Abstract
Burkholderia cenocepacia strain K56-2 typically has rough colony morphology on agar medium; however, shiny colony variants (shv) can appear spontaneously. These shv all had a minimum of 50% reduction in biomass formation and were generally avirulent in an alfalfa seedling infection model. Three shv—K56-2 S15, K56-2 S76, and K56-2 S86—were analyzed for virulence in a chronic agar bead model of respiratory infection and, although all shv were able to establish chronic infection, they produced significantly less lung histopathology than the rough K56-2. Transmission electron microscopy revealed that an extracellular matrix surrounding bacterial cells was absent or reduced in the shv compared to the rough wild type. Transposon mutagenesis was performed on the rough wild-type strain and a mutant with an insertion upstream of ORF BCAS0225, coding for a putative LysR-type regulator, exhibited shiny colony morphology, reduced biofilm production, increased N-acyl homoserine lactone production, and avirulence in alfalfa. The rough parental colony morphotype, biofilm formation, and virulence in alfalfa were restored by providing BCAS0225 in trans in the BCAS0225::pGSVTp-luxCDABF mutant. Introduction of BCAS0225 restored the rough morphotype in several shv which were determined to have spontaneous mutations in this gene. In the present study, we show that the conversion from rough wild type to shv in B. cenocepacia correlates with reduced biofilm formation and virulence, and we determined that BCAS0225 is one gene involved in the regulation of these phenotypes.
The Burkholderia cepacia complex (Bcc) is a group of closely related species that are ubiquitous in the environment and are also a particularly problematic source of respiratory infections in cystic fibrosis (CF) patients (33). To date, the Bcc comprises at least nine defined bacterial species that have been isolated from the sputa of CF patients, as well as from the environment (12, 13, 30, 31). Burkholderia cenocepacia is one of the most prevalent Bcc species in CF patients, and evidence suggests it is the most virulent (41, 50). The factors that allow B. cenocepacia to establish infection and persist in the lungs of CF patients remain relatively undefined.
Recently, it was shown that B. cenocepacia strain C1394 could undergo colony variation from a matte to a shiny colony morphotype through passage in mice (11). This colonial transformation was accompanied by hyperpiliation and increased production of exopolysaccharides (EPS) (11). The shiny variant was more persistent in a neutropenic mouse lung infection model than the original matte isolate (11). The switch in colony morphotypes may therefore represent a part of the adaptation mechanisms that are triggered by the conditions encountered in the host (11).
Variation in colony morphology, which may or may not be reversible, has been reported for many species of bacteria. Conversion from smooth to rugose in Vibrio cholerae (54, 58, 60) and Salmonella enterica serovar Typhimurium (2), from opaque to translucent in V. parahaemolyticus (20, 21, 34), from smooth to wrinkly and fuzzy spreaders in Pseudomonas fluorescens (39), from smooth to wrinkly in Pseudomonas aeruginosa (14), from rough to smooth in Actinobacillus actinomycetemcomitans (24), and the appearance of small colony variants in P. aeruginosa (19) are examples of modifications in colony morphology.
Other bacterial behaviors are associated with changes in colony morphology including motility, biofilm formation (19, 54, 60), and resistance to antibiotics (19), chlorine (60), and osmotic or oxidative stresses (54). However, these altered phenotypes are not always directly related to the change in colony morphology but rather result from the original mutation causing the colonial transformation, since these mutations often occur in transcriptional regulators. Colony morphology variation can also alter virulence as has been reported in Ralstonia solanacearum (38) and in V. cholerae (56, 62). V. cholerae has the ability to reversibly alter its colony morphology, switching between two forms, smooth and rugose. The rugose variant is more resistant to chlorine, acid, UV light, and complement-mediated bactericidal activities than the smooth wild-type form (36, 42, 60). Rugose strains are virulent, and disease symptoms have been demonstrated in rabbits and human volunteers infected with these strains (36, 42, 60).
In the present study, we describe the spontaneous appearance of shiny variants (shv) in B. cenocepacia strain K56-2. Since colony morphology variation has previously been shown to affect virulence in B. cenocepacia (11), selected shv were evaluated for virulence in both plant and animal infection models. A putative transcriptional regulator encoded by the BCAS0225 gene was identified that when mutated resulted in a shiny colony morphotype, as well as a defect in biofilm production and virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the present study are described in Table 1. The bacterial strains used here were routinely grown in LB broth or on 1.5% Lennox L agar (LB; Invitrogen, Burlington, Ontario, Canada) supplemented with antibiotics when appropriate and incubated at 37°C unless specified otherwise. Antibiotics were used at the following concentrations: 100 μg of trimethoprim (Tp)/ml, 200 μg of tetracycline (Tc)/ml for B. cenocepacia K56-2 and 100 μg of Tp/ml, 50 μg of gentamicin (Gm)/ml, and 50 μg of kanamycin (Km)/ml for Escherichia coli. Agrobacterium tumefaciens strains were grown at 28°C for 24 to 48 h with the addition of 25 μg of Km/ml and 4.5 μg of Tc/ml for A. tumefaciens A136(pMV26)(pCF218) (9) and 4.5 μg of Tc/ml and 50 μg of spectinomycin/ml for A. tumefaciens A136(pCF218)(pCF372) (61). For N-acyl homoserine lactone (AHL) extraction and detection assays, B. cenocepacia cultures were grown in Trypticase soy broth (TSB; Becton Dickinson, Sparks, MD). For protease assays, overnight cultures were grown in LB. For the Chrome Azurol S assay, cultures were grown at 32°C in succinate medium supplemented with 10 mM ornithine. For swarming and swimming motility assays, cultures were grown in nutrient broth (NB; Becton Dickinson) supplemented with 0.5% glucose. For alfalfa inoculation, overnight cultures were grown in LB with shaking (Invitrogen) in a 96-well plate. For animal experiments, cultures were grown in dialyzed and chelated TSB-DC (37) medium overnight. All chemicals were purchased from Sigma-Aldrich Canada, Ltd., (Oakville, Ontario, Canada).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| HB101 | supE44 hsdS20(rB mB) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 45 |
| TOP10 | F−mcrA Δ(mrr-hsdRMS mcrBC) φ80lacΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Smr) endA1 nupG | Invitrogen |
| A. tumefaciens A136 | Ti plasmidless host | C. Fuqua |
| B. cenocepacia | ||
| K56-2 | CF sputum isolate (Canada); ET12 lineage; cblA+recA subgroup A | 32 |
| K56-2 S1 to S93 | shv obtained in vitro | This study |
| K56-2 BCAS0225::Tn | BCAS0225::TnRhaBout derivative of K56-2; Tpr | This study |
| K56-2 BCAS0225::pGSVTp-luxCDABE | K56-2 with pGSVTp-luxCDABE inserted in BCAS0225; Tpr | This study |
| K56-2 BCAM1200::pGSVTp-luxCDABE | K56-2 with pGSVTp-luxCDABE inserted in BCAM1200; Tpr | This study |
| Plasmids | ||
| pCF218 | IncP plasmid expressing TraR; Tcr | 61 |
| pCF372 | pUCD2 with traI-lacZ fusion; Spra | 23 |
| pMV26 | traI-luxCDABE fusion; Kmr | 9, 49 |
| pRK2013 | Mobilizing vector, ColE1 Tra (RK2); Kmr | 22 |
| pUCP26 | Broad-host-range vector; Tcr | 57 |
| pSCrhaBout | pTnMod-OTp′, rhaR rhaS PrhaB; Tpr | 6 |
| pGSVTp-lux | Mobilizable suicide vector containing lux operon, derivative from pGSV3-lux (35); OriT; Tpr | C. Kooi |
| pGSVTp-lux-BCAS0225 | pGSVTp-lux with 563-bp EcoRI internal fragment of BCAS0225; Tpr | This study |
| pGSVTp-lux-BCAM1200 | pGSVTp-lux with 343-bp EcoRI fragment of BCAM1200; Tpr | This study |
| pUCP26-BCAS0225 | pUCP26 with 1.7-kb PstI-BamHI fragment containing BCAS0225 and upstream region; Tcr | This study |
Spr, spectinomycin resistance.
DNA manipulations.
Restriction endonucleases and T4 DNA polymerase were purchased from Invitrogen. T4 ligase was purchased from New England Biolabs (Mississauga, Ontario, Canada) or Invitrogen. Oligonucleotide primers (data not shown) were synthesized at the University of Calgary Core DNA and Protein Services (Calgary, Alberta, Canada). PCR was performed with either Platinum Taq DNA polymerase or Platinum Taq DNA Polymerase High Fidelity (Invitrogen). DNA fragments used for cloning were purified by using the QIAquick gel extraction kit (Qiagen, Mississauga, Ontario, Canada). The University of Calgary Core DNA and Protein Services performed nucleotide sequencing and sequence analysis was performed with the aid of DNAMAN sequence analysis software (Lynnon BioSoft, Vandreuil, Quebec, Canada).
Isolation of shv of B. cenocepacia K56-2.
One isolated K56-2 rough colony grown on LB agar was inoculated into 5 ml of fresh LB and incubated at 37°C for approximately 24 h with shaking (∼250 rpm). The following day, a 100-μl aliquot was serially diluted into fresh LB, plated onto LB agar plates (Invitrogen), and incubated at 37°C for 48 h. After 48 h, agar plates were put at room temperature for up to 24 h in order to distinguish the shv from the rough colonies. Isolated K56-2 shv were transferred into 100 μl of LB in a 96-well plate format, incubated at 37°C overnight with shaking, and frozen in glycerol to a final concentration of 15% (vol/vol).
In vitro stability of the colony morphotypes.
To evaluate the in vitro stability of both morphotypes, a single isolated colony from each of the rough colonies and shv of K56-2 grown on LB agar were inoculated into 5 ml of LB, followed by incubation for approximately 24 h (one passage) with shaking or for 72 h (one passage) with no shaking. After each passage, the cultures were thoroughly mixed, and a 100-μl aliquot was used to inoculate 5 ml of fresh medium, followed by incubation as described above. The process was repeated for periods of 14 days for the shaking cultures (14 passages) and 21 days for the static cultures (7 passages). After each passage, cultures were serially diluted and plated onto LB agar to determine the percentages of shv and rough colonies.
Congo red binding assays.
Cultures grown in 96-well microtiter plates were inoculated manually (2 μl) or with a 48-pin replicator onto LB agar plates containing Congo red dye at either 100 μg/ml or 0.01% (wt/vol). Plates were incubated at 37°C for 24 to 48 h, and the binding properties were determined by the color of the resulting colony. Red was indicative of high affinity, and pink and/or white was indicative of a decrease in binding affinity.
Phenotypic assays.
Protease activity was determined as previously described by using skim milk as a substrate (48). Briefly, overnight cultures were subcultured (1:100) into 10 ml of fresh LB medium until mid-log phase of growth, normalized to an optical density at 600 nm of 0.3, and spot inoculated (2 μl) in triplicate or with a 48-pin replicator onto skim milk agar. Plates were incubated for 24 h and analyzed for zones of clearing around the spotted colony. Siderophore activity present in culture supernatants was measured by using Chrome Azurol S assays as previously described (27). Motility assays were performed as previously described (4, 29). Assays were performed in triplicate and repeated at least twice with similar results.
AHL extraction, thin-layer chromatography, and luminescence bioassays.
AHLs were extracted from culture supernatants as previously described and AHL profiles were determined by thin-layer chromatography-AHL bioassays using A. tumefaciens A136(pCF218)(pCF372) as the reporter strain (28). Synthetic N-hexanoyl-l-homoserine lactone and N-octanoyl-l-homoserine lactone (Sigma-Aldrich) were used as reference standards. For the detection and monitoring of AHL production, a bioluminescence agar plate assay was developed using the A. tumefaciens strain A136(pCF218)(pMV26) as the reporter strain (9, 49). An overnight culture of the reporter strain was mixed in a ratio of 1:80 (vol/vol) with TSB containing 0.7% agar (wt/vol). Each plate was made with 20 ml of the TSB agar reporter strain mixture and allowed to dry for approximately 2 h at room temperature before use. Then, 2 μl of an overnight culture grown in TSB of the strains to be tested was spotted onto the plate, allowed to dry for 20 min, and incubated at 28°C for 24 h. Luminescence was detected by using a Fluorchem 8900 digital camera system.
Biofilm on abiotic surface and pellicle formation.
Biofilm assays on abiotic surfaces were performed as previously described (4) by growing bacteria in 96-well microtiter plates (Nunc, Roskilde, Denmark) covered with a 96-peg lid (Nunc) and quantitating the biomass formed on the polystyrene pegs. For pellicle formation, colonies were inoculated into a borosilicate glass tubes (16 by 150 mm) containing 5 ml of fresh LB and incubated statically at 37°C for 5 days. The pellicle at the air-liquid interface was qualitatively evaluated.
TEM.
For transmission electron microscopy (TEM), 3- to 5-day-old colonies grown on LB agar plates were removed from the agar surface by cutting them out with a thin layer of agar. TEM was performed by the University of Calgary Microscopy and Imaging Facility (http://microscopy.myweb.med.ucalgary.ca/). Briefly, colonies were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 1 h at room temperature. Samples were washed three times in 0.1 M cacodylate buffer (pH 7.4) postfixed in 1% osmium tetroxide buffered with 0.1 M cacodylate for 1 h at room temperature. Samples were then rinsed twice briefly with distilled water and stained en bloc for 30 min in 0.5% aqueous uranyl acetate. Samples were then dehydrated in an ethanol series and embedded in Spurr's resin. Thin sections were cut with a diamond knife on a Reichert Ultracut E and stained with uranyl acetate and lead citrate. Sample analysis was performed using a Hitachi H-7000 transmission electron microscope at 75 kV.
Alfalfa infection assay.
Briefly, the seedlings were germinated (3) and placed in 24-well plates (Becton Dickinson) containing 2 ml of water agar instead of petri dishes as previously described (3). A total of 30 μl of overnight cultures grown in LB at 37°C with shaking in a 96-well plate was used to surface inoculate three unwounded seedlings per well. For complementation experiments, Tc was included in the culture medium, and the bacteria were subsequently pelleted and resuspended in LB prior to inoculation onto seedlings. The 24-well plates containing seedlings were sealed with parafilm and incubated in a warm room (37°C) under a desk lamp producing artificial light. The seedlings were visually monitored for disease symptoms at 5 days postinfection (p.i.). Strains were tested with 12 seedlings in at least two assays with similar results, and the results presented are from one assay. The recovery of bacteria from infected plants was performed as previously described (3).
Animal studies.
Sprague-Dawley rats (150 to 175 g; Charles River Canada, Inc.) were tracheotomized under anesthesia and inoculated with approximately 104 CFU of the appropriate strain embedded in agar beads as previously described (7). At 14 days p.i., the lungs from four to five animals from each group were removed aseptically and homogenized (Polytron Homogenizer; Brinkman Instruments, Westbury, NY) in 3 ml of phosphate-buffered saline (PBS; 10 mM sodium phosphate, 150 mM NaCl [pH 7.5]). The homogenates were serially diluted in PBS and plated on B. cepacia selective agar (26). The lungs of four to five additional animals from each group were removed en bloc, fixed in 10% formalin, and examined for quantitative pathological changes. Infiltration of the lung with inflammatory cells and exudates was measured as previously described (3, 4).
Transposon mutagenesis and identification of insertion sites.
Rhamnose-dependent shv mutants of B. cenocepacia K56-2 were generated by using the plasmid pSCrhaBout as previously described (6). Briefly, the plasmid pSCrhaBout was conjugated into B. cenocepacia K56-2 by triparental mating using pRK2013 as the mobilizing plasmid (22). Transconjugants were selected on LB agar plates containing 100 μg of Tp/ml, 50 μg of Gm/ml, and 0.2% glucose. Colonies that exhibited a shiny colony morphology were selected, cultured in LB medium containing 100 μg of Tp/ml in 96-well format, and replica plated onto LB agar with 100 μg of Tp/ml and either 0.2% glucose or 0.2% rhamnose. Mutants that appeared shiny on LB agar plates containing glucose and rough on LB agar plates containing rhamnose were selected for further analysis. Genomic DNA was isolated from the mutants by using the Promega Wizard Genomic DNA purification kit, digested with NotI, self-ligated with T4 DNA ligase (Invitrogen), and transformed into E. coli DH5α competent cells. Transformants were selected on LB agar plates containing 100 μg of Tp/ml. The locations of the transposon insertion sites were determined by comparing the DNA sequences obtained by sequencing with primer 824 (6) with the genome of B. cenocepacia J2315 by BLAST (www.sanger.ac.uk/Projects/B_cenocepacia/). The annotated region of surrounding the insertion site was determined by using Artemis software (44).
Mutant construction (luxCDABE fusion construction).
The genes BCAS0225 and BCAM1200 in B. cenocepacia K56-2 were inactivated by the insertion of pGSVTp-lux. This vector was constructed from pGSV3-lux (35) by deleting the Gm resistance cassette at the SstI site and inserting a 708-bp SstI fragment containing the Tp resistance cassette from p34S-Tp (18). A 563-bp internal region of BCAS0225 was amplified by using the primers 5′-EcoRI-BCAS0225 and 3′-EcoRI-BCAS0225, while a 343-bp region of BCAM1200 was amplified with the primers 5′-EcoRI-BCAM1200 and 3′-EcoRI-BCAM1200. The amplification products were subcloned into pCR2.1-TOPO and transformed into TOP10F′ One-Shot E. coli DH5α competent cells (Invitrogen). The EcoRI fragment was subsequently cloned into pGSVTp-lux and mobilized into K56-2 (22). Transconjugants were selected on LB agar plates containing 100 μg of Tp/ml and 50 μg of Gm/ml. Insertions at the correct loci were verified by PCR using the primer combinations luxCR and 5′-EcoRI-BCAS0225 or 3′-EcoRI-BCAS0225 for BCAS0225 and with the primers 5′-EcoRI-BCAM1200 or 3′-EcoRI-BCAM1200 for BCAM1200.
Cloning of BCAS0225 and complementation of shv with BCAS0225.
A 1,740-bp fragment containing the BCAS0225 gene was amplified from K56-2 genomic DNA using Platinum Taq DNA Polymerase High Fidelity, with the primers 5′-PstI-BCAS0225 and 3′-BamHI-BCAS0225 and then subcloned into pCR2.1-TOPO and transformed into TOP10F′ One-Shot E. coli DH5α Competent Cells. A PstI-BamHI fragment containing BCAS0225 and the upstream region was then cloned into pUCP26 (57) digested with the same enzymes to generate pUCP26-BCAS0225. Competent cells were prepared from BCAS0225::pGSVTp-luxCDABE and K56-2 shv as previously described (17), and pUCP26-BCAS0225 was introduced by electroporation.
Statistical analysis.
Analysis of variance (ANOVA) was performed with INSTAT software (GraphPad Software). A P value of <0.05 was considered statistically significant.
RESULTS
Isolation of shv of K56-2.
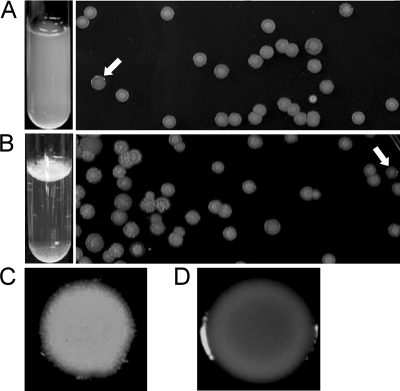
The colony morphology of K56-2 on agar is usually rough, but shv can often be detected (Fig. 1). Cultures have a homogenous distribution when grown in liquid medium with shaking and, when plated on agar medium, both rough and shv were identified (Fig. 1A). K56-2 cultures grown statically in liquid medium formed a pellicle at the air-liquid interface (Fig. 1B). When this pellicle was plated on agar medium, three colony morphotypes were identified: the typical rough colony, shv, and a larger flat rough colony (Fig. 1B). The K56-2 shv had a uniform, smooth, and shiny appearance (Fig. 1D), whereas rough colonies developed a pattern within the colony over time and had fuzzy edges (Fig. 1C).
FIG. 1.
Isolation of shv from shaken (A) and static (B) cultures of K56-2. The white arrows indicate the shv. Representative rough (C) and shv (D) colonies at higher magnification are also shown.
Stability of the colony morphotypes of K56-2.
To determine the frequency of the appearance of shv, the percentage of shv versus rough colonies obtained on agar medium was monitored after serial passages in both shaken and static liquid cultures. In shaken cultures, the rough morphotype was relatively stable over a period of 14 days, with <1% of the population consisting of shv (data not shown). In static cultures, 100% of the colonies obtained from the first three passages were rough, but the shv population increased with each of the following passages, suggesting that the shiny morphotype is more adapted for this growth environment (data not shown). The shv evaluated were stable in both shaken and static growth conditions over the same period of time as mentioned above (data not shown). The growth rate of representative rough colonies and shv were determined to be the same in shaking cultures (data not shown), indicating that differences in isolation frequency were not due to one morphotype outgrowing the other.
Phenotypic clustering of K56-2 shv.
A number of phenotypes were examined in isolated shv to determine whether they differed between shv and rough colony morphotypes. Ninety-three shv were characterized for Congo red binding, biofilm formation on polystyrene pegs, pellicle formation, protease activity, AHL production, swarming and swimming motility, and virulence in alfalfa. Variation in protease activity, AHL production, and virulence in alfalfa were used as parameters for clustering the shv into 15 distinct groups (data not shown). Features shared by all shv were the reduced ability to bind Congo red and reduction in biofilm formation, and 80% were avirulent in the alfalfa infection model (data not shown). Representative shv from seven of the phenotypic groups (data not shown) were selected for further analysis. These shv exhibited differences in protease activity, AHL production, virulence, siderophore production, and motility (data not shown).
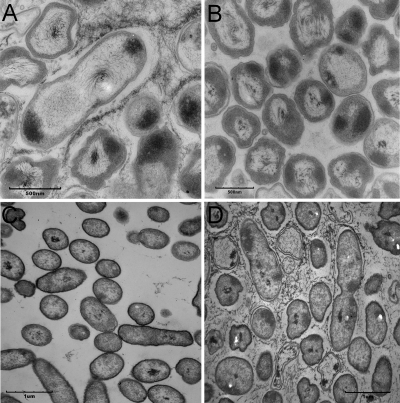
Smooth, flat colonies on agar surfaces generally have a reduction in the production of the extracellular matrix and are frequently biofilm defective (5). All seven shv had at least a 50% reduction in biofilm formation on polystyrene pegs (P < 0.01 [ANOVA]) compared to the rough parent strain (data not shown). Six of seven shv could form a pellicle-like structure at the air-liquid interface (data not shown). However, pellicles formed by the shv were not rigid as those formed by the rough K56-2 (data not shown). To determine whether reduction in biofilm formation correlated with a reduction in EPS-like matrix, representative rough colonies and shv colonies grown on agar were analyzed by TEM to identify ultrastructural differences. An extracellular matrix surrounded cells from rough colonies (Fig. 2A) but was absent in shv K56-2 S76 (Fig. 2B), as well as in K56-2 S15 and K56-2 S86 (data not shown).
FIG. 2.
Transmission electron micrographs of K56-2 rough colony showing the presence of an extracellular matrix that is absent in the shv (A), K56-2 S76 (B), K56-2 BCAS0225::pGSVTp-luxCDABE (pUCP26) (C), and K56-2 BCAS0225::pGSVTp-luxCDABE (pUCP26-BCAS0225) (D). Bars represent 500 nm in panels A and B and 1 μm in panels C and D.
Effect of colony morphology on virulence in plants and animals.
The selected shv, with the exception of K56-2 S15, were avirulent in the alfalfa seedling infection model, as evidenced by the absence of symptoms of disease (Table 2). The number of bacteria recovered 5 days p.i. from infected seedlings varied between the shv (Table 2) and was lower than K56-2 for K56-2 S60, K56-2 S62, K56-2 S76, K56-2 S86, and K56-2 S92 by almost 1 log unit (P < 0.05 [ANOVA]). The numbers of K56-2 S15 and K56 S39 recovered were similar to those for rough K56-2 (Table 2).
TABLE 2.
Virulence of the B. cenocepacia K56-2 shv in the alfalfa seedling infection model
| Strain | Symptoms of disease (%)a | Mean CFU ± SDb |
|---|---|---|
| K56-2 (rough parent) | 100 | 9.82 × 107 ± 5.44 × 107 |
| K56-2 S15 | 100 | 9.29 × 107 ± 1.54 × 107 |
| K56-2 S39 | 0 | 7.20 × 107 ± 3.37 × 107 |
| K56-2 S60 | 0 | 1.94 × 107 ± 2.89 × 106** |
| K56-2 S62 | 0 | 1.82 × 107 ± 3.92 × 106** |
| K56-2 S76 | 0 | 2.17 × 107 ± 3.50 × 106** |
| K56-2 S86 | 0 | 3.71 × 107 ± 2.09 × 107* |
| K56-2 S92 | 0 | 1.32 × 107 ± 1.29 × 106** |
That is, the percentage of seedlings demonstrating symptoms of disease 5 days p.i. as previously described (3).
Mean CFU recovered from three infected alfalfa seedlings. Experiments were repeated at least twice with similar results. Results significantly different than K56-2, as determined by ANOVA, are indicated by asterisks: *, P < 0.05; and **, P < 0.01.
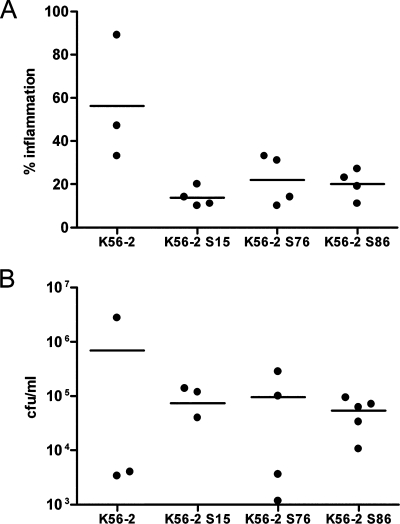
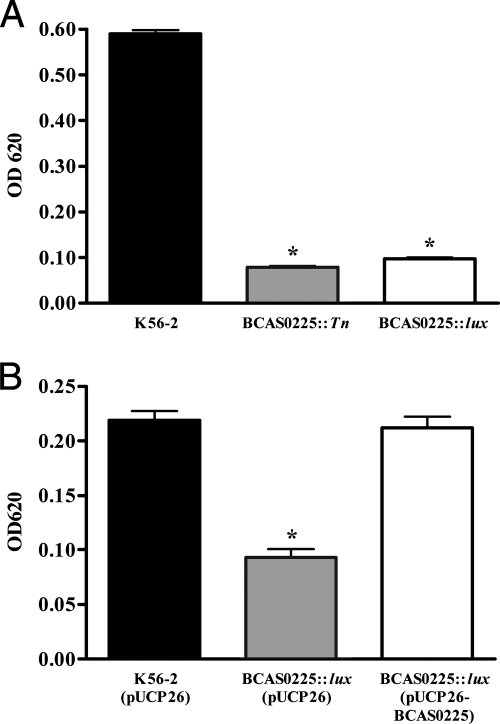
We evaluated the virulence of three shv—K56-2 S15, K56-2 S76, and K56-2 S86—using the rat agar bead model (7). K56-2 S15 was fully virulent in alfalfa (Table 2), but did not produce detectable AHLs or protease (data not shown). K56-2 S76 was representative of a group with parental protease activity but had increased AHL production (data not shown). K56-2 S86 had reduced expression of all virulence traits analyzed (data not shown). Rats infected with each of these shv had a significant decrease in quantitative lung histopathology compared to rough K56-2 (P < 0.05 [ANOVA]) (Fig. 3A). There was no significant difference in the numbers of bacteria recovered from the lungs of infected animals (Fig. 3B), although in most cases the number of CFU recovered from the rats infected with shv were higher than those infected with the wild type. No shv were recovered from rats infected with the rough K56-2, and no rough colonies were recovered from animals infected with shv, demonstrating that the morphotypes were stable in animals (data not shown).
FIG. 3.
Virulence of K56-2 shv in rats. (A) Scatter plot of the percentage of the lung with inflammatory exudates at 14 days p.i. detected in stained sections of the left lung from three to four animals per bacterial strain. K56-2 S15 and K56-2 S76 (P < 0.01 [ANOVA]) and K56-2 S86 (P < 0.05 [ANOVA]) were significantly different than K56-2. (B) Scatter plot of the CFU recovered 14 days p.i. from the lungs of four to five animals per bacterial strain. One animal each infected with K56-2 and K56-2 S15 cleared bacteria from their lungs.
Identification of a gene involved in the conversion from rough to shiny colony morphology.
We used a transposon mutagenesis approach with pSCrhaBout, which delivers an outward-oriented rhamnose-inducible promoter (6), to identify genes involved in the conversion from rough to shiny morphotypes. The insertion of the PrhaB promoter upstream of a gene involved in the colony morphotype conversion should result in shv that would be restored to the rough morphotype on medium with rhamnose and make it possible to differentiate between mutations that occurred spontaneously and those resulting from transposon insertions. Approximately 96,000 transconjugants were analyzed, and those that exhibited shv morphology on LB agar containing glucose were replica plated onto plates containing rhamnose or glucose. Of the 361 Tn-shv screened in this manner, 36 appeared shiny on glucose and rough on rhamnose (data not shown). To confirm that the Tn-shv mutants had properties similar to those of the spontaneously isolated shv previously characterized, the Tn-shv were analyzed for biofilm production and virulence on alfalfa seedlings. Twelve Tn-shv were found to exhibit phenotypes characteristic of the spontaneously isolated shv in that they were deficient in biofilm production and avirulent on alfalfa (data not shown).
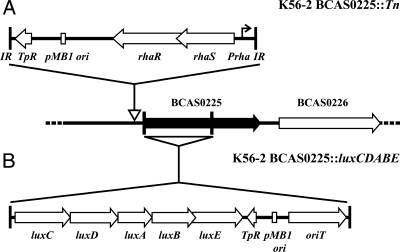
The location of the transposon insertion in these 12 mutants was determined by sequencing the flanking region and using the sequence to identify the location in the unpublished genome of B. cenocepacia J2315, which is an ET12 clone related to K56-2 (32). Since the 12 mutants selected for sequence analysis appeared to be rhamnose inducible, the insertion sites were expected to be located within promoter regions. Interestingly, only 2 of the 12 mutants analyzed contained insertions upstream of genes. One mutant had an insertion that occurred upstream of an open reading frame, BCAM1200, a putative membrane protein, while the other occurred upstream of BCAS0225, a putative transcriptional regulator that belongs to the LysR family (Fig. 4A).
FIG. 4.
Schematic diagrams of the K56-2 BCAS0225 transposon (A) and pGSVTp-lux insertion mutants (B). The site of insertion of the transposon is indicated by an open arrow upstream of BCAS0225. The site of the single crossover for insertion of pGSVTp-lux is indicated by the triangle below BCAS0225. The two vertical lines represent the fragment cloned into pGSVTp-lux.
The BCAS0225 mutant exhibits shiny colony morphology and is defective in biofilm formation, extracellular matrix formation, and virulence.
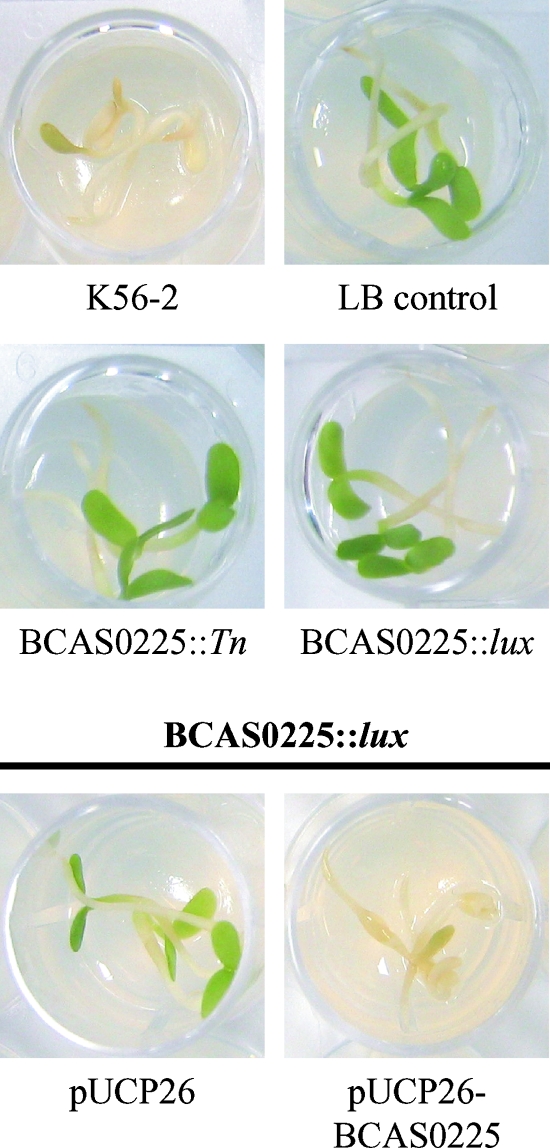
To investigate the involvement of BCAM1200 and BCAS0225 in the conversion from rough to shv in K56-2, insertional mutants were constructed by single recombination using pGSVTp-lux (BCAM1200::pGSVTp-luxCDABE and BCAS0225::pGSVTp-luxCDABE). Only inactivation of BCAS0225 resulted in the shv morphotype (Fig. 5A). Biofilm formation in both the BCAS0225 transposon mutant and the BCAS0225::pGSVTp-luxCDABE mutant were markedly reduced (P < 0.01 [ANOVA]) compared to K56-2 (Fig. 6A). To assess the role of BCAS0225 in virulence, the alfalfa seedling infection model was used (3). Neither of the BCAS0225 mutants caused disease symptoms in alfalfa seedlings, indicating that their phenotypes were consistent with the spontaneously isolated K56-2 shv (Fig. 7). The BCAS0225 mutants were most similar to the shv K56-2 S76 in that they produced parental levels of protease and slightly more AHL than K56-2 (data not shown).
FIG. 5.
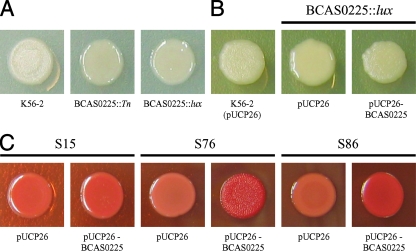
Colony morphology of K56-2 BCAS0225 mutants and complementation of shv. (A) K56-2, BCAS0225::Tn, and BCAS0225::pGSVTp-luxCDABE on LB agar; (B) complementation of BCAS0225::pGSVTp-luxCDABE with pUCP26-BCAS0225 on LB with 200 μg of Tc/ml; (C) effect of BCAS0225 in trans on colony morphology and Congo red binding of K56-2 S15, K56-2 S76, and K56-2 S86.
FIG. 6.
Biofilm formation of K56-2 BCAS0225 mutants. (A) Comparison of biofilm formation in K56-2, BCAS0225::Tn, and BCAS0225::pGSVTp-luxCDABE. (B) Restoration of biofilm formation in BCAS0225::pGSVTp-luxCDABE by complementation with pUCP26-BCAS0225. The results shown are the mean ± the standard deviation of three replicates. An asterisk indicates a value significantly different than K56-2 (ANOVA). The biomass obtained in the experiment shown in panel B is lower than that in panel A due to the presence of Tc in the culture medium required to maintain pUCP26.
FIG. 7.
Alfalfa seedling virulence of K56-2 BCAS0225 mutants. Three seedlings placed on agar in a 24-well plate were inoculated with each strain as described in Materials and Methods. Seedlings were visually inspected for disease symptoms characterized by necrosis and yellow or brown leaves on day 5 p.i.
To confirm that the shv morphotype observed in K56-2 BCAS0225::pGSVTp-luxCDABE was due to the disruption of BCAS0225 and not the result of polar effects caused by the insertion of pGSVTp-lux, we cloned the BCAS0225 gene along with the putative promoter region into pUCP26 (57). The resulting plasmid, pUCP26-BCAS0225, was electroporated into K56-2 BCAS0225::pGSVTp-luxCDABE. Complementation with pUCP26-BCAS0225, but not the vector alone, restored the rough colony morphology (Fig. 5B). Introduction of pUCP26-BCAS0225 also restored biofilm formation to levels observed in the rough K56-2 (Fig. 6B) and virulence in the alfalfa model (Fig. 7). The extracellular matrix observed in rough K56-2 (Fig. 2A) was absent in the BCAS0225 mutant (Fig. 2C) and restored when BCAS0225 was provided in trans (Fig. 2D). Taken together, these results confirm the involvement of BCAS0225 in colony morphology, biofilm formation, and virulence.
Complementation of spontaneous K56-2 shv and sequence analysis.
We examined the possibility that a defect in BCAS0225 was responsible for the shiny colony morphology observed in the seven shv representing different phenotypic clusters. First, we attempted to complement the shv morphotype by introducing pUCP26-BCAS0225 into these strains. The plasmid pUCP26-BCAS0225 restored the rough morphotype, Congo red binding, and AHL production to wild-type levels in K56-2 S76 (Fig. 5C, Table 3, and data not shown). The rough phenotype was also restored in three of five other shv (Table 3). BCAS0225 in trans increased Congo red binding in K56-2 S86 (Fig. 5C) and partially restored the rough morphotype, but this intermediate rough morphotype was only detectable on LB agar and not on any of the other media tested (data not shown). This is in contrast to shv K56-2 S76, whose colony morphology was consistently complemented with BCAS0225 on several types of culture medium (Fig. 5C and data not shown).
TABLE 3.
Characterization of BCAS0225 in representative K56-2 shv
| Shva | Effect of BCAS0225 in trans on colony morphotype | BCAS0225 genotypeb |
|---|---|---|
| K56-2 S15 | Shiny | WTc |
| K56-2 S39 | Rough | G insertion (710) |
| K56-2 S60 | Rough | G insertion (710) |
| K56-2 S62 | Rough | G insertion (710) |
| K56-2 S76 | Rough | G insertion (710) |
| K56-2 S86 | Intermediate | WT |
| K56-2 S92 | Shiny | WT |
| K56-2 S16* | Rough | A-to-G substitution (704) |
| K56-2 S71* | Rough | G insertion (710) |
| K56-2 S73* | Rough | A-to-G substitution (722) |
*, Shv from K56-2 S76 cluster as described elsewhere (data not shown).
The number in parentheses refers to the nucleotide position of the mutation. WT, wild type.
Sequence identical to K56-2.
To determine whether the spontaneous shv contained mutations in BCAS0225, this gene was amplified (Table 3) and sequenced. All of the shv that were complemented by BCAS0225 contained the same frameshift mutation in BCAS0225 (Table 3), whereas K56-2 S86 and the shv that were not complemented had the parental sequence. These data indicate that the shv morphotype is frequently due to mutations in BCAS0225 but that other unidentified mutations also may lead to shv colonies. The partial complementation of K56-2 S86 when pUCP26-BCAS0225 is introduced may be due to a copy number effect. BCAS0225 was sequenced from three additional shv from the same phenotypic cluster as K56-2 S76, and two additional mutations were identified in BCAS0225 (Table 3). These three shv were also restored to the rough morphotype when complemented with BCAS0225 (data not shown).
DISCUSSION
Bacteria often encounter many different environments, ranging from soil to mammalian hosts, and therefore must be able to adapt in order to survive (52). Many bacterial species, including V. cholerae (54, 58, 60), S. enterica serovar Typhimurium (2), and P. aeruginosa (19), are capable of existing in more than one form, which may provide an advantage for survival in different environments (39). Changes in colony morphology may be transient as in the case of phase variation, which results from the differential expression of certain genes or it may be sustained, if it is the result of a loss of function or deletion mutation.
In the present study, we demonstrated that B. cenocepacia K56-2 undergoes a conversion from a rough to a shiny colony morphotype that correlates with reduced biofilm production, reduced virulence in a chronic lung infection model, in most cases avirulence on alfalfa, and reduced or absent extracellular matrix. However, there is substantial diversity with regard to other phenotypes examined. The recovery of different colony morphotypes after growth of K56-2 in liquid cultures was similar to what has been described for P. fluorescens, where phenotypic variants were derived from a single smooth ancestor grown in microcosms (39). In our experiments, shv were recovered from both shaken and static cultures of K56-2 and were extremely stable (Fig. 1 and data not shown).
Using transposon mutagenesis we identified one regulatory gene, BCAS0225, that influences colony morphotype. BCAS0225 encodes a putative transcriptional regulator belonging to the LysR family. BCAS0225 mutants were shiny, defective in biofilm formation (Fig. 6A), and avirulent on alfalfa (Fig. 7) and had scant extracellular matrix (Fig. 2C). These phenotypes reverted to wild type upon complementation with an intact BCAS0225 gene. Several of the spontaneous shv had either frameshift or substitution mutations in this gene and were restored to the rough morphotype when complemented with BCAS0225 (Table 3).
LysR-type transcriptional regulators are among the most common types of positive regulators in prokaryotes (46). LysR-type transcriptional regulators contain an N-terminal DNA-binding domain, a central domain involved in coinducer recognition and/or response, and a C-terminal domain required for both DNA binding and coinducer response (46). The N-terminal and central regions are the most conserved. The sequence of BCAS0225 is highly conserved among other B. cenocepacia strains (>96% identity in amino acid sequences) and members of the Bcc (data not shown). The C-terminal amino acid sequence of BCAS0225 has the greatest homology to several LysR family regulatory proteins in Bordetella species, including BPP4163 from B. parapertussis 12822 and BB4633 from B. bronchiseptica RB50 (37% identity and 57% similarity and 37% identity and 56% similarity, respectively).
The identification of frameshift mutations in BCAS0225 in K56-2 S39, K56-2 S60, K56-2 S62, and K56-2 S76 indicates that mutations in this gene are responsible for at least some spontaneous shv, but clearly other mechanisms are also involved since three of the characterized shv did not contain BCAS0225 mutations, and the rough morphotype was not restored by the presence of BCAS0225 in trans (Table 3). The frameshift mutation resulting from an insertion of a G at position 710 in BCAS0225 was the most common mutation identified, but two additional mutations resulting from A-to-G substitutions were also identified in the shv K56-2 S76-like phenotypic cluster. All mutations identified were in the C terminus of BCAS0225. Studies are under way to determine other mechanisms that lead to the shiny colony morphotype.
Mutations in BCAS0225 are unlikely to account for all of the phenotypic variation in the shv since the four shv with mutations in BCAS0225 did not have the same AHL and protease phenotypes. The BCAS0225 mutants were most similar to K56-2 S76. It is possible that K56-2 S60 and K56-2 S39 contain additional mutations that would explain their AHL and protease phenotypes. The seemingly high frequency of spontaneous mutations in B. cenocepacia indicates the importance of performing complementation experiments in any comparative studies between mutants and the wild type in this organism.
Several regulatory genes have been identified in other species that, when mutated or overexpressed, alter colony morphotypes. Genes required for the synthesis of V. cholerae EPS (VPS) that is essential for the rugose phenotype are under a complex regulatory circuit comprising the negative regulator HapR (62) and the positive regulators VpsR (59), VpsT (8), and RocS (40). In Pseudomonas sp. strain PCL1171, colony morphology variation has been associated with random mutations occurring in the two-component regulatory system GacA/GacS that are caused by the inefficient repair by MutS, one of the central components of the mismatch repair system (52, 53), thereby affecting several of its biocontrol and colonizing traits (52). GGDEF and EAL domain proteins, responsible for the synthesis and degradation of cyclic-di-GMP, respectively, have been shown to influence cell surface properties, biofilm production, and colony morphotype in various organisms, including Vibrio, Yersinia, Salmonella, and Pseudomonas species (for reviews, see references 15 and 43). P. aeruginosa lasR mutants in CF isolates typically have an iridescent metallic sheen resulting from the accumulation of the intercellular signal 4-hydroxy-2-heptylquinoline (16). A LysR regulator, originally designated mor and subsequently shown to be oxyR, has been reported to control a switch in colony morphology in E. coli (25, 55).
Rugose variants of V. cholerae produce more biofilms on abiotic surfaces (54, 60), mainly due to the overproduction of VPS. These EPS comprise at least part of the extracellular matrix that can be visualized by TEM between bacteria of the rugose morphotype but was absent in the smooth morphotype colonies (1, 59). This has also been described for S. enterica serovar Typhimurium rugose variants (2). Similarly, our TEM micrographs revealed the presence of a similar extracellular matrix surrounding bacteria in the rough colonies that was absent in the shv and a BCAS0225 mutant (Fig. 2). These data suggest that BCAS0225 may regulate genes involved in the biosynthesis or production of this extracellular matrix. Straus et al. (51) described an extracellular toxic complex in P. cepacia (previous name for Bcc strains) composed of carbohydrates, lipopolysaccharides, and proteins that was toxic to mice and could result in lung histopathology when inoculated into rats. It is possible that the extracellular matrix observed in the rough colonies includes this extracellular toxic complex and that its absence in the shv would contribute to the decreased virulence observed.
Chung et al. (11) reported that there was more abundant EPS in the shiny derivative, C1394mp2, compared to the rough or matte strain. Although we have not specifically analyzed EPS in K56-2 rough and shv colonies, we would predict, based on our TEM data and studies in V. cholerae (1, 59) and S. enterica serovar Typhimurium (2), that the rough morphotypes would have more EPS. Further studies are needed to determine the differences in the extracellular matrix associated with biofilm formation and virulence between the rough and shv forms and the role of BCAS0225 in its production.
Changes in colony morphology have been associated with resistance to stress and increased survival in particular environment. P. aeruginosa has been shown to acquire a number of common mutations during the course of chronic infection in CF airways, particularly in virulence factors important in acute infections and antibiotic efflux pumps (47). One of the genes frequently mutated is lasR, which regulates a number of virulence factor genes. lasR gene mutants could be distinguished on agar plates after extended incubation by their characteristic metallic sheen (16). Spontaneous lasR mutants have also been isolated from in vitro cultures. Interestingly, loss-of-function mutations in lasR were shown to result in a growth advantage in a number of conditions, including in medium containing amino acids thought to be important in the lung environment (16). Therefore, mutations in lasR may provide a selective survival advantage in the CF lung, which would explain the frequency of isolation of lasR mutants.
The previously described shiny variant of strain C1394 was shown to persist better than the matte or rough morphotype after pulmonary challenge in a leukopenic mouse model (11). In this particular model, Bcc strains that are more virulent are typically cleared from the lungs, whereas less-virulent strains or species persist (10). The C1394 matte strain was cleared from the lungs, whereas the shiny variant C1394mp2 was maintained (11), suggesting that there is a selective advantage for this colony variant in this infection model. In our study we found no difference in survival or persistence between the shv and rough morphotypes of K56-2 in the rat agar bead chronic infection model (Fig. 3B). Both colony types had the same growth rates in liquid culture medium, and the BCAS0225 mutations did not affect in vitro growth rates (data not shown). We have not yet identified any shv from rat lung cultures, although shv are recovered from infected alfalfa seedlings at a relatively high frequency compared to liquid medium. Therefore, if conversion to shv does provide a selective advantage for K56-2, we have not yet determined the environmental conditions that would demonstrate this advantage or the factors that might trigger the colony switch.
In conclusion, we have demonstrated that shv arise spontaneously from strain K56-2 and that shv consistently produce less biofilm and less extracellular matrix and are less virulent. Other virulence phenotypes vary between isolated shv. Spontaneous mutations of the BCAS0225 gene resulting in a loss of function is one mechanism that leads to a shiny colony morphotype. Studies are under way to determine the function of BCAS0225, as well as to identify other mechanisms of conversion to the shiny morphotype.
Acknowledgments
This study was supported by grants from the Canadian Institutes of Health Research and the Canadian Cystic Fibrosis Foundation (CCFF) special initiative in memory of Michael O'Reilly. S.P.B was the recipient of a studentship from the CCFF.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 29 October 2007.
REFERENCES
- 1.Ali, A., M. H. Rashid, and D. K. Karaolis. 2002. High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl. Environ. Microbiol. 685773-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anriany, Y. A., R. M. Weiner, J. A. Johnson, C. E. De Rezende, and S. W. Joseph. 2001. Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl. Environ. Microbiol. 674048-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernier, S. P., L. Silo-Suh, D. E. Woods, D. E. Ohman, and P. A. Sokol. 2003. Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect. Immun. 715306-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernier, S. P., and P. A. Sokol. 2005. Use of suppression-subtractive hybridization to identify genes in the Burkholderia cepacia complex that are unique to Burkholderia cenocepacia. J. Bacteriol. 1875278-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 1320-26. [DOI] [PubMed] [Google Scholar]
- 6.Cardona, S. T., C. L. Mueller, and M. A. Valvano. 2006. Identification of essential operons with a rhamnose-inducible promoter in Burkholderia cenocepacia. Appl. Environ. Microbiol. 722547-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cash, H. A., D. E. Woods, B. McCullough, W. G. Johanson, Jr., and J. A. Bass. 1979. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am. Rev. Respir. Dis. 119453-459. [DOI] [PubMed] [Google Scholar]
- 8.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 1861574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers, C. E., M. B. Visser, U. Schwab, and P. A. Sokol. 2005. Identification of N-acylhomoserine lactones in mucopurulent respiratory secretions from cystic fibrosis patients. FEMS Microbiol. Lett. 244297-304. [DOI] [PubMed] [Google Scholar]
- 10.Chu, K. K., D. J. Davidson, T. K. Halsey, J. W. Chung, and D. P. Speert. 2002. Differential persistence among genomovars of the Burkholderia cepacia complex in a murine model of pulmonary infection. Infect. Immun. 702715-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, J. W., E. Altman, T. J. Beveridge, and D. P. Speert. 2003. Colonial morphology of Burkholderia cepacia complex genomovar III: implications in exopolysaccharide production, pilus expression, and persistence in the mouse. Infect. Immun. 71904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coenye, T., T. Spilker, A. Van Schoor, J. J. LiPuma, and P. Vandamme. 2004. Recovery of Burkholderia cenocepacia strain PHDC from cystic fibrosis patients in Europe. Thorax 59952-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5719-729. [DOI] [PubMed] [Google Scholar]
- 14.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 1846481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 1502497-2502. [DOI] [PubMed] [Google Scholar]
- 16.D'Argenio, D. A., M. Wu, L. R. Hoffman, H. D. Kulasekara, E. Deziel, E. E. Smith, H. Nguyen, R. K. Ernst, T. J. Larson Freeman, D. H. Spencer, M. Brittnacher, H. S. Hayden, S. Selgrade, M. Klausen, D. R. Goodlett, J. L. Burns, B. W. Ramsey, and S. I. Miller. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64512-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis, J. J., and P. A. Sokol. 1995. Electrotransformation of Pseudomonas. Methods Mol. Biol. 47125-133. [DOI] [PubMed] [Google Scholar]
- 18.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 642710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416740-743. [DOI] [PubMed] [Google Scholar]
- 20.Enos-Berlage, J. L., Z. T. Guvener, C. E. Keenan, and L. L. McCarter. 2005. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 551160-1182. [DOI] [PubMed] [Google Scholar]
- 21.Enos-Berlage, J. L., and L. L. McCarter. 2000. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J. Bacteriol. 1825513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haase, E. M., J. L. Zmuda, and F. A. Scannapieco. 1999. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 672901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson, I., and P. Owen. 1997. The autoregulatory protein Mor and OxyR are identical. Microbiology 143(Pt. 5)1482. [DOI] [PubMed] [Google Scholar]
- 26.Henry, D. A., M. E. Campbell, J. J. LiPuma, and D. P. Speert. 1997. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J. Clin. Microbiol. 35614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 1832212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewenza, S., M. B. Visser, and P. A. Sokol. 2002. Interspecies communication between Burkholderia cepacia and Pseudomonas aeruginosa. Can. J. Microbiol. 48707-716. [DOI] [PubMed] [Google Scholar]
- 30.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 16492-96. [DOI] [PubMed] [Google Scholar]
- 31.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51533-538. [DOI] [PubMed] [Google Scholar]
- 32.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3144-156. [DOI] [PubMed] [Google Scholar]
- 34.McCarter, L. L. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 1803166-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, R. A., S. Reckseidler-Zenteno, H. Kim, W. Nierman, Y. Yu, A. Tuanyok, J. Warawa, D. DeShazer, and D. E. Woods. 2004. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 724172-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris, J. G., Jr., M. B. Sztein, E. W. Rice, J. P. Nataro, G. A. Losonsky, P. Panigrahi, C. O. Tacket, and J. A. Johnson. 1996. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 1741364-1368. [DOI] [PubMed] [Google Scholar]
- 37.Ohman, D. E., J. C. Sadoff, and B. H. Iglewski. 1980. Toxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect. Immun. 28899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poussier, S., P. Thoquet, D. Trigalet-Demery, S. Barthet, D. Meyer, M. Arlat, and A. Trigalet. 2003. Host plant-dependent phenotypic reversion of Ralstonia solanacearum from non-pathogenic to pathogenic forms via alterations in the phcA gene. Mol. Microbiol. 49991-1003. [DOI] [PubMed] [Google Scholar]
- 39.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 39469-72. [DOI] [PubMed] [Google Scholar]
- 40.Rashid, M. H., C. Rajanna, A. Ali, and D. K. Karaolis. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 227113-119. [DOI] [PubMed] [Google Scholar]
- 41.Reik, R., T. Spilker, and J. J. Lipuma. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 432926-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, E. W., C. J. Johnson, R. M. Clark, K. R. Fox, D. J. Reasoner, M. E. Dunnigan, P. Panigrahi, J. A. Johnson, and J. G. Morris, Jr. 1992. Chlorine and survival of “rugose” Vibrio cholerae. Lancet 340740. [DOI] [PubMed] [Google Scholar]
- 43.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57629-639. [DOI] [PubMed] [Google Scholar]
- 44.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16944-945. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., Fritsch, E. F., and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47597-626. [DOI] [PubMed] [Google Scholar]
- 47.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 1038487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sokol, P. A., D. E. Ohman, and B. H. Iglewski. 1979. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J. Clin. Microbiol. 9538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 1493649-3658. [DOI] [PubMed] [Google Scholar]
- 50.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Straus, D. C., M. K. Lonon, D. E. Woods, and C. W. Garner. 1989. Production of an extracellular toxic complex by various strains of Pseudomonas cepacia. J. Med. Microbiol. 3017-22. [DOI] [PubMed] [Google Scholar]
- 52.van den Broek, D., G. V. Bloemberg, and B. Lugtenberg. 2005. The role of phenotypic variation in rhizosphere Pseudomonas bacteria. Environ. Microbiol. 71686-1697. [DOI] [PubMed] [Google Scholar]
- 53.van den Broek, D., A. W. T. F. Chin, G. V. Bloemberg, and B. J. Lugtenberg. 2005. Molecular nature of spontaneous modifications in gacS which cause colony phase variation in Pseudomonas sp. strain PCL1171. J. Bacteriol. 187593-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 643648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warne, S. R., J. M. Varley, G. J. Boulnois, and M. G. Norton. 1990. Identification and characterization of a gene that controls colony morphology and auto-aggregation in Escherichia coli K-12. J. Gen. Microbiol. 136455-462. [DOI] [PubMed] [Google Scholar]
- 56.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development, and virulence in Vibrio cholerae O139. Mol. Microbiol. 39223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 14881-86. [DOI] [PubMed] [Google Scholar]
- 58.White, P. B. 1938. The rugose variant of vibrios. J. Pathol. Bacteriol. 461-6. [Google Scholar]
- 59.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 1831716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 964028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, J., J. W. Beaber, M. I. More, C. Fuqua, A. Eberhard, and S. C. Winans. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 1805398-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5647-656. [DOI] [PubMed] [Google Scholar]