Abstract
Anaerobic ammonium oxidation (anammox) is an ecologically and industrially important process and is performed by a clade of deeply branching Planctomycetes. Anammox bacteria possess an intracytoplasmic membrane-bounded organelle, the anammoxosome. In the present study, the ultrastructures of four different genera of anammox bacteria were compared with transmission electron microscopy and electron tomography. The four anammox genera shared a common cell plan and contained glycogen granules. Differences between the four genera included cell size (from 800 to 1,100 nm in diameter), presence or absence of cytoplasmic particles, and presence or absence of pilus-like appendages. Furthermore, cytochrome c proteins were detected exclusively inside the anammoxosome. This detection provides further support for the hypothesis that this organelle is the locus of anammox catabolism.
Bacteria performing anaerobic ammonium oxidation (anammox) are key players in the global nitrogen cycle and have been applied in wastewater treatment for the removal of ammonium (9, 20, 40). Anammox is the anaerobic oxidation of ammonium with nitrite as the electron acceptor and dinitrogen gas as the product (43). Anammox bacteria form a monophyletic group, or clade, deeply branching inside the phylum Planctomycetes (30, 31, 38). So far, four anammox genera have been described. “Candidatus Kuenenia” (29), “Candidatus Brocadia” (16, 19, 38), and “Candidatus Anammoxoglobus” (14) have all been enriched from activated sludge. Of these three anammox genera, “Candidatus Anammoxoglobus propionicus” and “Candidatus Brocadia fulgida” have also been shown to be very efficient at the oxidation of small organic acids, such as propionate, acetate, and formate, next to ammonium (14, 15). The fourth genus, “Candidatus Scalindua” (20, 28), has often been detected in natural habitats, especially in marine sediments and oxygen minimum zones (23, 31, 47).
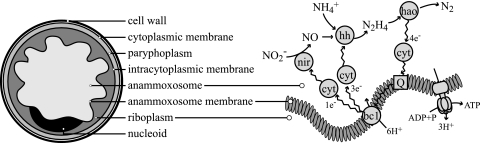
As is the case for all other Planctomycetes, the anammox bacteria have an ultrastructure atypical for bacteria, with a compartmentalized cytoplasm and apparently no real periplasmic space (10, 21). Based on transmission electron microscopy (TEM) studies of “Candidatus Brocadia anammoxidans,” the anammox cell plan was described as follows (21). The cytoplasm is divided into three separate compartments bounded by individual bilayer membranes (Fig. 1). The innermost cytoplasmic compartment, the anammoxosome, is bounded by the anammoxosome membrane, contains iron particles and tubule-like structures (21, 44, 45), and occupies most of the volume of the cell. The second cytoplasmic compartment, the riboplasm, contains ribosomes and the nucleoid and is bounded on the outside by an intracytoplasmic membrane. The third and outermost cytoplasmic compartment, the paryphoplasm, is bounded on the outside by the cytoplasmic membrane. Outside the cytoplasmic membrane resides a putative peptidoglycanless cell wall with no outer membrane. However, it should be noted that the exact organization of the anammox cell envelope and the status of the paryphyoplasm compartment remain uncertain. The recently published metagenome of “Candidatus Kuenenia stuttgartiensis” revealed several characteristics that do suggest the presence of a gram-negative-like cell wall with an outer membrane and periplasmic space (37).
FIG. 1.
Schematic drawing (left), representing the ultrastructure of anammox bacteria and the postulated coupling of the anaerobic oxidation of ammonium (37) to the anammoxosome membrane (right) for the buildup of a proton motive force and subsequent ATP synthesis. nir, nitrite reductase (cytochrome cd1); hh, hydrazine hydrolase; hao, hydrazine/hydroxylamine oxidoreductase (octaheme cyrochrome c) (27, 35); cyt, mono- or diheme cytochrome c electron carriers (8, 11); bc1, cytochrome bc1 complex (complex III); Q, coenzyme Q (ubiquinone).
The function of the anammoxosome has been hypothesized to be the production of energy, which is analogous to the function of mitochondria in eukaryotes (21, 44). In this hypothesis, energy from the anaerobic oxidation of ammonium is conserved in the form of a proton motive force over the anammoxosome membrane. This process has been postulated to be catalyzed by several cytochrome c proteins, as shown in Fig. 1 (37). In this model, nitrite is first reduced to nitric oxide by a cytochrome c- and cytochrome d1-containing nitrite reductase (NirS). Nitric oxide and ammonium are then assumed to be combined into hydrazine by hydrazine hydrolase, and the latter is finally oxidized to dinitrogen gas by hydrazine/hydroxylamine oxidoreductase, an octaheme cytochrome c (27, 35). The four electrons derived from this oxidation are transferred first to soluble cytochrome c electron carriers (8, 11) and then to ubiquinone, followed by the cytochrome bc1 complex (complex III), other soluble cytochrome c electron carriers, and finally to nitrite reductase and hydrazine hydrolase. In theory, this results in the buildup of a proton motive force that could be used to drive ATP synthesis. The immunogold localization of one of the key enzymes (hydrazine/hydroxylamine oxidoreductase [Fig. 1]) to the anammoxosome provided the first (and only) proof that the mechanism described in Fig. 1 might occur inside the anammoxosome (21). However, the anammoxosome membrane is predominantly in a curved configuration (45) and the function of this curvature could be to enlarge the membrane area available for the membrane-associated catabolic process just described.
In this study, we used TEM and electron tomography to investigate whether the anammox cell plan was conserved in all four anammox genera described so far: “Candidatus Kuenenia stuttgartiensis,” “Candidatus Brocadia fulgida,” “Candidatus Anammoxoglobus propionicus,” and “Candidatus Scalindua spp.” Additional ultrastructural features were also observed. For all genera, riboplasmic granules were identified as glycogen by glycogen staining. “Candidatus Brocadia fulgida” and “Candidatus Anammoxoglobus propionicus” contained additional riboplasmic particles of unknown identity. Further, TEM showed the presence of anammox cells with pilus-like appendages among “Candidatus Scalindua spp.” Finally, we used the intrinsic peroxidase activity of cytochrome c to investigate the hypothesis that the anaerobic ammonium oxidation takes place within the anammoxosome. Staining for peroxidase activity occurred only inside the anammoxosome, with profound accumulation of reaction product along a 150-nm rim on the inside of the anammoxosome membrane. This result provided further indication that the anammoxosome might be functioning as an energy-generating bacterial “organelle.”
MATERIALS AND METHODS
Anammox enrichment cultures.
Samples containing enrichment cultures of “Candidatus Kuenenia stuttgartiensis,” “Candidatus Brocadia fulgida” (15, 16), “Candidatus Anammoxoglobus propionicus” (14), and “Candidatus Scalindua spp.” (1:1 coculture of two “Candidatus Scalindua” species) (J. van de Vossenberg, unpublished data) were taken from continuous or sequencing batch reactors (39).
Sample preparation for TEM.
Small aggregates of “Candidatus Kuenenia stuttgartiensis,” “Candidatus Brocadia fulgida,” “Candidatus Anammoxoglobus propionicus,” and “Candidatus Scalindua spp.” cells were transferred into a 100-μm cavity of a planchette (3-mm diameter, 0.1/0.2-mm depth; Engineering Office M. Wohlwend GmbH, CH-9466 Sennwald, Switzerland) containing 1-hexadecene (41), closed with the flat side of a lecithin-coated planchette (3 mm, 0.3-mm depth), and cryofixed by high-pressure freezing (Leica EMHPF, Leica Microsystems, Vienna, Austria). Freeze-substitution was performed in acetone containing 2% osmium tetroxide, 0.2% uranyl acetate, and 1% H2O (46). Samples were kept at −90°C for 47 h, brought to −60°C at 2°C per hour, kept at −60°C for 8 h, brought to −30°C at 2°C per hour, and kept at −30°C for 8 h in a freeze-substitution unit (AFS, Leica Microsystems, Vienna, Austria). Uranyl acetate was removed by washing the samples four times for 30 min in the AFS device at −30°C with acetone containing 2% osmium tetroxide and 1% H2O. Fixation was then continued for 1 h on ice. Osmium tetroxide and H2O were removed by washing two times for 30 min on ice with anhydrous acetone. Samples were gradually infiltrated with Epon resin (22). Epon was polymerized for 72 h at 60°C.
The “Candidatus Brocadia fulgida” sample (see Fig. 3C and D) was processed as described above, with the following changes. Cells were cryofixed by high-pressure freezing in a HPM010 HPF (BAL-TEC, Inc., Balzers, Liechtenstein) and freeze-substituted in anhydrous acetone containing 2% osmium tetroxide and 0.5% uranyl acetate. Samples were kept at −90°C for 24 h and −80°C for 24 h, brought to −45°C at 2°C per hour, kept at −45°C for 2 h, brought to 0°C at 22.5°C per hour, and brought to 20°C at 10°C per hour. Samples were washed four times for 20 min with anhydrous acetone. Epon was polymerized for 24 h at 60°C.
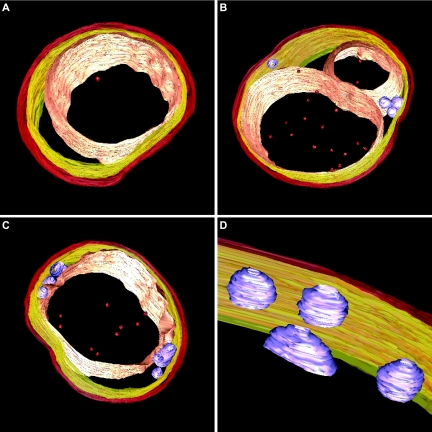
FIG. 3.
Snapshots of electron tomography models showing the conserved anammox cell plan in three anammox genera. (A) “Candidatus Kuenenia stuttgartiensis” cell. (B) “Candidatus Anammoxoglobus propionicus” cell with riboplasmic particles and with the anammoxosome compartment being divided or lying in a bent configuration in the cell. (C) “Candidatus Brocadia fulgida” cell with (D) riboplasmic particles. Models show (from outside to inside) cell boundary (red), intracytoplasmic membrane (yellow), riboplasmic particles (purple), anammoxosome membrane (pink), and putative intra-anammoxosome iron particles (red). Electron tomography movies are available at http://www.microbiology.science.ruonl/niftrik.
All sections were cut using a Reichert Ultracut E microtome (Leica Microsystems, Vienna, Austria). Ultrathin (for TEM, ca. 70-nm) and semithin (for electron tomography, 250- to 400-nm) sections were collected on carbon-Formvar-coated 50-mesh copper square grids.
Ultrathin sections of Epon-embedded cells (ca. 70 nm) were poststained with 20% uranyl acetate in 70% methanol for 4 min and Reynolds lead citrate staining (26) for 2 min and investigated at 80 to 120 kV by using a transmission electron microscope (Tecnai 10 or Tecnai 12; FEI Company, Eindhoven, The Netherlands). Images were recorded using a charge-coupled device camera (MegaView II, analySIS).
Electron tomography.
The resolution of a tomogram is determined by the thickness of the section and the number of projections (17). So, to obtain an optimal combination of both resolution and volume of the cell imaged, we used sections with a maximum thickness of 400 nm. Ten-nanometer colloidal gold particles were applied to one surface of semithin sections (200 to 400 nm) of high-pressure frozen, freeze-substituted, and Epon-embedded “Candidatus Kuenenia stuttgartiensis” “Candidatus Brocadia fulgida,” and “Candidatus Anammoxoglobus propionicus” cells to function as fiducial markers by incubating sections on a protein A-10-nm gold solution. The sections were poststained with 2% uranyl acetate in water for 10 minutes. Specimens were placed in a high-tilt specimen holder, and dual axis datasets were recorded at 200 kV (Tecnai 20 LaB6; FEI Company, Eindhoven, The Netherlands) by either manually rotating the grid 90° (Fischione 2020 advanced tomography holder; Fischione Instruments, Pittsburgh, PA) or rotating the grid 90° inside the microscope (Fischione rotation holder; Fischione Instruments, Pittsburgh, PA). The angular tilt range was from −65° to 65°, with an increment of 1°. Binned (2 by 2) images (1,024 by 1,024 pixels) were recorded using a charge-coupled device camera (TemCam F214; TVIPS GmbH, Gauting, Germany). Automated data acquisition of the tilt series was carried out using Xplore 3D (FEI Company, Eindhoven, The Netherlands). Tomograms from each tilt axis were computed with the R-weighted back-projection algorithm and combined into one double-tilt tomogram using the IMOD software package (18). Double-tilt tomograms were analyzed and modeled with the IMOD software package. Features of interest were contoured manually in serial optical slices extracted from the tomogram.
Polysaccharide (glycogen) stain.
The glycogen stain, modified after the method of Thiery (42), was performed on four different anammox genera: “Candidatus Kuenenia stuttgartiensis” “Candidatus Brocadia fulgida,” “Candidatus Anammoxoglobus propionicus,” and “Candidatus Scalindua spp.” Ultrathin sections of high-pressure frozen, freeze-substituted, and Epon-embedded cells were collected on carbon-Formvar-coated 100-mesh hexagonal golden grids. All incubations and washing steps were performed on drops. Sections were incubated for 30 min with 1% periodic acid and washed for 60 min with water. Negative controls were incubated with water instead of periodic acid. Sections were incubated overnight with 0.2% thiocarbohydrazide in 20% acetic acid. Sections were washed with 10% acetic acid (two times for 15 min), 5% acetic acid (two times for 5 min), 2% acetic acid (5 min), and water (3 times for 5 min). Sections were incubated for 30 min with 1% silver albumin and washed with water (three times for 5 min). Finally, sections were incubated for 5 min with 5% sodium thiosulfate and washed with water (three times for 5 min each). The sections were investigated unstained. In this method, periodic acid breaks open the polysaccharide molecules, after which thiocarbohydrazide can attach. This complex is then stained black by the addition of silver albumin.
Cytochrome peroxidase reaction.
The cytochrome peroxidase reaction was modified from the method of Seligman et al. (33). “Candidatus Kuenenia stuttgartiensis” cells were fixed for 30 min in 0.1 M cacodylate buffer, pH 7.2, containing 1.5% glutaraldehyde and 4% formaldehyde and washed with buffer. Cells were incubated for 15 min with 2.5 mM diaminobenzidine tetrahydrochloride (DAB) and 0.02% H2O2 in 0.1 M cacodylate buffer, pH 6.5, and washed with cold buffer, pH 7.2, containing 0.15 M sucrose (to prevent the relocation of precipitates). Negative controls were preincubated for 15 min with 0.01 M potassium cyanide in 0.1 M cacodylate buffer, pH 6.5, and then incubated with DAB and H2O2 in the presence of potassium cyanide. Cells were postfixed for 90 min in 2% osmium tetroxide at 4°C, washed with water, and embedded in 2% low-melting-point agarose. Cells were dehydrated in a graded ethanol series (70, 80, 90, 96, and 100% ethanol) and then in 100% propylene oxide and subsequently embedded in Epon resin. Epon was polymerized for 48 h at 60°C. Ultrathin sections were investigated unstained, poststained for 1 min with Reynolds lead citrate, and poststained for 4 min with 20% uranyl acetate in 70% methanol and 2 min with Reynolds lead citrate.
Energy dispersive X-ray analysis.
“Candidatus Brocadia fulgida” cells were cryofixed by high-pressure freezing, freeze-substituted in anhydrous acetone containing 2% osmium tetroxide, and embedded in Epon resin as described above. Semithin sections (∼250 nm) were collected on carbon-Formvar-coated 50-mesh nickel square grids and analyzed with TEM-energy dispersive X-ray analysis (EDX) for their elemental compositions. Spot measurements were performed with the following settings: acceleration voltage, 200 kV; 5 eV/17 μs; spot size, 8; and dwell time, 60 s (Tecnai 20 FEG; FEI Company, Eindhoven, The Netherlands) using ES vision software (Emispec Systems, Inc., Tempe, AZ).
Comparing anammox cell and anammoxosome volume.
To compare the four anammox genera with respect to size (diameter) and what percentage of the cell was occupied by the anammoxosome compartment, we deducted the specific cell and anammoxosome areas (nm2) from transmission electron micrographs by using the Scion Image Software (Scion Corporation, Frederick, MD).
Genome and sequence analysis.
In our search for specific genes, protein sequences of known genes were blasted against the “Candidatus Kuenenia stuttgartiensis” genome by using the NCBI blastp program (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Putative homologs were subsequently aligned to the known genes by using the PIR pairwise alignment tool (http://pir.georgetown.edu/pirwww/search/pairwise.shtml).
RESULTS
The ultrastructures of the four anammox genera “Candidatus Kuenenia stuttgartiensis,” “Candidatus Brocadia fulgida,” “Candidatus Anammoxoglobus propionicus,” and “Candidatus Scalindua spp.” were investigated and compared using TEM. “Candidatus Kuenenia stuttgartiensis,” “Candidatus Brocadia fulgida,” and “Candidatus Anammoxoglobus propionicus” were also investigated using electron tomography. We observed that the anammox cell plan was conserved in all four anammox genera (Fig. 2 and 3). The cytoplasm was divided into three compartments separated by individual membranes. The cell wall was lined on the inside by a membrane (the cytoplasmic membrane, consistent with data of Lindsay et al. [21]). A second membrane (the intracytoplasmic membrane) separated the first and outermost cytoplasmic compartment (the paryphoplasm) (Fig. 2) from the second electron-dense cytoplasmic compartment (the riboplasm) (Fig. 2). A third membrane was in a predominantly curved configuration and separated the riboplasm from the third and largest electron-light, cytoplasmic compartment (the anammoxosome) (Fig. 2). All four anammox genera contained electron-dense particles (Fig. 2 and 3) and tubule-like structures in the anammoxosome (21, 44, 45). In “Candidatus Brocadia fulgida,” the electron-dense particles were previously shown to contain iron (45).
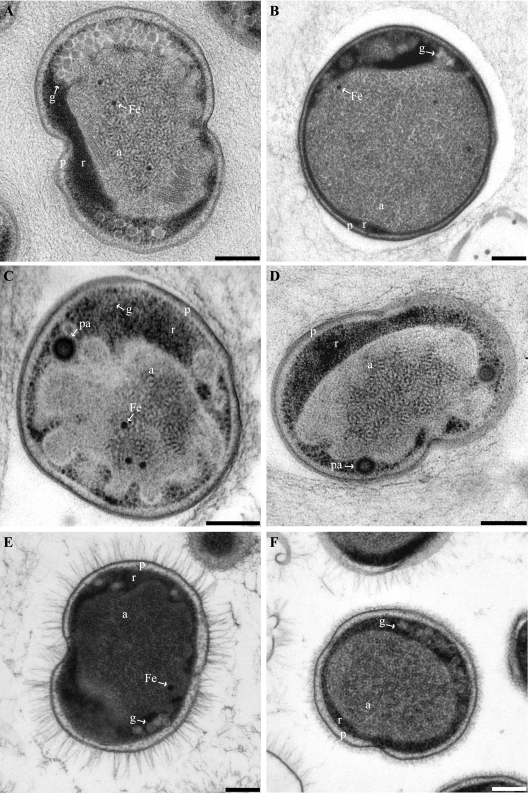
FIG. 2.
Transmission electron micrographs of high-pressure frozen, freeze-substituted, and Epon-embedded thin sections of four anammox genera. All cells are divided into three separate compartments by individual membranes: the paryphoplasm (p), riboplasm (r), and anammoxosome (a) compartments. (A) Dividing “Candidatus Kuenenia stuttgartiensis” cell. (B) “Candidatus Anammoxoglobus propionicus” cell. (C and D) “Candidatus Brocadia fulgida” cells showing riboplasmic particles (pa). (D) Dividing cell. (E and F) Dividing “Candidatus Scalindua spp.” cells with (E) and without (F) pilus-like appendages. Micrographs further show glycogen (g) and putative intra-anammoxosome iron particles (Fe). Scale bars, 200 nm.
The cells of the genera “Candidatus Kuenenia stuttgartiensis” and “Candidatus Brocadia fulgida” both had an average diameter of 800 nm, with the anammoxosome compartment occupying 61% ± 5% of the cell volume. The “Candidatus Scalindua spp.” were both slightly larger, with an average cell diameter of 950 nm. However, the anammoxosome compartment remained the same size as those in “Candidatus Kuenenia stuttgartiensis” and “Candidatus Brocadia fulgida,” resulting in the anammoxosome compartment occupying 51% ± 8% of the cell volume in “Candidatus Scalindua spp.” The genus “Candidatus Anammoxoglobus propionicus” was the largest cell type, with a typical cell diameter of 1,100 nm. In this genus, the anammoxosome was also relatively larger than that in cells of the other genera and occupied on average 66% ± 6% of the cell volume.
In addition to the finding that the anammox cell plan established in previous studies was conserved among the four anammox genera, we also observed as-yet-undescribed ultrastructural features. The TEM study of the four anammox genera showed numerous spherical, aggregated granules in all four genera (Fig. 2). These granules occurred only in the riboplasmic compartment, and each had a diameter of approximately 55 nm. We identified the granules as polysaccharide (glycogen) storage granules by performing a glycogen stain on thin sections of all anammox genera (Fig. 4). “Candidatus Kuenenia stuttgartiensis” cells especially stored large amounts of glycogen (Fig. 4A). The negative controls (Fig. 4B), incubated with water instead of periodic acid, showed no staining at all; instead, the glycogen granules appeared bright white. An analysis of the “Candidatus Kuenenia stuttgartiensis” genome revealed the presence of the glycogen synthesis genes glgA (bacterial glycogen synthase, EC 2.4.1.21), glgB (glycogen branching enzyme, EC 2.4.1.18), and glgC (glucose-1-phosphate adenylyltransferase, EC 2.7.7.27) (12, 24). Open reading frames were found with 30 to 55% protein sequence identity to the full-length Escherichia coli (4) glycogen genes: one copy of glgA (kuste2463), one copy of glgB (kuste3063), and two copies of glgC (kuste2466 and kustb0247). Glycolysis and gluconeogenesis were also completely encoded in the genome of “Candidatus Kuenenia stuttgartiensis” (37).
FIG. 4.
Transmission electron micrographs showing glycogen staining of high-pressure frozen, freeze-substituted, and Epon-embedded thin sections of four anammox genera. All investigated anammox genera show glycogen staining in the riboplasmic compartment. (A and B) “Candidatus Kuenenia stuttgartiensis.” (B) Negative control incubated with water instead of periodic acid. (C) “Candidatus Anammoxoglobus propionicus.” (D) “Candidatus Brocadia fulgida.” (E and F) “Candidatus Scalindua spp.” cells with (E) and without (F) pilus-like appendages. Scale bars, 200 nm.
Another observation was that some “Candidatus Anammoxoglobus propionicus” and “Candidatus Brocadia fulgida” cells contained additional particles in the riboplasmic compartment (Fig. 2C and D and Fig. 3B to D). The appearance of these particles was different from the appearance of the glycogen granules. The spherical and sometimes tubular particles were on average 83 nm in diameter, and the lining of these particles appeared more electron dense than the interior. This could mean that the particles were membrane bound, though their electron density prohibited a definite interpretation. Electron tomography suggested that these particles were separate entities; no connections to any of the other membranes were observed (Fig. 3B to D). EDX analysis was performed on “Candidatus Brocadia fulgida” cells (data not shown), and no differences in elemental composition between the riboplasmic particles and the riboplasmic compartment were detected.
For the reactor sample containing a 1:1 coculture of two “Candidatus Scalindua” species (J. van de Vossenberg, unpublished data), two morphotypes were observed: one with pilus-like appendages and one without (Fig. 2E and F). When present, these appendages were evenly spread over the whole cell surface and had an average length of 131 nm. The “Candidatus Scalindua” cells without appendages had an irregular layer outside the cell wall (Fig. 2F) that was not observed for the other genera examined.
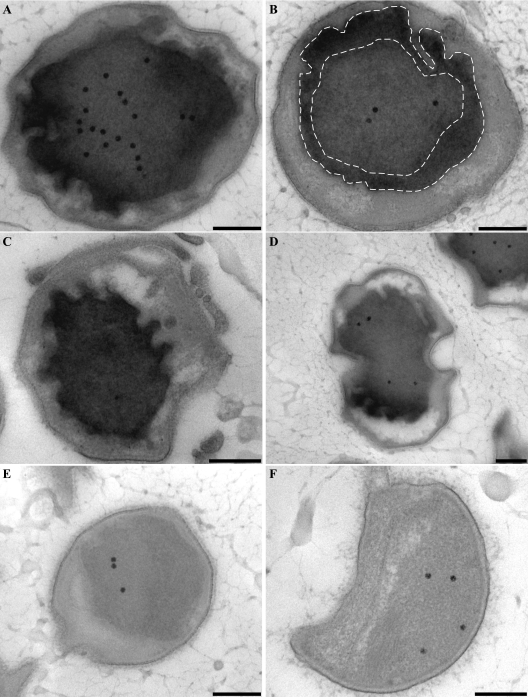
To further investigate the hypothesis that the anammoxosome compartment is the locus of all catabolic reactions, a cytochrome peroxidase stain was performed on “Candidatus Kuenenia stuttgartiensis” cells. This was done to investigate the location of the cytochrome c proteins involved in the anaerobic oxidation of ammonium (Fig. 1). The oxidation of DAB resulted in the accumulation of an electron-dense product inside the anammoxosome, especially along the inside of the anammoxosome membrane (Fig. 5). The staining was most intense up to approximately 150 nm from the anammoxosome membrane (Fig. 5B) and in membrane protrusions (Fig. 5D). This result indicated that anammox cytochrome c proteins were predominantly located at or in close proximity to the inside of the anammoxosome membrane. The intensity of the staining can be explained by the fact that cytochrome c proteins constitute up to 30% of the total cell protein of anammox bacteria (8, 27) and that the genome of “Candidatus Kuenenia stuttgartiensis” encodes at least 59 different cytochrome c proteins (37). Staining was completely absent in the negative controls (Fig. 5E and F), which were preincubated with KCN and then incubated with DAB and H2O2 in the presence of KCN. No staining was observed in any other parts of the cell, including the paryphoplasm and the outside of the anammoxosome membrane.
FIG. 5.
Transmission electron micrographs of chemically fixed and Epon-embedded thin sections of “Candidatus Kuenenia stuttgartiensis” cells showing cytochrome peroxidase staining. This figure is best viewed on screen to avoid change of contrast by printer settings. (A to D) Cytochrome peroxidase staining is observed solely in the anammoxosome compartment. Intense staining occurs within close proximity to the anammoxosome membrane, as outlined by the dashed lines (B) and in places where the membrane is curved (D). (E and F) Negative controls preincubated with KCN and incubated with DAB and H2O2 in the presence of KCN. All sections were poststained for 1 min with Reynolds lead citrate. Scale bars, 200 nm.
DISCUSSION
Since their discovery in the 1990s in a Gist-Brocades pilot plant (19), anammox bacteria have been found in many different environments, such as marine suboxic zones, coastal sediments, lakes, and wastewater treatment plants (31). So far, four anammox genera have been described, with 16S rRNA gene sequence identities between the species ranging between 87 and 99% (31). Despite this relatively large phylogenetic distance, all anammox organisms belong to the same family, Anammoxaceae (13) and are all capable of performing anaerobic ammonium oxidation. The feature of a differentiated cytoplasm is also shared by a phylogenetically larger group to which the anammox bacteria belong, the phylum Planctomycetes. Most genus-level groups within the Planctomycetes have different types of compartmentalization. This study showed that the anammox ultrastructure is conserved among all anammox genera described so far. The cytoplasms of all four genera were divided into three separate compartments by individual membranes. The innermost anammoxosome compartment made up most of the volume of the cell and was bounded by a predominantly curved membrane. Furthermore, all genera contained putative iron particles and tubule-like structures inside the anammoxosome (21, 44, 45).
It is likely that the function of the anammox cell plan is linked to a specific function in this group. The hypothesis that the anammoxosome compartment is dedicated to the generation of energy is supported by the cytochrome peroxidase stain that was performed on “Candidatus Kuenenia stuttgartiensis” cells. Heme-c containing proteins, cytochromes, exhibit a wide variety of biological functions (7). One of these functions is the reduction of peroxides (peroxidase activity), which, among other proteins, is performed by heme-c cytochromes (6). The anammox cytochromes proposed to be involved in the anaerobic oxidation of ammonium to dinitrogen gas all contain heme-c groups and can thus be stained by the DAB method (33). This stain is not specific for these cytochromes alone and will stain all other proteins with peroxidase activity as well. Staining was observed only inside the anammoxosome and was most intense in curvatures and in a 150-nm rim along the inside of the anammoxosome membrane. These results suggest that the anammox enzymes are indeed either attached to or associated with the anammoxosome membrane and that they reside on the anammoxosome side of this membrane as was proposed previously (Fig. 1). The specific location of enzymes in places where the membrane was curved further strengthens the idea of an energy-generating organelle, where the membrane is folded to enhance catabolic activity. In bacteria, cytochromes with heme-c (such as hydroxylamine oxidoreductase and nitrite reductase) have so far been found only in the periplasmic space (for a review, see references 2 and 3). The absence of peroxidase staining in the paryphoplasm compartment is consistent with the idea that this compartment is a cytoplasmic and not a periplasmic compartment (21). The absence of staining in the paryphoplasm also supports the idea that the anammoxosome is a separate entity and not a simple invagination of the intracytoplasmic membrane. If this were the case, staining would have been observed inside the paryphoplasm. Other evidence that supports the status of the anammoxosome as a separate compartment consists of the successful purification of intact anammoxosomes from anammox cells after lysis (36), the exclusive localization of hydrazine/hydroxylamine oxidoreductase to the anammoxosome with immunogold labeling (21), the apparent absence of membrane links between the anammoxosome and the paryphoplasm as observed by electron tomography and the observed vertical inheritance of the anammoxosome during cell division (L. van Niftrik, W. J. C. Geerts, E. G. van Donselaar, B. M. Humbel, R. I. Webb, H. R. Harhangi, H. J. M. Op den Camp, J. A. Fuerst, A. J. Verkleij, M. S. M. Jetten, and M. Strous, submitted for publication). Still, we cannot rule out the fact that the biogenesis of new anammoxosome matrix and membrane somehow proceeds via the paryphoplasm, i.e., via vesicular transport.
All examined anammox genera were found to store glycogen in their riboplasmic compartment. The exact role of glycogen in bacteria is not entirely clear, though evidence indicates that glycogen functions as an energy and carbon storage compound, providing energy and carbon for cell survival under stress or starvation (12). Also, glycogen storage is linked to excess carbon and/or the lack of a required nutrient (especially nitrogen) in the medium. In stationary-phase Salmonella enterica serovar Enteritidis cells, glycogen stores are used for the formation of a biofilm (5). Anammox bacteria can also grow in a dense biofilm (the extracellular matrix can be seen in Fig. 2A to D and Fig. 4C). This property is selected for during the enrichment of anammox bacteria in sequencing batch or continuous reactors; single cells will be washed or pumped out of the reactor. Therefore, it could be that anammox bacteria also store and use glycogen for biofilm formation. However, the storage of glycogen is not likely to be triggered solely by biofilm formation since the anammox cells in the predominantly single-cell “Candidatus Scalindua spp.” culture also stored glycogen, though to a lesser extent. It would be interesting to further investigate the effect of the culture “phase” (start-up or steady state), medium (carbon or nutrients), and biofilm versus that of single cells on anammox glycogen storage.
The nature of the riboplasmic particles that were observed in “Candidatus Brocadia fulgida” and “Candidatus Anammoxoglobus propionicus” remains unclear. EDX analysis showed no differences in elemental composition between the riboplasm and these particles. This means there was no accumulation of a specific element, as was the case for the anammoxosomal iron particles (45). The riboplasmic particles also did not stain in the glycogen stain and were ultrastructurally different from the glycogen granules. It would be interesting to investigate whether they represent another form of carbon and energy storage material, namely polyhydroxyalkanoates (PHA) (for a review, see reference 1). If so, it will probably be a property evolved by these specific anammox genera alone, since a search through the genome of “Candidatus Kuenenia stuttgartiensis” showed that it does not contain any homologs to the PHA synthase gene, phaC, necessary for synthesis of PHAs (34), which is consistent with the absence of the particles in “Candidatus Kuenenia stuttgartiensis.” If “Candidatus Brocadia fulgida” and “Candidatus Anammoxoglobus propionicus” store PHAs, it is very likely that this storage is linked to their specific property of being very efficient at cooxidizing small organic acids (14, 15); PHA, like glycogen, is known to be accumulated when a carbon source, such as propionate, is provided in excess and a growth nutrient is limited (25). A final possibility is that the particles are really vesicles that are derived from the intracytoplasmic or anammoxosome membrane; electron tomography never showed a connection with any of these two membranes, but such connections might occur only periodically.
In the sample containing two “Candidatus Scalindua” species, cells with pilus-like appendages were observed. Pili, also named fimbriae, are hairlike extensions from the bacterial cell surface for adherence to the environment (32). The “Candidatus Scalindua” cells without pilus-like appendages still had an irregular layer on the outer side of the cell wall. This layer could mean that these cells had lost their pilus-like appendages instead of having had none at all. However, the possibility remains that the pilus-like appendages are not actual pili but are instead composed of the extracellular polymeric substance that anammox bacteria produce to attach to each other.
This research has shown that the anammox cell plan is conserved among the four anammox genera known so far and suggests that the hypothesis that the anammoxosome compartment is specifically used for energy generation is plausible. This supports the view that these prokaryotes have indeed evolved a specialized membrane-bounded organelle, a feature before ascribed only to eukaryotes. Further investigation of the pilus-like appendages, the identification of the riboplasmic particles in “Candidatus Brocadia fulgida” and “Candidatus Anammoxoglobus propionicus,” and a possible relationship between glycogen (and possibly PHA) accumulation and anammox growth conditions should be productive for new insights into how anammox cells function during their unique metabolism.
Acknowledgments
We thank Boran Kartal, Katinka van de Pas-Schoonen, and Jack van de Vossenberg for the anammox enrichment cultures, Chris Schneijdenberg and Hans Meeldijk for technical assistance, John Geus and Alya Yakushevska for their assistance with the EDX analysis, and Gijs Kuenen for discussions and feedback.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arp, D. J., and L. Y. Stein. 2003. Metabolism of inorganic N compounds by ammonium-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 38471-495. [DOI] [PubMed] [Google Scholar]
- 3.Averill, B. A. 1996. Dissimilatory nitrite and nitric oxide reductases. Chem. Rev. 962951-2964. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1462. [DOI] [PubMed] [Google Scholar]
- 5.Bonafonte, M. A., C. Solano, B. Sesma, M. Alvarez, L. Montuenga, D. García-Ros, and C. Gamazo. 2000. The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 19131-36. [DOI] [PubMed] [Google Scholar]
- 6.Braun, M., and L. Thöny-Meyer. 2004. Biosynthesis of artificial microperoxidases by exploiting the secretion and cytochrome c maturation apparatuses of Escherichia coli. Proc. Natl. Acad. Sci. USA 10112830-12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapmann, S. K., S. Daff, and A. W. Munro. 1997. Heme: the most versatile redox centre in biology? Struct. Bond. 8839-70. [Google Scholar]
- 8.Cirpus, I. E. Y., M. de Been, H. J. M. Op den Camp, M. Strous, D. Le Paslier, J. G. Kuenen, and M. S. M. Jetten. 2005. A new soluble 10 kDa monoheme cytochrome c-552 from the anammox bacterium Candidatus “Kuenenia stuttgartiensis. ” FEMS Microbiol. Lett. 252273-278. [DOI] [PubMed] [Google Scholar]
- 9.Francis, C. A., J. M. Beman, and M. M. M. Kuypers. 2007. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 119-27. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst, J. A. 1995. The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology 1411493-1506. [DOI] [PubMed] [Google Scholar]
- 11.Huston, W. M., H. R. Harhangi, A. P. Leech, C. S. Butler, M. S. M. Jetten, H. J. M. Op den Camp, and J. W. B. Moir. 2007. Expression and characterisation of a major c-type cytochrome encoded by gene kustc0563 from Kuenenia stuttgartiensis as a recombinant protein in Escherichia coli. Protein Expr. Purif. 5128-33. [DOI] [PubMed] [Google Scholar]
- 12.Iglesias, A. A., and J. Preiss. 1992. Bacterial glycogen and plant starch biosynthesis. Biochem. Educ. 20196-203. [Google Scholar]
- 13.Jetten, M. S. M., H. J. M. Op den Camp, J. G. Kuenen, and M. Strous. Taxonomic description of the family Anammoxaceae. In B. Hedlund, N. R. Krieg, B. J. Paster, K.-H. Schleifer, J. T. Staley, N. Ward, and W. B. Whitman (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 4, in press. Springer, New York, NY.
- 14.Kartal, B., J. Rattray, L. A. van Niftrik, J. van de Vossenberg, M. C. Schmid, R. I. Webb, S. Schouten, J. A. Fuerst, J. Sinninghe Damsté, M. S. M. Jetten, and M. Strous. 2007. Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 3039-49. [DOI] [PubMed] [Google Scholar]
- 15.Kartal, B., L. van Niftrik, J. Rattray, J. L. C. M. van de Vossenberg, M. C. Schmid, J. Sinninghe Damsté, M. S. M. Jetten, and M. Strous. Candidatus “Brocadia fulgida”: an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol. Ecol., in press. doi: 10.1111/j.1574-6941.2007.00408.x. [DOI] [PubMed]
- 16.Kartal, B., L. van Niftrik, O. Sliekers, M. C. Schmid, I. Schmidt, K. van de Pas-Schoonen, I. Cirpus, W. van der Star, M. van Loosdrecht, W. Abma, J. G. Kuenen, J.-W. Mulder, M. S. M. Jetten, H. Op den Camp, M. Strous, and J. van de Vossenberg. 2004. Application, eco-physiology and biodiversity of anaerobic ammonium-oxidizing bacteria. Rev. Environ. Sci. Biotechnol. 3255-264. [Google Scholar]
- 17.Koster, A. J., R. Grimm, D. Typke, R. Hegerl, A. Stoschek, J. Walz, and W. Baumeister. 1997. Perspectives of molecular and cellular electron tomography. J. Struct. Biol. 120276-308. [DOI] [PubMed] [Google Scholar]
- 18.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 11671-76. [DOI] [PubMed] [Google Scholar]
- 19.Kuenen, J. G., and M. S. M. Jetten. 2001. Extraordinary anaerobic ammonium-oxidizing bacteria. ASM News 67456-463. [Google Scholar]
- 20.Kuypers, M. M. M., A. O. Sliekers, G. Lavik, M. Schmid, B. Barker Jørgensen, J. G. Kuenen, J. S. Sinninghe Damsté, M. Strous, and M. S. M. Jetten. 2003. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422608-611. [DOI] [PubMed] [Google Scholar]
- 21.Lindsay, M. R., R. I. Webb, M. Strous, M. S. M. Jetten, M. K. Butler, R. J. Forde, and J. A. Fuerst. 2001. Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175413-429. [DOI] [PubMed] [Google Scholar]
- 22.Mollenhauer, H. H. 1964. Plastic embedding mixtures for use in electron microscopy. Stain Technol. 39111-114. [PubMed] [Google Scholar]
- 23.Penton, C. R., A. H. Devol, and J. M. Tiedje. 2006. Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl. Environ. Microbiol. 726829-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preiss, J., S.-G. Yung, and P. A. Baecker. 1983. Regulation of bacterial glycogen synthesis. Mol. Cell. Biochem. 5761-80. [DOI] [PubMed] [Google Scholar]
- 25.Ramsay, B. A., K. Lomaliza, C. Chavarie, B. Dubé, P. Bataille, and J. A. Ramsay. 1990. Production of poly-(β-hydroxybutyric-co-β-hydroxyvaleric) acids. Appl. Environ. Microbiol. 562093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schalk, J., S. de Vries, J. G. Kuenen, and M. S. M. Jetten. 2000. Involvement of a novel hydroxylamine oxidoreductase in anaerobic ammonium oxidation. Biochemistry 395405-5412. [DOI] [PubMed] [Google Scholar]
- 28.Schmid, M., K. Walsh, R. Webb, W. I. C. Rijpstra, K. van de Pas-Schoonen, M. J. Verbruggen, T. Hill, B. Moffett, J. Fuerst, S. Schouten, J. S. Sinninghe Damsté, J. Harris, P. Shaw, M. Jetten, and M. Strous. 2003. Candidatus “Scalindua brodae,” sp. nov., Candidatus “Scalindua wagneri,” sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 26529-538. [DOI] [PubMed] [Google Scholar]
- 29.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. Jetten, J. W. Metzger, K. H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 2393-106. [DOI] [PubMed] [Google Scholar]
- 30.Schmid, M. C., B. Maas, A. Dapena, K. van de Pas-Schoonen, J. van de Vossenberg, B. Kartal, L. van Niftrik, I. Schmidt, I. Cirpus, J. G. Kuenen, M. Wagner, J. S. Sinninghe Damsté, M. Kuypers, N. P. Revsbech, R. Mendez, M. S. M. Jetten, and M. Strous. 2005. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 711677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid, M. C., N. Risgaard-Petersen, J. van de Vossenberg, M. M. M. Kuypers, G. Lavik, J. Petersen, S. Hulth, B. Thamdrup, D. Canfield, T. Dalsgaard, S. Rysgaard, M. K. Seir, M. Strous, H. J. M. Op den Camp, and M. S. M. Jetten. 2007. Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ. Microbiol. 91476-1484. [DOI] [PubMed] [Google Scholar]
- 32.Scott, J. R., and D. Zähner. 2006. Pili with strong attachments: gram-positive bacteria do it differently. Mol. Microbiol. 62320-330. [DOI] [PubMed] [Google Scholar]
- 33.Seligman, A. M., M. J. Karnovsky, H. L. Wasserkrug, and J. S. Hanker. 1968. Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB). J. Cell Biol. 381-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheu, D.-S., Y.-T. Wang, and C.-Y. Lee. 2000. Rapid detection of polyhydroxyalkanoate-accumulating bacteria isolated from the environment by colony PCR. Microbiology 1462019-2025. [DOI] [PubMed] [Google Scholar]
- 35.Shimamura, M., T. Nishiyama, H. Shigetomo, T. Toyomoto, Y. Kawahara, K. Furukawa, and T. Fujii. 2007. Isolation of a multiheme protein with features of a hydrazine-oxidizing enzyme from an anaerobic ammonium-oxidizing enrichment culture. Appl. Environ. Microbiol. 781065-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinninghe Damsté, J. S., M. Strous, W. I. C. Rijpstra, E. C. Hopmans, J. A. J. Geenevasen, A. C. T. van Duin, L. A. van Niftrik, and M. S. M. Jetten. 2002. Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nature 419708-712. [DOI] [PubMed] [Google Scholar]
- 37.Strous, M., E. Pelletier, S. Mangenot, T. Rattei, A. Lehner, M. W. Taylor, M. Horn, H. Daims, D. Bartol-Mavel, P. Wincker, V. Barbe, N. Fonknechten, D. Vallenet, B. Segurens, C. Schenowitz-Truong, C. Médigue, A. Collingro, B. Snel, B. E. Dutilh, H. J. M. Op den Camp, C. van der Drift, I. Cirpus, K. T. van de Pas-Schoonen, H. R. Harhangi, L. van Niftrik, M. Schmid, J. Keltjens, J. van de Vossenberg, B. Kartal, H. Meier, D. Frishman, M. A. Huynen, H.-W. Mewes, J. Weissenbach, M. S. M. Jetten, M. Wagner, and D. Le Paslier. 2006. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440790-794. [DOI] [PubMed] [Google Scholar]
- 38.Strous, M., J. A. Fuerst, E. H. M. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. M. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400446-449. [DOI] [PubMed] [Google Scholar]
- 39.Strous, M., J. J. Heijnen, J. G. Kuenen, and M. S. M. Jetten. 1998. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl. Microbiol. Biotechnol. 50589-596. [Google Scholar]
- 40.Strous, M., and M. S. M. Jetten. 2004. Anaerobic oxidation of methane and ammonium. Annu. Rev. Microbiol. 5899-117. [DOI] [PubMed] [Google Scholar]
- 41.Studer, D., M. Michel, and M. Müller. 1989. High-pressure freezing comes of age. Scan. Microsc. Suppl. 3253-269. [PubMed] [Google Scholar]
- 42.Thiery, J. P. 1967. Demonstration of polysaccharides in thin sections by electron microscopy. J. Microsc. 6987-1018. [Google Scholar]
- 43.van de Graaf, A. A., P. de Bruijn, L. A. Robertson, M. S. M. Jetten, and J. G. Kuenen. 1997. Metabolic pathway of anaerobic ammonium oxidation on basis of N-15 studies in a fluidized bed reactor. Microbiology 1432415-2421. [DOI] [PubMed] [Google Scholar]
- 44.van Niftrik, L. A., J. A. Fuerst, J. S. Sinninghe Damsté, J. G. Kuenen, M. S. M. Jetten, and M. Strous. 2004. The anammoxosome: an intracytoplasmic compartment in anammox bacteria. FEMS Microbiol. Lett. 2337-13. [DOI] [PubMed] [Google Scholar]
- 45.van Niftrik, L., W. J. C. Geerts, E. G. van Donselaar, B. M. Humbel, A. Yakushevska, A. J. Verkleij, M. S. M. Jetten, and M. Strous. 2007. Combined structural and chemical analysis of the anammoxosome: a membrane-bounded intracytoplasmic compartment in anammox bacteria. J. Struct. Biol. [Epub ahead of print] doi: 10.1016/j.jsb.2007.05.005. [DOI] [PubMed]
- 46.Walther, P., and A. Ziegler. 2002. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J. Microsc. 2083-10. [DOI] [PubMed] [Google Scholar]
- 47.Woebken, D., B. M. Fuchs, M. M. M. Kuypers, and R. Amann. 2007. Potential interactions of particle-associated anammox bacteria with bacterial and archaeal partners in the Namibian upwelling system. Appl. Environ. Microbiol. 734648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]