Abstract
The saeRS two-component regulatory system regulates transcription of multiple virulence factors in Staphylococcus aureus. In the present study, we demonstrated that the saePQRS region in Staphylococcus epidermidis is transcriptionally regulated in a temporal manner and is arranged in a manner similar to that previously described for S. aureus. Studies using a mouse foreign body infection model demonstrated that the virulence of strain 1457 and the virulence of a mutant, strain 1457 saeR, were statistically equivalent. However, histological analyses suggested that the polymorphonuclear neutrophil response at 2 days postinfection was significantly greater in 1457-infected mice than in 1457 saeR-infected mice, demonstrating that SaeR influences the early, acute phases of infection. Microarray analysis demonstrated that a saeR mutation affected the transcription of 65 genes (37 genes were upregulated and 28 genes were downregulated); in particular, 8 genes that facilitate growth under anaerobic conditions were downregulated in 1457 saeR. Analysis of growth under anaerobic conditions demonstrated that 1457 saeR had a decreased growth rate compared to 1457. Further metabolic experiments demonstrated that 1457 saeR had a reduced capacity to utilize nitrate as a terminal electron acceptor and exhibited increased production of lactic acid in comparison to 1457. These data suggest that in S. epidermidis SaeR functions to regulate the transition between aerobic growth and anaerobic growth. In addition, when grown anaerobically, 1457 saeR appeared to compensate for the redox imbalance created by the lack of electron transport-mediated oxidation of NADH to NAD+ by increasing lactate dehydrogenase activity and the subsequent oxidation of NADH.
Prokaryotic two-component regulatory systems modulate gene expression in response to different environmental signals. Analyses of the available Staphylococcus aureus genomes have revealed that S. aureus encodes 16 putative two-component regulatory systems (14), 6 of which have been characterized (3, 10, 13, 31, 36, 43, 53). One two-component regulatory system, encoded by saeRS, was originally identified as a transposon mutant system deficient in the synthesis of several exoproteins, including α- and β-hemolysin, and coagulase (21). SaeRS has subsequently been shown to regulate staphylococcal immune evasion proteins (48), adhesins (26), and γ-hemolysin (58). Importantly, sae inactivation reduces invasion of S. aureus in several animal models and cell lines (20, 35, 52, 57).
The sae operon in S. aureus is comprised of four genes, saePQRS, with saeR encoding the response regulator and saeS encoding the sensor kinase (18). The functions of SaeP and SaeQ are unknown, but it has been postulated that they have important roles in sae-dependent regulation (44, 52). Transcriptional studies revealed that the sae genes in S. aureus are translated from three or four different transcripts and that saeRS is present in three of these transcripts (44, 47, 52). Transcription of the sae mRNAs is growth phase dependent, activated by agr (19, 44, 52), and regulated by SaeR autoactivation (44). Although several environmental conditions affect sae expression, including low pH, salt, glucose, or subinhibitory concentrations of antibiotics (44), the specific activating signal remains unknown.
In this work, we demonstrated that the sae locus is present in the closely related, opportunistic pathogen Staphylococcus epidermidis, the preeminent cause of biomaterial-related infections in hospital settings. In contrast to S. aureus, which produces a large array of toxins and adherence factors, S. epidermidis produces very few virulence factors. Of the virulence factors that have been described (32, 54, 60, 61), a major virulence mechanism associated with S. epidermidis is its ability to form a biofilm, which is largely composed of polysaccharide intercellular adhesin (PIA) (38). PIA, which is synthesized by enzymes encoded in the four-gene icaADBC operon (27), dramatically reduces the effectiveness of antibiotics and the host immune system (55, 56). Importantly, S. epidermidis icaADBC operon mutants are less virulent in animal models of biomaterial infection (49, 50). SaeRS is required for transcription of several virulence genes in S. aureus, which led us to speculate that the SaeRS system has a similar function in S. epidermidis.
MATERIALS AND METHODS
Bacterial strains and plasmids used in study.
Bacterial strains and plasmid constructs used in the current study are listed in Table 1.
TABLE 1.
Bacterial strains, bacteriophage, and plasmids used in the current study
| Bacterial strain, bacteriophage, or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Chemically competent plasmid cloning host | Invitrogen |
| S. epidermidis 1457 | Biofilm positive strain, transducible with phage 71 | 39 |
| S. epidermidis 1457 NR | 1457 resistant to novobiocin (1 μg/ml) and rifampin (10 μg/ml) | This study |
| S. carnosus TM300 | ica-negative strain, negative control in the biofilm assay | 24 |
| S. aureus RN4220 | Restriction-negative, modification-positive isolate | 34 |
| 1457 saeR | saeR knockout mutant of strain 1457, Tmpr | This study |
| 1457 saeR/pNF155 | 1457 saeR complemented with saeRS | This study |
| Bacteriophage | ||
| Phage 71 | S. epidermidis transducing phage | 40 |
| Plasmids | ||
| pUC19 | High-copy-number plasmid with multiple cloning site, Ampr | Invitrogen |
| pGO558 | dhfr cloned into the SalI site of pUC19, Ampr Tmpr | G. Archer |
| pROJ6448 | pE194 with pC221 nick site for mobilization, temperature-sensitive replicon, Ermr | 46 |
| pNF38 | sae allelic replacement vector, Ampr Tmpr Ermr | This study |
| pNF41 | RNAIII allelic replacement vector, Ampr Tmpr Ermr | This study |
| pGO1 | Conjugative plasmid, Genr | 2 |
| pC221 | Mobilizeable plasmid, Camr | 46 |
Abbreviations: Amp, ampicillin; Tmp, trimethoprim; Gen, gentamicin; Cam, chloramphenicol; Erm, erythromycin.
Culture media and conditions.
Staphylococcal strains were cultured on tryptic soy agar (TSA) (Difco, Detroit, MI) or in tryptic soy broth (TSB) (Difco) at 37°C except where indicated otherwise. Luria-Bertani Miller broth or agar (Difco) was used to propagate Escherichia coli strain DH5α. Antibiotics and medium additives were purchased from Sigma (St. Louis, MO) and used at the following concentrations: ampicillin, 50 μg/ml; erythromycin, 10 μg/ml for staphylococci and 500 μg/ml for E. coli; trimethoprim, 10 μg/ml; and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 50 μg/ml. Aerobic growth studies of strains 1457 and 1457 saeR were performed by diluting overnight cultures 1/200 (normalized for growth) into TSB, which was incubated at 37°C at 225 rpm with a flask-to-volume ratio of 10:1. Anaerobic growth studies were performed by diluting an overnight culture (grown in prereduced TSB) 1/50 in TSB containing 3 mg/liter resazurin (Acros Organics, Geel, Belgium), 1.0 g/liter l-cysteine hydrochloride monohydrate (MP Biomedicals, Solon, OH), and 3 mM potassium nitrate (Sigma). Growth was monitored in an anaerobic chamber (model 1029; Thermo Scientific, Waltham, MA) at 37°C.
Molecular analyses and assembly of saeR allelic replacement vectors.
PCR primers for amplification of saePQRS from S. epidermidis 1457 were designed using the genome sequence of S. epidermidis RP62A (GenBank accession number CP000029.) The cloning host for the saeR allelic replacement experiments was E. coli DH5α (Invitrogen, Carlsbad, CA). First, a 965-bp piece of DNA encompassing saeQ and the 5′ end of saeR was amplified using forward primer 373 (5′-GGAATTCGAAAGAGAGTGTTAATCATGAAG-3′) and reverse primer 374 (5′-CGGGATCCGAAGCAAGATACCATAGCAATTC-3′) (Fig. 1). Primers 373 and 374 contained EcoRI and BamHI restriction sites, respectively (underlined). This piece of DNA was subsequently ligated to the EcoRI and BamHI sites of pUC19 (59), which was renamed pNF30. Next, a 949-bp piece of DNA encompassing the 3′ end of saeR and the 5′ end of saeS was amplified using forward primer 359 (5′-ACGCGTCGACCCCTACACTATTACAACTGTG-3′) and reverse primer 360 (5′-AACTGCAGCTAAACGTTCTTTCAAAGATACG-3′) (Fig. 1). Primers 359 and 360 contained SalI and PstI sites, respectively (underlined). This piece of DNA was subsequently ligated into the SalI and PstI sites of pNF30, which was then called pNF32. dhfr, encoding trimethoprim resistance, was excised from pGO558 using SalI and cloned into the SalI site of pNF32, yielding pNF34. Finally, pROJ6448 (46), a temperature-sensitive derivative of pE194 (28), was ligated into the PstI site of pNF34, yielding pNF38.
FIG. 1.
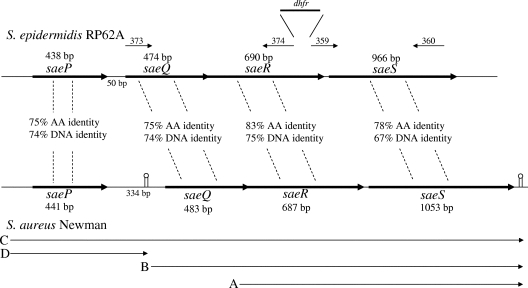
Comparison of sae nucleotide and protein sequences from S. aureus Newman (GenBank accession number AJ556794) and S. epidermidis RP62A. The DNA identity percentages indicate the level of nucleotide sequence identity for each of the sae genes, as well as the saePQ intergenic region. Percentages of amino acid (AA) identity are also indicated. The positions of primers 373, 374, 359, and 360 (see text) are indicated by arrows. sae transcripts, as defined by Novick and Jiang (44) and Steinhuber et al. (52), are labeled A, B, C, and D.
pNF38 was subsequently electroporated into RN4220, a restriction-deficient, modification-proficient strain of S. aureus (34), using previously described protocols (51). Modified pNF38 DNA was isolated from RN4220 using a Wizard plus MidiPrep kit (Promega, Madison, WI) and subsequently electroporated into S. epidermidis 1457 (39) using the electroporation protocol developed by Augustin and Götz (4). To detect chromosomal integration of pNF38 into saeR, strain 1457 containing pNF38 was grown to mid-exponential phase in 10 ml TSB (Difco, Sparks, MD) containing 10 μg/ml erythromycin (Sigma, St. Louis, MO) at 30°C. The culture was then diluted 1:100 in fresh TSB (100 ml) and was incubated at 43°C (nonpermissive temperature) overnight with shaking at 100 rpm. This step was repeated the following day. The culture was then plated (with incubation at 43°C) onto TSA containing trimethoprim (10 μg/ml; Sigma) and replica plated onto TSA containing 10 μg/ml erythromycin to count colonies that were trimethoprim resistant and erythromycin susceptible (i.e., colonies with double recombination in saeR and subsequent loss of erythromycin resistance). Following confirmation of gene knockouts by PCR and Southern blot hybridization, knockout alleles were backcrossed into wild-type S. epidermidis 1457 using transducing phage 71, as described previously (40). The allelic replacement of saeR with dhfr resulted in the loss of 134 bp.
Mouse foreign body infection model.
The mouse foreign body infection model was used to evaluate the virulence of S. epidermidis 1457 and 1457 saeR (50). Both strains were inoculated into 100 ml TSB and were incubated for 16 h at 37°C with shaking at 250 rpm. Bacteria were harvested by centrifugation and suspended in sterile saline (0.9% NaCl) to obtain concentrations of 1 × 107, 1 × 108, and 1 × 109 CFU/ml. Following ketamine/xylazine administration, the skin on an animal's back and flank was shaved with surgical clippers and disinfected with povidone-iodine. One-centimeter catheter segments (Jelco 16G FEP catheter; Johnson and Johnson, New Brunswick, NJ) were implanted into the subcutaneous space in 25- to 30-g male Swiss-Webster mice. One hundred microliters of saline (0.9%) containing either 106, 107, or 108 CFU of 1457 or 1457 saeR was inoculated into the flank of each mouse containing an implanted catheter. The mice were housed for 7 days with unrestricted access to water and rodent diet. Animals were euthanized by carbon dioxide inhalation, and the catheters were explanted.
To obtain S. epidermidis cell counts, the catheter was first removed from a mouse flank and placed into 1 ml of saline. Next, a standardized amount or pericatheter tissue, excluding the overlying cutaneous tissue, was removed from the animal and placed into 1 ml of saline. Both samples were mixed with a vortex mixer to dislodge adherent cells, and the cells were enumerated after dilution and plating on TSA. Tissue sample results were standardized by determining the number of cells per gram of tissue. A statistical analysis (Mann-Whitney test) was performed with GraphPad Prism version 4.03 for Windows (GraphPad Software, San Diego, CA).
Tissues used for histological examination were harvested at 2 and 7 days postinfection and were stored in 10% formalin. Embedding, cutting, and eosin and hematoxylin staining of these tissues were performed by the Eppley Histology Core Laboratory at the University of Nebraska Medical Center. For enumeration of the mononuclear cell populations at the site of infection, tissue from the catheterized site was collected at 2 and 7 days postinfection and was placed in a 15-ml conical centrifuge tube containing 10 ml RPMI 1640 supplemented with 5% heat-inactivated fetal bovine serum (Mediatech, Herndon, VA), 400 IU/ml collagenase IV, and 30 μg/ml DNase I (Sigma). Tissue was placed on a GyroMini nutating mixer (Labnet, Edison, NJ) and incubated at 37°C for 2 h. To obtain a suspension of single cells, tissue was passed through a stainless steel mesh, and the entire cell fraction was centrifuged at 300 × g for 5 min at 4°C. Cells were washed in phosphate-buffered saline, and the pellet was resuspended in Hanks balanced salt solution (Mediatech, Herndon, VA). The mononuclear cell population was enriched by using a density gradient (Lympholyte-M; Cedarlane Labs, Burlington, Canada) as directed by the manufacturer. Briefly, the cell pellet was suspended in 5 ml Hanks balanced salt solution, underlaid with 5 ml Lympholyte-M, and centrifuged at 1,000 × g for 20 min at room temperature. Dead cells and red blood cells were pelleted, and mononuclear cells were collected at the interface. Cytocentrifuge preparations of the isolated mononuclear cell populations were obtained with a Cytopro 7620 (Wescor, Logan, UT). Differential cell counts were obtained by staining slides with a Diff-Quick kit (ThermoFisher Scientific, Waltham, MA) as directed by the manufacturer. A minimum of 200 cells per slide were counted.
RNA isolation and Northern analysis.
Cultures of S. epidermidis 1457 were grown overnight in TSB, diluted 1:200 into fresh TSB (flask-to-volume ratio, 5:1; shaking at 200 rpm [microaerobic conditions]), and grown at 37°C to optical densities at 600 nm (OD600) of 0.25 (early exponential growth), 3.0 (mid-exponential growth), and 8.2 (late exponential growth). RNA was then isolated as described by Luong et al. (37). The following primer sets were used to amplify regions of the sae locus that were used as DNA probes for the Northern blots: for saeP (379 bp), forward primer AGCGCCACCAAAAATTACAT and reverse primer TTACATTAGGCGCATGTGGA; and for saeS (285 bp), forward primer CGATTGGAGGTCGATACTGG and reverse primer TCTGAAGGTTTACGGGATGG. DNA probes were labeled using digoxigenin-labeled dUTP (Roche, Indianapolis, IN).
Transcriptional profiling.
Overnight cultures of S. epidermidis 1457 and 1457 saeR were diluted 1:100 into fresh TSB and grown at 37°C to an OD600 of 2.7 (flask-to-volume ratio, 5:1; shaking at 200 rpm [microaerobic conditions]). RNA was converted to cDNA, and microarray analysis was performed according to the manufacturer's instructions (Affymetrix expression analysis technical manual; Affymetrix, Inc., Santa Clara, CA) for antisense prokaryotic arrays essentially as described previously by Beenken and colleagues (7). To ensure reproducibility, two cDNA samples from each strain were prepared from two separate experiments. Each cDNA sample was hybridized to an S. epidermidis GeneChip. Signal intensity values for each qualifier (predicted open reading frame [ORF] and intergenic region) were normalized to the median signal intensity value for each GeneChip. Sample values were then averaged. Genes for which there was at least a twofold difference (P ≤0.05, t test) in RNA titer between 1457 and 1457 saeR were considered differentially expressed in a saeR-dependent manner.
Reverse transcriptase PCR (RT-PCR) was used to confirm the microarray data for genes involved in anaerobic metabolism (Table 2). RNA was isolated as described above, and 10 ng was used in each reaction mixture. A One Tube RT-PCR kit (Roche Diagnostics) was used according to the manufacturer's recommendations. Oligonucleotides used for detection of specific genes are listed in Table 2. All reactions were allowed to proceed for 22 cycles with an annealing temperature of 50°C. gyrA was used as an internal standard as described previously by Conlon et al. (16). Amplified products were visualized on a 1.5% agarose (Sigma, St. Louis, MO) gel. Five of the seven genes (SE214, SE1977, SE2170, SE2171, and SE2172) were further confirmed by Northern blot analysis. Primer sets described in Table 2 were used to amplify regions of each gene that were used as DNA probes.
TABLE 2.
RT-PCR primers
| Gene | Forward primer | Reverse primer |
|---|---|---|
| SE1977 | 1285 (TTTTTGATCGTGTAACGGACAG) | 1286 (TGAAATGATTCCTCAAATAGGC) |
| SE2170 | 1287 (AATTGCGGCATGAGATTAGG) | 1288 (AAACGCGCATTAACTGCTTC) |
| SE2171 | 1289 (CGCATCGTCTTGCTGTTTC) | 1290 (TGAAGGTGTTCGATGCAGTG) |
| SE2172 | 1291 (TCACCAAGCAGTTCCACAAG) | 1292 (TGCAAACGTTTCAAGCAGTC) |
| SE0640 | 1293 (TGCTTCATATCCTCTTTAATTTCATC) | 1294 (TTTGTTGTTCTTGGAGCTTTG) |
| SE0227 | 1295 (AAGGCCCACTCATATGTTGC) | 1296 (CGCTTTTCGTCTTACCGTTC) |
| SE0214 | 1297 (TTAGGTCCAGCACCAGAACC) | 1298 (CGATACCAGTTGCCATTGTG) |
| SE0215 | 1299 (TGAAATCTTGCCGTACAAACC) | 1300 (TCAGGCGGTTCAACATTTTC) |
| gyrA | 5 (GGGTAAATATCACCCTCATGG) | 6 (GCAGTTGGGAAATCAGGACC) |
Biofilm and PIA assay.
Assays for the production of biofilm were performed using the method of Christensen et al. (15). Cells for PIA immunoblot assays were cultured in TSB overnight with shaking. The next day, cells were diluted 1:100 into TSB and grown to an OD600 of 1.0. Cell extracts from 3 × 108 CFU were prepared, spotted on 0.45-μm-pore-size polyvinylidene difluoride (Immobilon-P; Millipore, Bedford, MA) membranes, and developed as previously described (17). The anti-PIA antibody used in these assays was diluted 1:250,000 (the antibody was a kind gift from D. Mack).
Metabolic analyses.
Both 1457 and 1457 saeR were grown anaerobically in TSB as described above. Nitrate and lactic acid in the culture medium (TSB; after centrifugation and collection of the supernatant) were quantified using an R-Biopharm metabolic assay kit (R-Biopharm, Marshall, MI). Nitrite in the culture medium was quantified using the Griess reagent (Invitrogen, Carlsbad, CA).
RESULTS
sae is present in S. epidermidis.
BLAST searches (1) performed with the sequences of saeR and saeS from S. aureus strain Newman (GenBank accession number AJ556795 [52]) indicated that genes encoding SaeR and SaeS homologs were present in the genomes of S. epidermidis strains RP62A and ATCC 12228 (GenBank accession numbers CP000029 and AE015929, respectively.) In addition, two ORFs (annotated as SERP0367 and SERP0366 in the RP62A genome sequence and as SE_0481 and SE_0480 in the ATCC 12228 genome sequence) were found upstream of saeR. Comparison of the S. epidermidis genomic sequence data with the S. aureus strain Newman genome indicated that the farthest upstream of these ORFs shared 74% nucleotide sequence identity with S. aureus saeP (designated ORF4) (Fig. 1). Similarly, saeQ from Newman (designated ORF3) shared 75% nucleotide identity with the second ORF in S. epidermidis 1457. The saeR and saeS genes were also similar in the two species, sharing 75 and 67% nucleotide identity, respectively. As a result, we designated the farthest upstream gene in S. epidermidis 1457 saeP and the second gene saeQ (per Novick and Jiang [44]). Primers designed using the S. epidermidis RP62A genome sequence amplified products of the appropriate size (from saeP, saeQ, saeR, and saeS) from S. epidermidis 1457 (data not shown) (39). Therefore, S. epidermidis 1457 was used in further studies as it is amenable to genetic manipulation (25).
Transcriptional analysis of the saePQRS region.
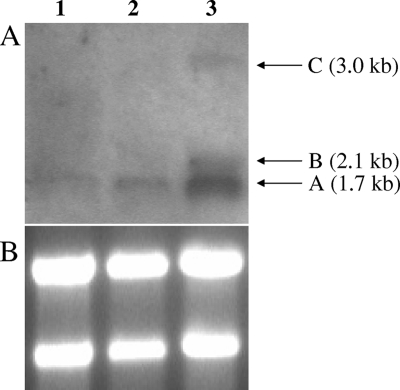
Previous Northern blot analysis of S. aureus saePQRS demonstrated that this region was comprised of four transcriptional units, transcripts A, B, C, and D (44, 52) (Fig. 1). Transcript A is produced early in the exponential phase of growth, whereas transcripts B, C, and D are transcribed late in the exponential growth phase. The transition from producing transcript A to producing transcripts B, C, and D is under the control of agr (44). Northern blot analysis of S. epidermidis 1457 demonstrated that the transcriptional regulation of the saePQRS region was similar to that in S. aureus. Using saeS as a DNA probe, a single transcript that was approximately 1.7 kb long (corresponding to transcript A) was detected during early exponential growth, whereas two additional transcripts that were approximately 2.1 and 3.0 kb long (corresponding to transcripts B and C) were detected later in the exponential growth phase (Fig. 2). In contrast to the results for S. aureus, using saeP as a probe, a smaller 0.5-kb transcript corresponding to transcript D was never detected at any phase of growth (Fig. 1) (data not shown).
FIG. 2.
Northern blot analysis of the sae operon hybridized with a saeS DNA probe. (A) RNA isolated from 1457 at the following times: lane 1, early exponential growth (OD600, 0.25); lane 2, 1457 mid-exponential growth (OD600, 3.0); lane 3, late exponential growth (OD600, 8.2). The arrows indicate the positions of transcripts A, B, and C as discussed in text. (B) RNA gel hybridized with saeS in panel A. Note the equal loading of RNA based on 16S-23S rRNA ethidium bromide staining intensity.
Construction and characterization of a saeR mutant in S. epidermidis.
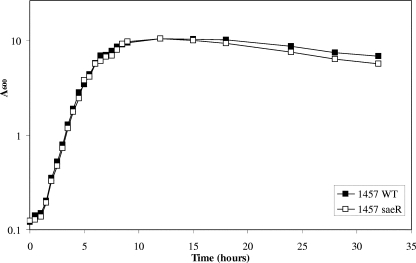
Because sae has been shown to be a regulator of virulence in S. aureus, we speculated that sae controls virulence factor expression in S. epidermidis. To test this possibility, we constructed a sae mutant strain of S. epidermidis 1457 by insertion of the dhfr gene into the predicted effector domain of saeR. Following plasmid integration and vector excision by secondary recombination, disruption of saeR was confirmed by PCR and Southern blot hybridization (data not shown). To ensure that observations made with this mutant strain were due to sae interruption and not due to secondary mutations in other genes, the disrupted saeR allele was backcrossed into wild-type strain 1457 with phage 71. Similar to the results for S. aureus, disruption of saeR did not result in a significant aerobic growth defect (Fig. 3).
FIG. 3.
Comparison of growth of 1457 (WT) and 1457 saeR.
In contrast to S. aureus, biofilm formation is the primary virulence mechanism in S. epidermidis; hence, we assessed the effect of saeR inactivation on biofilm formation. Average A650 values from six biofilm assays indicated that the saeR mutant produced slightly more biofilm than its wild-type counterpart (A650, 2.19 versus 1.90). As expected, two negative control strains, strains 1457 ica (25) and ica-negative Staphylococcus carnosus strain TM300 (24), failed to produce biofilm (A650, 0.33 and 0.28, respectively.) PIA immunoblots indicated that the levels of PIA production were equivalent in 1457 and 1457 saeR (data not shown).
Virulence of an S. epidermidis saeR mutant in a mouse foreign body infection model.
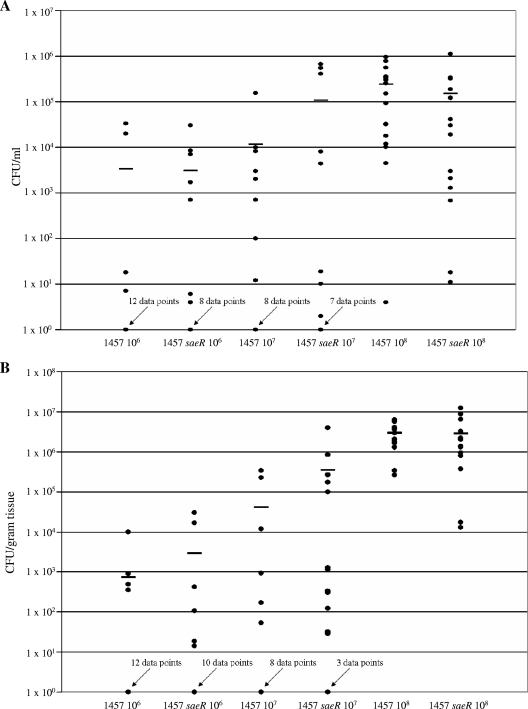
Animal models of infection have demonstrated that sae is required for virulence of S. aureus (5, 8, 20, 22, 23, 57); therefore, we hypothesized that loss of SaeR in S. epidermidis would result in a decrease in virulence. To examine this hypothesis, wild-type strain 1457 and its isogenic saeR mutant were compared using the mouse foreign body infection model. For these experiments, infectious doses of 106, 107, and 108 bacteria were inoculated into catheter segments implanted in the subcutaneous space of mice. The number of catheters analyzed for each inoculum was as follows: for 106 CFU of 1457 and 1457 saeR, 16 catheters each; for 107 CFU of 1457 and 1457 saeR, 15 catheters each; and for 108 CFU of 1457 and 1457 saeR, 16 catheters each. Catheters were explanted 7 days postinfection, and the numbers of S. epidermidis cells associated with the catheters were determined following dilution and plating. The numbers of bacteria isolated from the catheters inoculated with strain 1457 and the numbers of bacteria isolated from the catheters inoculated with strain 1457 saeR were statistically equivalent for all infectious doses (Fig. 4A) (for 106 CFU, P = 0.24; for 107 CFU, P = 1.00; for 108 CFU, P = 0.21). In addition, tissues directly surrounding the catheters were explanted, and the numbers of bacteria associated with these tissues were also determined. Similar to the bacterial loads on the catheters, the numbers of bacteria per gram of tissue were equivalent for strains 1457 and 1457 saeR for each inoculum (Fig. 4B) (for 106 CFU, P = 0.63; for 107 CFU, P = 0.06; for 108 CFU, P = 0.40). Taken together, these data demonstrate that S. epidermidis can establish an infection independent of SaeR.
FIG. 4.
Animal studies with 1457 and 1457 saeR. (A) Numbers of cells obtained from catheters inoculated with 1457 and 1457 saeR. The mean values are indicated by bars. The numbers of data points where no bacteria were detected in the samples are indicated above the arrows. (B) Numbers of cells obtained from tissues surrounding catheters inoculated with 1457 and 1457 saeR. The mean values are indicated by bars. The numbers of data points where no bacteria were detected in the samples are indicated above the arrows.
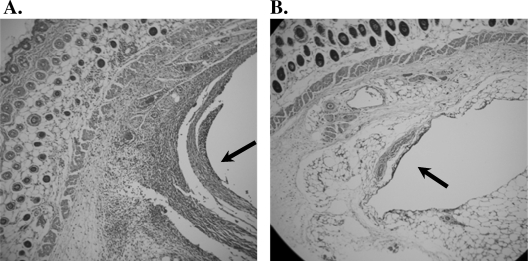
Despite the in vivo growth similarities of the wild-type and saeR mutant strains, we chose to perform a more thorough histological examination of catheter beds to determine if saeR inactivation affected the host response. Indeed, differences in the number of polymorphonuclear neutrophils (PMNs) present at the site of infection were observed for wild-type and saeR mutant cells (Fig. 5A and 5B). Eosin and hematoxylin staining of the tissues revealed that darkly staining immune cells were more prevalent adjacent to catheters infected with wild-type strain 1457. Specifically, 2 days after infection, the proportion of cells with morphological characteristics consistent with those of PMNs was higher in the 1457-infected mice (49.3% ± 3%) than in the 1457 saeR-infected mice (26.7% ± 3%) (P < 0.05). The numbers of PMNs were similar for the two groups of mice after 7 days of infection (P ≥ 0.05) (data not shown). These data demonstrate that SaeR directly or indirectly influences the early, acute phase(s) of an infection.
FIG. 5.
Hematoxylin- and eosin-stained tissues surrounding catheters infected with 1457 (A) or 1457 saeR (B). Note the darkly staining granulocytes in catheter beds (indicated by an arrow) in 1457 (A) compared to 1457 saeR (B).
Transcriptional profiling.
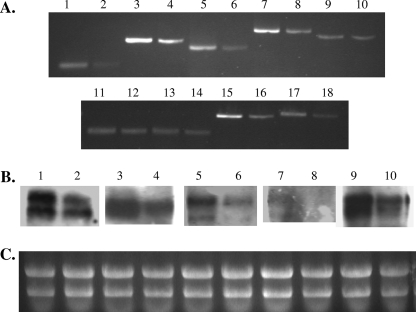
To ascertain possible causes of the alteration in the innate immune response to S. epidermidis with saeR inactivated, we performed a transcriptional profile analysis of the wild-type and saeR mutant strains. We determined that 65 genes were affected by SaeR (37 genes were upregulated and 28 genes were downregulated) (Table 3). In contrast to the findings for S. aureus, in which the large majority of the genes regulated by SaeR code for extracellular toxins and cellular adherence factors (35, 47), genes affected by the loss of SaeR in S. epidermidis had multiple functions, including pyruvate metabolism (e.g., pflA, pflB, ackA, and butA), amino acid synthesis and metabolism (e.g., hisG, hisD, hisH, argG, argJ, and argC), and redox regulation (putative REX regulator, trxA, and trxB). In a comparison to microarray analyses of three S. aureus strains (COL, Newman, and WCUH29), only four genes were identified in S. epidermidis that were also regulated by saeR in S. aureus (35, 47). These four genes were geh (lipase), esp (S. epidermidis)/sspA (S. aureus) (V8 protease), lrgA (holin protein), and the gene encoding a LysM/CHAP domain protein. Interestingly, geh was negatively regulated by saeR in S. epidermidis, but it is positively regulated in S. aureus. Of particular interest were a number of genes that were positively regulated by SaeR that may facilitate growth under anaerobic conditions. These genes included the genes encoding the putative NADH:flavin oxidoreductase/fumarate reductase (SE0195), the putative C-4 dicarboxylate anaerobic carrier (SE0227), complex I NADH:ubiquinone oxidoreductase (snoG; SE0640), assimilatory nitrite reductase (nasE; SE1977), the putative anaerobic C-4 dicarboxylate transporter (SE2170), the putative anaerobic ribonucleoside reductase activator (SE2171), and the class III anaerobic reductase (nrdD; SE2172). Due to the close proximity of the SE2170, SE2171, and SE2172 genes, these genes may be members of an operon. Taken together, these data led us to speculate that SaeRS may be involved in sensing and responding to anaerobiosis. RT-PCR was used to confirm that expression of eight genes (SE1977, SE2170, SE2171, SE2172, SE0640, SE0227, SE0214, and SE0215 [Tables 2 and 3]) possibly involved in anaerobic growth was indeed downregulated in 1457 saeR compared to 1457 (Fig. 6). As shown in Fig. 6, we were unable to consistently demonstrate that expression of two of the genes identified by microarray analysis (SE0640 or snoG [Fig. 6A, lanes 11 and 12] and SE0227 [Fig. 6A, lanes 13 and 14]) was indeed downregulated in 1457 saeR. As the proteins encoded by both of these genes presumably function under anaerobic conditions, the inconsistency in expression levels may be related to variable oxygen tensions in repeat microaerobic cultures. The expression of all other genes tested was indeed downregulated in 1457 saeR compared to 1457 as assessed by RT-PCR (Fig. 6A) or Northern analysis (Fig. 6B).
TABLE 3.
Microarray analysis of wild-type strain S. epidermidis 1457 compared to strain 1457 saeR
| Gene | Fold changea | Locusb |
|---|---|---|
| Genes upregulated in wild-type strain 1457 compared to strain 1457 saeR | ||
| Thiamine biosynthesis lipoprotein (apbE) | 4.7↑ | SE0194 |
| Putative NADH:flavin oxidoreductase/fumarate reductase | 5.2↑ | SE0195 |
| Putative transmembrane efflux pump protein | 3.7↑ | SE0196 |
| Formate acetyltransferase (pflB) | 7.1↑ | SE0214 |
| Formate acetyltransferase-activating enzyme (pflA) | 5.4↑ | SE0215 |
| Putative C-4 dicarboxylate anaerobic carrier | 2.2↑ | SE0227 |
| Putative LysM/CHAP domain protein | 2.2↑ | SE0433 |
| Staphylococcal cell division protein (scdA) | 2.3↑ | SE0439 |
| Staphylococcus aureus exoprotein (saeR) | 8.2↑ | SE0479 |
| Staphylococcus aureus exoprotein (saeP) | 7.0↑ | SE0480 |
| Staphylococcus aureus exoprotein (saeQ) | 47.3↑ | SE0481 |
| K+-H+/Zn-Co-Cd antiporter (czcD) | 2.1↑ | SE0577 |
| Complex I NADH:ubiquinone oxidoreductase (snoG) | 2.4↑ | SE0640 |
| Peptide binding protein (oppA) | 2.1↑ | SE0684 |
| Putative pyruvate ferrodoxin oxidoreductase, alpha subunit | 2.4↑ | SE0967 |
| Glycyl tRNA synthetase | 4.0↑ | SE1252 |
| Acetate kinase (ackA) | 2.0↑ | SE1387 |
| Putative REX redox regulator | 2.7↑ | SE1647 |
| Putative alanine racemase | 2.2↑ | SE1769 |
| Putative SIR2-NAD-dependent deacetylase | 2.4↑ | SE1789 |
| Putative d-octopine dehydrogenase | 2.1↑ | SE1874 |
| Phosphotransferase system arbutin-like II BC component (glvC) | 2.4↑ | SE1897 |
| Assimilatory nitrite reductase (nasE) | 5.3↑ | SE1977 |
| Anti-holin (lrgA) | 2.4↑ | SE2013 |
| Anti-holin (lrgB) | 3.3↑ | SE2014 |
| Glucose-1-dehydrogenase | 2.6↑ | SE2032 |
| Putative Gcn5-related N-acetyltransferase | 2.2↑ | SE2069 |
| Putative alcohol dehydrogenase | 5.3↑ | SE2098 |
| Putative anaerobic C-4 dicarboxylate transporter | 2.2↑ | SE2170 |
| Putative anaerobic ribonucleoside reductase activator | 3.1↑ | SE2171 |
| Class III anaerobic reductase (nrdD) | 3.5↑ | SE2172 |
| Acetoin reductase (butA) | 8.8↑ | SE2225 |
| Putative autolysin/LysM/CHAP domain protein | 2.3↑ | SE2319 |
| 5-Methyltetrahydropteroyltriglutamate-homosysteine 5-methyltransferase (metE) | 2.2↑ | SE2382 |
| Genes which are downregulated in wild-type strain 1457 compared to strain 1457 saeR | ||
| ATP phosphoribosyltransferase (hisG) | 3.1↓ | SE0271 |
| Histidinol dehydrogenase (hisD) | 3.3↓ | SE0272 |
| Amidotransferase (hisH) | 2.4↓ | SE0274 |
| Lipase (gehC) | 3.1↓ | SE0281 |
| Argininosuccinate synthase (argG) | 2.7↓ | SE0657 |
| Putative ABC transporter | 2.6↓ | SE0998 |
| RecA regulon repressor (lexA) | 2.6↓ | SE1022 |
| Thymidylate synthase (thyA) | 3.5↓ | SE1120 |
| Thioredoxin reductase (trxB) | 2.1↓ | SE1168 |
| Arginine acetyltransferase (argJ) | 2.3↓ | SE1211 |
| N-Acetylglutamate gamma-semialdehyde dehydrogenase (argC) | 2.8↓ | SE1212 |
| Riboflavin synthase alpha chain (ribB) | 2.4↓ | SE1440 |
| Riboflavin-specific deiminase (ribD) | 2.1↓ | SE1441 |
| Putative amino acid (possible glutamine) ABC transporter ATP binding protein | 3.2↓ | SE1540 |
| Putative amino acid (possible glutamine) binding periplasmic protein | 2.5↓ | SE1541 |
| V8 protease (esp) | 2.3↓ | SE1543 |
| Putative thioredoxin peroxidase-bacterioferritin | 2.0↓ | SE1547 |
| Putative cationic (Mg2+, Co2+) transporter, CorA family | 2.0↓ | SE1958 |
| Thioredoxin (trxA) | 2.2↓ | SE2097 |
| Other genes | ||
| Hypothetical protein | 3.6↓ | SE0280 |
| Hypothetical protein | 2.2↓ | SE0366 |
| Conserved hypothetical protein | 2.1↓ | SE0392 |
| Hypothetical protein | 2.4↓ | SE0570 |
| Hypothetical protein (possible Na+/K+ antiporter) | 2.0↓ | SE0637 |
| Hypothetical protein | 2.5↓ | SE1024 |
| Hypothetical protein | 2.1↓ | SE1174 |
| Hypothetical protein | 2.0↓ | SE1448 |
| Putative integrase | 2.2↓ | SE1472 |
| Hypothetical protein | 2.6↑ | SE1903 |
| Hypothetical protein | 2.8↑ | SE2173 |
| Hypothetical protein | 5.4↑ | SE2331 |
An arrow pointing up indicates a gene which is upregulated in wild-type strain 1457 compared to strain 1457 saeR. An arrow pointing down indicates a gene which is downregulated in wild-type strain 1457 compared to strain 1457 saeR.
Locus in the S. epidermidis ATCC 12228 genome sequence (GenBank accession number NC 004461).
FIG. 6.
(A) RT-PCR of genes involved in anaerobic growth for confirmation of transcriptional profiling. Lanes 1 and 2, 1457 (lane 1) and 1457 saeR (lane 2) amplified with primers 1285 and 1286 (SE1977); lanes 3 and 4, 1457 (lane 3) and 1457 saeR (lane 4) amplified with primers 1287 and 1288 (SE2170); lanes 5 and 6, 1457 (lane 5) and 1457 saeR (lane 6) amplified with primers 1289 and 1290 (SE2171); lanes 7 and 8, 1457 (lane 7) and 1457 saeR (lane 8) amplified with primers 1291 and 1292 (SE2172); lanes 9 and 10, 1457 (lane 9) and 1457 saeR (lane 10) amplified with primers 5 and 6 (gyrA); lanes 11 and 12, 1457 (lane 11) and 1457 saeR (lane 12) amplified with primers 1293 and 1294 (SE0640); lanes 13 and 14, 1457 (lane 13) and 1457 saeR (lane 14) amplified with primers 1295 and 1296 (SE0227); lanes 15 and 16, 1457 (lane 15) and 1457 saeR (lane 16) amplified with primers 1297 and 1298 (SE0214); lanes 17 and 18, 1457 (lane 17) and 1457 saeR (lane 18) amplified with primers 1299 and 1300 (SE0215). (B) Northern blot analysis of genes involved in anaerobic growth. Lanes 1 and 2, 1457 (lane 1) and 1457 saeR (lane 2) hybridized with an SE2170 DNA probe; lanes 3 and 4, 1457 (lane 3) and 1457 saeR (lane 4) hybridized with an SE2171 DNA probe; lanes 5 and 6, 1457 (lane 5) and 1457 saeR (lane 6) hybridized with an SE1977 DNA probe; lanes 7 and 8, 1457 (lane 7) and 1457 saeR (lane 8) hybridized with an SE2172 DNA probe; lanes 9 and 10, 1457 (lane 9) and 1457 saeR (lane 10) hybridized with an SE0214 DNA probe. (C) RNA gel hybridized with various probes described above for panel B. Note the equal loading of RNA based on 16S-23S rRNA ethidium bromide staining intensity.
Growth of 1457 and 1457 saeR under anoxic growth conditions.
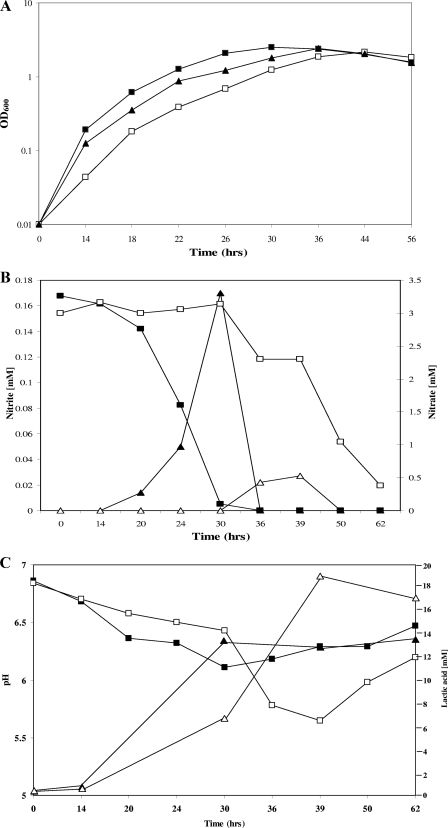
To assess if SaeR alters anaerobic physiology, strains 1457 and 1457 saeR were grown at 37°C in an anaerobic chamber, and the growth was monitored. As expected, the results demonstrated that 1457 saeR had a lower growth rate than 1457, although the growth yields were similar (Fig. 7A).
FIG. 7.
(A) Effect of anaerobiosis on growth of 1457 (▪), 1457 saeR (□), and 1457 saeR/pNF115 (▴). The results are representative of three separate experiments. (B) Production of nitrite by 1457 (▴) and 1457 saeR (▵) superimposed on curves showing the utilization of nitrate by 1457 (▪) and 1457 saeR (□). (C) Production of lactic acid by 1457 (▴) and 1457 saeR (▵) superimposed on curves showing the pH values of the 1457 growth medium (▪) and the 1457 saeR growth medium (□). The data in panels B and C are representative of five separate experiments.
Staphylococci can use nitrate as a terminal electron acceptor under anoxic conditions (11, 12, 41, 45). Since snoG is postulated to be an important oxidoreductase that is linked to nitrate reductase activity in electron transport (6), we hypothesized that the growth defect detected in 1457 saeR is due to a decreased ability to utilize nitrate as a terminal electron acceptor. To test this possibility, strains 1457 and 1457 saeR were grown anaerobically in TSB supplemented with 3 mM potassium nitrate. The results demonstrated that 1457 depleted the nitrate in the culture medium; however, strain 1457 saeR had a significantly reduced rate of nitrate utilization (Fig. 7B). Strain 1457 exhausted the nitrate in the culture medium by 30 h after the beginning of of growth; however, strain 1457 saeR never depleted the nitrate. Furthermore, after 39 h of growth, when the OD600 of 1457 and 1457 saeR were similar, there was still approximately 2.3 mM nitrate left in the culture supernatant of 1457 saeR, whereas no nitrate was detected in the culture supernatant of 1457. Anaerobiosis reduces nitrate to nitrite; therefore, our data demonstrating that nitrate utilization was significantly decreased in strain 1457 saeR compared to 1457 led us to speculate that the accumulation of nitrite in the culture medium would be greater in 1457 cultures than in 1457 saeR cultures (41). As predicted, nitrite began accumulating coincident with a decrease in the nitrate concentration in the 1457 culture medium. In contrast, the concentration of nitrite in the culture medium of 1457 saeR was at the detection limit of the assay (Fig. 7B). Consistent with previous observations, nitrite was extracted from the culture medium only after the concentration of nitrate became limited (41) (Fig. 7B).
The impaired anaerobic growth of strain 1457 saeR results in decreased utilization of nitrate as a terminal electron acceptor. This situation can affect redox homeostasis by limiting the ability of the bacterium to oxidize NADH to NAD+. We hypothesized that to compensate for the lack of electron transport-mediated oxidation of NADH, more pyruvate would be shunted through lactate dehydrogenase and result in increased lactic acid accumulation in the culture medium of strain1457 saeR. Increasing lactate dehydrogenase (EC 1.1.1.27) activity would help mitigate a redox imbalance by coupling the dismutation of pyruvate to lactic acid with the oxidation of NADH to NAD+. As predicted, the pH minimum of the 1457 saeR growth medium was significantly lower than the pH minimum of the 1457 culture medium (Fig. 7C), suggesting that more organic acids were produced by 1457 saeR than by 1457. Consistent with the pH data, we found a higher concentration of lactic acid in the culture medium of strain 1457 saeR than in the culture medium of the isogenic wild-type strain (Fig. 7C). Collectively, these data suggest that the decreased pH observed in the 1457 saeR culture medium was due to increased production of lactic acid.
Complementation of 1457 saeR with pNF155.
1457 saeR/pNF155, which contained full-length saeRS on a multicopy plasmid, partially complemented the growth defect observed in 1457 saeR grown under anoxic conditions (Fig. 7A). Consistent with these data, pNF155 did not completely restore 1457 saeR to the 1457 phenotype with regard to nitrate utilization and lactic acid production. For instance, after 39 h of growth, 1.0 mM nitrate and 11.2 mM lactic acid (mean concentrations) remained in the culture supernatant of 1457 saeR/pNF115 (n = 3; data not shown).
DISCUSSION
The sae two-component regulatory system in S. aureus has emerged as an important regulator of virulence. The research described here is the first step in characterization of the sae operon in S. epidermidis and the first attempt to identify the environmental conditions that modulate the regulatory response of sae. The S. epidermidis sae operon was found to be organized in a manner similar to the S. aureus operon. Each gene in strain RP62A was found to share at least 67% nucleotide identity to a gene from S. aureus strain Newman, and each protein was found to share at least 75% amino acid identity. In addition, Northern analysis demonstrated that the transcriptional regulation of sae in S. epidermidis strain 1457 was similar to that in S. aureus (44, 47, 52) (Fig. 1). Relative to S. aureus, S. epidermidis has very few defined virulence factors. Of the known virulence factors and mechanisms in S. epidermidis, the most extensively studied are the production of PIA/biofilm and the poly-γ-dl-glutamic acid capsule (32, 49, 50). Our data demonstrate that transcription of the genes encoding these virulence factors occurs independent of SaeR in S. epidermidis. This observation provided an excellent opportunity to examine the physiological function of sae in S. epidermidis. Nevertheless, since there is a clear association between virulence and the sae operon in S. aureus, we utilized a mouse foreign body infection model, a highly pertinent model of S. epidermidis-mediated infection, to determine the level of virulence of the saeR mutant compared to that of the wild type. Furthermore, this model has been used to demonstrate differences in the virulence of strains that produce similar amounts of PIA/biofilm (32). Interestingly, even though strains 1457 and 1457 saeR were equally able to establish an infection in the mouse foreign body infection model, the host responses to these two strains differed dramatically (Fig. 5A and 5B). At 2 days postinoculation, the PMN response was significantly greater in the tissue of mice infected with 1457 than in the tissue of mice infected with 1457 saeR. Our first inclination was to determine whether SaeR regulates the production of phenol-soluble modulins, which are known immune mediators (54). However, no difference in the production of phenol-soluble modulins was detected between the saeR mutant and the wild type (data not shown). Further studies are needed to determine which immune modulators are regulated by SaeR during the acute phase of infection.
Transcriptional profiling revealed that SaeR affects metabolic processes, including redox activity, pyruvate metabolism, and amino acid synthesis and metabolism; of particular interest were several genes predicted to be involved in anaerobic metabolism. Two of the genes (SE0227 and SE2170) encode putative anaerobic C-4 dicarboxylate transporters, which facilitate transport of alternative electron acceptors (e.g., fumarate or succinate) (29). In addition, a putative NADH:flavin oxidoreductase/fumarate reductase (SE0195) was also positively regulated by SaeR. The class III anaerobic reductase (nrdD; SE2172) is an anaerobic ribonucleotide reductase that catalyzes the formation of DNA from RNA building blocks. NrdD is inactivated by molecular oxygen and appears to be essential for anaerobic growth (30, 42). Lastly, snoG, encoding part of the complex I NADH:ubiquinone oxidoreductase, is important in anaerobic respiration linked to nitrate reduction. Taken together, these data suggest that one function of SaeR is to regulate the transition from aerobic growth to anaerobic growth. The fact that saeR inactivation did not alter aerobic growth but did result in an anaerobic growth impairment strongly supports this suggestion (Fig. 7A).
Glycolysis generates two molecules of pyruvate for every molecule of glucose consumed; however, in the process it reduces two molecules of NAD+ to NADH. Reduction of NAD+ to NADH without an equivalent means to oxidize NADH can create a redox imbalance and inhibit growth. Staphylococci primarily reduce pyruvate to lactic acid during anaerobic growth, a process that regenerates NAD+ (9, 33). Under anaerobic or microaerobic growth conditions, NAD+ can be regenerated from NADH by both the membrane-bound nitrate reductase and the NADH:ubiquinone oxidoreductase (SnoA-G) (6). However, saeR inactivation decreased transcription of snoG, slowing the utilization of nitrate and likely resulting in the decreased growth rate. To compensate for the redox imbalance, strain 1457 saeR shunts pyruvate into lactate dehydrogenase, a reaction allowing for the stoichiometric oxidation of NADH produced during glycolysis (Fig. 7C). In agreement with these data, microarray analysis demonstrated that transcription of both trxA and trxB is downregulated in the saeR mutant, whereas REX, a redox-responsive regulator, is upregulated in the saeR mutant (Table 3). The result of this compensation by 1457 saeR is a much more acidic extracellular milieu; the consequence of this metabolic switch, especially within a biofilm, is currently being studied in our laboratories.
It is not known why pNF115 is unable to completely restore the wild-type phenotype to 1457 saeR. However, the explanation may be related to the fact that the entire two-component regulator (saeRS) is present on a multicopy plasmid. Unfortunately, unlike the situation in S. aureus, in S. epidermidis there is no genetic system to complement mutants in single copy.
In contrast to S. aureus, our data suggest that in S. epidermidis the sae two-component regulatory system functions to control basic metabolic processes but few of the limited number of virulence genes (i.e., lipase and V8 protease genes). Based on the genomic similarities between S. aureus and S. epidermidis, we speculate that saeRS may function to indirectly regulate virulence gene expression by altering the metabolic status of the bacteria, particularly under conditions in which oxygen availability is low, such as the conditions in a deep abscess or biofilm. This hypothesis is actively being tested in our laboratories.
Acknowledgments
This work was supported by Public Health Service grant AI49311 from the National Institute of Allergy and Infectious Diseases to P.D.F.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 22 October 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Archer, G. L., and J. L. Johnston. 1983. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob. Agents Chemother. 2470-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291159-170. [DOI] [PubMed] [Google Scholar]
- 4.Augustin, J., and F. Götz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54203-207. [DOI] [PubMed] [Google Scholar]
- 5.Bae, T., A. K. Banger, A. Wallace, E. M. Glass, F. Aslund, O. Schneewind, and D. M. Missiakas. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA 10112312-12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer, A. S., P. McNamara, M. R. Yeaman, N. Lucindo, T. Jones, A. L. Cheung, H. G. Sahl, and R. A. Proctor. 2006. Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1. J. Bacteriol. 188211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 1864665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benton, B. M., J. P. Zhang, S. Bond, C. Pope, T. Christian, L. Lee, K. M. Winterberg, M. B. Schmid, and J. M. Buysse. 2004. Large-scale identification of genes required for full virulence of Staphylococcus aureus. J. Bacteriol. 1868478-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumenthal, H. J. 1972. Glucose metabolism in staphylococci, p. 111-135. In J. O. Cohen (ed.), The staphylococci. Wiley-Interscience, New York, NY.
- 10.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28183-200. [DOI] [PubMed] [Google Scholar]
- 11.Burke, K. A., A. E. Brown, and J. Lascelles. 1981. Membrane and cytoplasmic nitrate reductase of Staphylococcus aureus and application of crossed immunoelectrophoresis. J. Bacteriol. 148724-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burke, K. A., and J. Lascelles. 1979. Partial purification and some properties of the Staphylococcus aureus cytoplasmic nitrate reductase. J. Bacteriol. 139120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, P. R., T. Bae, W. A. Williams, E. M. Duguid, P. A. Rice, O. Schneewind, and C. He. 2006. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2591-595. [DOI] [PubMed] [Google Scholar]
- 14.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 401-9. [DOI] [PubMed] [Google Scholar]
- 15.Christensen, G. D., L. M. Baddour, B. M. Madison, J. T. Parisi, S. N. Abraham, D. L. Hasty, J. H. Lowrance, J. A. Josephs, and W. A. Simpson. 1990. Colonial morphology of staphylococci on memphis agar: phase variation of slime production, resistance to beta-lactam antibiotics, and virulence. J. Infect. Dis. 1611153-1169. [DOI] [PubMed] [Google Scholar]
- 16.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 1844400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cramton, S. E., M. Ulrich, F. Gotz, and G. Doring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 694079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraudo, A. T., A. Calzolari, A. A. Cataldi, C. Bogni, and R. Nagel. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 17715-22. [DOI] [PubMed] [Google Scholar]
- 19.Giraudo, A. T., C. Mansilla, A. Chan, C. Raspanti, and R. Nagel. 2003. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol. 46246-250. [DOI] [PubMed] [Google Scholar]
- 20.Giraudo, A. T., H. Rampone, A. Calzolari, and R. Nagel. 1996. Phenotypic characterization and virulence of a sae− agr− mutant of Staphylococcus aureus. Can. J Microbiol. 42120-123. [DOI] [PubMed] [Google Scholar]
- 21.Giraudo, A. T., C. G. Raspanti, A. Calzolari, and R. Nagel. 1994. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 40677-681. [DOI] [PubMed] [Google Scholar]
- 22.Goerke, C., U. Fluckiger, A. Steinhuber, V. Bisanzio, M. Ulrich, M. Bischoff, J. M. Patti, and C. Wolz. 2005. Role of Staphylococcus aureus global regulators sae and σB in virulence gene expression during device-related infection. Infect. Immun. 733415-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 401439-1447. [DOI] [PubMed] [Google Scholar]
- 24.Gotz, F., and B. Schumacher. 1987. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40285-288. [Google Scholar]
- 25.Handke, L., S. Slater, K. Conlon, S. O'Donnell, M. Olson, K. Bryant, M. Rupp, J. O'Gara, and P. Fey. 2007. σB and SarA act at the level of icaADBC transcription to regulate PIA production in Staphylococcus epidermidis. Can. J. Microbiol. 5382-91. [DOI] [PubMed] [Google Scholar]
- 26.Harraghy, N., J. Kormanec, C. Wolz, D. Homerova, C. Goerke, K. Ohlsen, S. Qazi, P. Hill, and M. Herrmann. 2005. sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology 1511789-1800. [DOI] [PubMed] [Google Scholar]
- 27.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 201083-1091. [DOI] [PubMed] [Google Scholar]
- 28.Iordanescu, S. 1976. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch. Roum. Pathol. Exp. Microbiol. 35111-118. [PubMed] [Google Scholar]
- 29.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kroger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 155339-56. [DOI] [PubMed] [Google Scholar]
- 30.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 6771-98. [DOI] [PubMed] [Google Scholar]
- 31.Kaito, C., K. Kurokawa, Y. Matsumoto, Y. Terao, S. Kawabata, S. Hamada, and K. Sekimizu. 2005. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol. Microbiol. 56934-944. [DOI] [PubMed] [Google Scholar]
- 32.Kocianova, S., C. Vuong, Y. Yao, J. M. Voyich, E. R. Fischer, F. R. DeLeo, and M. Otto. 2005. Key role of poly-gamma-dl-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Investig. 115688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krebs, H. A. 1937. Dismutation of pyruvic acid in Gonococcus and Staphylococcus. Biochem. J. 31661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 35.Liang, X., C. Yu, J. Sun, H. Liu, C. Landwehr, D. Holmes, and Y. Ji. 2006. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 744655-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luong, T. T., P. M. Dunman, E. Murphy, S. J. Projan, and C. Y. Lee. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 1881899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mack, D., M. Haeder, N. Siemssen, and R. Laufs. 1996. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J. Infect. Dis. 174881-884. [DOI] [PubMed] [Google Scholar]
- 39.Mack, D., N. Siemssen, and R. Laurs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 602048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nedelmann, M., A. Sabottke, R. Laufs, and D. Mack. 1998. Generalized transduction for genetic linkage analysis and transfer of transposon insertions in different Staphylococcus epidermidis strains. Zentbl. Bakteriol. 28785-92. [DOI] [PubMed] [Google Scholar]
- 41.Neubauer, H., and F. Gotz. 1996. Physiology and interaction of nitrate and nitrite reduction in Staphylococcus carnosus. J. Bacteriol. 1782005-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nordlund, P., and P. Reichard. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75681-706. [DOI] [PubMed] [Google Scholar]
- 43.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 44.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 1492709-2717. [DOI] [PubMed] [Google Scholar]
- 45.Pantel, I., P. E. Lindgren, H. Neubauer, and F. Gotz. 1998. Identification and characterization of the Staphylococcus carnosus nitrate reductase operon. Mol. Gen. Genet. 259105-114. [DOI] [PubMed] [Google Scholar]
- 46.Projan, S. J., and G. L. Archer. 1989. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J. Bacteriol. 1711841-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogasch, K., V. Ruhmling, J. Pane-Farre, D. Hoper, C. Weinberg, S. Fuchs, M. Schmudde, B. M. Broker, C. Wolz, M. Hecker, and S. Engelmann. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 1887742-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rooijakkers, S. H., M. Ruyken, J. van Roon, K. P. van Kessel, J. A. van Strijp, and W. J. van Wamel. 2006. Early expression of SCIN and CHIPS drives instant immune evasion by Staphylococcus aureus. Cell. Microbiol. 81282-1293. [DOI] [PubMed] [Google Scholar]
- 49.Rupp, M. E., J. S. Ulphani, P. D. Fey, and D. Mack. 1999. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 672656-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rupp, M. E., J. S. Ulphani, P. D. Fey, and D. Mack. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body model. Infect. Immun. 672627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schenk, S., and R. A. Laddiga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94133-138. [DOI] [PubMed] [Google Scholar]
- 52.Steinhuber, A., C. Goerke, M. G. Bayer, G. Doring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 1856278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, J., L. Zheng, C. Landwehr, J. Yang, and Y. Ji. 2005. Identification of a novel essential two-component signal transduction system, YhcSR, in Staphylococcus aureus. J. Bacteriol. 1877876-7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vuong, C., M. Durr, A. B. Carmody, A. Peschel, S. J. Klebanoff, and M. Otto. 2004. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell. Microbiol. 6753-759. [DOI] [PubMed] [Google Scholar]
- 55.Vuong, C., S. Kocianova, J. M. Voyich, Y. Yao, E. R. Fischer, F. R. DeLeo, and M. Otto. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 27954881-54886. [DOI] [PubMed] [Google Scholar]
- 56.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6269-275. [DOI] [PubMed] [Google Scholar]
- 57.Xiong, Y. Q., J. Willard, M. R. Yeaman, A. L. Cheung, and A. S. Bayer. 2006. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J. Infect. Dis. 1941267-1275. [DOI] [PubMed] [Google Scholar]
- 58.Yamazaki, K., F. Kato, Y. Kamio, and J. Kaneko. 2006. Expression of gamma-hemolysin regulated by sae in Staphylococcus aureus strain Smith 5R. FEMS Microbiol. Lett. 259174-180. [DOI] [PubMed] [Google Scholar]
- 59.Yanisch-Perron, D., J. Vierra, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 60.Yao, Y., D. E. Sturdevant, and M. Otto. 2005. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191289-298. [DOI] [PubMed] [Google Scholar]
- 61.Yao, Y., D. E. Sturdevant, A. Villaruz, L. Xu, Q. Gao, and M. Otto. 2005. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect. Immun. 731856-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]