Abstract
Typical enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) employ either Nck, TccP/TccP2, or Nck and TccP/TccP2 pathways to activate the neuronal Wiskott-Aldrich syndrome protein (N-WASP) and to trigger actin polymerization in cultured cells. This phenotype is used as a marker for the pathogenic potential of EPEC and EHEC strains. In this paper we report that EPEC O125:H6, which represents a large category of strains, lacks the ability to utilize either Nck or TccP/TccP2 and hence triggers actin polymerization in vitro only inefficiently. However, we show that infection of human intestinal biopsies with EPEC O125:H6 results in formation of typical attaching and effacing lesions. Expression of TccP in EPEC O125:H6, which harbors an EHEC O157-like Tir, resulted in efficient actin polymerization in vitro and enhanced colonization of human intestinal in vitro organ cultures with detectable N-WASP and electron-dense material at the site of bacterial adhesion. These results show the existence of a natural category of EPEC that colonizes the gut mucosa using Nck- and TccP-independent mechanisms. Importantly, the results highlight yet again the fact that conclusions made on the basis of in vitro cell culture models cannot be extrapolated wholesale to infection of mucosal surfaces and that the ability to induce actin polymerization on cultured cells should not be used as a definitive marker for EPEC and EHEC virulence.
Enteropathogenic Escherichia coli (EPEC) comprises a category of diarrheagenic E. coli that was the first to be implicated in human disease. In 1987 the World Health Organization assigned EPEC to serogroups O26, O55, O86, O111, O114, O119, O125, O126, O127, O128, O142, and O158 (reviewed in reference 7). EPEC is divided into two evolutionary related lineages termed EPEC 1 (typified by expression of flagellar antigen H6 and intimin α) and EPEC 2 (typified by expression of flagellar antigen H2, intimin β, and TccP2) (10, 34, 35). Enterohemorrhagic E. coli (EHEC) constitutes a subgroup of Shiga toxin-producing E. coli that can cause bloody diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome. EHEC O157:H7 is the most common and virulent serotype that is implicated worldwide in human disease (reviewed in reference 17).
While colonizing the gut mucosa, EPEC and EHEC trigger widespread ultrastructural changes which are characterized by localized disintegration of the brush border microvilli and close association of the bacteria with the enterocyte plasma membrane, termed attaching and effacing (A/E) lesions (27). Formation of A/E lesions can be reproduced ex vivo by infection of cultured human intestinal explants (in vitro organ culture [IVOC]) with EPEC (22). The genes necessary for EPEC A/E lesion formation in vitro are carried on the locus of enterocyte effacement (26), which encodes a type III secretion system (15), the adhesin intimin (16), chaperones, translocator, and six effector proteins, including Tir (translocated intimin receptor) (20).
Once translocated, Tir is integrated into the host cell plasma membrane in a hairpin loop topology (13). The extracellular loop, present above the plasma membrane, serves as a receptor for the bacterial adhesin intimin. Results from EPEC infection of cultured epithelial cells in vitro have shown that clustering of Tir by intimin (4) leads to phosphorylation of a Tir tyrosine residue (19) which is present in the context of a consensus binding site (YPDEP/D/V) for the mammalian adaptor proteins Nck1 and -2 (referred to collectively as Nck throughout this report). Binding of Nck to phosphorylated Tir leads to recruitment and activation of the neuronal Wiskott-Aldrich syndrome protein (N-WASP), initiating actin polymerization via the actin-related protein 2/3 (Arp2/3) complex (reviewed in reference 6).
Strains belonging to EPEC 1, commonly represented by O127:H6 strain E2348/69, trigger actin polymerization predominantly via the Nck actin polymerization pathway, while strains belonging to EPEC 2, commonly represented by O111:NM strain B171, can trigger actin polymerization in vitro by redundant mechanisms involving either Nck or TccP2, which is functionally interchangeable with TccP of EHEC O157:H7 (34). TccP/EspFU is a bacterial effector protein that, although it has not been shown to bind Tir directly, binds directly to N-WASP, leading to recruitment of the Arp2/3 complex and localized actin polymerization (5, 11). Recent studies have shown that a conserved NPY carboxy-terminal Tir motif in EPEC and EHEC is involved in Nck-independent actin polymerization in the former and TccP-dependent actin polymerization pathway in the latter (1). Importantly, the Nck, TccP, and TccP2 actin polymerization pathways are all dispensable for A/E lesion formation on human IVOC (11, 32, 34) and in mouse gut following infection with Citrobacter rodentium (9).
Recently while screening for the presence of tccP and tccP2 in clinical EPEC isolates, we discovered that strains belonging to EPEC O125:H6 naturally encode non-tyrosine-phosphorylated Tir and yet are tccP and tccP2 gene negative (30). The aim of this study was to investigate whether EPEC O125:H6 triggered actin polymerization in vitro and ex vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown for 8 h in Luria-Bertani medium before being diluted into Dulbecco's modified Eagle's medium and incubated at 37°C in 5% CO2 statically overnight. Growth medium was supplemented with ampicillin (50 μg/ml) or kanamycin (50 μg/ml), when necessary.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| TUV 93-0 | EHEC O157:H7 strain EDL933, stx-negative (intimin γ) | ATCC |
| E2348/69 | Wild-type EPEC O127:H6 (intimin α) | 25 |
| ICC223 | E. coli O125:H6 isolated in Brazil from a diarrheagenic case (intimin α) | This study |
| CPG35 | E. coli O125:H6 isolated in the United Kingdom from a diarrheagenic case (intimin α) | This study |
| N67 | E. coli O125:H6 isolated in the United Kingdom from a diarrheagenic case (intimin α) | This study |
| 2741-5 | E. coli O125:H6 isolated in Brazil from a diarrheagenic case (intimin α) | This study |
| ICC224 | ICC223Δtir::Km | This study |
| ICC225 | Δtir in EPEC O127:H6 strain E2398/69 | This study |
| TUV 93-0Δtir | Δtir in EHEC O157:H7 | 22 |
| Plasmids | ||
| pACYC-P | pACYC derivative encoding mtc (map tir cesT 5′ eae) from E2348/69 | 18 |
| pKD46 | Helper plasmid encoding λ red recombinase | 8 |
| pKD4 | Template | 10 |
| pSA10 | pKK177-3 derivative containing lacQ | 31 |
| pICC368 | pSA10 derivative encoding TirICC223 | This study |
| pICC369 | pSA10 derivative encoding TUV 93-0 TccP-HA fusion protein | This study |
Infection of cultured cells.
Bacterial cultures were used to infect HeLa cells grown on coverslips in 24-well plates for 3 h (E2348/69), 6 h (TUV 93-0), or 5 h (all other strains) as described elsewhere (34). Cell monolayers were then fixed in 3.7% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 4 min. EHEC O157:H7 and EPEC E2348/69 were visualized with goat anti-O157 antiserum (Fitzgerald Industries) and anti-O127 antiserum (a gift from Roberto La Ragione, Veterinary Laboratories Agency [VLA], United Kingdom), respectively. O125 bacteria were visualized with Hoechst 33342 DNA stain (Molecular Probes) or anti-O125 antiserum (a gift from Roberto La Ragione, VLA, United Kingdom), actin was stained with Oregon Green-phalloidin (Invitrogen), and hemagglutinin (HA)-tagged TccP was detected by anti-HA as described elsewhere (21, 34). TirEPEC and TirEHEC were detected as described in references 34 and 11, respectively. Samples were analyzed using a Zeiss Axioimager fluorescence microscope, and images were processed using Axiovision and Adobe Photoshop software.
Recombinant DNA.
The tir gene of ICC223 was amplified by PCR using primers 125tir-F2 and 125tir-R2 (Table 2), cloned into pSA10 (generating plasmid pICC368), and sequenced (accession number AB355659). A tir deletion mutant in strain ICC223 was made using the lambda red system (8) with primers 125tir-F1 and 125tir-R1 and pKD4 as template, generating strain ICC224. Primers flanking the deleted region and inside the kanamycin cassette were used in a PCR to verify the deletion (primer pairs kt with tir-flank-F and k2 with tir-flank-R) (Table 2). E2348/69Δtir (ICC225) (Table 1) was generated using the lambda red system (8) with primers EPEC FRT and Tir EPEC FRT rev; the deletion was confirmed by PCR using the primer pairs Map303 with kt and 3CesT rev with k2 (Table 2).
TABLE 2.
Primers used in this study
| Name | Nucleotide sequence (5′-3′) |
|---|---|
| kt | CGGCCACAGTCGATGAATCC |
| k2 | CGGTGCCCTGAATGAACTGC |
| 125tir-F1(m) | ATCCCAATGTGAATAATTCAATTCCTCCTGCACCTCCATTACCTTCACAAGTGTAGGCTGGAGCTGCTTC |
| 125tir-R1 | TTAGACGAAACGATGGGATCCCGGCGCTGGTGGGTTATTCGAAGTATTCACATATGAATATCCTCCTTAG |
| 125tir-F2(k) | ATGCCTATTGGTAATCTTGGT |
| 125tir-R2 | TAGCTGCAGTTAACGAAACGATGGGATCC |
| tir-flank-F(detec) | ATGCCTATTGGTAATCTTGGT |
| tir-flank-R | TAAAAGTTCAGATCTTGATGACAT |
| tir EPEC FRT for | ATGCCTATTGGTAACCTTGGTAATAATGTAAATGGCAATCATTTAATTCCTGTGTAGGCTGGAGCTGCTTCG |
| tir EPEC FRT rev | TTAAACGAAACGTACTGGTCCCGGCGTTGGTGCGGCATTTACAGAACTTACATATGAATATCCTCCTTAG |
| map303 for | AAGAATTCATTAGTAAGGAGACTAAATGT |
| 3cesT rev | AAAAGATCTTTATCTTCCGGCGTAATAATG |
| pkk-tccP-F1 | CCGGAATTCATGATTAACAATGTTTCTTCA |
| pkk-tccP-R1 | AAACTGCAGTCAAGCGTAGTCTGGGACGTCGTATGGGTAAGCGTAGTCTGGGACGTCGTATGGGTACGAGCGCTTAGATGTATTAATGCC |
To create TccP-HA, the coding sequence for tccP was amplified (primers pkk-tccP-F1 and pkk-tccP-R1 [Table 2]) and cloned into pSA10 as described previously (11).
Sequence comparison of Tir proteins.
Clustal W was used for making a multiple alignment of the TirEPEC_O125:H6 sequences with 14 known Tir sequences which were retrieved from the GenBank database. A phylogenic tree was constructed with the neighbor-joining algorithm of the MEGA 3.1 software (24). Poisson correction with the complete deletion of gaps was used to calculate protein distances. Bootstrap analysis with 1,000 replicates was performed to evaluate the significance of the internal branches.
In vitro organ cultures.
Pediatric tissue was obtained with fully informed parental consent and local ethical committee approval using grasp forceps during routine endoscopic investigation of intestinal disorders. Small intestinal mucosal biopsies which appeared macroscopically normal were taken for organ culture experiments as described previously (14). Adherence was examined using tissue from six patients (ages between 139 and 201 months) by scanning electron microscopy and five further cases (ages between 141 and 200 months) by cryosectioning, immunostaining, and transmission electron microscopy as described elsewhere (32). IVOC infected with EHEC O157:H7 strain TUV 93-0 was used as a positive control. In each experiment, a noninfected sample was included to exclude endogenous bacterial adhesion.
For immunofluorescence, samples were embedded in optimal cutting temperature compound (Sakura), snap-frozen in liquid nitrogen, and stored at −70°C until use. Serial sections of 8 μm were cut with an MTE cryostat (SLEE Technik), picked up on poly-l-lysine-coated slides, and air dried. Tissue sections were fixed in formalin for 10 min and blocked with 0.5% bovine serum albumin, 2% normal goat serum in phosphate-buffered saline for 20 min at room temperature. Slides were incubated with rabbit anti-TirEHEC_O157:H7, anti-TccP, or anti-N-WASP (kindly provided by Silvia Lommel, Institute for Cell Biology, University of Bonn, Bonn, Germany) for 60 min at room temperature, washed, and incubated in Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G (Molecular Probes) for 30 min. Counterstaining of bacteria and cell nuclei was performed using propidium iodide (Sigma). Epithelial cells were stained with mouse anti-cytokeratin (Dako) and Alexa Fluor 647-conjugated goat anti-mouse immunoglobulin G (Molecular Probes). Sections were analyzed with a Radiance 2100 confocal laser scanning microscope (Bio-Rad, United Kingdom).
Nucleotide sequence accession number.
The nucleotide sequence for the tir gene of ICC223 was deposited in the GenBank database under accession no. AB355659.
RESULTS
EPEC O125:H6 cannot trigger efficient actin polymerization in vitro.
In order to characterize the ability of EPEC O125:H6 to trigger actin polymerization in vitro, HeLa cells were infected with four different strains isolated in Brazil and the United Kingdom and with controls EPEC 1 O127:H6 (E2348/69) and EHEC O157:H7 (EDL933 stx-negative strain TUV 93-0) (Table 1). All four O125:H6 strains failed to trigger detectable actin polymerization under attached bacteria (data not shown). We selected one of the O125:H6 EPEC strains isolated in Brazil (ICC223) for further detailed analysis.
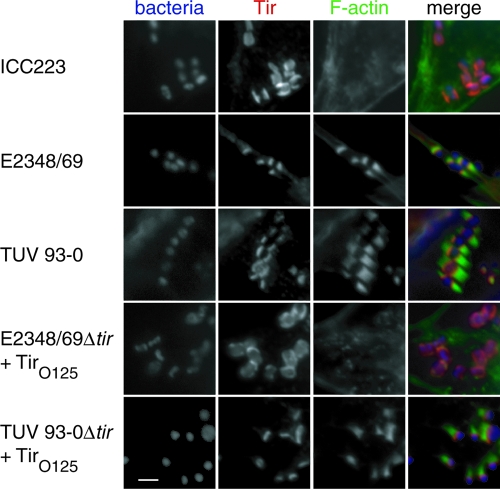
The competence of ICC223-induced actin remodeling was further quantified by counting the percentage of cell-associated bacteria which were also associated with intense F-actin staining. Regions of 5 to 20 bacteria per cell were examined for each strain in three separate experiments carried out in duplicate. One hundred bacteria per coverslip were examined. Quantifying the efficiency of actin polymerization revealed weak actin aggregation under only 3% of adherent ICC223 cells despite efficient Tir translocation (Fig. 1; Table 3). This was in sharp contrast to cells infected with controls EPEC 1 O127:H6 (E2348/69) and EHEC O157:H7 (TUV 93-0), which induced efficient actin polymerization under 75% and 70% of attached bacteria, respectively (Fig. 1; Table 3). The 3% of adherent ICC223 cells showing weak actin accretion is comparable to the frequency seen after infection with E2348/69 expressing Tir Y474F (3).
FIG. 1.
EPEC O125:H6 strain ICC223 cannot efficiently induce actin polymerization in infected HeLa cells, while the controls EPEC O127:H6 strain E2348/69 and EHEC O157:H7 strain TUV 93-0 trigger efficient actin polymerization. Expression of TirEPEC_O125:H6 in EPEC E2348/69Δtir did not restore actin polymerization, while expressing TirEPEC_O125:H6 in EHEC TUV 93-0Δtir resulted in strong actin polymerization. Bacterial DNA was visualized in blue using Hoechst 33342. Tir is labeled red with anti-TirEHEC or TirEPEC (for strain E2348/69) antiserum. Actin was labeled in green using Oregon Green-conjugated phalloidin. Separate monochrome images of the UV, red, and green fluorescence channels and a merged color image are shown. Bar, 5 μm.
TABLE 3.
Quantification of efficiency of actin polymerization triggered during bacterial infection
| Strain | Actin accretion beneath attached bacteria (% of adherent bacteria)a |
|---|---|
| ICC223 (O125:H6) | 3.8 ± 1b |
| E2348/69 (O127:H6) | 75 ± 4 |
| TUV 93-0 (O157:H7) | 70.6 ± 5 |
| ICC224(pICC368) | 3.7 ± 1.5b |
| E2348/69Δtir(pICC368) | 3.1 ± 0.6b |
| TUV 93-0Δtir(pICC368) | 73.3 ± 4.7 |
| ICC223(pICC369) | 81 ± 3.1 |
Data are means ± standard deviations of three independent experiments carried out in duplicate. The efficiency of actin accretion was quantified by counting the percentage of cell-associated bacteria which were also associated with aggregated F-actin staining. Regions of 5 to 20 attached bacteria per cell from 100 bacteria per coverslip were examined.
Weak actin recruitment was detected beneath the adherent bacteria but was distinct from the intensely stained F-actin pedestals triggered by strains E2348/69, TUV 93-0, TUV 93-0Δtir(pICC368), and ICC223(pICC369).
Sequence and functional analyses of TirEPEC_O125:H6.
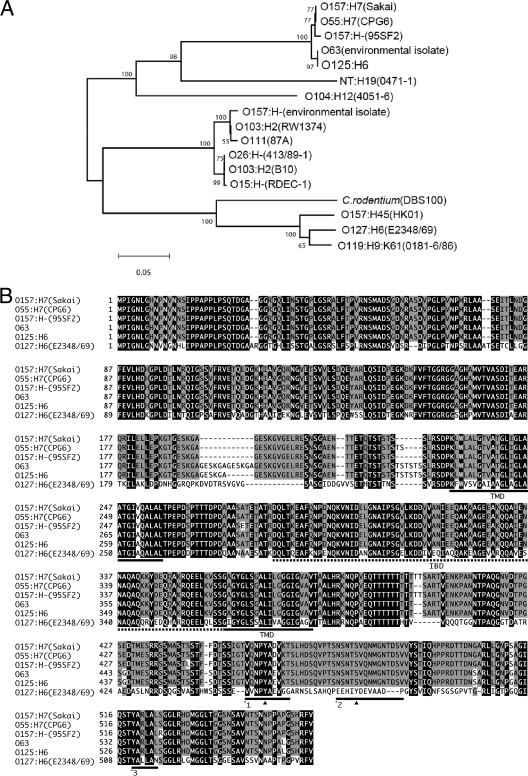
In order to characterize Tir of ICC223, the tir gene was amplified by PCR, cloned into pSA10 (generating plasmid pICC368), and sequenced (accession number AB355659). Multiple sequence alignment of the 569 amino acids of TirICC223 with representative Tir sequences in the database revealed that the two transmembrane domains and the intimin-binding domain were highly conserved. The phylogenetic relationship of TirEPEC_O125:H6 with available Tir molecules revealed that it is on the same main branch as TirEHEC_O157, TirEPEC_O55:H7, and TirEPEC_O63 (Fig. 2A). Moreover, TirICC223 shares 96.8% and 56.9% sequence identity with TirEHEC_O157:H7 and TirEPEC_O127:H6, respectively, and 100% sequence identity (with the exception of deletion of one of the GESKGA repeats) with TirEPEC_O63 of an environmental EPEC isolate belonging to serogroup O63 (Fig. 2B). In particular, the sequence confirms that TirEPEC_O125:H6 lacks the Y474 equivalent of TirEPEC_O127:H6 (Fig. 2B), which is consistent with our inability to detect phosphorylated TirEPEC_O125:H6 during infection (30). The NPY motif that is implicated in the inefficient Nck-independent actin polymerization pathway in EPEC O127:H6 and the TccP-mediated actin polymerization pathway in EHEC O157:H7 (1) is conserved in TirEPEC_O125:H6.
FIG. 2.
A. The phylogenetic relationship of the tir genes from EPEC O125:H6 with selected previously published tir genes. B. The amino acid sequence of Tir proteins of EPEC O125:H6 strain ICC223 was aligned with previously published Tir sequences as described elsewhere (30). The tir genes of amino acid residues identical in all the proteins are indicated in black, and the residues shared by no less than 50% identity within all proteins are gray. The intimin-binding domain and two predicted transmembrane domains are indicated by a dashed line and underlining, respectively. Black triangles indicate the tyrosine residues phosphorylated by a host cell kinase(s). Underlining with *1 indicates the regions containing Y454 that are involved in pedestal formation via the TirEHEC_O157:H7-TccP/EspFu pathway and the alternative TirEPEC_O127:H6-Nck-independent pathway. Underlining with *2 indicates the TirEPEC_O127:H6 Y474 involved in the Nck pedestal formation pathway. The underlining with *3 indicates the region corresponding to the O157 EHEC Tir residues 519 to 524 that may be related to the type III secretion system-dependent secretion efficiency.
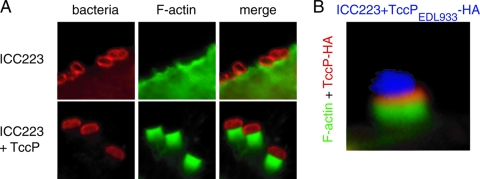
We next generated a tir deletion mutant in strain ICC223, generating strain ICC224. Complementing the ICC224 mutant with pICC368, which overexpresses TirEPEC_O125:H6, did not reveal Tir tyrosine phosphorylation, recruitment of Nck (data not shown), or conferred actin polymerization activity following infection of HeLa cells (Table 3). In contrast, when the mutant was complemented with a plasmid encoding TirEPEC_O127:H6 (pACYC-P) (18), we detected Tir tyrosine phosphorylation, recruitment of Nck, and efficient actin polymerization under attached ICC224 bacteria (data not shown). In a reciprocal experiment, pICC368 encoding TirEPEC_O125:H6 was used to complement TUV 93-0Δtir (22) and E2348/69Δtir (ICC225) (Table 1). While TUV 93-0Δtir was deficient in actin polymerization (data not shown), expression of TirEPEC_O125:H6 restored actin polymerization activity to the wild-type level (73.3%) (Fig. 1; Table 3). In contrast, only background actin staining was observed under E2348/69Δtir expressing TirEPEC_O125:H6 (Fig. 1; Table 3). Finally we transformed wild-type EPEC O125:H6 strain ICC223 with a plasmid encoding HA-tagged TccPEHEC_O157:H7 (pICC369). Infection of HeLa cells showed that ICC223 (stained with anti-O125 antiserum) expressing TccP can effectively trigger actin polymerization; anti-HA staining confirmed that TccP was concentrated at the tip of the pedestal (Fig. 3). Taken together these results show that EPEC O125:H6 strains have the potential to trigger actin polymerization in HeLa cells, provided that they are equipped with either TccP or with Tir that can undergo tyrosine phosphorylation. In this respect, wild-type EPEC O125:H6 exhibits a similar phenotype to that of EHEC O157:H7ΔtccP (11).
FIG. 3.
A. ICC223 binds to HeLa cells but cannot trigger efficient actin polymerization. Expression of TccPEHEC_O157:H7 confers strong actin polymerization activity. B. HA staining shows that TccP is concentrated under attached ICC223 bacteria.
EPEC O125:H6 induces A/E lesions on human intestinal in vitro organ cultures.
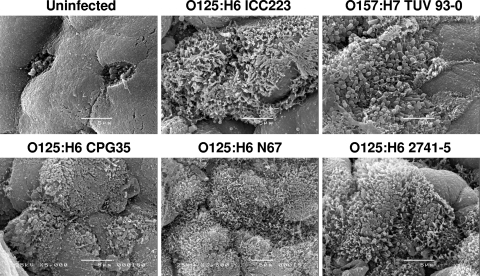
We previously showed that EPEC O127:H6 (E2348/69) expressing Tir-Y454F/Y474F (32) and EHEC O157:H7ΔtccP (11) can induce A/E lesions during infection of human IVOC. Accordingly, we tested if EPEC O125:H6 strain ICC223, which naturally lacks tccP/tccP2 and carries a non-tyrosine-phosphorylated Tir, can infect human IVOC ex vivo. Scanning electron microscopy analysis of 8-h IVOC samples showed that all EPEC O125:H6 strains adhered to human terminal ileum. Adherence patterns were similar to the EHEC strain TUV 93-0 control, with intimate bacterial attachment and microvillous elongation of the IVOC tissue between adhering bacteria (Fig. 4).
FIG. 4.
Scanning electron micrographs of EPEC O125:H6 strains (ICC223, 35, N67, and 2741-5) and EHEC O157:H7 (TUV 93-0) on human terminal ileum after 8 h of IVOC. All strains show intimate adherence to the mucosa, and microvillous elongation between attaching bacteria is evident. An uninfected sample was included as a negative control. Bar, 5 μm.
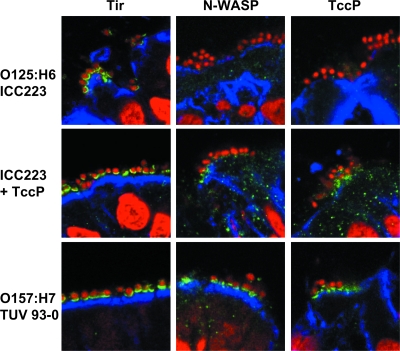
Immunofluorescence staining of cryosections revealed localization of translocated Tir underneath adherent ICC223 and TUV 93-0 (Fig. 5). In contrast, efficient N-WASP recruitment could be observed beneath adherent TUV 93-0, whereas only a minority of ICC223 bacteria showed a weak positive reaction. This phenotype is reminiscent of TUV 93-0ΔtccP, which also shows intimate adherence to the terminal ileum in the absence of N-WASP recruitment (11). Importantly, expression of TccP in ICC223 resulted in efficient recruitment of TccP and N-WASP at the site of bacterial attachment (Fig. 5). In addition, colonization of terminal ileum by ICC223 expressing TccP appeared to be enhanced compared to ICC223, as intimately adhering bacteria were detected on four of four biopsies infected with ICC223 expressing TccP, compared to two of four biopsies infected with ICC223. This result may suggest that expression of TccP, although not essential, may increase colonization efficiency.
FIG. 5.
Immunofluorescence staining of cryosections of human terminal ileum infected with EPEC O125:H6 (ICC223), its TccP-expressing derivative (ICC223 + TccP), and EHEC O157:H7 (TUV 93-0). Whereas all strains show Tir translocation (green) into the host cell membrane, N-WASP staining (green) can be observed underneath TccP-expressing TUV 93-0 and ICC223 + TccP but is only very weakly recruited beneath a minority of ICC223 bacteria. Sections were counterstained with propidium iodide (red) and anticytokeratin (blue) to visualize bacteria/cell nuclei and epithelial cells, respectively. Shown are merged images of all fluorescence channels.
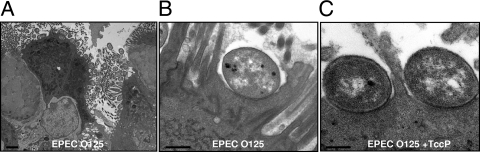
Finally, infected IVOC were analyzed by transmission electron microscopy. This revealed that while ICC223 can efficiently trigger A/E lesions (Fig. 6), the intensity of electron-dense staining under attached bacteria, indicating actin polymerization, was variable and much less profound than that seen after infection with wild-type EPEC O127:H6 or EPEC O127:H6 expressing Tir Y474S (32). ICC223 expressing TccP was associated with an increase in electron-dense material compared to ICC223 (Fig. 6C). These results suggest that EPEC O125:H6, like EHEC O157:H7ΔtccP, can cause intimate attachment and microvillous effacement without efficient recruitment of N-WASP or F-actin beneath adhering bacteria. Expression of TccP restores efficient recruitment of N-WASP and actin polymerization and increases the colonization efficiency of human intestinal IVOC.
FIG. 6.
Transmission electron microscopy of human IVOC infected with ICC223. A. ICC223 efficiently colonizes the gut mucosa. B. Typical A/E lesion with intimate bacterial attachment and effacement of brush border microvilli; increased electron density at the site of bacterial attachment (representing accumulated actin) is not apparent. C. ICC223 expressing a TccP A/E lesion, showing increased electron density in the epithelium at the site of attachment. Bars, 2 μm (A) or 0.5 μm (B and C).
DISCUSSION
The ability of typical EHEC O157:H7 and EPEC strains to trigger efficient actin polymerization on cultured cells is linked to activation of N-WASP. However, while typical EHEC O157:H7 strains use the effector protein TccP (5, 11), EPEC strains use the host adaptor protein Nck, which binds tyrosine-phosphorylated Tir (2, 12). Interestingly, non-O157 EHEC (29) and EPEC strains belonging to lineage 2 (34) can use both the Nck and TccP2 actin polymerization pathways, while atypical sorbitol-fermenting EHEC O157 expresses both TccP and TccP2 (29). The fact that EPEC and EHEC express what seem to be redundant mechanisms to efficiently trigger actin polymerization is suggestive of an essential role and of selective pressure to maintain this capability.
In this paper we have shown that EPEC O125:H6 expresses a Tir which naturally lacks a Y474 equivalent (19) and hence cannot trigger actin polymerization using the Nck pathway (2, 12). All the tested EPEC O125:H6 strains also naturally lack TccP and TccP2 (30) and hence cannot use these effector proteins to trigger efficient actin polymerization. Accordingly, as we observed during infection of HeLa cells, EPEC O125:H6 can only trigger inefficient actin polymerization, presumably by using the NPY motif, which is conserved in EPEC and EHEC strains (1). Expressing TccP in EPEC O125:H6 enabled the strain to efficiently trigger actin polymerization in infected HeLa cells. Consistent with these findings, we found that TirEPEC_O125:H6 is phylogenetically clustered with, and functionally interchangeable with, TirEHEC_O157:H7.
Importantly, we have demonstrated that in spite of their inability to trigger efficient actin polymerization on cultured HeLa cells, EPEC O125:H6 strains can infect human intestinal explants, intimately attach to the enterocytes, and trigger effacement of the brush border microvilli. These IVOC phenotypes parallel those reported for EHEC O157:H7ΔtccP (11), EPEC 1 strain E2348/69 O127:H6 expressing TirY474S (32), and C. rodentium (9). It therefore appears that during infection of mucosal surfaces neither Nck nor TccP is needed for A/E lesion formation. However, although the number of IVOC used was relatively small, we observed that EPEC O125:H6 expressing TccP colonizes the mucosa of the terminal ileum (four IVOC out of four) more efficiently than wild-type O125:H6 (two IVOC out of four). Moreover, infection of IVOC with O125:H6 expressing TccP resulted in detection of N-WASP at the site of bacterial attachment and accumulation of electron-dense material under attached bacteria, believed to be actin. A possible interpretation of the data is that colonization and A/E lesion formation can be achieved by EPEC and EHEC in the absence of efficient actin polymerization activity. However, the ability to efficiently polymerize actin might stabilize initial adhesion and increase the long-term colonization potential. Indeed, interfering with actin cell signaling seems to modulate the ability of EPEC to remain attached to IVOC, as EPECΔmap and EPECΔespH mutants detach from IVOC at a high frequency, leaving behind pedestal footprints (33). As Map and EspH cooperate with Tir in coordinating actin dynamics, our results suggest that timely and efficient polymerization of actin, although not essential for colonization, might provide a subtle advantage over EPEC and EHEC strains lacking this capability. In order to address this hypothesis experimentally, we are currently engineering site-directed Tir mutants in Citrobacter rodentium, the mouse pathogen equivalent of EPEC and EHEC (28), that will be used in competitive index studies with the wild-type strain.
The ability of EPEC and EHEC to trigger actin polymerization in cultured cells has been used for many years as the main virulence marker for EPEC and EHEC since Knutton et al. (21) developed the fluorescent actin staining (FAS) test. The current study shows that relying on the FAS test alone is not sufficient. While FAS-positive strains are likely to be pathogenic, locus of enterocyte effacement-positive strains that fail to trigger actin polymerization in vitro cannot be classified as nonpathogenic, and alternative assays should be employed. Indeed, the phenotype we described for EPEC O125:H6 is not uncommon. A previous study showed that 29% of eae-positive strains isolated from children in the United Kingdom were FAS negative on HEp-2 cells but produced typical A/E lesions on human IVOC (23). These findings reinforce the important differences in signal transduction between cultured epithelial cells and mucosal surfaces (32) and suggest the existence of an important subgroup of EPEC strains that utilize a TccP- and Nck-independent pathway to adhere and trigger A/E lesion formation on mucosal surfaces.
Acknowledgments
We thank Junkal Garmendia for the HA-TccP construct, Silvia Lommel, Institute for Cell Biology, University of Bonn, Bonn, Germany, for the anti-N-WASP antibody, and Roberto La Ragione, VLA, United Kingdom, for anti-O125.
Work in the laboratory of A.D.P. was supported by the NIH (grant R37AI21657 to J. B. Kaper). The work in the laboratory of G.F. was supported by a BBSRC China partnering award and the Wellcome Trust.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 5 November 2007.
REFERENCES
- 1.Brady, M. J., K. G. Campellone, M. Ghildiyal, and J. M. Leong. 23 May 2007. Enterohaemorrhagic and enteropathogenic Escherichia coli Tir proteins trigger a common Nck-independent actin assembly pathway. Cell. Microbiol. doi: 10.1111/j.1462-5822.2007.00954.x. [DOI] [PubMed]
- 2.Campellone, K. G., A. Giese, D. J. Tipper, and J. M. Leong. 2002. A tyrosine-phosphorylated 12-amino-acid sequence of enteropathogenic Escherichia coli Tir binds the host adaptor protein Nck and is required for Nck localization to actin pedestals. Mol. Microbiol. 431227-1241. [DOI] [PubMed] [Google Scholar]
- 3.Campellone, K. G., and J. M. Leong. 2005. Nck-independent actin assembly is mediated by two phosphorylated tyrosines within enteropathogenic Escherichia coli Tir. Mol. Microbiol. 56416-432. [DOI] [PubMed] [Google Scholar]
- 4.Campellone, K. G., S. Rankin, T. Pawson, M. W. Kirschner, D. J. Tipper, and J. M. Leong. 2004. Clustering of Nck by a 12-residue Tir phosphopeptide is sufficient to trigger localized actin assembly. J. Cell Biol. 164406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campellone, K. G., D. Robbins, and J. M. Leong. 2004. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7217-228. [DOI] [PubMed] [Google Scholar]
- 6.Caron, E., V. F. Crepin, N. Simpson, J. Garmendia, and G. Frankel. 2006. Subversion of actin dynamics by EPEC and EHEC. Curr. Opin. Microbiol. 940-45. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H. D., and G. Frankel. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 2983-98. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, W., B. A. Vallance, Y. Li, J. L. Puente, and B. B. Finlay. 2003. Citrobacter rodentium translocated intimin receptor (Tir) is an essential virulence factor needed for actin condensation, intestinal colonization and colonic hyperplasia in mice. Mol. Microbiol. 4895-115. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Investig. 107539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garmendia, J., A. Phillips, Y. Chong, S. Schuller, O. Marches, S. Dahan, E. Oswald, R. K. Shaw, S. Knutton, and G. Frankel. 2004. TccP is an enterohaemorrhagic E. coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell. Microbiol. 61167-1183. [DOI] [PubMed] [Google Scholar]
- 12.Gruenheid, S., R. DeVinney, F. Bladt, D. Goosney, S. Gelkop, G. D. Gish, T. Pawson, and B. B. Finlay. 2001. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3856-859. [DOI] [PubMed] [Google Scholar]
- 13.Hartland, E. L., M. Batchelor, R. M. Delahay, C. Hale, S. Matthews, G. Dougan, S. Knutton, I. Connerton, and G. Frankel. 1999. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 32151-158. [DOI] [PubMed] [Google Scholar]
- 14.Hicks, S., G. Frankel, J. B. Kaper, G. Dougan, and A. D. Phillips. 1998. Role of intimin and bundle foming pili in enteropathgenic Escherichia coli adhesion to paediatric intestine in vitro. Infect. Immun. 661570-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 927996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 877839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karch, H., P. I. Tarr, and M. Bielaszewska. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295405-418. [DOI] [PubMed] [Google Scholar]
- 18.Kenny, B. 2001. The enterohaemorrhagic Escherichia coli (serotype O157:H7) Tir molecule is not functionally interchangeable for its enteropathogenic E. coli (serotype O127:H6) homologue. Cell. Microbiol. 3499-510. [DOI] [PubMed] [Google Scholar]
- 19.Kenny, B. 1999. Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol. 311229-1241. [DOI] [PubMed] [Google Scholar]
- 20.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91511-520. [DOI] [PubMed] [Google Scholar]
- 21.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 571290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutton, S., D. R. Lloyd, and A. S. McNeish. 1987. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect. Immun. 5569-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knutton, S., R. Shaw, A. D. Phillips, H. R. Smith, G. A. Willshaw, P. Watson, and E. Price. 2001. Phenotypic and genetic analysis of diarrhea-associated Escherichia coli isolated from children in the United Kingdom. J. Pediatr. Gastroenterol. Nutr. 3332-40. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 25.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, and S. Sotman. 1978. Escherichia coli that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:119-122. [DOI] [PubMed] [Google Scholar]
- 26.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 921664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 411340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mundy, R., T. T. MacDonald, G. Dougan, G. Frankel, and S. Wiles. 2005. Citrobacter rodentium of mice and man. Cell. Microbiol. 71697-1706. [DOI] [PubMed] [Google Scholar]
- 29.Ogura, Y., T. Ooka, A. Whale, J. Garmendia, L. Beutin, S. Tennant, G. Krause, S. Morabito, I. Chinen, T. Tobe, H. Abe, R. Tozzoli, A. Caprioli, M. Rivas, R. Robins Browne, T. Hayashi, and G. Frankel. 2007. TccP2 of O157:H7 and non-O157 enterohemorrhagic Escherichia coli (EHEC): challenging the dogma of EHEC-induced actin polymerization. Infect. Immun. 75604-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ooka, T., M. A. M. Vieira, Y. Ogura, L. Beutin, R. L. Ragione, P. M. van Diemen, M. P. Stevens, I. Aktan, S. Cawthraw, A. Best, R. T. Hernandes, G. Krause, T. A. T. Gomes, T. Hayashi, and G. Frankel. 2007. Characterisation of tccP2 carried by atypical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 271126-135. [DOI] [PubMed] [Google Scholar]
- 31.Schlosser-Silverman, E., M. Elgrably-Weiss, I. Rosenshine, R. Kohen, and S. Altuvia. 2000. Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J. Bacteriol. 1825225-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schüller, S., Y. Chong, J. Lewin, B. Kenny, G. Frankel, and A. D. Phillips. 2007. Tir phosphorylation and Nck/N-WASP recruitment by enteropathogenic and enterohaemorrhagic Escherichia coli during ex vivo colonization of human intestinal mucosa is different to cell culture models. Cell. Microbiol. 91352-1364. [DOI] [PubMed] [Google Scholar]
- 33.Shaw, R. K., J. Cleary, G. Frankel, and S. Knutton. 2005. Enteropathogenic Escherichia coli interaction with human intestinal mucosa: role of effector proteins in brush border remodelling and attaching and effacing lesion formation. Infect. Immun. 731243-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whale, A. D., R. T. Hernandes, O. Tadasuke, B. L., S. Schüller, J. Garmendia, L. Crowther, M. A. M. Vieira, Y. Ogura, G. Krause, A. D. Phillips, T. A. T. Gomes, T. Hayashi, and G. Frankel. 2007. TccP2-mediated subversion of actin dynamics by EPEC 2a distinct evolutionary lineage of enteropathogenic Escherichia coli. Microbiology 1531743-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whittam, T. S., and E. A. McGraw. 1996. Clonal analysis of EPEC serogroups. Rev. Microbiol. Sao Paulo 27(Suppl. l)7-16. [Google Scholar]