Abstract
The life cycle of the apicomplexan parasite Toxoplasma gondii requires that an infectious cyst develop and be maintained throughout the life of the host. The molecules displayed on the parasite surface are important in controlling the immune response to the parasite. T. gondii has a superfamily of glycosylphosphatidylinositol (GPI)-anchored surface antigens, termed the surface antigen (SAG) and SAG-related surface antigens, that are developmentally regulated during infection. Using a clustering algorithm, we identified a new family of 31 surface proteins that are predicted to be GPI anchored but are unrelated to the SAG proteins, and thus we named these proteins SAG-unrelated surface antigens (SUSA). Analysis of the single nucleotide polymorphism density showed that the members of this family are the most polymorphic genes within the T. gondii genome. Immunofluorescence of SUSA1 and SUSA2, two members of the family, revealed that they are found on the parasite surface. We confirmed that SUSA1 and SUSA2 are GPI anchored by phospholipase cleavage. Analysis of expressed sequence tags (ESTs) revealed that SUSA1 had 22 of 23 ESTs from chronic infection. Analysis of mRNA and protein confirmed that SUSA1 is highly expressed in the chronic form of the parasite. Sera from mice with chronic T. gondii infection reacted to SUSA1, indicating that SUSA1 interacts with the host immune system during infection. This group of proteins likely represents a new family of polymorphic GPI-anchored surface antigens that are recognized by the host's immune system and whose expression is regulated during infection.
Toxoplasma gondii is an obligate intracellular parasite and a member of the phylum Apicomplexa, which also includes Plasmodium, Cryptosporidium, Cyclospora, Eimeria, and Sarcocystis (3). T. gondii can reproduce both sexually and asexually. Sexual reproduction occurs only in the feline intestine and results in oocysts being shed in the feces. Within the environment, oocysts develop into infectious sporozoites. The asexual cycle has two developmental stages, namely, a rapidly replicating form called the tachyzoite and a slow-growing stage called the bradyzoite. Bradyzoites form tissue cysts in the central nervous system and muscle tissue and represent the chronic stage of infection. T. gondii is acquired orally either by ingestion of oocyst-contaminated foods or by eating undercooked, bradyzoite-harboring meat products. T. gondii infections in immunocompetent individuals are generally asymptomatic, but infections in immunocompromised individuals and fetuses are life-threatening (9).
Despite its ability to reproduce sexually and its broad geographic range, T. gondii has a largely clonal population structure comprised principally of three lines (15, 25). Analysis of genetic polymorphisms indicates that these three lines emerged from a single genetic cross approximately 10,000 years ago (27), establishing two major alleles for each locus (12). The type I lineage is highly virulent, with injection of one viable parasite being lethal to a mouse, whereas types II and III are relatively avirulent. Type II and III lines readily establish chronic infections in mice and humans, whereas type I strains do not. In this study, we used strain RH as a typical type I strain and PRU as a typical type II strain.
Each developmental stage of T. gondii interacts with the host immune system. A delicate balance between elicitation and suppression of the host responses must be achieved to ensure survival of both the host and the parasite (7). Initial interaction with the host is accomplished primarily through parasite surface antigens. Extensive research has been performed to identify and define the functions of the superfamily of glycosylphosphatidylinositol (GPI)-anchored T. gondii surface antigens, which includes surface antigen (SAG) and SAG-related sequence (SRS) proteins (4, 17, 20). Analysis of the T. gondii genome predicts 161 unique SRS proteins (17). Most characterized members of this family are found exclusively on the surfaces of either tachyzoites or bradyzoites. The exact role of each family member is unknown, but it has been determined that SAG1 and SAG3 have roles in attachment and/or invasion (13, 21). It has been speculated that the presence of numerous SAG proteins on the surfaces of tachyzoites could regulate virulence by controlling the elicitation of the immune response. Likewise, the bradyzoite-specific SAG proteins could then be involved in immune evasion. Finally, the numerous SAG proteins could also be responsible for allowing T. gondii to invade a wide variety of host cells (20).
To identify new surface-exposed proteins, we developed an in silico screen based on the assumption that immune pressure may lead to the evolution of antigenically variant proteins at the parasite surface. We combined this with the knowledge that an endoplasmic reticulum (ER)-type signal sequence is often required for export across the cell membrane and screened the annotated T. gondii genome (www.toxodb.org) for polymorphic families containing a conserved signal sequence. This screen revealed a new family of 31 predicted GPI-anchored proteins. This study determined the location of and GPI anchor addition to this protein family and then examined the developmental regulation and immune reaction against one member during infection.
MATERIALS AND METHODS
Screen for polymorphic families and the presence of ER-type signal sequences.
To search for polymorphic protein clusters, we used TRIBE-MCL, a clustering algorithm based on stimulation of Markov matrices in flow (11). We searched the approximately 7,800 predicted proteins annotated in the T. gondii genome (http://toxodb.org/download/release-4.2/Tgondii/TgondiiAnnotatedProteins_toxoDB-4.2.fasta). The MCL analysis is a three-step process; the BLAST step was run with an E value cutoff of 1 and the filter on, the MCL Markov matrix map was created with an E value cutoff of 1e−4 and heavyweight, and clustering was done with an inflation value of 2.8 and scheme −5. Because our focus was secreted proteins, we analyzed only clusters with proteins predicted to contain an ER-type signal sequence, based on SignalP 2.0 (22).
Cell culture and parasite strains.
Strains were maintained as tachyzoites by serial passage on monolayers of human foreskin fibroblasts. Tachyzoite conditions were Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin (Gibco), and 2 mM l-glutamine, with incubation at 37°C with 5% CO2. Bradyzoite conditions were RPMI 1640 supplemented with 1% fetal bovine serum and 1% penicillin-streptomycin, buffered with 50 mM HEPES to pH 8, with incubation at 37°C with ambient CO2. T. gondii RHΔHXGPRT was used for expression of green fluorescent protein (GFP) fusions from the α-tubulin promoter, and strain PRU was used for Northern blot analysis and expression of the 65.m01148 GFP fusion from its native promoter.
Generation of protein fusion constructs and their expression in T. gondii.
The open reading frames (ORFs) of 65.m01148 and 65.m01173 were amplified using primers 65-48-Nsi (5′-ATGCATGCTGTTGCACTCAGAGGCTT-3′) and 65-48-Pac (5′-TTAATTAAAATGTGGAGAACAGTGCAGCT-3′) for 65.m01148 and primers 65-73-Nsi (5′-ATGCATGGACACATTTTTCGAGCATT-3′) and 65-73-Pac (5′-TTAATTAATAGATCAGCAGAGACGCA-3′) for 65.m01173. NsiI and PacI sites were engineered into the primers for subcloning into pT/230 (26), which will express the ORFs from the α-tubulin promoter. The GFP sequence was inserted in frame into each ORF near the 5′ end, using BsiWI sites for 65.m01148 and BstEII sites for 65.m01173. In order to insert a hemagglutinin (HA) tag, a region of the ORF was amplified using primers that would incorporate an HA tag near the 3′ end of the ORF (primers for 65.m01148 were 48HAF [5′-GCAAATACCATAAGCAAGG-3′] and 48HAR [5′-GTGCACTCGCGTAGTCTGGGACGTCGTATGGGTATAGTTCCGGAGTGACTGT GTC] and primers for 65.m01173 were 73HAF [5′-GGTTACCCAACACCGCTG-3′] and 73HAR [5′-GTGCACCGCGTAGTCTGGGACGTCGTATGGGTAGACAATCGTCAGGTATGCCTC]). The PCR products were cloned into pCR 2.1 (Invitrogen) according to the manufacturer's protocols and verified by sequencing with M13F and M13R vector primers. The resulting plasmids were digested with ApaLI and XcmI to excise a 344-bp band for 65.m01148 and a 634-bp band for 65.m01173. These were subcloned into pT/230 65.m01148 or 65.m01173 that was digested with ApaLI and XcmI to remove the identical region to be replaced with the HA version. The native promoter of 65.m01148 was PCR amplified using primers 48endogpromF (5′-AAGCTTGAACATGAGAGTGAGCTC-3′) and 48endogpromR (5′-CTGTCTCGTCGATGACAC-3′). This 1.4-kb region includes 1.2 kb upstream of the predicted start methionine of 65.m01148 and a HindIII site at the 5′ end of 48endogpromF for subcloning. The α-tubulin 65.m01148 GFP construct was digested with HindIII and BstXI to remove the α-tubulin promoter and replace it with the native promoter. The dihydrofolate reductase-thymidylate synthetase sequence (8) was subcloned into all plasmids. Twenty-five milligrams of each linearized construct was electroporated with 1 × 107 T. gondii RHΔHXGPRT or PRU parasites, and stable transformants were selected as resistant to 1 μM pyrimethamine.
IFA.
Confluent human foreskin fibroblasts on coverslips were infected with recently lysed parasites for 24 h under tachyzoite conditions or for 5 days under bradyzoite conditions, as described above. For extracellular parasites, recently lysed parasites in Dulbecco's modified Eagle medium were applied to coverslips and allowed to attach for 30 min. The monolayer or extracellular parasites were fixed with 3% formaldehyde and then permeabilized and blocked in 3% bovine serum albumin-0.2% Triton X-100 (for permeabilized immunofluorescence assay [IFA]) or in 3% bovine serum albumin (for nonpermeabilized IFA). Primary antibodies for fusion proteins were rabbit anti-GFP (A11122; Invitrogen), mouse anti-GFP (sc-9996; Santa Cruz Biotechnology, Inc.), and mouse anti-HA (AFC-101P; Covance Innovative Antibodies) and colocalized with SAG1, P36, ROP1, IMC1, GRA4, and BIP. Secondary antibodies were Alexa Fluor 633-conjugated goat anti-mouse or anti-rabbit and Alexa Fluor 488-conjugated goat anti-mouse or anti-rabbit (Molecular Probes). Coverslips were mounted onto slides by using VectaShield mounting medium containing 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Laboratories). Samples were examined using a motorized Zeiss Axioplan IIi microscope equipped with a rear-mounted excitation filter wheel, a triple-pass (DAPI-fluorescein isothiocyanate-Texas Red) emission cube, differential interference contrast optics, and a Hamamatsu ORCA-AG charge-coupled device camera. Serial image stacks (0.2-μm z increments) were collected at a magnification of ×100 (PlanApo oil immersion objective; 1.4 numerical aperture) and were deconvolved and pseudocolored using OpenLabs 4.0 software (Improvision).
Analysis of GPI anchor addition.
Parasites expressing HA epitope-tagged versions of 65.m01148 and 65.m01173 were digested with phosphatidylinositol-specific phospholipase C (PI-PLC) as previously described (14). Briefly, lysed parasites were washed three times with phosphate-buffered saline and once with PI-PLC buffer (10 mM Tris-HCl, pH 7.5, 0.75 M sucrose, and 10 mM glucose). Parasites were resuspended in 20 μl of PI-PLC buffer and treated with 1 U of PI-PLC (Molecular Probes) at 37°C for 2 hours. Treated parasites were collected by centrifugation at 11,600 × g for 1 minute. The supernatant was collected for Western analysis, and the cells were washed three times in PI-PLC buffer, applied to coverslips, and processed for IFA as stated above. Supernatants were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon-P membranes (Millipore). Membranes were incubated with primary antibody (anti-HA, 1:2,500; anti-SAG1, 1:1,000) for 1 hour and with the secondary antibody (mouse anti-rabbit immunoglobulin G [IgG] conjugated to horseradish peroxidase; 1:4,000) for 1 hour. Detection was performed with the Amersham Bioscience ECL Western blotting system for chemiluminescence according to the manufacturer's protocol.
RNA isolation and Northern hybridization.
Parasites were harvested for RNA isolation by syringing (27 gauge for tachyzoites and 30 gauge for bradyzoites) to release parasites. Total RNA was isolated from equal numbers of tachyzoites and bradyzoites, using Ultraspec RNA (Biotecx Laboratories, Inc.) according to the manufacturer's protocol. Harvested total RNA was separated in a formaldehyde-agarose gel and transferred to Zeta-Probe blotting membranes (Bio-Rad). A region of the 5′-untranslated region of 65.m01148 was PCR amplified from T. gondii genomic DNA, using primers 48probeF (5′-GTAAGGTTGAACTTCAGCC-3′) and 48probeR (5′-GAACAACGGCATTTGCAG-3′), and was used as a probe. Primers TgTubulinF (5′-CCTGTCTGTTGACTACGGCAAG-3′) and TgTubulinR (5′-CGTCACCATAGCCCTCCTC-3′) were used to amplify the T. gondii α-tubulin probe. Hybridization and stringent washes were performed as previously described (2).
Peptide production and purification.
The variable region of 65.m01148 was amplified with primers 48protF (5′-CCATGGGCCCGAAGGGTGGACCCGGTC-3′) and 48protR (5′-CTCGAGCTCTTCAGCGTTATGTGC-3′), which added an NcoI site on the 5′ end and an XhoI site on the 3′ end. The PCR product was cloned into pCR 2.1 according to the manufacturer's protocols and verified by sequencing with M13F and M13R vector primers. The plasmid was then digested with NcoI and XhoI to obtain a 425-bp fragment, which was subcloned into pET-28a(+) (Novagen) that was linearized with NcoI and XhoI. The construct was transformed into the Rosetta strain; the peptide was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C; and the soluble fraction, a 16-kDa protein with a C-terminal six-His tag, was purified using His-Bind Quick 900 cartridges (Novagen) according to the manufacturer's protocol.
Immunoblots with chronic sera.
The 65.m01148 peptide was separated in a nondenaturing 20% acrylamide gel without sodium dodecyl sulfate and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). Ponceau S staining (0.1% [wt/vol] Ponceau S in 5% acetic acid) was done to confirm that transfer was complete. Sera collected from CBA/J mice infected with 2 × 104 PRU tachyzoites for 22 days were used as the primary antibody. The secondary antibody was donkey anti-mouse IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, Inc.). Chemiluminescence detection was done with an Amersham Bioscience ECL Western blotting system.
RESULTS
Identification and sequence analysis of a predicted GPI-anchored protein family.
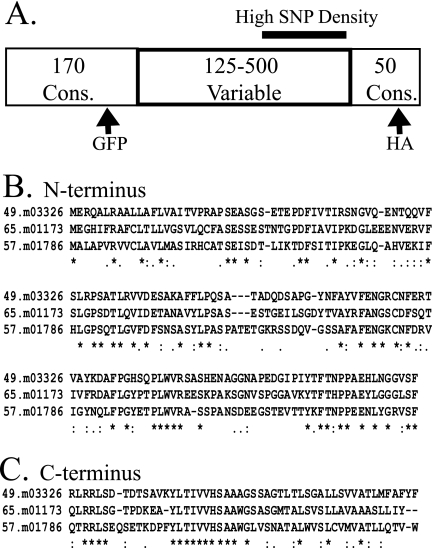
We hypothesized that since host-exposed proteins are under antigenic pressure, they are likely to belong to polymorphic families. Because these proteins are targeted to the parasite surface, they frequently contain an ER-type secretion signal. We used TRIBE-MCL to search the annotated T. gondii database for polymorphic families containing a secretion signal and identified a predicted protein family with 31 members that contain a GPI anchor addition signal (Table 1). Analysis of ToxoDB with any one of the family member sequences will list the other 30 members as paralogs. The amino acid sequences of the family members were analyzed using the multiple sequence alignment program T-Coffee (http://www.ebi.ac.uk/t-coffee/) to find the regions of sequence conservation. The optimal global alignment revealed that the family contains a variable region of 125 to 500 amino acids flanked by conserved domains at the N and C termini of approximately 170 and 50 amino acids, respectively (Fig. 1A). It was clear from this alignment that family members on the same chromosome had greater sequence identity to each other than they did to family members on the other two chromosomes, highlighting errors in homologous recombination as the most common method of gene duplication to expand the family. Figure 1 shows an alignment of the conserved domains for a representative family member from each chromosome (Fig. 1B shows the N-terminal 170 amino acids, and Fig. 1C shows the C-terminal 50 amino acids).
TABLE 1.
Members of the polymorphic protein familya
| Annotation | Chromosome | EST(s) (no. of ESTs) | SNP density (no. of SNPs/kb of coding sequence)
|
SNPs at 3′ end | ||
|---|---|---|---|---|---|---|
| I vs II | I vs III | II vs III | ||||
| 49.m03322 | VI | CAST (2) | 14 | 14 | 0 | ✓ |
| 49.m03323 | VI | 35 | 35 | 0 | ||
| 49.m03324 | VI | CAST (1) | 22 | 23 | 0.81 | ✓ |
| 49.m03325 | VI | 30 | 30 | 0 | ||
| 49.m03326 | VI | 44 | 45 | 0.8 | ✓ | |
| 49.m03327 | VI | CAST (1) | 56 | 56 | 0 | |
| 49.m03328 | VI | 46 | 46 | 0 | ✓ | |
| 49.m03329 | VI | 30 | 30 | 0 | ✓ | |
| 49.m03330 | VI | 10 | 11 | 0.72 | ||
| 49.m03331 | VI | 27 | 27 | 0 | ✓ | |
| 57.m01785 | IX | ND | ND | ND | ✓ | |
| 57.m01786 | IX | ND | ND | ND | ||
| 57.m01787 | IX | ND | ND | ND | ||
| 65.m01148 (SUSA1) | XII | In vivo bradyzoites (22), RH (1) | 7.3 | 13 | 15 | ✓ |
| 65.m01149 | XII | Coug (1) | 6.3 | 51 | 52 | ✓ |
| 65.m01150 | XII | Coug (1) | 5.4 | 40 | 43 | ✓ |
| 65.m02531 | XII | 39 | 4.6 | 37 | ✓ | |
| 65.m01162 | XII | 36 | 36 | 17 | ||
| 65.m02532 | XII | 21 | 25 | 25 | ||
| 65.m01163 | XII | Coug (1) | 29 | 45 | 42 | ✓ |
| 65.m01164 | XII | Coug (1) | 32 | 31 | 33 | |
| 65.m01165 | XII | 12 | 45 | 47 | ||
| 65.m01166 | XII | 4.8 | 32 | 34 | ||
| 65.m02533 | XII | 2.2 | 46 | 47 | ||
| 65.m01167 | XII | RH (1), Coug (1), VEG (1) | 35 | 51 | 44 | ✓ |
| 65.m01168 | XII | Coug (1) | 26 | 36 | 19 | ✓ |
| 65.m01169 | XII | RH (1), Coug (1), VEG (1) | 5.7 | 45 | 43 | ✓ |
| 65.m01170 | XII | RH (3), Coug (5) | 0 | 30 | 30 | ✓ |
| 65.m01171 | XII | 2.5 | 21 | 22 | ✓ | |
| 65.m01172 | XII | 4.6 | 26 | 24 | ✓ | |
| 65.m01173 (SUSA2) | XII | ME49 (1), Coug (1) | 27 | 35 | 41 | ✓ |
Members of the polymorphic family are identified by their annotations from ToxoDB. 65.m01148 and 65.m01173 were further characterized in the present study. All ESTs are from the tachyzoite form, unless stated otherwise. The SNP densities listed in ToxoDB were generated by sequence comparisons of the three lineages (I versus II, I versus III, and II versus III). Family members that have a highly polymorphic region at the 3′ end of the variable region are designated with a check in the final column.
FIG. 1.
Sequence comparison of the GPI-anchored protein family. (A) Representation of a consensus sequence of the protein family members on chromosome XII. The sizes of variable and conserved (Cons) regions of the family are given as numbers of amino acids. Placement of the GFP and HA tags is designated by arrows. The region of high SNP density is shown as a bar in the 3′ end of the variable region. Alignments were performed with the N-terminal 170 amino acids (B) and the C-terminal 50 amino acids (C) of 65.m01173, 49.m03325 (chromosome VI), and 57.m01786 (chromosome IX). Symbols underneath the alignments represent identical amino acids in all sequences (*), conserved substitutions (:), and semiconserved substitutions (.), as defined by T-Coffee (http://www.ebi.ac.uk/t-coffee/).
Family members are highly polymorphic.
We examined the density of single nucleotide polymorphisms (SNPs) within this family of predicted GPI-anchored proteins. While the genome-wide polymorphism rate between the three lines is 0.65%, those for the new family range from 1.1 to 5.6% (Table 1). While all currently characterized genes from T. gondii have only two major allelic types for each locus, several family members on chromosome XII have distinct alleles for each lineage. This highlights that this region of the chromosome may contain a higher than average mutation rate or a recombination hot spot. Additionally, several of the proteins contain a region of high SNP density of approximately 80 amino acids long within the variable region (Fig. 1A). Taken together, these results suggest that these family members are under immune pressure to vary their sequence.
The largest cluster of highly polymorphic genes is located on chromosome XII, and therefore we chose the genes on the opposite ends of this cluster, 65.m01148 and 65.m01173, for further examination. We also wanted to focus our efforts on family members that were expressed at different parasite stages, based on expressed sequence tag (EST) profiles (Table 1). Of the 23 ESTs from the 65.m01148 gene, 22 are from in vivo ME49 bradyzoites. 65.m01173 has two ESTs of tachyzoite origin, one from ME49 and the other from the COUG strain. The 65.m01173 predicted protein is highly polymorphic, with a unique allele in each of the three major lines, and is a representative consensus sequence for the members on chromosome XII.
Localization of 65.m01148 and 65.01173.
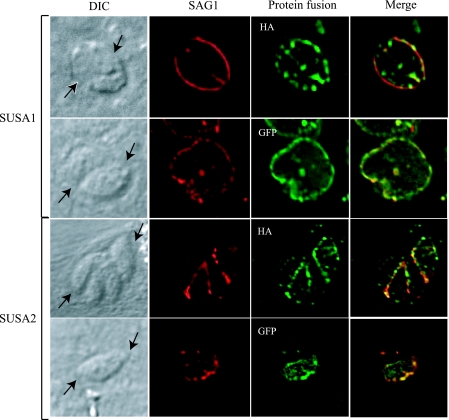
To examine the locations of 65.m01148 and 65.m01173, we created GFP and HA protein fusions with the predicted proteins expressed constitutively from the α-tubulin promoter. GFP was added approximately 80 amino acids downstream of the signal sequence, and HA was inserted upstream of the predicated GPI cleavage site (15 amino acids upstream for 65.m01148 and 2 amino acids upstream for 65.m01173). T. gondii clones engineered to express 65.m01148 and 65.m01173 as GFP or HA protein fusions were analyzed by immunofluorescence to determine the locations of the proteins in tachyzoites. Colocalization with antibodies to the T. gondii surface (SAG1), inner membrane complex (IMC1), rhoptries (ROP1), and dense granules (GRA4) showed that 65.m01148 and 65.m01173 fused with GFP and HA were located on the parasite surface (Fig. 2 and data not shown). Staining with antibodies to HA and GFP was not observed in wild-type parasites (data not shown). Due to their surface location, we named these proteins SAG-unrelated surface antigen 1 (SUSA1; 65.m01148) and SUSA2 (65.m01173).
FIG. 2.
Colocalization of SUSA1 (65.m01148) and SUSA2 (65.m01173) with SAG1. The images show immunofluorescence of intracellular parasites expressing SUSA1 and SUSA2 protein fusions with GFP or HA (green) and colocalized with SAG1 (red). Arrows indicate the locations of the parasitophorous vacuole that contains multiple tachyzoites. DIC, differential interference contrast.
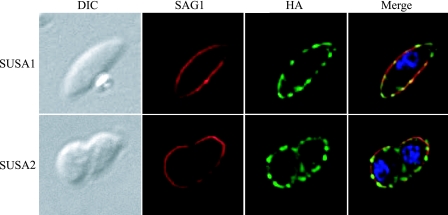
To verify that SUSA1 and SUSA2 are located on the surface of the parasite, we performed colocation studies with SAG1 antibodies and extracellular parasites, with and without permeabilization. Similar to SAG1, SUSA1- and SUSA2-HA protein fusions were detectable without permeabilization (Fig. 3). No staining of IMC1 was seen in nonpermeabilized parasites (data not shown). This confirms that SUSA1 and SUSA2 proteins are on the parasite surface.
FIG. 3.
Nonpermeabilized IFA showing the surface location of SUSA1 and SUSA2. Extracellular parasites expressing the HA-tagged versions of SUSA1 (one tachyzoite pictured) and SUSA2 (two tachyzoites pictured) were fixed and blocked without permeabilization and then stained for SAG1 (red) and HA (green). DIC, differential interference contrast.
SUSA-HA protein fusions are GPI-anchored proteins.
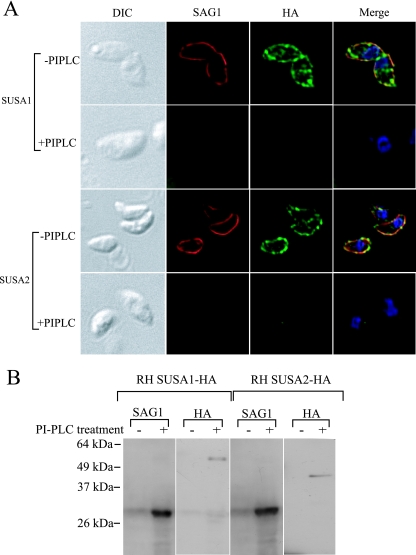
The surface location of SUSA1 and SUSA2, along with their predicted GPI anchor addition signal, indicated that they were GPI-anchored proteins. For a definitive answer, we treated free parasites expressing SUSA1-HA or SUSA2-HA with PI-PLC and examined the parasites by IFA. SAG1, SUSA1-HA, and SUSA2-HA were no longer detected by antibodies in parasites digested with PI-PLC, while the surface proteins were detected in untreated parasites (Fig. 4A). Additionally, Western immunoblotting of supernatants from parasites digested with PI-PLC showed protein bands of the appropriate size for SUSA1 and SUSA2 reacting with anti-HA (Fig. 4B). No reactive proteins were detected in the supernatants of untreated parasites (Fig. 4B). Taken together, these data provide evidence that SUSA1 and SUSA2 are GPI-anchored proteins.
FIG. 4.
PI-PLC treatment of parasites expressing HA-tagged versions of SUSA1 and SUSA2. (A) Free parasites were treated with PI-PLC for 2 hours, fixed on coverslips, and visualized with antibodies to SAG1 and HA. The first column consists of differential interference contrast images that show extracellular parasites (two, one, three, and two tachyzoites per picture, from the top to the bottom of the column). The first row of each set shows untreated parasites, and the second row shows PI-PLC-treated parasites. Merged images include SAG1 (red), HA (green), and DAPI (blue). Images for untreated and treated parasites were captured with identical exposure times. (B) Western hybridization using antibody to HA as the primary antibody was performed on supernatants from the PI-PLC treatments to detect released SUSA1 and SUSA2. SAG1, a known GPI-anchored protein used as a control.
Transcriptional regulation of SUSA1.
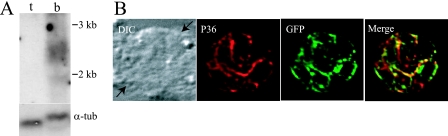
Because SUSA1 had multiple ESTs from the in vivo bradyzoite library, we examined its transcript abundance in tachyzoites and bradyzoites. Previous microarray analysis showed that SUSA1 was one of the most up-regulated genes in bradyzoites (SUSA1 is ctoxoqual 4192 and 4416 [6]). We confirmed this microarray result by using Northern blot hybridizations. A probe for the SUSA1 5′-untranslated region showed a 2.4-kb transcript in PRU bradyzoites that was not detected in tachyzoites (Fig. 5A). Reprobing of this blot with an α-tubulin probe showed that each lane had an approximately equal quantity of T. gondii RNA. Thus, SUSA1 is transcriptionally up-regulated in the bradyzoite stage.
FIG. 5.
Developmental regulation of SUSA1. (A) Expression of SUSA1 was examined by Northern hybridization with tachyzoites (t) and bradyzoites (b). α-tubulin (α-tub) is shown as a loading control. The shift in the α-tubulin band was due to more host cell RNA being present in the bradyzoite sample, causing the band to be slightly displaced. (B) Bradyzoites expressing a GFP protein fusion of SUSA1 driven by its native promoter were visualized by antibodies against P36 (red) and GFP (green). Arrows indicate the location of the parasitophorous vacuole. DIC, differential interference contrast.
Regulation and location of SUSA1 protein in bradyzoites.
The SUSA1 transcript is up-regulated in the bradyzoite stage of T. gondii. We therefore wanted to examine the regulation and location of the SUSA1 protein in bradyzoites by immunofluorescence. In order to observe endogenous expression of SUSA1, we subcloned the native SUSA1 promoter into the SUSA1-GFP fusion and selected for transformants in PRU. By IFA, SUSA1-GFP driven by the native promoter was expressed robustly and colocalized with SRS9, a bradyzoite-specific SAG1 family surface antigen that is the main interactive protein with the P36 monoclonal antibody (Fig. 5B) (28). These results show that the expression of at least one member of this new family of surface antigens is controlled during infection.
Chronic infection sera are reactive against SUSA1.
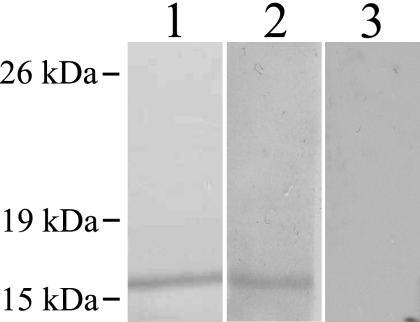
Because SUSA1 is present on the surfaces of bradyzoites, we wanted to determine if the protein was an antigen recognized during natural infection. We collected sera from mice that were infected with T. gondii PRU for 22 days, which represents an early chronic infection, when cysts are readily detectable in the brain. Sera from infected mice were reactive with a polypeptide generated from the variable region of SUSA1 (Fig. 6). Chronic infection sera were not reactive with purified lysate from the Rosetta strain (data not shown). Serum reactivity indicates that SUSA1 interacts with the immune system of the host.
FIG. 6.
Detection of SUSA1 peptide with chronic sera. A 16-kDa polypeptide of SUSA1 was visualized by Ponceau S staining following transfer (lane 1). Sera from mice infected with T. gondii detected the SUSA1 polypeptide on a nondenaturing gel (lane 2). A no primary antibody control is shown in lane 3.
DISCUSSION
The current study has identified a new family of polymorphic proteins whose genes are clustered on three chromosomes. Two members were shown to be on the parasite surface and attached by a GPI anchor. While this study focused on the two family members encoded on both ends of the largest gene cluster on chromosome XII, further characterization must be performed on members from chromosomes VI and IX to ensure that they are also on the parasite surface and are true paralogs. It is noteworthy that ToxoDB contains no ESTs for several of the family members and thus that some of the genes may be pseudogenes, not expressed genes. It could be that most of the family members are expressed at a low level, as even members with ESTs contain just one or two; only SUSA1 contains numerous ESTs in the database. An alternative explanation for the lack of ESTs is that the genes are expressed in parasite forms that are not represented in the EST libraries (e.g., merozoites). Likewise, because the in vivo bradyzoite library represents a single time point during a mouse infection, other family members may be expressed at different time points during infection or within different animal hosts. Further analysis will examine if and when the other family members are expressed.
The life cycle of T. gondii requires that this parasite form an infectious bradyzoite cyst that will be maintained throughout the life of the host. T. gondii meets this challenge and sustains itself within a host that is armed with several ways to eliminate nonself particles. Other pathogens circumvent host efforts to eradicate them through alteration of their cell surfaces. Some bacterial pathogens are able to evade the immune system through phase variation of phosphorylcholine, a host-like molecule, on the surfaces of their membranes (24, 29). The variant surface glycoprotein coat of Trypanosoma brucei has been shown to undergo antigenic variation, allowing parasites expressing variants not recognized by the immune system to escape destruction (1). Plasmodium falciparum has a similar system with which it alters proteins expressed on the surfaces of infected erythrocytes to prevent it from being destroyed (23). Frequent antigenic variation aids in evasion of the immune response and forces the cellular immune components to restart the process of identifying the pathogen as a foreign particle.
How T. gondii bradyzoite cysts evade host immune surveillance is still largely a mystery. Few members of the SAG1 family have been identified as bradyzoite specific, and even fewer have been characterized functionally. Constitutive expression of SRS9, a bradyzoite-specific surface antigen, resulted in greater clearance of parasites during infection, with fewer parasites persisting as cysts (19). Additionally, a T. gondii SRS9 knockout strain was shown to have no effect on replication or dissemination during acute infection, but cyst burdens resulting from chronic infection were diminished compared to those for the wild type (18). These findings emphasize the importance of T. gondii varying its surface presentation throughout infection to ensure immune evasion and persistence (18, 19). The up-regulation of SUSA1 in the bradyzoite stage highlights the potential role of this protein in infection persistence.
In addition to assisting T. gondii in immune response evasion, bradyzoite-specific surface antigens may serve as a protective barrier to ensure parasite survival within the stomach. Bradyzoites have been shown to be more stable than tachyzoites when exposed to gastric digestion, as measured by infectivity of parasites in mice following digestion with acid pepsin (16). Because this stability has been attributed to the surface of the bradyzoite, SUSA1 may be important for survival in pepsin acid (9). The reliability of acid pepsin digestion stability as a method to differentiate between tachyzoites and bradyzoites has been questioned, since tachyzoites are occasionally infectious orally (10). The conflicting evidence could be due to the methods used to infect the mice. Oral gavage administration of luciferase-expressing parasites resulted in an inconsistent pattern of dissemination, with parasites initially observed in the chest area. Natural feeding of brain cyst homogenate showed infection initiating in the abdominal area (5). Further experiments with SUSA1 will examine if gene deletions affect infection persistence as well as infectivity in the next host by using natural feeding.
The great variety of surface antigens is likely necessary for attachment and invasion of a wide range of host cells. Surface antigens have been shown to be involved in attachment to and invasion of host cells (20); however, all proteins involved in these processes have not been identified. As components of the parasite surface, SUSA1 and SUSA2 may interact with the host and could serve roles that are similar or redundant to those of the SAG family of surface antigens. Attachment and invasion of T. gondii tachyzoites have been diminished following pretreatment of the parasites with antibodies to SAG1 (13, 21). Somewhat surprisingly, parasites with a deletion of SAG1 were shown to have a twofold increase in attachment and invasion efficiency compared to that of the wild type (15). Functional analysis of SUSA1 and SUSA2 will reveal if this family shares function as well as location with the SAG family.
The distribution of the protein family genes in the genome and the high degree of polymorphisms indicate the possibility of gene duplication and immune pressure. The family is clustered together on three chromosomes, VI, IX, and XII, in groups of 3 to 13 genes. This gene arrangement led to the hypothesis that the various genes (i) are expressed at different times to allow for phase variation at the surface to evade the immune system and allow for persistence or (ii) are specific for invasion of a certain type of host cell. The bradyzoite-specific expression of SUSA1 supports the idea of the family being involved in phase variation of the parasite surface. SUSA1 could be expressed only in the early progression of chronic infection, followed by the expression of a different member of the family later in chronic infection. Continual modification of the surfaces of bradyzoites could allow for persistence in the chronic stage of infection and for reactivation of bradyzoites by evasion of the immune system. Monitoring the expression of the SUSA family throughout the course of chronic infection will allow us to define its role in persistence and possible antigenic variation.
Acknowledgments
We sincerely thank Travis Harrison for help with TRIBE-MCL, Jay Bangs for the use of his microscope, Jon Boyle for help analyzing the microarray and polymorphism data, John Boothroyd, John Mansfield, and Donna Paulnock for helpful discussions, ToxoDB (www.toxodb.org), and the following individuals for antibodies: Gary Ward (IMC1), John Boothroyd (SAG1), Joe Schwartzman (ROP1), Jay Bangs (BIP), Jean-François Dubremetz (P36), and David Sibley (GRA4).
This research was supported by National Institutes of Health (NIH) award A1054603 (L.J.K.), Regional Center of Excellence V Great Lakes award 1-U54-AI-057153 (K.H.), and NIH National Research Service award T32 AI007414 (K.N.O.).
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 15 October 2007.
REFERENCES
- 1.Aitcheson, N., S. Talbot, J. Shapiro, K. Hughes, C. Adkin, T. Butt, K. Sheader, and G. Rudenko. 2005. VSG switching in Trypanosoma brucei: antigenic variation analysed using RNAi in the absence of immune selection. Mol. Microbiol. 571608-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2002. Analysis of RNA by Northern and slot blot analysis, p. 4.9.1-4.9.19. In Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 3.Black, M. W., and J. C. Boothroyd. 2000. Lytic cycle of Toxoplasma gondii. Microbiol. Mol. Biol. Rev. 64607-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boothroyd, J. C., A. Hehl, L. J. Knoll, and I. D. Manger. 1997. The surface of Toxoplasma: more or less. Int. J. Parasitol. 283-9. [DOI] [PubMed] [Google Scholar]
- 5.Boyle, J. P., J. P. J. Saeij, and J. C. Boothroyd. 2007. Toxoplasma gondii: inconsistent dissemination patterns following oral infection in mice. Exp. Parasitol. 116302-305. [DOI] [PubMed] [Google Scholar]
- 6.Cleary, M. D., U. Singh, I. J. Blader, J. L. Brewer, and J. C. Boothroyd. 2002. Toxoplasma gondii asexual development: identification of developmentally regulated genes and distinct patterns of gene expression. Eukaryot. Cell 1329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denkers, E. Y., and R. T. Gazzinelli. 1998. Regulation and function of T-cell-mediated immunity during Toxoplasma gondii infection. Clin. Microbiol. Rev. 11569-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donald, R. G., and D. S. Roos. 1993. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc. Natl. Acad. Sci. USA 9011703-11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubey, J. P. 1994. Toxoplasmosis. JAMA 2051593-1598. [PubMed] [Google Scholar]
- 10.Dubey, J. P. 1998. Re-examination of resistance of Toxoplasma gondii tachyzoites and bradyzoites to pepsin and trypsin digestion. Parasitology 11643-50. [DOI] [PubMed] [Google Scholar]
- 11.Enright, A. J., S. Van Dongen, and C. A. Ouzounis. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 301575-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grigg, M. E., S. Bonnefoy, A. B. Hehl, Y. Suzuki, and J. C. Boothroyd. 2001. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294161-165. [DOI] [PubMed] [Google Scholar]
- 13.Grimwood, J., and J. E. Smith. 1996. Toxoplasma gondii: the role of parasite surface and secreted proteins in host cell invasion. Int. J. Parasitol. 26169-173. [DOI] [PubMed] [Google Scholar]
- 14.Howard, A. D., J. Berger, L. Gerber, P. Familletti, and S. Udenfriend. 1987. Characterization of the phosphatidylinositol-glycan membrane anchor of human placental alkaline phosphatase. Proc. Natl. Acad. Sci. USA 846055-6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howe, D. K., and L. D. Sibley. 1995. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J. Infect. Dis. 1721561-1566. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, L., J. S. Remington, and M. L. Melton. 1960. The resistance of the encysted form of Toxoplasma gondii. J. Parasitol. 4611-21. [PubMed] [Google Scholar]
- 17.Jung, C., C. Y.-F. Lee, and M. E. Grigg. 2004. The SRS superfamily of Toxoplasma surface proteins. Int. J. Parasitol. 34285-296. [DOI] [PubMed] [Google Scholar]
- 18.Kim, S.-K., A. Karasov, and J. C. Boothroyd. 2007. Bradyzoite-specific surface antigen SRS9 plays a role in maintaining Toxoplasma gondii persistence in the brain and in host control of parasite replication in the intestine. Infect. Immun. 751626-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, S. K., and J. C. Boothroyd. 2005. Stage-specific expression of surface antigens by Toxoplasma gondii as a mechanism to facilitate parasite persistence. J. Immunol. 1748038-8048. [DOI] [PubMed] [Google Scholar]
- 20.Lekutis, C., D. J. P. Ferguson, M. E. Grigg, M. Camps, and J. C. Boothroyd. 2001. Surface antigens of Toxoplasma gondii: variations on a theme. Int. J. Parasitol. 311285-1292. [DOI] [PubMed] [Google Scholar]
- 21.Mineo, J. R., and L. H. Kasper. 1994. Attachment of Toxoplasma gondii to host cells involves major surface protein, SAG-1 (P30). Exp. Parasitol. 7911-20. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 101-6. [DOI] [PubMed] [Google Scholar]
- 23.Peters, J., E. Fowler, M. Gatton, N. Chen, A. Saul, and Q. Cheng. 2002. High diversity and rapid changeover of expressed var genes during the acute phase of Plasmodium falciparum infections in human volunteers. Proc. Natl. Acad. Sci. USA 9910689-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serino, L., and M. Virji. 2000. Phosphorylcholine decoration of lipopolysaccharide differentiates commensal Neisseriae from pathogenic strains: identification of licA-type genes in commensal Neisseriae. Mol. Microbiol. 351550-1559. [DOI] [PubMed] [Google Scholar]
- 25.Sibley, L. D., and J. C. Boothroyd. 1992. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature 35982-85. [DOI] [PubMed] [Google Scholar]
- 26.Soldati, D., and J. C. Boothroyd. 1995. A selector of transcription initiation in the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 1587-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su, C., D. Evans, R. H. Cole, J. C. Kissinger, J. W. Ajioka, and L. D. Sibley. 2003. Recent expansion of Toxoplasma through enhanced oral transmission. Science 299414-416. [DOI] [PubMed] [Google Scholar]
- 28.Van, T. T., S. K. Kim, M. Camps, J. C. Boothroyd, and L. J. Knoll. 2007. The BSR4 protein is up-regulated in Toxoplasma gondii bradyzoites, however the dominant surface antigen recognized by the P36 monoclonal antibody is SRS9. Int. J. Parasitol. 37877-885. [DOI] [PubMed] [Google Scholar]
- 29.Weiser, J. N., M. Shchepetov, and S. T. H. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic. Infect. Immun. 65943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]