Abstract
Activation of the innate immune system by bacterial DNA and DNA of other invertebrates represents a pathogen recognition mechanism. In this study we investigated macrophage responses to DNA from the intestinal protozoan parasite Entamoeba histolytica. E. histolytica genomic DNA was purified from log-phase trophozoites and tested with the mouse macrophage cell line RAW 264.7. RAW cells treated with E. histolytica DNA demonstrated an increase in levels of tumor necrosis factor alpha (TNF-α) mRNA and protein production. TNF-α production was blocked by pretreatment with chloroquine or monensin. In fact, an NF-κB luciferase reporter assay in HEK cells transfected with human TLR9 demonstrated that E. histolytica DNA signaled through Toll-like receptor 9 (TLR9) in a manner similar to that seen with CpG-ODN. Immunofluorescence assays confirmed NF-κB activation in RAW cells, as seen by nuclear translocation of the p65 subunit. Western blot analysis demonstrated mitogen-activated protein kinase activation by E. histolytica DNA. E. histolytica DNA effects were abolished in MYD88−/− mouse-derived macrophages. In the context of disease, immunization with E. histolytica DNA protected gerbils from an E. histolytica challenge infection. Taken together, these results demonstrate that E. histolytica DNA is recognized by TLR9 to activate macrophages and may provide an innate defense mechanism characterized by the induction of the inflammatory mediator TNF-α.
The vertebrate immune system has evolved to recognize pathogen-associated molecular patterns (PAMP) in order to counteract bacterial, viral, and parasitic infections (18). The Toll-like receptor (TLR) family of proteins specifically recognizes these PAMPs and activates antipathogen responses characterized by Th1 cytokines (1). The role of TLRs is to discriminate self from “non-self” molecules, such as unmethylated CpG DNA. Mammalian DNA is highly methylated at the CG dinucleotide whereas bacterial DNA and DNA of other invertebrates is not, and this feature is key to its immunostimulatory activity (27, 40). CpG DNA must first be endocytosed, and upon endosomal acidification it is specifically engaged by TLR 9 (TLR9) (17, 28). The activated receptor initiates a signaling cascade, starting with the recruitment of MYD88, IRAK, and TRAF6, which ultimately leads to the downstream activation of MAPK and NF-κB (10, 24). CpG DNA demonstrates mitogenic activity with respect to B cells (16, 27) and activates dendritic cells and macrophages to produce proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α), interleukin-1 (IL-1), and IL-6 (25, 26).
The intestinal protozoan parasite Entamoeba histolytica is the etiological agent of amebiasis in humans. Although amebiasis remains a prevalent disease in developing countries, only 10 to 14% of infected individuals progress to invasive amebiasis characterized by amebic colitis. The pathological events or immune events leading to amebic invasion are still unknown; however, recent epidemiological studies revealed acquired resistance and inherited susceptibility to amebiasis (2, 13, 14). Many studies have also established that cell-mediated or Th1 immunity is of importance in host defense against amebiasis. Lymphocytes from individuals who have recovered from invasive amebiasis display amebicidal activity and secrete high amounts of IL-2 and gamma interferon (IFN-γ) (33, 34). Other cells demonstrate amebicidal activity such as activated neutrophils and macrophages (35-37), and TNF-α as well as nitric oxide has a crucial role in initiating this effect.
Th1 polarization in response to amebic infection could be due to innate activation of TLR. It is obvious that NF-κB must be involved in the observed inflammation response, which can lead to tissue damage; however, it may also be required for disease resolution. It has been shown that inhibition of NF-κB blocks the inflammatory response to ameba in human intestinal xenograft models (38), whereas patients given corticosteroids have increased disease severity (21). Several parasite molecules could be involved in early immune activation events. E. histolytica lipopeptidophosphoglycan has been shown to signal through TLR2 and TLR4, inducing Th1 cytokines in monocytes (30). The parasite cell surface lectin galactose-N-acetyl-d-galactosamine-inhibitable lectin (Gal-lectin) is well characterized and known to induce direct dendritic cell and macrophage activation (19, 35, 36). The role of other parasite molecules in innate immunity and Th1 polarization remains unclear. Given the immunogenicity of foreign DNA and its role in Th1 induction via TLR9, there is a potential role for E. histolytica DNA in the activation of innate immunity.
In this study, we determined whether E. histolytica DNA could activate naive murine macrophages. Herein we demonstrate that E. histolytica DNA activates macrophages to produce TNF-α and that this activation is mediated through TLR9. Blocking endosomal maturation greatly reduced TNF-α expression; furthermore, this expression was lost in MYD88−/− bone marrow-derived macrophages. Repeated injection of E. histolytica DNA in gerbils conferred protection from amebic challenge infection and was characterized by IFN-γ expression. Thus, we identified another parasite molecule capable of activating an innate immune response which may contribute to host defense against amebiasis.
MATERIALS AND METHODS
Reagents and parasites.
E. histolytica strain HM1:IMSS trophozoites were grown in axenic culture in TYI-S-33 medium (20). Parasites were grown for 72 h (log phase) for use in all experiments. The native Gal-lectin was purified from amebae on an immunoaffinity column as previously described (32). E. histolytica DNA was extracted from trophozoites as previously described (3). Briefly, the parasite pellet was frozen in liquid nitrogen and ground to a fine powder with mortar and pestle. The pellet was resuspended in lysis buffer containing 1 mg/ml proteinase K (Invitrogen) and incubated for 30 min at 65°C. The lysate was subjected to a series of phenol:chloroform extractions, and DNA was precipitated with ethanol. DNA was treated with RNase A (Qiagen) for 30 min at 37°C before use in experiments. Lipopolysaccharide (LPS) (Escherichia coli serotype 0111:B4; Sigma-Aldrich, St. Louis, MO) was used at 100 ng/ml or 1 μg/ml as indicated. CpG-ODN 10103 (TCG TCG TTT CGT CGT TTT GTC GTT) with a full phosphorothioate backbone was purchased from Coley Pharmaceutical Group (Kanata, Canada) and used at 2 μM. Calf thymus DNA (Sigma) was used as a negative control at 30 μg/ml. Salmonella genomic DNA was purified as previously described (31) and used as a positive control at 30 μg/ml. Chloroquine and monensin (Sigma) were used for blocking experiments, and DNase I (Qiagen) was used to digest DNA overnight. Polymyxin B sulfate was purchased from Sigma.
Cell culture.
The mouse macrophage cell line RAW 264.7 and HEK 293 cells were cultivated in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, 10 mM HEPES, and 5 mg/ml penicillin-streptomycin. Cells from the L929 mouse fibroblast cell line were cultivated in minimal essential medium (Invitrogen) supplemented as described above. Bone marrow-derived macrophages were propagated from wild-type (C57BL/6) or MYD88−/− mice as previously described (19). Briefly, femurs and tibias were removed, and the bone marrow was flushed using a 27-gauge syringe. Red blood cells were lysed with red blood cell lysing buffer (Sigma) and washed three times in RPMI 1640 medium (Invitrogen). Cells were cultured in six-well plates (2.5 × 105 cells/ml) in RPMI 1640 media supplemented with 5% fetal bovine serum, 10 mM HEPES, 50 μM 2-mercaptoethanol, 2 mM glutamine, 5 mg/ml penicillin-streptomycin sulfate and 10 ng/ml recombinant moose GM-CSF (R&D Systems, Minneapolis, MN). Nonadherent cells were removed after 5 days, and remaining cells were cultured another 48 h and then subcultured into new plates to enrich the macrophage population.
Real-time PCR.
RAW cells or bone marrow-derived macrophages were stimulated with the indicated antigens or media alone for 3 h. Total mRNA was extracted using Trizol (Invitrogen) following the manufacturer's instructions. DNase-treated total mRNA (1 to 2 μg) was reverse transcribed with Moloney murine leukemia virus enzyme and random hexamer primers (Invitrogen). Real-time PCR analysis using SYBR green (Qiagen) and mouse-specific primers (9) was used to determine relative cytokine gene expression levels. Cytokine gene expression levels are represented as severalfold increases over RPMI-stimulated control results corrected for the presence of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) housekeeping gene as determined by the 2ΔΔct method (9). Real-time DNA amplification assays were conducted in triplicate using a Rotor-Gene 3000 system (Corbett Research). For gerbil cytokine expression determinations, spleen cells were stimulated in vitro for 18 h and mRNA was extracted and cDNA prepared as described above. Real-time PCR was performed as described above but with gerbil-specific Taqman probes (20).
TNF-α bioassay and Western blot analysis.
The L929 mouse fibroblast cell line was used in a TNF-α bioassay with treated RAW cell supernatants. L929 cells were trypsinized and resuspended at a cell density of 3.5 × 105 cell/ml. A 100-μl cell suspension was added to each well in a 96-well plate, and cells were grown overnight. The following day the media was removed and replaced with minimal essential medium containing 50 μg/ml cycloheximide and either TNF-α standards or 24-h-stimulated RAW cell supernatants. Plates were incubated for 18 h at 37°C and then analyzed for cell viability with crystal violet stain. Cell supernatants were also concentrated and separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred to nitrocellulose and probed for TNF-α protein (Santa Cruz).
Griess reaction.
NO production in RAW cell culture supernatants (96 h stimulation) was measured as nitrite accumulation in a Griess reaction assay. Cell supernatants (50 μl/well) were transferred to a 96-well plate and mixed with equal volumes of 0.1% N-(1-naphtyl)-ethylenediamine dihydrochloride (Sigma) and 1% sulfanilamide (Sigma) in 2.5% H3PO4. Absorbance was read at 540 nm and compared to a sodium nitrite (NaNO2) standard curve.
Mitogen-activated protein kinase (MAPK) Western blotting.
RAW cells were stimulated with the indicated antigens for 30 min. The cells were washed, and the cell pellet was collected. Cytoplasmic extracts were prepared with NE-PER nuclear and cytoplasmic extraction reagents (Pierce) following the manufacturer's instructions. Cytoplasmic proteins (30 μg) were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. The membrane was blocked with a 5% milk solution, washed with Tween-Tris-buffered saline, and incubated with primary antibody against phosphorylated p38, Jun N-terminal protein kinase (JNK), and extracellular signal-regulated kinase (ERK) (Santa Cruz). Bound antibody was visualized using secondary horseradish peroxidase-conjugated antibody and enhanced chemiluminescence (ECL Plus reagent; Amersham).
Immunofluorescence.
RAW cells (1.5 × 106 cell/ml) were grown on glass coverslips overnight for use in an immunofluorescence assay. The following day, cells were washed and treated with LPS (1 μg/ml), E. histolytica DNA (30 μg/ml), or control DNA (30 μg/ml) for 45 min at 37°C. After incubation, cells were washed and fixed with 100% methanol at −20°C for 5 min. Cells were washed again and permeabilized with 2% Igepal (Sigma) for 5 min at room temperature. Cells were washed again and then blocked with 1% bovine serum albumin-phosphate-buffered saline (bovine serum albumin-PBS) for 30 min at room temperature and washed thoroughly. To visualize NF-κB translocation, cells were stained with p65 antibody (Santa Cruz 8008) diluted 1:200 in PBS-0.1% bovine serum albumin for 1 h at room temperature in a humidified chamber. Cells were washed and stained with anti-mouse fluorescein isothiocyanate-conjugated secondary antibody (Chemicon) for 30 min as described above. After incubation the cells were washed and mounted with VectaShield onto glass slides. Slides were visualized using a Nikon Eclipse E400 fluorescence microscope.
Transient transfection and reporter assay.
HEK 293 cells were seeded at 6.0 × 105 cell/well and transfected overnight in six-well plates by a CaCl2 precipitation method with 1 μg pNFKB-Luc (Stratagene) and 1 μg pcDNA human TLR9 or control plasmid. After overnight culture growth the transfection medium was removed and cells were stimulated with various stimuli for 12 h. Cell lysates were assayed for luciferase activity by use of a luciferase report assay system (Promega). Luciferase activity was measured with a luminometer (Victor II; Wallac) and was normalized to control transfection results and is expressed as severalfold increases over matched control results.
Animal immunizations.
Male gerbils (Meriones unguiculatus) (Charles River, St. Constant, Canada) (six to nine weeks old) were injected intramuscularly in the hind legs with 50 μg E. histolytica DNA only, 10 μg of Gal-lectin and 50 μg E. histolytica DNA, or 10 μg Gal-lectin only in 100 μl PBS. Gerbils received identical booster injections at 7 and 14 days after the initial injection. At day 20, gerbils were anesthetized and challenged via intrahepatic injection of 106 E. histolytica amebae (HM1:IMSS) into the left liver lobe as previously described (6). Gerbils were sacrificed at 10 days postchallenge, and spleens and serum samples were collected. Livers were removed, and amebic liver abscess (ALA) weight relative to total liver weight was measured. All protocols in this study were carried out with the approval of the University of Calgary Animal Care Committee.
Statistical analysis.
Differences between experimental groups in the mean values were compared using Student's unpaired t test. P values < 0.05 are considered significant.
RESULTS
Induction of TNF-α by E. histolytica DNA.
CpG DNA is known to induce a range of cytokines in macrophages, including IL-12, TNF-α, IL-1β, and IL-6 (26). We first evaluated the ability of E. histolytica DNA to activate macrophages by determining TNF-α induction. RAW mouse macrophages were stimulated with increasing amounts of E. histolytica DNA, and TNF-α mRNA expression was measured by real-time PCR. As shown in Fig. 1A, E. histolytica DNA induces increased TNF-α mRNA expression in a dose-dependent manner. Treatment of E. histolytica DNA with polymyxin B did not alter the TNF-α response in macrophages, indicating that the activation was not due to LPS contamination (data not shown). We also compared TNF-α induction by E. histolytica DNA to results obtained with CpG-ODN and LPS, both of which represent PAMPs known to activate macrophages. E. histolytica DNA-induced TNF-α expression was not as high as LPS- and CpG-ODN-positive control results; however, it was significantly higher than that seen with the nonstimulatory control GpC-ODN (Fig. 1B). Additionally, E. histolytica DNA was able to stimulate Il-1β and Il-6 mRNA expression (data not shown).
FIG. 1.

E. histolytica DNA activates macrophage TNF-α production. (A) Real-time PCR analysis was performed on RAW 264.7 macrophage mRNA collected after 3 h of in vitro incubation with increasing amounts of E. histolytica (E.h) DNA (1 to 25 μg/ml). Data are represented as severalfold increases over medium-only control results for mRNA expression. (B) TNF-α mRNA levels of RAW 264.7 macrophages treated with CpG-ODN or GpC-ODN (2 μM), E. histolytica DNA (30 μg/ml), or LPS (1 μg/ml) for 3 h. Data points represent means ± standard errors of the means of the results obtained from three experiments.
In order to confirm that the observed increase in TNF-α mRNA corresponds to an increase in cytokine release from macrophages we performed a bioassay with mouse L929 fibroblasts which are sensitive to TNF-α. RAW cells were treated for 24 h and cell culture supernatants added to the responder cells for 18 h. In agreement with the real-time PCR data, LPS and E. histolytica DNA were both positive (P < 0.05) for the production of TNF-α protein, as was Salmonella enterica serovar Typhimurium DNA (Fig. 2). In contrast, calf thymus DNA and DNase-treated E. histolytica DNA did not induce significant amounts of TNF-α secretion. These data implied that macrophages were activated by E. histolytica DNA and thus we focused our attention on the characterization of this response.
FIG. 2.
E. histolytica DNA induces TNF-α protein production. RAW 264.7 macrophages were treated in vitro for 24 h with media only, LPS (1 μg/ml), E. histolytica (E.h) DNA (30 μg/ml), S. enterica serovar Typhimurium (S.t) DNA (30 μg/ml), calf thymus (C. thymus) DNA (30 μg/ml), or DNase-treated E. histolytica DNA (30 μg/ml). Culture supernatants were used in an L929 TNF-α bioassay. Data points represent means ± standard errors of the means of the results obtained from three experiments. An asterisk indicates a significant increase in TNF-α protein level compared to results seen with cells treated with media only (P < 0.05). The blot represents the corresponding bands for TNF-α protein (17 kDa) in culture supernatants.
Effect on nitric oxide production.
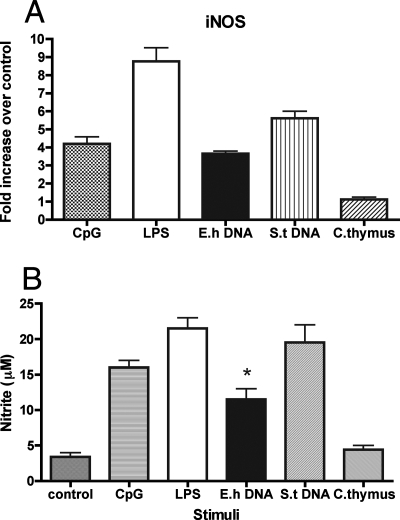
Activated macrophages are a source of nitric oxide, which helps generate effective innate immunity against viruses, bacteria, and parasites. Nitric oxide is also the main cytotoxic molecule produced by activated macrophages for cytotoxicity against E. histolytica. We evaluated the effect of various stimuli on macrophage nitric oxide production. In the experiment shown in Fig. 3 A, we measured iNOS mRNA expression in control and treated macrophages. The data show that E. histolytica DNA could increase inducible nitric oxide synthase (iNOS) mRNA to an extent similar to that seen with CpG-ODN, while LPS and S. enterica serovar Typhimurium DNA induced higher mRNA expression levels. Calf thymus DNA, however, could not induce an increase in iNOS mRNA levels. We used 96-h culture supernatants in a Griess reaction to indirectly measure nitric oxide production by stimulated macrophages. E. histolytica DNA was able to induce nitric oxide production significantly more than control and calf thymus DNA (P < 0.05). The amount of NO measured, however, was less than that seen with LPS, CpG-ODN, or S. enterica serovar Typhimurium DNA stimulation (Fig. 3B).
FIG. 3.
E. histolytica DNA activates macrophage iNOS production. (A) Real-time PCR analysis was performed on IFN-γ (100 U/ml)-primed RAW 264.7 macrophage mRNA collected after 6 h of in vitro incubation with CpG-ODN (2 μM), LPS (1 μg/ml), E. histolytica (E.h) DNA (30 μg/ml), S. enterica serovar Typhimurium (S.t) DNA (30 μg/ml), and calf thymus (C. thymus) DNA (30 μg/ml). Data are represented as severalfold increases over medium-only control results for mRNA expression. (B) Macrophages were stimulated as described above for 24 h, and culture supernatants were used in a Griess reaction to measure nitrite levels. Data points represent means ± standard errors of the means of the results obtained from three experiments. An asterisk indicates a significant increase in nitrite level compared to the results obtained with cells treated with media only (P < 0.05).
Inhibition of acidification.
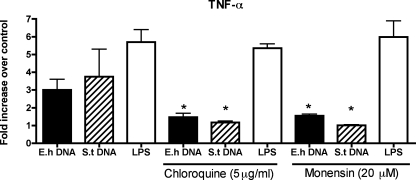
TLR9 is activated intracellularly by bacterial DNA and CpG-ODNs by translocation from the endoplasmic reticulum to endosomes containing the CpG DNA (28). Endosomal acidification and maturation seem to be required for proper TLR9 signaling. We tested the effects of chloroquine (5 μg/ml) and monensin (20 μM), both inhibitors of endosomal acidification, on macrophage TNF-α expression after exposure to various stimuli. Pretreatment of RAW macrophages with chloroquine or monensin reduced the potency of both E. histolytica DNA and S. enterica serovar Typhimurium DNA (P < 0.05) but not that of LPS (Fig. 4). In fact, retained activity with LPS demonstrated specificity for TLR9 signaling and also demonstrated that the concentration of inhibitors used, which could have accounted for reduced mRNA levels, did not cause cytotoxicity.
FIG. 4.
TNF-α expression by E. histolytica DNA-stimulated macrophages requires endosomal acidification. RAW 264.7 macrophages were pretreated for 30 min with or without chloroquine (5 μg/ml) or monensin (20 μM) and subsequently stimulated for 3 h with E. histolytica (E.h) DNA (30 μg/ml), S. enterica serovar Typhimurium (S.t) DNA (30 μg/ml), or LPS (1 μg/ml). mRNA was collected, and the level of TNF-α expression was measured by real-time PCR. Data points represent means ± standard errors of the means of the results obtained from three experiments. An asterisk indicates a significant decrease in level compared to the results obtained with cells not treated with inhibitors (P < 0.05).
HEK-TLR9 NF-κB reporter assay.
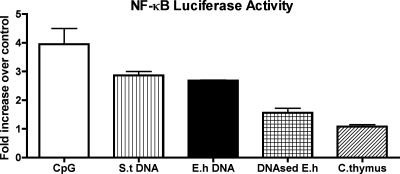
We suspected that TLR9 might be involved in E. histolytica DNA macrophage activation, in which case NF-κB would be involved in proinflammatory cytokine production. HEK 293 cells were transiently transfected with human TLR9 and an NF-κB-luciferase reporter plasmid and treated with various stimuli. The most effective NF-κB activator was CpG-ODN followed by S. enterica serovar Typhimurium DNA and E. histolytica DNA (Fig. 5). In contrast, DNase-treated E. histolytica DNA had reduced NF-κB activity whereas calf thymus DNA did not significantly induce any activation.
FIG. 5.
TLR9-dependent NF-κB activation by E. histolytica DNA. HEK 293 cells were transiently transfected with pcDNA-human TLR9 (pcDNA-hTLR9) and NF-κB luciferase reporter plasmid and treated with CpG-ODN (2 μM), S. enterica serovar Typhimurium (S.t) DNA (30 μg/ml), E. histolytica (E.h) DNA (30 μg/ml), DNAse-treated E. histolytica DNA (30 μg/ml), or calf thymus (C. thymus) DNA (30 μg/ml). Luciferase induction was measured and is reported as severalfold increases in level compared to the results obtained with cells transfected with empty pcDNA plasmid. Data points represent means ± standard errors of the means of the results obtained from three experiments.
Impaired response to E. histolytica DNA in MYD88−/− cells.
MYD88 is an adaptor protein in the signal transduction pathway mediated by Toll receptors, and it is essential for mounting an innate immune response. We propagated bone marrow-derived macrophages from wild-type and MYD88−/− mice to assess the biological function of TLR signaling in response to E. histolytica DNA. In wild-type macrophages both LPS and E. histolytica DNA induced high TNF-α mRNA expression levels (Fig. 6A and B). In contrast, MYD88−/− macrophages showed significantly reduced TNF-α expression in response to E. histolytica DNA. LPS also had reduced potency in the MYD88−/− macrophages but to a lesser extent than E. histolytica DNA. Thus, E. histolytica DNA-mediated signal transduction is dependent on MYD88, a key adaptor protein in TLR9 signaling.
FIG. 6.
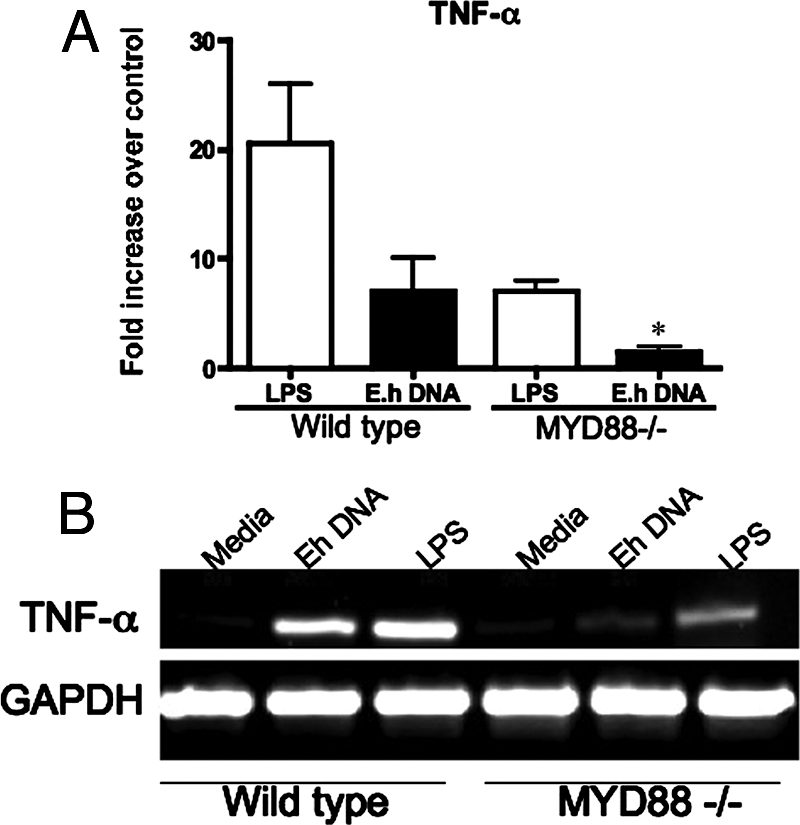
MYD88 is required for E. histolytica DNA-induced TNF-α expression. Bone marrow-derived macrophages were propagated from wild-type and MYD88−/− mice and stimulated in vitro for 3 h with media, LPS (1 μg/ml), or E. histolytica (E.h) DNA (30 μg/ml). TNF-α mRNA expression was measured by real-time PCR (A), and PCR products were processed on 1.5% agarose gels (B). Data points represent means ± standard errors of the means of the results obtained from three experiments. An asterisk indicates a significant decrease in TNF-α expression compared to wild-type macrophage results (P < 0.05).
MAPK and NF-κB activation by E. histolytica DNA.
MYD88 is known to be involved in the activation of all three MAPK pathways in TLR signaling. Therefore, we analyzed MAPK activation in response to various stimuli known to require TLR signaling. Cells were treated for 30 min, and cellular extracts were probed in Western blot analyses using phosphorylated MAPK antibodies. CpG-ODN, LPS, and S. enterica serovar Typhimurium DNA as well as E. histolytica DNA all induced phosphorylation of p38, ERK, and JNK whereas calf thymus DNA and medium alone did not (Fig. 7). Next, we confirmed the activation of NF-κB by detecting p65 subunit translocation into the nucleus by immunofluorescence. Treated and untreated cells were stained with a p65 antibody and visualized for fluorescence. RAW macrophages that were not treated or were treated with calf thymus DNA showed p65 staining in the cytoplasm, indicating no NF-κB activation (Fig. 8a and b). However, macrophages treated with S. enterica serovar Typhimurium DNA, LPS, or E. histolytica DNA exhibited strong p65 staining in the nucleus indicative of p65 translocation and NF-κB activation (Fig. 8c, d, and e). Thus, E. histolytica DNA activates NF-κB in a manner similar to that seen with LPS and bacterial DNA.
FIG. 7.
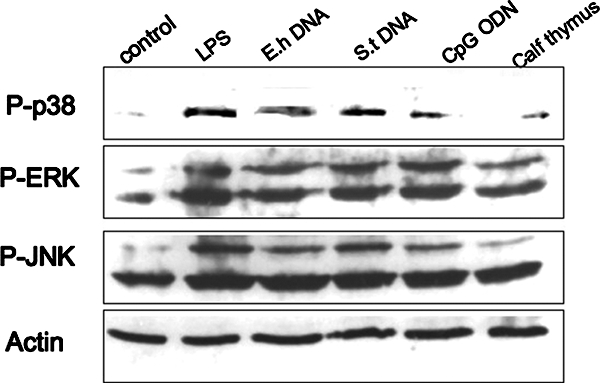
Macrophage activation by E. histolytica DNA is mediated by MAPK. RAW 264.7 macrophages were stimulated for 30 min with medium only (control), LPS (1 μg/ml), E. histolytica (E.h) DNA (30 μg/ml), S. enterica serovar Typhimurium (S.t) DNA (30 μg/ml), CpG-ODN (2 μM), and calf thymus DNA (30 μg/ml). Cellular extracts were separated by electrophoresis and then blotted and probed with antibodies specific for phosphorylated ERK, JNK, or p38. Total actin antibody was used as a loading control. Data shown are from one experiment and are representative of the results of three independent experiments.
FIG. 8.
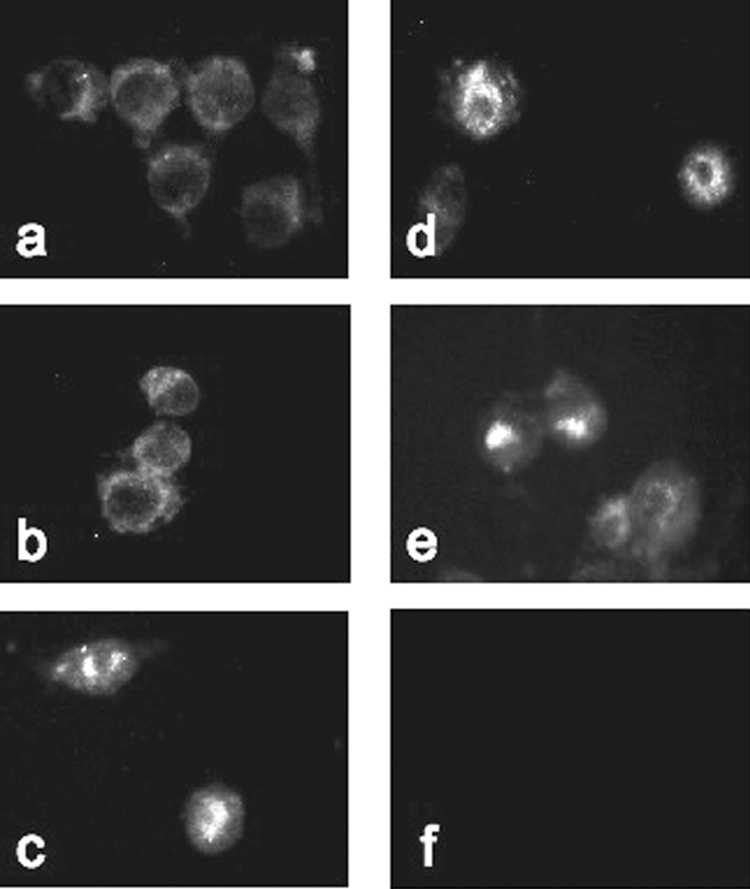
Immunofluorescence analysis of NF-κB activation. RAW 264.7 cells were treated in vitro for 45 min with medium (a), calf thymus DNA (b), S. enterica serovar Typhimurium DNA (c), LPS (d), or E. histolytica DNA (e) and assessed for p65 subunit localization. Isotype controls were stimulated with LPS and probed with normal mouse immunoglobulin G (f). Images are representative of the results of three independent experiments.
Interplay between LPS and E. histolytica DNA.
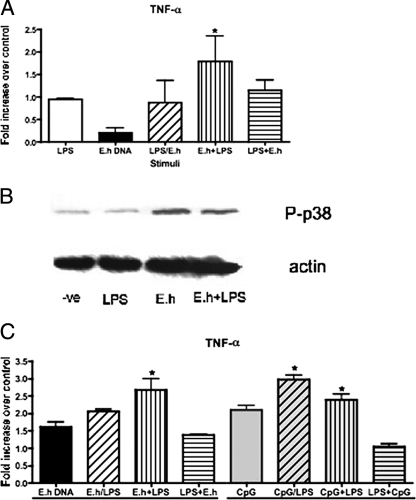
Desensitization, or tolerance, can occur in vitro with submaximal doses of LPS that reduce macrophage effector functions in response to subsequent stimulation with other activators. Similarly, activation through MYD88-dependent pathways can prime cells to respond to LPS in a MYD88-independent way (4). In the case of amebiasis, not all patients develop invasive disease and the factors involved in susceptibility are yet unknown. To consider the possibility that E. histolytica DNA could alter immune activation or deactivation, we examined the interplay with LPS. We determined the submaximal doses of LPS and E. histolytica DNA (data not shown) and used those concentrations to evaluate the interplay between LPS and E. histolytica DNA macrophage activation. Submaximal doses of LPS and E. histolytica DNA alone did not induce TNF-α expression (Fig. 9A); neither did treatment with both submaximal doses simultaneously. Pretreatment of cells with E. histolytica DNA, however, increased the subsequent response to submaximal LPS stimulation. On the other hand, treatment with LPS tolerized the response to E. histolytica DNA, as this order of stimulation induced less TNF-α expression. In fact, Western blot analysis demonstrated that submaximal LPS doses do not induce p38 phosphorylation, while pretreatment with E. histolytica DNA could increase this phosphorylation event (Fig. 9B). We compared the priming effects of submaximal E. histolytica DNA to those of CpG-ODN and found that TNF-α expression patterns were similar with both treatments (Fig. 9C). In both cases, pretreatment with CpG DNA (ODN or E. histolytica) increased the response to LPS (P < 0.05) whereas pretreatment with LPS reduced the response to CpG DNA.
FIG. 9.
E. histolytica DNA pretreatment enhances subsequent activation of macrophages by submaximal LPS. (A) RAW 264.7 cells were left untreated or pretreated for 6 h with submaximal LPS (0.1 ng/ml) or E. histolytica (E.h) DNA (1 μg/ml). After pretreatment, cells were washed and treated for 3 h with medium or with LPS or E. histolytica DNA or with both simultaneously (LPS/E.h) at the concentrations specified above. TNF-α mRNA expression was measured by real-time PCR. (B) Western blot analysis of p38 phosphorylation in cells treated with medium (-ve), LPS (0.1 ng/ml), or E. histolytica DNA (1 μg/ml) or pretreated with E. histolytica DNA as described above and subsequently stimulated with LPS. (C) TNF-α mRNA expression in cells treated as described above or treated with CpG-ODN (2 μM) instead of E. histolytica DNA. Data points represent means ± standard errors of the means of the results obtained from three experiments. An asterisk indicates a significant increase in TNF-α expression compared to the results seen with cells treated with submaximal doses alone (P < 0.05).
In vivo proinflammatory effect of E. histolytica DNA.
Finally, we addressed in vivo response to E. histolytica DNA in the gerbil model of amebiasis. We evaluated the protective potential of E. histolytica DNA in comparison to that of Gal-lectin, a known parasite immunogen. Animals were immunized with 50 μg E. histolytica DNA or 10 μg Gal-lectin or both E. histolytica DNA and Gal-lectin. Upon challenge infection with live amebic trophozoites, E. histolytica DNA alone and Gal-lectin alone demonstrated equal levels of protective immunity against amebic liver abscess formation (Table 1). Immunization with both E. histolytica DNA and Gal-lectin, however, conferred complete protection. Spleen cells were isolated from immunized animals, and cytokine expression was analyzed by real-time PCR. Figure 10 shows that both E. histolytica DNA and Gal-lectin induced the expression of IL-4 and IFN-γ but that animals immunized with both antigens had significantly higher levels of cytokine expression, specifically, of IFN-γ expression (P < 0.05).
TABLE 1.
Prevention of ALA in gerbils by immunization with E. histolytica DNA
| Immunization group | % (± SEM) ALA of total liver wt | Viable parasitesa | % Efficacyb |
|---|---|---|---|
| PBS | 12 ± 4 | + | 0 |
| E. histolytica DNA | 26 ± 5 | + | 33 |
| Gal-lectin | 3 ± 3 | + | 25 |
| E. histolytica DNA + Gal-lectin | 0 | − | 100c |
ALA contents were aspirated, and viable trophozoite levels were determined by in vitro culture. +, presence; −, absence.
Treatment efficacy to protect against ALA = 100 × (protected gerbil results/infected gerbil results); n = 6/group.
P < 0.05.
FIG. 10.
Taqman real-time PCR analysis of spleen cytokine gene expression. Gene expression levels were normalized to 18S rRNA and are represented as severalfold increases over the results seen with normal nontreated gerbil mRNA. There were higher levels of IFN-γ and IL-4 in the protected gerbils (*, P < 0.05). Data points represent means ± standard errors of the means of results from three independent PCRs. Eh, E. histolytica.
DISCUSSION
The mammalian immune system recognizes foreign DNA through TLR9 expressed on immune cells. This recognition is ascribed to unmethylated CpG motifs, which are present at a high frequency in DNA of bacteria and other pathogens. In the event of an E. histolytica infection, the lysis of trophozoites may allow the intestinal immune system to sample amebic DNA and induce an inflammatory response. In this report we show that E. histolytica DNA stimulates macrophage activation in a TLR9-dependent manner similar to that seen with CpG DNA. Our studies clearly demonstrate that E. histolytica DNA is a stimulus for macrophage activation. As is consistent with the activation potential of CpG DNA, E. histolytica DNA induced TNF-α expression in murine macrophages. This cytokine expression was blocked by the presence of chloroquine and monensin, indicating signaling via TLR9. Moreover, we found that E. histolytica DNA could induce NF-κB activation in HEK-TLR9-transfected cells. TLR signaling was confirmed by the requirement for the presence of MYD88 for E. histolytica DNA activity. Taken together, these results demonstrate that E. histolytica DNA can trigger innate immune responses.
Monocytes-macrophages, dendritic cells, and B cells recognize unmethylated CpG dinucleotides through TLR9 (28). It is well established that bacterial DNA is a potent immune stimulator due to the high frequency of these unmethylated CpG motifs. In contrast to bacterial DNA, E. histolytica DNA has an AT-rich (75.3%) genome and CG dinucleotides are underrepresented (22). It has been reported that E. histolytica has a DNA methyltransferase of the DNMT2 family which targets rRNA (29). Methylation seems to occur mostly at AT dinucleotides and not at the common CpG site as in vertebrate systems (5). It is interesting that despite the differences between bacteria and this parasite in DNA composition characteristics, E. histolytica DNA was still able to induce some activation in macrophages. However, the variation in the magnitude of responses could be attributed to these innate differences in DNA composition, mainly, the degree of difference in CpG frequency methylation patterns. The observation that S. enterica serovar Typhimurium DNA and CpG-ODN had greater activation potential than E. histolytica DNA was consistent with this theory. Other protozoan parasite DNAs such as that of Trypanosoma brucei have also been reported to activate immune cells; however, the extent of activation is lower than that seen with bacterial DNA due to differential unmethylated CpG frequencies (11).
Prominent inflammation is a major component of amebiasis, and NF-κB activation is known to be required for initiation of this response in cases of amebic infection (38). NF-κB controls the gene expression of proinflammatory cytokines such as IL-1, IL-6, and TNF-α. Here we report that E. histolytica DNA can activate NF-κB via TLR9 signaling and can induce the production of the proinflammatory cytokine TNF-α as well as nitric oxide in macrophages. It has been shown that activation of macrophages with Th1 cytokines results in amebicial activity mediated by nitric oxide production and that this activation may be essential for innate immunity against the parasite (35, 36, 39). Since macrophages have an important role in disease protection, it was important to identify a parasite molecule that was able to stimulate a cell-mediated response. As is consistent with the crucial role of cellular responses, our immunization studies demonstrated resistance to amebic infection mediated by IFN-γ production. In humans, higher IFN-γ production has been correlated with a reduced risk of amebic disease (12). IFN-γ is a key Th1 cytokine which could prime macrophages at the site of infection to respond to amebic DNA and become cytotoxic against the invading parasite. Previous studies have demonstrated that repeated administration of CpG-ODN in mice could protect against lethal bacterial or parasite challenge infections and that this protection is associated with high-level IL-6 and IFN-γ production (15, 23). In this way, E. histolytica DNA injections could have elicited immune activation sufficient to provide some protection against parasite challenge infections. However, combining E. histolytica DNA with another strong Th1 immunogen, Gal-lectin, conferred complete protection mediated by higher Th1 responses.
The majority of cases of E. histolytica infection do not result in invasive disease; however, the nature of the initiation of the invasion process remains unknown. Evidence suggests that modulation of host response may result in the establishment and persistence of amebic infection. It is possible that amebic colitis may be a result of immune responses to direct parasite insult, but it could also be due to a combination of gut immune events that could facilitate invasion. It has been increasingly observed that different TLR agonists can have an impact on the immune response to other TLR ligands, resulting in altered inflammatory responses (7, 8). During infection, the immune system is exposed to various stimuli, and the interplay between them could potentiate disease or attenuate it. In this study we evaluated the priming and tolerance effects of LPS and E. histolytica DNA. LPS is recognized by TLR4, which can signal via MYD88-dependent and MYD88-independent pathways (4), while CpG DNA can only signal in a MYD88-dependent manner (24). Our results demonstrated that E. histolytica DNA could prime macrophages to be responsive to otherwise nonstimulating LPS doses. On the other hand, LPS rendered macrophages less responsive to E. histolytica DNA, causing a tolerance effect. This effect could be due to the MYD88 pathways involved in TLR4 signaling, whereby pretreatment with LPS would down-regulate the potential to subsequently signal through TLR9, as MYD88 would have been activated. The reverse addition of E. histolytica DNA followed by LPS, however, allows subsequent stimulation with LPS to signal through an alternative MYD88-independent pathway. The nature of host susceptibility to amebic infection is unknown; however, one could postulate that previous, concurrent, and subsequent intestinal infections with other pathogens could alter the outcome of amebic infection.
In conclusion, the results reported herein indicate that E. histolytica DNA could represent a PAMP that is recognized by the innate immune system through TLR9. This finding further elucidates the initial proinflammatory host response to amebic infection and identifies another parasite molecule involved in the multifactorial process of amebiasis. Future studies will need to evaluate the activation potential of E. histolytica DNA with other cell types and identify its role in pathogenesis and or protection from disease.
Acknowledgments
This research was funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada. C. P. A. Ivory was the recipient of a Ph.D. scholarship from McGill University, and M. Prystajecky was supported by a Canadian Association of Gastroenterology summer scholarship award.
We thank Douglas Golenbock (University of Massachusetts Medical School) for kindly providing the pcDNA-hTLR9 plasmid and Brigitte Allard (University of North Carolina—Chapel Hill) for breeding the Myd88−/− mice and preparing the marrow.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 5 November 2007.
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406782-787. [DOI] [PubMed] [Google Scholar]
- 2.Arellano, J., M. Perez-Rodriguez, M. Lopez-Osuna, J. R. Velazquez, J. Granados, N. Justiniani, J. I. Santos, A. Madrazo, L. Munoz, and R. Kretschmer. 1996. Increased frequency of HLA-DR3 and complotype SCO1 in Mexican mestizo children with amoebic abscess of the liver. Parasite Immunol. 18491-498. [DOI] [PubMed] [Google Scholar]
- 3.Arteaga-Nieto, P., A. Alvarez-Vargas, C. Cano-Canchola, A. Flores-Carreron, E. Lopez-Romerro, and C. Calvo-Mendez. 2000. Improved purification of DNA from Entamoeba histolytica. Focus 2214-15. [Google Scholar]
- 4.Bagchi, A., E. A. Herrup, H. S. Warren, J. Trigilio, H. S. Shin, C. Valentine, and J. Hellman. 2007. MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J. Immunol. 1781164-1171. [DOI] [PubMed] [Google Scholar]
- 5.Bernes, S., R. Siman-Tov, and S. Ankri. 2005. Epigenetic and classical activation of Entamoeba histolytica heat shock protein 100 (EHsp100) expression. FEBS Lett. 5796395-6402. [DOI] [PubMed] [Google Scholar]
- 6.Chadee, K., and E. Meerovitch. 1984. The pathogenesis of experimentally induced amebic liver abscess in the gerbil (Meriones unguiculatus). Am. J. Pathol. 11771-80. [PMC free article] [PubMed] [Google Scholar]
- 7.Dalpke, A. H., M. D. Lehner, T. Hartung, and K. Heeg. 2005. Differential effects of CpG-DNA in Toll-like receptor-2/-4/-9 tolerance and cross-tolerance. Immunology 116203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrovolskaia, M. A., A. E. Medvedev, K. E. Thomas, N. Cuesta, V. Toshchakov, T. Ren, M. J. Cody, S. M. Michalek, N. R. Rice, and S. N. Vogel. 2003. Induction of in vitro reprogramming by Toll-like receptor (TLR)2 and TLR4 agonists in murine macrophages: effects of TLR “homotolerance” versus “heterotolerance” on NF-kappa B signaling pathway components. J. Immunol. 170508-519. [DOI] [PubMed] [Google Scholar]
- 9.Giulietti, A., L. Overbergh, D. Valckx, B. Decallonne, R. Bouillon, and C. Mathieu. 2001. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25386-401. [DOI] [PubMed] [Google Scholar]
- 10.Häcker, H. 2000. Signal transduction pathways activated by CpG-DNA. Curr. Top. Microbiol. Immunol. 24777-92. [DOI] [PubMed] [Google Scholar]
- 11.Haddad, E. B., M. Birrell, K. McCluskie, A. Ling, S. E. Webber, M. L. Foster, and M. G. Belvisi. 2001. Role of p38 MAP kinase in LPS-induced airway inflammation in the rat. Br. J. Pharmacol. 1321715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haque, R., D. Mondal, J. Shu, S. Roy, M. Kabir, A. N. Davis, P. Duggal, and W. A. Petri, Jr. 2007. Correlation of interferon-gamma production by peripheral blood mononuclear cells with childhood malnutrition and susceptibility to amebiasis. Am. J. Trop. Med. Hyg. 76340-344. [PubMed] [Google Scholar]
- 13.Haque, R., D. Mondal, P. Duggal, M. Kabir, S. Roy, B. M. Farr, R. B. Sack, and W. A. Petri, Jr. 2006. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect. Immun. 74904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haque, R., I. M. Ali, R. B. Sack, B. M. Farr, G. Ramakrishnan, and W. A. Petri, Jr. 2001. Amebiasis and mucosal IgA antibody against the Entamoeba histolytica adherence lectin in Bangladeshi children. J. Infect. Dis. 1831787-1793. [DOI] [PubMed] [Google Scholar]
- 15.Harris, T. H., J. M. Mansfield, and D. M. Paulnock. 2007. CpG ODN treatment enhances innate resistance and acquired immunity to African trypanosomes. Infect. Immun. 755658-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartmann, G., and A. M. Krieg. 2000. Mechanism and function of a newly identified CpG DNA motif in human primary B cells. J. Immunol. 164944-953. [DOI] [PubMed] [Google Scholar]
- 17.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408740-745. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, and R. A. Ezekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 2841313-1318. [DOI] [PubMed] [Google Scholar]
- 19.Ivory, C. P., and K. Chadee. 2007. Activation of dendritic cells by the Gal-lectin of Entamoeba histolytica drives Th1 responses in vitro and in vivo. Eur. J. Immunol. 37385-394. [DOI] [PubMed] [Google Scholar]
- 20.Ivory, C. P., K. Keller, and K. Chadee. 2006. CpG-oligodeoxynucleotide is a potent adjuvant with an Entamoeba histolytica Gal-inhibitable lectin vaccine against amoebic liver abscess in gerbils. Infect. Immun. 74528-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanani, S. R., and R. Knight. 1969. Amoebic dysentery precipitated by corticosteroids. Br. Med. J. 3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlin, S., I. Ladunga, and B. E. Blaisdell. 1994. Heterogeneity of genomes: measures and values. Proc. Natl. Acad. Sci. USA 9112837-12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinman, D. M., J. Conover, and C. Coban. 1999. Repeated administration of synthetic oligodeoxynucleotides expressing CpG motifs provides long-term protection against bacterial infection. Infect. Immun. 675658-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieg, A. M. 2000. Signal transduction induced by immunostimulatory CpG DNA. Springer Semin. Immunopathol. 2297-105. [DOI] [PubMed] [Google Scholar]
- 25.Krieg, A. M. 2000. Immune effects and mechanisms of action of CpG motifs. Vaccine 19618-622. [DOI] [PubMed] [Google Scholar]
- 26.Krieg, A. M. 2002. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 20709-760. [DOI] [PubMed] [Google Scholar]
- 27.Krieg, A. M., A. K. Yi, S. Matson, T. J. Waldschmidt, G. A. Bishop, R. Teasdale, G. A. Koretzky, and D. M. Klinman. 1995. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374546-549. [DOI] [PubMed] [Google Scholar]
- 28.Latz, E., A. Schoenemeyer, A. Visintin, K. A. Fitzgerald, B. G. Monks, C. F. Knetter, E. Lien, N. J. Nilsen, T. Espevik, and D. T. Golenbock. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5190-198. [DOI] [PubMed] [Google Scholar]
- 29.Lavi, T., E. Isakov, H. Harony, O. Fisher, R. Siman-Tov, and S. Ankri. 2006. Sensing DNA methylation in the protozoan parasite Entamoeba histolytica. Mol. Microbiol. 621373-1386. [DOI] [PubMed] [Google Scholar]
- 30.Maldonado-Bernal, C., C. J. Kirschning, Y. Rosenstein, L. M. Rocha, N. Rios-Sarabia, M. Espinosa-Cantellano, I. Becker, I. Estrada, R. M. Salazar-Gonzalez, C. Lopez-Macias, H. Wagner, J. Sanchez, and A. Isibasi. 2005. The innate immune response to Entamoeba histolytica lipopeptidophosphoglycan is mediated by toll-like receptors 2 and 4. Parasite Immunol. 27127-137. [DOI] [PubMed] [Google Scholar]
- 31.Marmur, J., and P. Doty. 1961. Thermal renaturation of deoxyribonucleic acids. J. Mol. Biol. 3585-594. [DOI] [PubMed] [Google Scholar]
- 32.Ravdin, J. I., C. F. Murphy, R. A. Salata, R. L. Guerrant, and E. L. Hewlett. 1985. N-Acetyl-d-galactosamine-inhibitable adherence lectin of Entamoeba histolytica. I. Partial purification and relation to amoebic virulence in vitro. J. Infect. Dis. 151804-815. [DOI] [PubMed] [Google Scholar]
- 33.Salata, R. A., A. Martinez-Palomo, H. W. Murray, L. Conales, N. Trevino, E. Segovia, C. F. Murphy, and J. I. Ravdin. 1986. Patients treated for amebic liver abscess develop cell-mediated immune responses effective in vitro against Entamoeba histolytica. J. Immunol. 1362633-2639. [PubMed] [Google Scholar]
- 34.Schain, D. C., R. A. Salata, and J. I. Ravdin. 1992. Human T-lymphocyte proliferation, lymphokine production, and amebicidal activity elicited by the galactose-inhibitable adherence protein of Entamoeba histolytica. Infect. Immun. 602143-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Séguin, R., B. J. Mann, K. Keller, and K. Chadee. 1995. Identification of the galactose-adherence lectin epitopes of Entamoeba histolytica that stimulate tumor necrosis factor-alpha production by macrophages. Proc. Natl. Acad. Sci. USA 9212175-12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Séguin, R., B. J. Mann, K. Keller, and K. Chadee. 1997. The tumor necrosis factor alpha-stimulating region of galactose-inhibitable lectin of Entamoeba histolytica activates gamma interferon-primed macrophages for amebicidal activity mediated by nitric oxide. Infect. Immun. 652522-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seydel, K. B., T. Zhang, and S. L. Stanley, Jr. 1997. Neutrophils play a critical role in early resistance to amebic liver abscesses in severe combined immunodeficient mice. Infect. Immun. 653951-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seydel, K. B., E. Li, Z. Zhang, and S. L. Stanley, Jr. 1998. Epithelial cell-initiated inflammation plays a crucial role in early tissue damage in amebic infection of human intestine. Gastroenterology 1151446-1453. [DOI] [PubMed] [Google Scholar]
- 39.Seydel, K. B., S. J. Smith, and S. L. Stanley, Jr. 2000. Innate immunity to amebic liver abscess is dependent on gamma interferon and nitric oxide in a murine model of disease. Infect. Immun. 68400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner, H. 1999. Bacterial CpG DNA activates immune cells to signal infectious danger. Adv. Immunol. 73329-368. [DOI] [PubMed] [Google Scholar]