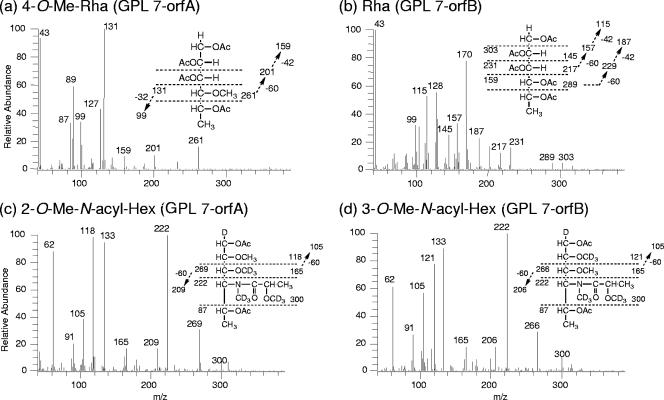
The GPLs produced in the transformants were purified using preparative TLC; their molecular weights were measured using MALDI-TOF MS (Fig. 4). The main molecularly related ions of GPL 7 and GPL 12 were detected as m/z 1,897 and 1,911, respectively, for [M + Na]+ (Fig. 4a and b). The predominant m/z values were 1,911 for GPL 7-orfA, 1,897 for GPL 7-orfB, and 1,911 for GPL7-orfAB (Fig. 4c to e). The molecular weight of GPL 7-orfB was the same as that of GPL 7, and those of GPL 7-orfA and GPL 7-orfAB were equal to that of GPL 12. Next, MALDI-TOF/TOF MS analysis was performed to determine the glycosyl pattern, using fragment ions of glycosyl cleavage. The fragment ions of the GPL vector (equal to GPL 7) showed m/z 254, 400, 546, and 692 for cleavage in turn from terminal 4N-acyl-hexose (Hex) and 336, 482, and 628 for cleavage in the opposite direction from 6-d-Tal (Fig. 5a). The fragment ions of GPL 7-orfA, m/z 414 and 642, were different from those of GPL 7, i.e., m/z 400 and 628, respectively; they demonstrated that the mass number of the sugar next to the terminal Hex increased 14 mass units (Fig. 5b). This result suggests that the second sugar from the terminal one was changed from Rha to O-methyl rhamnose (O-Me-Rha). Similarly, the fragment pattern of GPL 7-orfAB was identical to that of GPL 7-orfA, although that of GPL 7-orfB was the same as that of GPL 7 (Fig. 5c and d). Altogether, GPL 7-orfAB was predicted to have a modification of the O-Me position in the terminal Hex along with the substitution of O-Me-Rha for Rha in the sugar next to the terminal Hex; GPL 7-orfB was modified only at the O-Me position in the terminal Hex.
FIG. 4.
MALDI-TOF MS spectra of GPLs derived from M. intracellulare serotype 7, serotype 12, and serotype 7 transformants. a.u., absorbance units.
FIG. 5.
Fragment patterns of MALDI-TOF/TOF MS spectra of OSEs in GPLs derived from M. intracellulare serotype 7, serotype 12, and serotype 7 transformants. The MALDI-TOF/TOF MS spectra were acquired using 10 mg/ml 2,5-dihydroxybenzoic acid in ethanol-water (3:7 [vol/vol]) as the matrix; the molecularly related ions were detected as [M + Na]+ in lift-lift mode. The assigned fragment patterns of glycosyl residues are depicted. a.u., absorbance units.
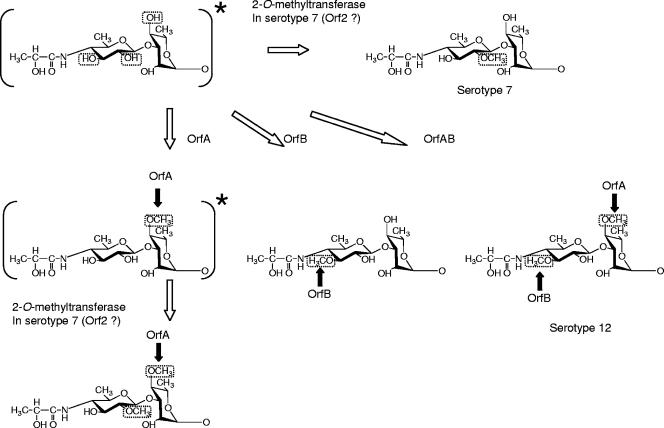
GC-MS analyses of alditol acetate and perdeuteromethyl alditol acetate derivatives were performed to assign the linkage position of O-Me. As portrayed in Fig. 6a and b, the alditol acetate derivatives of the second sugar from the terminal 4N-acyl-Hex in GPL 7-orfA and GPL 7-orfB were assigned to 1,2,3,5-tetra-acetyl-4-O-methyl-rhaminitol (m/z 99, 131, 159, 201, and 261) and 1,2,3,4,5-penta-acetyl-rhaminitol (m/z 115, 157, 187, 217, 231, 289, and 303), respectively. The perdeuteromethyl alditol acetate derivatives of the terminal sugar in GPL 7-orfA and GPL 7-orfB were assigned to 3-O-deuteromethyl-1,5-di-O-acetyl-4-2′-O-deuteromethyl-propanoyl-deu-teromethylamido-4,6-dideoxy-2-O-methyl-hexitol (m/z 105, 118, 165, 209, 222, 269, and 300) and 2-O-deuteromethyl-1,5-di-O-acetyl-4-2′-O-deuteromethyl-propanoyl-deuteromethyl-amido-4,6-dideoxy-3-O-methyl-hexitol (m/z 105, 121, 165, 206, 222, 266, and 300), respectively (Fig. 6c and d). In particular, the fragment ions of m/z 118 and 269 (Fig. 6c) versus m/z 121 and 266 (Fig. 6d) strongly indicated the different positions of linkages 2-O-Me and 3-O-Me. The alditol acetate and perdeuteromethyl alditol acetate derivatives in GPL 7-orfAB were detected with the same patterns of 4N-acyl-4,6-dideoxy-3-O-Me-Hex and 4-O-Me-Rha. According to these results, all OSE structures in GPLs derived from three serotype 7 transformants were assigned as listed in Table 2. Altogether, the functions of the two genes were defined. The orfA product transfers a methyl group to the C-4 position of Rha next to the terminal sugar, and the orfB product transfers a methyl group to the C-3 position of the terminal sugar (Fig. 7). The results demonstrated that GPL 7 in the serotype 7 strain was changed completely to GPL 12 by introduction of the orfA-orfB gene cluster.
FIG. 6.
Preparative GC-MS spectra of alditol acetate (a and b) and perdeuteromethylated alditol acetate (c and d) derivatives. The patterns of prominent fragment ions are presented. An SP-2380 column was used for the analysis of alditol acetate derivatives. The temperature program was started at 60°C, with an increase of 40°C/min to 260°C and a hold at 260°C for 25 min. An Equity-1 column was used for the perdeuteromethylated alditol acetate derivatives. The temperature program was 80°C for 1 min, with an increase of 20°C/min to 180°C followed by an increase of 8°C/min to 280°C.
TABLE 2.
Summarized structures of OSEs derived from serotype 7 transformants
| GPL | Molecular weight of OSE | Fragment ions in MALDI-TOF/TOF MS |
O-Methyl group
|
Structure of OSE | |
|---|---|---|---|---|---|
| Terminal sugar | Residue next to terminal sugar | ||||
| GPL 7 vector | 859 | 254, 400, 546, 692 | 2-O-Met | 4N-acyl-4,6-dideoxy-2-O-Me-Hex→Rha→Rha→Rha→6-d-Tal | |
| GPL 7-orfA | 873 | 254, 414, 560, 706 | 2-O-Met | 4-O-Met | 4N-acyl-4,6-dideoxy-2-O-Me-Hex→4-O-Me-Rha→Rha→Rha→6-d-Tal |
| GPL 7-orfB | 859 | 254, 400, 546, 692 | 3-O-Met | 4N-acyl-4,6-dideoxy-3-O-Me-Hex→Rha→Rha→Rha→6-d-Tal | |
| GPL 7-orfAB | 873 | 254, 414, 560, 706 | 3-O-Met | 4-O-Met | 4N-acyl-4,6-dideoxy-3-O-Me-Hex→4-O-Me-Rha→Rha→Rha→6-d-Tal |
FIG. 7.
Synthesis of O-methyl groups specific for GPL 7 and GPL 12 in the terminal disaccharide. The structures asterisked in the figure were not detected in this study. Serotype 12-specific O methylations and ORFs responsible for their syntheses are indicated by black arrows.
DISCUSSION
Nontuberculous mycobacteria, including the pathogenic species belonging to the MAC, have serotype-specific GPLs that are important components of the outer layer of the lipid-rich cell walls (5). Structural analyses of some serotype-specific GPLs derived from predominant clinical isolates have been reported (20). We recently determined the complete structure of serotype 7 GPL and the nucleotide sequence of the serotype 7-specific GPL biosynthesis cluster (10). In this cluster, Orfs 1, 3, and 9 might engender transfer of the two molecules of l-Rha and the terminal Hex of serotype 7 GPL (10). Orfs 4, 5, 7, and 8 are homologous to an aminotransferase, a carbamoyl phosphate synthase protein, a metallophosphoesterase, and an acyltransferase, respectively, and possibly relate to the biosynthesis of 2′-hydroxypropanoylamido in the terminal Hex. Based on analysis of sequence homology, these ORFs are probably responsible for the glycosylation of serotype 7 GPL. Serotype 12 GPL has a similar structure to that of serotype 7 GPL, except for O methylation (Fig. 1). In the present study, we cloned the serotype 12 GPL biosynthesis cluster and analyzed its sequence. Although the genetic organization of the gtfB-to-drrC region of the serotype 12 GPL biosynthetic cluster closely resembled that of serotype 7, significant differences were found in three loci (Fig. 2). The M. intracellulare serotype 12 strain NF 103 had one ORF encoding a transposase between orf7 and orf8 and had an orf6 homologous sequence with frameshift inactivation. Orf6 in M. intracellulare serotype 7 exhibits sequence similarity to nucleotide sugar epimerases/dehydrogenases, but NF 112, one of the M. intracellulare serotype 7 isolates, had an interrupted orf6 (10). These findings suggest that orf6 is not involved in biosynthesis of either serotype 7 GPL or serotype 12 GPL. The most important difference between the two serotypes is that M. intracellulare serotype 12 had two unique ORFs, orfA and orfB, instead of orf2 in M. intracellulare serotype 7. Actually, Orf2 in M. intracellulare serotype 7 was assigned to a methyltransferase and might be responsible for synthesis of the O-methyl group at the C-2 position in the terminal Hex. That possibility suggests that the two unique ORFs for serotype 12 encode O-methyltransferases that produce the serotype 12-specific structure. NF 027 (serotype 7) transformed with orfA produced 4N-acyl-4,6-dideoxy-2-O-Me-Hex→4-O-Me-Rha→Rha→Rha→6-d-Tal, indicating that the product from orfA had activity to synthesize an O-methyl group at C-4 in l-Rha next to the terminal Hex (Table 2 and Fig. 7). NF 027 transformed with orfB produced 4N-acyl-4,6-dideoxy-3-O-Me-Hex→Rha→Rha→Rha→6-d-Tal, indicating that the product from orfB had activity to synthesize an O-methyl group at C-3 in the terminal Hex. NF 027 transformed with orfA and orfB produced serotype 12-specific GPL, indicating that these two ORFs were responsible for producing the serotype 12-specific structure. The TLC patterns showed that the migration of GPL 7-orfB was different from that of GPL 7, although the MS data showed that they had the same molecular weight and the same number of methyl groups. A possible explanation for this is that a difference in the position of O methylation could influence hydrogen bond formation and the polarity of the whole molecule and consequently result in a different TLC migration pattern. GPL 7-orfB had an O-methyl group at C-3 but not at C-2 in the terminal Hex, indicating that the reaction of O methylation at C-2 by the 2-O-methyltransferase in serotype 7 is strongly inhibited by O-methylation at C-3. In addition, NF 027 transformed with orfA produced a trace of serotype 7-specifc GPL (Fig. 3, lane 4), and NF 027 transformed with orfA and orfB produced only serotype 12 GPL (Fig. 3, lane 6), suggesting that O methylation at C-2 in the terminal Hex might hinder the reaction of O methylation at C-4 in Rha next to the terminal Hex or that O methylation at C-3 in the terminal Hex might promote the reaction of O methylation at C-4 in Rha.
Because it is not likely that M. intracellulare serotypes 7 and 12 independently acquired different methyltransferase genes in the same genetic location between orf1 and orf3, the common ancestor for these two serotypes possibly had all three genes and activated them as the occasion demanded. However, our results showed that reactions of O methylation at C-3 and C-2 in the terminal Hex were competitive (Fig. 3, lane 5, and Table 2). Tsang et al. (26) reported that the frequency of isolation of MAC organisms from AIDS or non-AIDS patients varied among serotypes and that M. intracellulare serotype 12 was isolated more often than serotype 7. These two serotypes of M. intracellulare might have evolved to adapt to certain environments by losing orf2 or orfA-orfB.
Actually, GPLs are among the immunogenic molecules of the MAC. Tassel et al. reported that the core GPL seems to play a role in suppression of a mitogen-induced blastogenic response of spleen cells (25); furthermore, our previous study showed that sera of patients with MAC disease contain antibodies against GPLs and that the antibody level reflects disease activity (17). In addition, the immunomodulating activity of GPLs on macrophage functions is serotype dependent (13, 24). Elucidation of the structure-activity relationship of GPLs is necessary to better understand the pathogenesis of MAC infection.
Acknowledgments
This work was supported by grants from the Ministry of Health, Labor and Welfare (Emerging and Re-Emerging Infectious Diseases), the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Japan Health Sciences Foundation.
N.N. is grateful to M. Kai and M. Makino for helpful discussions.
Footnotes
Published ahead of print on 16 November 2007.
REFERENCES
- 1.Aspinall, G. O., D. Chatterjee, and P. J. Brennan. 1995. The variable surface glycolipids of mycobacteria: structures, synthesis of epitopes, and biological properties. Adv. Carbohydr. Chem. Biochem. 51169-242. [DOI] [PubMed] [Google Scholar]
- 2.Barrow, W. W., T. L. Davis, E. L. Wright, V. Labrousse, M. Bachelet, and N. Rastogi. 1995. Immunomodulatory spectrum of lipids associated with Mycobacterium avium serovar 8. Infect. Immun. 63126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt, A., N. Fujiwara, K. Bhatt, S. S. Gurcha, L. Kremer, B. Chen, J. Chan, S. A. Porcelli, K. Kobayashi, G. S. Besra, and W. R. Jacobs. 2007. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. USA 1045157-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan, P. J., and M. B. Goren. 1979. Structural studies on the type-specific antigens and lipids of the Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum serocomplex. Mycobacterium intracellulare serotype 9. J. Biol. Chem. 2544205-4211. [PubMed] [Google Scholar]
- 5.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 6429-63. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee, D., G. O. Aspinall, and P. J. Brennan. 1987. The presence of novel glucuronic acid-containing, type-specific glycolipid antigens within Mycobacterium spp. Revision of earlier structures. J. Biol. Chem. 2623528-3533. [PubMed] [Google Scholar]
- 7.Chatterjee, D., and K. H. Khoo. 2001. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell. Mol. Life Sci. 582018-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckstein, T. M., J. T. Belisle, and J. M. Inamine. 2003. Proposed pathway for the biosynthesis of serovar-specific glycopeptidolipids in Mycobacterium avium serovar 2. Microbiology 1492797-2807. [DOI] [PubMed] [Google Scholar]
- 9.Falkinham, J. O. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara, N., N. Nakata, S. Maeda, T. Naka, M. Doe, I. Yano, and K. Kobayashi. 2007. Structural characterization of a specific glycopeptidolipid containing a novel N-acyl-deoxy sugar from Mycobacterium intracellulare serotype 7 and genetic analysis of its glycosylation pathway. J. Bacteriol. 1891099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakomori, S. 1964. A rapid permethylation of glycolipid, and polysaccharide catalyzed by methylsulfinyl carbanion in dimethyl sulfoxide. J. Biochem. (Tokyo) 55205-208. [PubMed] [Google Scholar]
- 12.Heidelberg, T., and O. R. Martin. 2004. Synthesis of the glycopeptidolipid of Mycobacterium avium serovar 4: first example of a fully synthetic C-mycoside GPL. J. Org. Chem. 692290-2301. [DOI] [PubMed] [Google Scholar]
- 13.Kano, H., T. Doi, Y. Fujita, H. Takimoto, I. Yano, and Y. Kumazawa. 2005. Serotype-specific modulation of human monocyte functions by glycopeptidolipid (GPL) isolated from Mycobacterium avium complex. Biol. Pharm. Bull. 28335-339. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann, S. H. 2001. How can immunology contribute to the control of tuberculosis? Nat. Rev. Immunol. 120-30. [DOI] [PubMed] [Google Scholar]
- 15.Khoo, K. H., D. Chatterjee, A. Dell, H. R. Morris, P. J. Brennan, and P. Draper. 1996. Novel O-methylated terminal glucuronic acid characterizes the polar glycopeptidolipids of Mycobacterium habana strain TMC 5135. J. Biol. Chem. 27112333-12342. [DOI] [PubMed] [Google Scholar]
- 16.Khoo, K. H., E. Jarboe, A. Barker, J. Torrelles, C. W. Kuo, and D. Chatterjee. 1999. Altered expression profile of the surface glycopeptidolipids in drug-resistant clinical isolates of Mycobacterium avium complex. J. Biol. Chem. 2749778-9785. [DOI] [PubMed] [Google Scholar]
- 17.Kitada, S., R. Maekura, N. Toyoshima, N. Fujiwara, I. Yano, T. Ogura, M. Ito, and K. Kobayashi. 2002. Serodiagnosis of pulmonary disease due to Mycobacterium avium complex with an enzyme immunoassay that uses a mixture of glycopeptidolipid antigens. Clin. Infect. Dis. 351328-1335. [DOI] [PubMed] [Google Scholar]
- 18.Krzywinska, E., S. Bhatnagar, L. Sweet, D. Chatterjee, and J. S. Schorey. 2005. Mycobacterium avium 104 deleted of the methyltransferase D gene by allelic replacement lacks serotype-specific glycopeptidolipids and shows attenuated virulence in mice. Mol. Microbiol. 561262-1273. [DOI] [PubMed] [Google Scholar]
- 19.Maslow, J. N., V. R. Irani, S. H. Lee, T. M. Eckstein, J. M. Inamine, and J. T. Belisle. 2003. Biosynthetic specificity of the rhamnosyltransferase gene of Mycobacterium avium serovar 2 as determined by allelic exchange mutagenesis. Microbiology 1493193-3202. [DOI] [PubMed] [Google Scholar]
- 20.McNeil, M., A. Y. Tsang, and P. J. Brennan. 1987. Structure and antigenicity of the specific oligosaccharide hapten from the glycopeptidolipid antigen of Mycobacterium avium serotype 4, the dominant mycobacterium isolated from patients with acquired immune deficiency syndrome. J. Biol. Chem. 2622630-2635. [PubMed] [Google Scholar]
- 21.Miyamoto, Y., T. Mukai, N. Nakata, Y. Maeda, M. Kai, T. Naka, I. Yano, and M. Makino. 2006. Identification and characterization of the genes involved in glycosylation pathways of mycobacterial glycopeptidolipid biosynthesis. J. Bacteriol. 18886-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porcelli, S. A., and R. L. Modlin. 1999. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu. Rev. Immunol. 17297-329. [DOI] [PubMed] [Google Scholar]
- 23.Smith, I. 2003. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16463-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takegaki, Y. 2000. Effect of serotype specific glycopeptidolipid (GPL) isolated from Mycobacterium avium complex (MAC) on phagocytosis and phagosome-lysosome fusion of human peripheral blood monocytes. Kekkaku 759-18. [PubMed] [Google Scholar]
- 25.Tassell, S. K., M. Pourshafie, E. L. Wright, M. G. Richmond, and W. W. Barrow. 1992. Modified lymphocyte response to mitogens induced by the lipopeptide fragment derived from Mycobacterium avium serovar-specific glycopeptidolipids. Infect. Immun. 60706-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsang, A. Y., J. C. Denner, P. J. Brennan, and J. K. McClatchy. 1992. Clinical and epidemiological importance of typing of Mycobacterium avium complex isolates. J. Clin. Microbiol. 30479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]