Flagella are complex organelles whose synthesis depends upon approximately 50 gene products. Not surprisingly, multiple mechanisms control and direct flagellar biogenesis (reviewed in references 1, 15, and 55). Perhaps best understood is the control exerted at the level of transcription: hierarchies exist, such that the flagellar structural proteins are synthesized only under proper conditions and at the time each is needed for assembly. Many bacteria also possess posttranscriptional mechanisms that control translation or protein stability. Assembly itself also occurs in a highly ordered fashion, with the insertion of one component building on the insertion of a previous subunit. In the best-characterized bacteria, flagellar biogenesis begins with the insertion of an inner membrane protein called FliF (Fig. 1). Building on the assembled FliF subunits are proteins that comprise the flagellar type III secretory (TTS) apparatus. This device exports the protein subunits required to build the basal body, which includes both a cytoplasmic switching device and a complex of rod and ring proteins that spans the two membranes of the gram-negative bacterium. The completed basal body provides a narrow channel through which the more external components exit the cell for assembly. These external components include the hook (a flexible linker comprised of over 100 identical subunits) and the filament (a semirigid propeller built from thousands of flagellin subunits). Once assembled, the flagellum rotates, a process powered by the proton (or sodium) motive force via motor proteins localized in the cytoplasmic membrane in close proximity to the switching device. The direction of rotation is dictated by the switching device, which interfaces with a signaling pathway that delivers information about the cell's physicochemical environment. Chemotaxis, the resulting behavior, permits cells to migrate toward favorable environments. Finally, some bacteria eject their fully functional flagellum in coordination with the cell cycle.
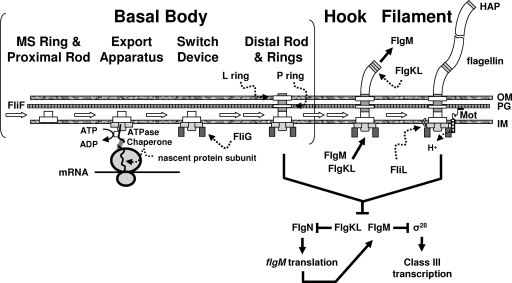
FIG. 1.
Flagellar assembly. Best studied in the enterics (Escherichia coli and Salmonella enterica serovar Typhimurium), the flagellum is comprised of three major substructures: the basal body, the hook, and the filament. These substructures are assembled in order (reviewed in reference 55). The first step in assembly appears to be the insertion into the inner membrane (IM) of the MS ring and proximal rod (composed entirely of FliF subunits) because this insertion can occur independently of all other flagellar components. The second and third steps, assembly of a TSS export apparatus and the switching device, including FliG, occur within the IM and at the cytoplasmic face of the MS ring, respectively. The fourth step, assembly first of the rod and then of the P- and L-ring components, depends upon the presence of both the assembled export apparatus and the switch. Assembly of the distal rod (as well as the hook and filament) depends upon the TSS export apparatus. Secretion of nascent subunits is assisted by a subunit-specific chaperone and depends upon a flagellum-specific ATPase. The first subunits secreted through this TTS apparatus complete the rod, which now bridges the IM, peptidoglycan (PG), and outer membrane (OM). In contrast, assembly of the P ring into the PG and the L ring into the OM depends upon the general secretory apparatus. Following this assembly process, the completed basal body can now serve as a channel through which subsequent proteins travel across the envelope to their assembly site located distal to the OM. First secreted through this envelope-spanning channel are dozens of hook subunits, which are polymerized at the distal end of the basal body. Next secreted and assembled at the distal end of the hook are three HAPs, which serve as adaptors between the larger hook and smaller filament diameters. Two of these HAPs, FlgK and FlgL, bind to their chaperone FlgN and inhibit its ability to activate flgM translation; their secretion relieves this inhibition. This permits increased synthesis of FlgM, the anti-sigma factor for the flagellum-specific sigma factor σ28. Secretion of FlgM relieves inhibition of σ28, which now becomes free to transcribe class III genes, including fliC, which encodes flagellin. Thousands of these flagellin subunits are secreted and assembled between the HAPs to produce the filament. Once the filament is assembled, the HAPs and FlgM are no longer secreted and they resume their inhibitory roles. The result is a semirigid helix strong enough to enable propulsion through a viscous environment. Propulsion occurs because the rod, hook, and filament rotate. Rotation is driven by energy generated by proton (H+) flow across the IM via a PG-anchored proton channel (Mot) that interacts with the switching device. Although the role of FliL is not understood, this protein is membrane bound and associated with the basal body complex (3, 55).
In recent years, researchers have discovered a new layer of regulation on top of the already sophisticated control of flagellar biogenesis and function. This layer depends on bis-(3′,5′)-cyclic diguanylic acid (also known as cyclic diguanylate, or c-di-GMP), a novel second messenger that apparently is unique to bacteria. This newly appreciated second messenger modulates diverse cellular processes, including virulence (40, 82). First identified as a positive effector of cellulose synthase in Gluconacetobacter xylinus (reviewed in reference 84), c-di-GMP regulates the transition from the motile, planktonic state to sessile, community-based behaviors, such as biofilm development. It tends to enhance biosynthesis of capsular and fimbrial components required by developing biofilms, while inhibiting flagella and pili that permit movement (33, 38, 81, 83, 92, 107).
Diguanylate cyclases (DGCs) synthesize c-di-GMP, while phosphodiesterases (PDEs) degrade it. Together, these activities maintain the steady-state concentration of c-di-GMP (84) (Fig. 2). DGC activity depends upon the highly conserved GGDEF (formerly DUF 1) domain (73, 74, 89, 92, 96), while PDE activity requires either the EAL (formerly DUF 2) domain (9, 19, 98) or the HD-GYP domain (26, 85). Such domains have been found associated with many diverse input and output domains (reviewed in reference 83), suggesting that they receive many different types of stimuli and respond through a variety of mechanisms. Intriguingly, hybrid proteins that possess both GGDEF and EAL domains exist. Those that have been tested, however, exhibit either DGC or PDE activity but not both (19, 45). This appears to be the case especially when one of the domains is poorly conserved. It has been proposed that these noncatalytic domains function in a regulatory capacity (19). On the basis of sheer abundance in diverse bacterial genomes, GGDEF, EAL, and HD-GYP domains represent a major family of signaling pathways that use c-di-GMP as their second messenger (28). Many bacteria possess multiple proteins with these domains (29). For example, Shewanella oneidensis possesses 98 of these proteins (101), Pseudomonas aeruginosa has 38 (50), Vibrio cholerae has 53 (54), V. fischeri has 48 (K. L. Visick, unpublished), and Escherichia coli has 29 (107). This abundance suggests that these bacteria possess a network of pathways that feeds c-di-GMP into a global pool or, alternatively, an array of unlinked pathways that produce small, localized, and thus highly specific concentrations of c-di-GMP (82, 83, 107).
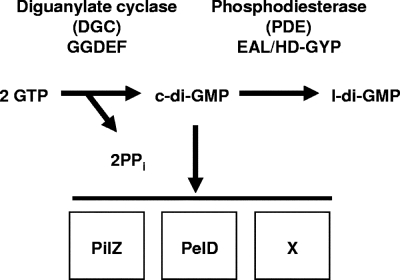
FIG. 2.
Fundamental c-di-GMP pathway. A DGC containing the conserved GGDEF domain catalyzes the synthesis of c-di-GMP from two GTP molecules with the release of pyrophosphate (PPi). A PDE containing the conserved EAL or HD-GYP domain catalyzes the degradation of c-di-GMP to linear diguanylate (l-di-GMP). Together, these two catalytic activities set the steady-state level of c-di-GMP, which can bind to a protein with the conserved PilZ or PelD domain or another as-yet-unidentified domain (X) (21).
Although it remains unclear how c-di-GMP influences behavior, some clues exist. Recently, Amikam and Galperin (5) used bioinformatics to identify a c-di-GMP-binding domain, termed PilZ after a protein that regulates type IV pilus assembly in P. aeruginosa (60). Subsequently, Ryjenkov and coworkers (88) showed that c-di-GMP binds the PilZ domain with a 1:1 stoichiometry. Typically, PilZ is only one domain of a multidomain protein. For example, the BcsA subunit of the cellulose synthesis complex contains a PilZ domain linked to a glycosyltransferase. The docking of c-di-GMP to this PilZ domain likely activates cellulose synthesis (5, 88). Multiple additional studies support the hypothesis that PilZ domain proteins bind c-di-GMP (18, 64, 76, 80). However, PilZ domains may not be the only c-di-GMP-binding motif, as some organisms that contain DGCs and PDEs do not appear to carry PilZ domain genes (5). Indeed, Lee and coworkers have recently identified PelD as a non-PilZ domain c-di-GMP-binding protein (51).
Many studies have suggested a role for c-di-GMP in controlling flagellum-based motility. However, most have utilized overexpression constructs that may lead to artifactual results due to unnaturally high (or low) levels of c-di-GMP. Furthermore, relatively few studies have investigated the level at which c-di-GMP exerts its control. However, those few studies have revealed a variety of ways in which c-di-GMP can impact motility. These putative or established mechanisms can be classified on the basis of the affected level: (i) transcription, (ii) posttranscription, (iii) function, or (iv) ejection. Although c-di-GMP impacts pilus-based twitching motility, biofilm formation, and related processes, a number of recent reviews have already covered these topics (12, 20, 40, 82, 83, 86, 97). Thus, they remain outside the scope of this review. Also outside this review are mechanisms that utilize homologs of the chemotaxis system to control non-flagellum-based processes, such as the WspR pathway of Pseudomonas (34). Instead, this minireview focuses strictly on mechanistic flagellum-based studies. At each level, we first briefly summarize the previously established mechanisms of control and then describe the predicted role of c-di-GMP.
Impact at the level of transcription.
Ordered assembly is maintained by two general mechanisms (reviewed in reference 25). First, transcription of flagellar genes occurs in a hierarchical manner. This is true for all bacteria studied to date, regardless of whether they assemble a single flagellum or many and whether they possess polar or peritrichous flagella (for a review, see reference 78). This ordering permits cells to couple gene expression to the flagellar assembly process, such that each protein is synthesized only when needed. Second, the flagellar export (TTS) apparatus switches substrates from components of the rod and hook subassemblies to components of the filament, a process that is described in more detail in the next section (for reviews, see references 1, 15, 25, and 94). In this section, we first describe the hierarchical process by which cells transcribe flagellar genes and then relate the evidence that c-di-GMP impacts this regulatory scheme.
Three major hierarchical schemes are known (reviewed in reference 78). The first (Fig. 3A), developed from studies of the enterics (primarily E. coli K-12 and Salmonella enterica serovar Typhimurium), relies on a class I master regulator (FlhDC) that acts in conjunction with the general sigma factor (σ70) to direct RNA polymerase (E) to transcribe class II genes. These class II genes encode components of the switch, export apparatus, basal body, and hook. They also include the gene for the flagellum-specific sigma factor FliA (σ28), which subsequently directs transcription of class III genes encoding later-assembled components, such as hook-associated proteins (HAPs), flagellin, the chemotactic signaling pathway (Che), and motor proteins (Mot). These genes are divided into two subsets on the basis of the regulators required for their transcription: class IIIA requires both σ28 and FlhDC, while class IIIB requires only σ28. One of the class IIIA genes encodes the σ28-specific anti-sigma factor FlgM, which provides feedback to σ28 regarding the state of flagellar assembly (reviewed in references 1, 15, and 55), while class IIIB includes two proteins (YcgR and YhjH) known to be associated with c-di-GMP (27, 47, 105).
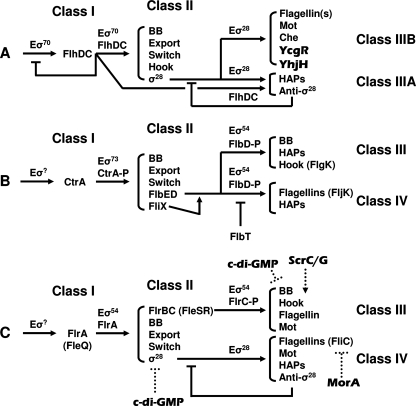
FIG. 3.
Schematic of transcription hierarchies, as determined for (A) the Enterobacteriaceae (1, 15, 25, 78, 94), (B) Caulobacter (24, 68, 109), and (C) the vibrios (and pseudomonads) (22, 62, 77). Arrows indicate induction, while a straight line in place of the arrowhead indicates inhibition. Note that the hierarchy for the Enterobacteriaceae includes c-di-GMP-associated proteins YcgR and YhjH, which are known to be encoded by genes within the flagellar regulon in Salmonella. The sigma factors at the top of the hierarchies for Caulobacter and vibrio/pseudomonad species are as yet undefined. The levels at which c-di-GMP impacts transcription for V. cholerae (cdgF overexpression; written as c-di-GMP in the figure), V. parahaemolyticus (ScrC/G), and P. putida (MorA) are not known, but the protein that is affected is indicated as an aid for the reader. BB, basal body; Mot, motor proteins; anti-σ28, FlgM; -P, phosphorylated.
The second and third schemes arose, respectively, from studies of Caulobacter crescentus (Fig. 3B) (24, 68, 109) and investigations of several vibrio and pseudomonad species (Fig. 3C) (22, 62, 77). Like the enterics, both are constructed in a hierarchical manner, each also with four major groupings of genes. The major difference lies in the regulators: the sigma factors, the master regulators, and the secondary regulators. Whereas the enterics use FlhDC, σ70, and σ28 to coordinate hierarchical control, C. crescentus uses its σ70 homolog (σ73) in conjunction with CtrA (a two-component response regulator) and σ54 in conjunction with FlbD (also a response regulator). In contrast, the vibrio/pseudomonad group uses σ54, two σ54 regulators (FlrA/FleQ and the response regulator FlrC/FleR), and σ28 to perform similar tasks.
A number of reports have documented cases in which overexpression of a DGC or a PDE impacts motility (8, 48, 54, 63, 66, 70, 79, 92). Although in many cases the levels impacted by overexpression have not been characterized, it is not unlikely that some of these proteins influence transcription of flagellar genes. Indeed, the hypothesis that c-di-GMP production affects transcription is supported by a couple of recent microarray studies (8, 34, 63). In particular, one study performed with V. cholerae, which assembles a single polar flagellum, revealed changes in transcript levels of numerous flagellar genes. Overexpression of the DGC CdgF (8) in V. cholerae caused a 2.1- to 2.5-fold decrease in the transcript levels of many class III and IV genes, including many of the flagellin and basal body proteins (Fig. 3C). Other flagellar transcripts were reduced between 1.5- and 1.9-fold. Importantly, the transcript level for the gene encoding σ28 also was reduced, a result that could easily account for the observed decrease in class IV transcripts. Some of these decreases could be seen as early as 15 min following induction of CdgF, timing that is consistent with control at the level of transcription. Not surprisingly, overexpression of CdgF caused a decrease in swimming migration through soft agar (0.3%) plates, while the corresponding overexpression of a PDE increased migration. A null mutation in cdgF, however, did not cause defects in motility or other c-di-GMP-associated phenotypes. This suggests that the influence of cdgF overexpression on motility is due to elevated levels of total intracellular c-di-GMP and that the natural role for cdgF does not include the control of flagellar gene transcription.
ScrC/ScrG.
A clearer example of transcriptional control by c-di-GMP has been documented for V. parahaemolyticus, an organism that serves as a model for bacterial differentiation (61). When grown in liquid media, this organism exists as a swimmer cell that assembles a single flagellum at one of its poles. This swimmer cell differentiates into a swarmer cell in response to environmental conditions that are thought to increase drag and/or decrease function of the polar flagelllum, such as increased viscosity or exposure to a surface. The swarmer cell is elongated and elaborates numerous lateral flagella arranged randomly about the cell surface, permitting the cell to migrate across wetted surfaces, a process called swarming (32, 65). A recent study (10) searched for genes that could inappropriately induce expression in liquid culture of a lateral flagellar gene, flgEL. One cosmid that conferred high levels of transcription to the flgEL reporter construct contained the scrABC operon (swarmer and capsular polysaccharide regulation) (10). The first gene (scrA) encodes a putative pyridoxal phosphate-dependent enzyme, while the second (scrB) encodes a putative periplasmic solute-binding protein. The third gene (scrC) encodes a membrane-associated GGDEF-EAL hybrid protein. Each of these genes appears to be involved in the observed phenotype, as chromosomal disruptions of scrA, scrB, or scrC substantially decreased the activity of the flgEL reporter fusion and significantly decreased swarming. Specifically, disruption of scrA by two different insertions, one predicted to be polar and a second predicted to be nonpolar, resulted in decreases of 75- and 7.5-fold, respectively, while disruption of either scrB or scrC reduced expression about 30-fold. Not surprisingly, decreases in motility and lateral gene transcription corresponded to significant decreases in lateral flagellin, the abundant subunit that comprises the external filament (Fig. 1). While complementation experiments verified the role of scrC, they also demonstrated that scrA and scrB are critical to operon function; thus, a full understanding of the individual roles of the last two genes awaits further characterization. However, it does appear that the activity of ScrC can be modulated by ScrB, as cooverexpression of these proteins increased flgEL expression while overexpression of ScrC alone appeared to decrease it. The authors proposed a model (Fig. 4A) in which ScrB, localized to the periplasm, receives a signal. In turn, ScrB communicates this information to ScrC via the latter protein's periplasmic domain. In response, ScrC controls the production of lateral flagella. Although V. parahaemolyticus can express either a single polar flagellum or multiple lateral flagella, the ability of the scrABC operon to impact motility appears to be limited to lateral flagella: swimming motility was unaltered by scr mutations. Furthermore, these genes do not appear to be required for the surface sensing that is mediated by the polar flagellum, as scr mutations do not substantially alter the induction of swarming that occurs through the loss of polar flagellum. These data thus suggest a novel function for the scr genes (10).
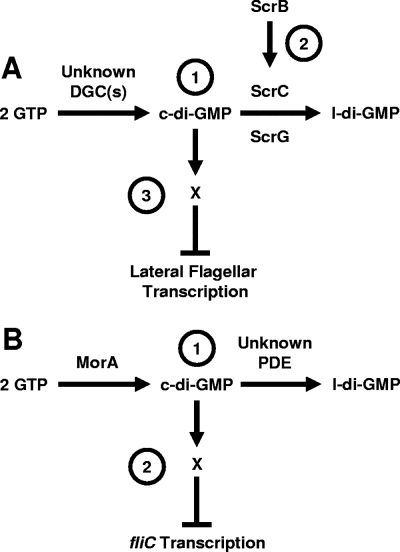
FIG. 4.
Working model for how c-di-GMP influences flagellar transcription. (A) Scr pathway of V. parahaemolyticus. (Label 1) The PDEs ScrC and ScrG and unidentified DGCs set the steady-state levels of c-di-GMP. (Label 2) ScrB, localized to the periplasm, receives a signal that causes ScrB to modulate ScrA activity. (Label 3) c-di-GMP binds to an unknown c-di-GMP-binding protein, X, to directly or indirectly inhibit lateral flagellar gene transcription. The role of ScrA is as yet unknown and is not included here. The net effect of Scr activity is to decrease c-di-GMP levels and, thus, increase lateral gene transcription. (B) P. putida MorA model. (Label 1) The levels of c-di-GMP are set through the activities of the putative DGC MorA and a hypothetical PDE. (Label 2) c-di-GMP together with a c-di-GMP-binding protein may directly or indirectly inhibit fliC transcription. Thus, the consequence of MorA activity is to decrease fliC transcription. l-di-GMP, linear diguanylate.
A second cosmid also substantially increased activity of the flgEL reporter (46). This second cosmid encoded ScrG, a protein whose domains include not only both GGDEF and EAL (like ScrC) but also PAS, which binds cofactors such as FAD or FMN and can respond to a variety of signals, such as light, oxygen, or redox (99). The GGDEF signature, however, is poorly conserved (HDDDF), while the EAL motif is ESL. Overexpression of ScrG alone was sufficient to induce both flgEL expression and production of lateral flagellins. Importantly, disruption of scrG resulted in both decreased lateral flagellin protein (to about 60% of WT) and decreased swarming, thus suggesting that the overexpression phenotype reflects a natural function of the protein. These overexpression phenotypes depended upon the EAL domain, as the truncated version ScrGΔEAL produced stable protein but failed to induce either transcription or flagellin production. Disruption of the EAL domain by substitution of ESL to ASL similarly abrogated activation. Surprisingly, both the deletion and the substitution converted ScrG into an inhibitor of transcription, as both variants caused flgEL transcription to be diminished relative to the level for the vector control. In contrast, substitutions in the GGDEF signature motif had little impact on protein function, even when combined with the EAL deletion; thus, single amino acids in the poorly conserved signature motif are not critical for the inhibitory activity (46). This genetic evidence suggests that ScrG functions as a PDE. Two-dimensional thin-layer chromatography supports this assignment; overexpression of ScrG decreases the steady-state levels of c-di-GMP (46). Given the similarities in domain structure and behavior, this is probably also the case for ScrC. For example, overexpression of either ScrABC or ScrG decreased transcription of capsular polysaccharide (cps) genes, probably as a consequence of elevated intracellular c-di-GMP (10, 46). Furthermore, the two sets of regulators suppressed each other: overexpression of ScrG restored swarming and flgEL expression to the scrABC operon mutants in an EAL-dependent manner, while a plasmid containing scrABC likewise restored those phenotypes to a scrG mutant. The interaction between these systems was investigated through the construction of double mutants. These analyses suggested that the scrABC locus played the greater role in controlling swarming motility; however, loss of both scrG and the scrA locus resulted in cumulative effects on both flgEL transcription and swarming motility (46).
To date, the level at which these regulators impact transcription of flgEL is unknown. Although the impact of scrA disruption on cps expression can be attributed to a newly discovered regulator (CpsR), its impact on motility cannot (31). Perhaps it depends upon one of the newly identified regulators of swarming (37).
MorA.
Another example of a GGDEF/EAL protein that impacts transcription of flagellar genes is the MorA protein of P. putida (Fig. 4B) (16). Disruption of morA led to enhanced swimming in soft agar (0.4%) motility plates, a phenotype that was largely restored by expression of morA from a plasmid and, in the context of numerous other studies, suggests that MorA acts as a DGC. Examination by transmission electron microscopy revealed that morA mutants contained substantially greater numbers of flagella. Furthermore, virtually all morA mutant cells contained flagella at all growth phases evaluated (early exponential, mid-exponential, and the exponential-to-stationary transition). This contrasts with wild-type cells, which restrict flagellar production largely to the exponential-to-stationary phase transition. These differences were reflected in the amounts of flagellin (FliC) protein produced by the two strains: the morA mutant contained substantially larger amounts of FliC protein. Similarly, the levels of fliC transcripts, as evaluated by Northern blot analysis, were greatly enhanced in the morA mutant, suggesting that MorA impacts flagellar biogenesis by controlling transcription. This effect appeared limited to P. putida, as a disruption of morA in P. aeruginosa failed to impact motility or levels of the fliC transcript. The level within the transcriptional hierarchy impacted by MorA remains to be determined.
Impact between transcription and assembly.
Bacteria also exert posttranscriptional control of flagellar biogenesis (1, 15, 25, 55). The ultimate goal of this regulation is to irreversibly switch the export substrate specificity from that of the subunits that comprise the rod and hook subassemblies to that of the subunits that form the filament (25) (Fig. 1). This control is coupled to the status of flagellar assembly and can occur at the level of translation, protein stability, and/or export and assembly. For example, translation of late (class III) genes has been shown to be positively regulated by a protein with weak similarity to translation initiation factors and by TTS chaperones. The latter proteins also protect their substrates (i.e., the products of these class III genes) from degradation and/or aggregation. Furthermore, the chaperone-substrate complexes appear to dock with the ATPase complex of the TTS export apparatus (3, 100) (Fig. 1). These observations have led to the hypothesis that translation of class III gene products might be localized to the export apparatus of actively assembling flagella (reviewed in reference 1), where the ability of subunits to be assembled into the growing flagellum dictates whether the protein will be translated and, if so, whether it is stable (3, 100). In this section, we describe two examples in which c-di-GMP appears to control motility at levels beyond transcription but prior to the production of fully functional flagella. The first example shows that c-di-GMP can impact both translation and secretion of late gene products, perhaps by interacting with one of these previously characterized mechanisms. The second example implicates c-di-GMP in a very early step in the export/assembly process (Fig. 1).
TipF.
The first example revolves around the TipF PDE of C. crescentus, an organism that has been studied extensively as a model for bacterial development and cell cycle control (14, 39, 53, 57, 95). This organism undergoes an asymmetric division that results in a motile swarmer cell and a nonmotile stalked cell. After a brief motile period, the swarmer cell ejects its single polar flagellum and grows a stalk at the pole previously occupied by the flagellum.
tipF was uncovered in a recent search for regulators that impact C. crescentus motility (36). Disruption of tipF resulted in nonmotile cells that lacked the hook and filament of the polar flagellum, as monitored by transmission electron microscopy. The nonmotile phenotype could be complemented by plasmid-borne wild-type TipF but not by a derivative (TipFE211A) with a disrupted EAL domain: this behavior supports the hypothesis that this protein indeed acts as a PDE (Fig. 5A). Although the tipF mutant exhibited near-normal levels (58% of wild type) of fljK flagellin transcription, it displayed little translation (2.5% of wild type) and highly reduced steady-state levels of the FljK protein. Similarly, the tipF mutant possessed decreased levels of the FlgE hook protein. In C. crescentus, the FlbT protein serves as a negative regulator of flagellin translation (Fig. 3B). When a mutation in flbT was combined with a tipF mutation, FljK levels increased, but the protein was not properly secreted, suggesting that TipF also controls motility at an additional level.
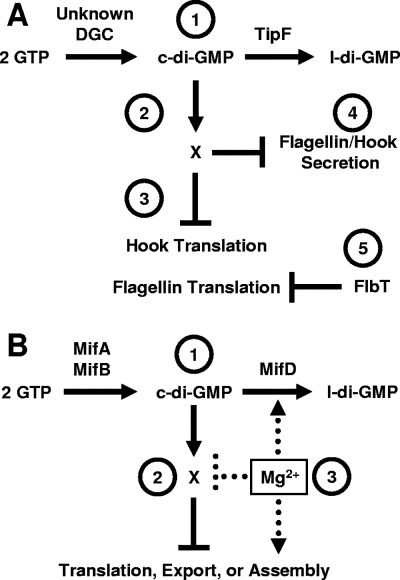
FIG. 5.
Working models for how c-di-GMP influences flagellar assembly. (A) C. crescentus TipF. (Label 1) An unknown DGC and the PDE TipF control the level of c-di-GMP. (Label 2) c-di-GMP binds to a c-di-GMP-binding protein, X, to directly or indirectly inhibit hook translation (or stability) and flagellin translation (label 3) and the secretion of both (label 4). FlbT independently inhibits flagellin translation (label 5). The net effect of TipF activity is to enhance hook and flagellin translation and secretion. (B) Mif model in V. fischeri. (Label 1) The DGCs MifA and MifB and the PDE MifD set the steady-state levels of c-di-GMP, which binds to an unknown c-di-GMP-binding protein, X (label 2). This complex interferes with the translation, export, and/or assembly of very early flagellar components. (Label 3) Mg2+ inhibits this process at some step downstream of c-di-GMP synthesis, e.g., activation of MifD activity or inhibition of the c-di-GMP-binding protein. l-di-GMP, linear diguanylate.
Investigations into the localization of TipF revealed that it could be found at the division plane approximately 30 min after the appearance of TipN, a presumptive “birth scar” protein that is thought to mark the new pole (36). It is at this new pole that the flagellum is eventually assembled, a process that clearly depends on TipF. Indeed, investigations of the localization of the switch component FliG (Fig. 1) revealed that while wild-type cells contained fluorescent foci of FliG-green fluorescent protein at both poles, the tipF mutant contained few such foci (19% of cells), the majority of which were mislocalized. Although the function of TipN does not appear to be related to the control of c-di-GMP levels, this protein is also required for correct localization of the flagellum: disruption of tipN resulted in cells with mislocalized flagella. Interestingly, a tipF tipN double mutant appeared similar to the tipF mutant in that no flagella were made, indicating that TipF acts in the pathway downstream of TipN and controls flagellar assembly regardless of flagellar placement (Fig. 5A).
Mif.
The second example of posttranscriptional control of motility comes from studies of V. fischeri. To assemble its polar tuft of flagella, this marine symbiont requires magnesium (Mg2+) at concentrations similar to those found in seawater. Under Mg2+-limited conditions, the vast majority of cells do not assemble flagella, apparently due to a paucity of flagellar proteins: Mg2+-limited cells possess very little of the highly abundant flagellin subunits that normally comprise the filament (69). Yet, Mg2+ does not substantially impact transcription of a variety of flagellar genes, including those for flagellin (70). Because Mg2+ does not seem to affect transcription but clearly influences steady-state protein levels (69, 70), it has been proposed that Mg2+ exerts its influence somewhere after transcript synthesis and stability. Because transmission electron microscopy found no obvious basal body-like structures at the poles of the nonflagellated Mg2+-limited cells (69), the block likely occurs just before or during an early stage of assembly, e.g., translation, protein stability, and/or export of basal body components (Fig. 5B).
Mutant screens to identify components of this novel magnesium-dependent induction of flagellation (Mif) pathway led to the identification of two genes that contribute to the inhibition of motility: in the absence of Mg2+, mutants defective for either gene migrated in soft (0.25%) agar substantially sooner and faster than the wild-type control (70). In the presence of Mg2+, however, both mutants migrated at a rate indistinguishable from that of the parent, suggesting that they were not simply hypermotile but instead no longer responded properly to the absence of Mg2+. The mifA gene encodes a DGC: when overexpressed in E. coli, it promotes synthesis of c-di-GMP. The mifB gene likely also encodes a DGC for it shares extensive homology with MifA and other DGCs. Like Mg2+, the impact of MifA and MifB appears to be posttranscriptional. The steady-state levels of flagellin subunits increased in mifA, mifB, or mifAB mutants and decreased when either protein was overexpressed, and yet none of these genetic manipulations exerted a substantial effect upon flagellar gene transcription (70).
Surprisingly, in the absence of Mg2+, a double mifA mifB mutant did not exhibit the full Mg2+-induced phenotype. Such behavior suggests the existence of an additional Mif component. It also supports a model in which Mg2+ does not feed into the system solely through MifA or MifB but leaves open the possibility that Mg2+ signals through another, more downstream component (Fig. 5B). Given the involvement of two DGCs, it seemed reasonable to hypothesize the existence of a gene that encodes a PDE that would specifically degrade the c-di-GMP synthesized by MifA and MifB. Indeed, such a gene has been identified: loss of MifD (a GGDEF-EAL hybrid with a nonconsensus GGDEF domain) results in decreased migration (even in the presence of Mg2+) and restores wild-type-like behavior to a mifB mutant (A. J. Wolfe and K. L. Visick, unpublished data). Whether other PDEs also participate in this pathway and whether PDE activity is modulated by Mg2+ remain to be determined.
Impact on function.
Upon completion of flagellar assembly, motor (Mot) proteins pump protons (H+) across the cytoplasmic membrane, providing energy to drive rotation (Fig. 1), while chemotaxis components control the direction of that rotation in response to physicochemical gradients (55). For the enterics E. coli and S. enterica serovar Typhimurium (YhjH/YcgR), for C. crescentus (DgcA/DgrAB), and for P. aeruginosa (SadB/BifA), evidence that c-di-GMP inhibits motility at the level of flagellar rotation exists.
YhjH/YcgR.
For E. coli, the evidence that c-di-GMP-related components might be involved came long before their biochemical functions became clear. An early study (47) revealed related roles in motility for two proteins: YcgR and YhjH, now known to be a PilZ domain protein (5, 18) and a PDE (90), respectively. These linked roles were identified through characterization of hns mutants, which exhibit motility defects (47). At the time, H-NS was known to impact motility at two levels: (i) a DNA-binding-dependent enhancement of flagellar biogenesis by promoting transcription of flhDC, which encode the master flagellar transcriptional regulator (Fig. 3A) (7), and (ii) a DNA-binding-independent modulation of flagellar rotation through interaction with the flagellar switch component FliG (23, 58). In an effort to separate these two functions, Ko and Park constructed hns mutants that transcribed flhDC in an H-NS-independent manner and found that they remained largely nonmotile (47). Careful phenotypic analyses of these mutants suggested that the absence of H-NS results in paralyzed flagella. Overexpression of the motor components FliG, MotB, and both MotB and MotA failed to restore motor function to the flhDC-constitutive hns mutant. In contrast, overexpression of MotA alone increased motility, albeit to a fraction of that of the wild-type parent. Since the levels of these motor components were not reduced in the flhDC-constitutive hns mutant relative to the wild type, Ko and Park argued against the lack of expression or protein instability as the cause of the H-NS-dependent motor defect. A search for transposon-based suppressors of this motor defect revealed ycgR, while a search for multicopy suppressors yielded yhjH. Although disruption of ycgR alone in an otherwise wild-type background did not cause a motility phenotype, loss of this gene in the flhDC-constitutive hns mutant restored flagellar rotation and, thus, promoted motility. Similar results were seen when multicopy yhjH was introduced into the flhDC-constitutive hns mutant. Bioinformatic analysis of the ycgR and yhjH promoter regions revealed sequences that suggested that both genes were σ28-dependent (class III) members of the flagellar regulon (Fig. 3A); indeed, transcription of both genes depended on flhDC (47).
Recently, many of these observations were confirmed for S. enterica serovar Typhimurium. As with E. coli, both yhjH and ycgR were shown to be class III members of the flagellar regulon (27, 105). Moreover, disruptions of yhjH led to a decreased migration rate, while disruptions in ycgR did not (27, 87, 88). Importantly, however, the decreased migration rate of the yhjH mutant could be reversed by a second disruption, one in ycgR, thus providing strong genetic evidence that the PilZ domain protein YcgR functions downstream of the PDE YhjH. In support of this hypothesis, biochemical and genetic analyses showed that YcgR must be able to bind c-di-GMP to antagonize the action of YhjH. Furthermore, the inability to observe any other phenotypes associated with c-di-GMP in Salmonella suggests that YhjH and YcgR are dedicated to motor control and provides evidence that c-di-GMP can be very specific (88).
Taken together, these studies suggest the following model (Fig. 6A). YhjH and an unknown DGC set the levels of c-di-GMP, which binds to YcgR. In a mechanism yet to be determined, this complex interferes with the proper association of the Mot proteins with FliG and the rest of the switching device. The result is a paralyzed flagellum. If the interaction of the Mot complex with FliG is a regulated process (104), then one could easily imagine that H-NS could enhance the process while the c-di-GMP-YcgR complex could inhibit it (47).
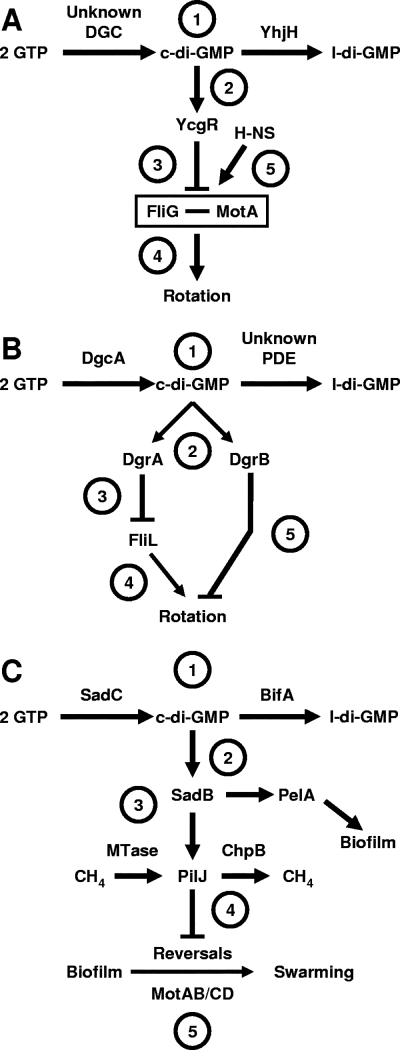
FIG. 6.
Working models for how c-di-GMP influences flagellar function. (A) YhjH/YcgR model for E. coli and S. enterica. (Label 1) The PDE YhjH and an unidentified DGC set the steady-state levels of c-di-GMP, which (label 2) binds to YcgR. (Label 3) This complex interferes with the ability of cells to properly insert the MotAB energy transduction complex next to the switching device component FliG in the completed basal body, resulting in (label 4) paralyzed flagella. (Label 5) The nucleoid protein H-NS antagonizes this effect by stabilizing the FliG-MotA interaction. The net effect of YcgR activity is to impair rotation, an activity opposed by YhjH. (B) DgcA/DgrAB model for C. crescentus. (Label 1) The DGC DgcA and an unidentified PDE set the steady-state levels of c-di-GMP, which (label 2) binds to DgrA and DgrB. (Label 3) Overexpression of DgrA decreases the steady-state levels of FliL. (Label 4) Since fliL mutants assemble paralyzed flagella, FliL may be integral to DgrA-dependent inhibition of rotation. (Label 5) Like DgrA, overexpression of DgrB inhibits flagellar rotation although it has no effect on FliL. The net effect of DgcA, DgrA, and DgrB is to impair rotation. (C) SadC-BifA model for P. aeruginosa. (Label 1) The DGC SadC and the PDE BifA set the steady-state levels of c-di-GMP, which (label 2) modulates the activity of SadB via a presently unknown mechanism. Also not understood is how SadB influences the activity of (label 3) PelA and the rest of the EPS biosynthetic pathway or (label 4) the methylation state of the chemoreceptor PilJ or (label 5) how PilJ controls the reversal rate, a process that also includes the energy transduction complexes MotAB and MotCD. The net result of SadC and SadB activity is to decrease the reversals necessary for swarming. l-di-GMP, linear diguanylate.
DgcA/DgrAB.
Recent work by Christen and colleagues (18) revealed that two other PilZ domain proteins (DgrA and DgrB; diguanylate receptors A and B) modulate flagellar function in response to high c-di-GMP levels. Overexpression of DgcA, a highly active C. crescentus DGC, disrupted motility (17, 18). However, overexpression of DgcA had no effect upon the levels of flagellin protein and the cells remained flagellated (18). This paralysis depended upon DgrA and DgrB: although disruption of dgrA or dgrB had no effect in wild-type cells, disruption of either alleviated the motility defect caused by DgcA overexpression. Thus, the DgcA/DgrAB story resembles that of YhjH/YcgR: in response to high levels of c-di-GMP, the PilZ domain protein appears to interfere with flagellar function rather than with the expression or assembly of the core flagellar components.
Like overexpression of dgcA, overexpression of either dgrA or dgrB resulted in cells with paralyzed flagella. Such cells contained normal levels of flagellar proteins chosen as representatives of each level of the flagellar hierarchy (Fig. 3). Furthermore, they elaborated seemingly complete flagella, and yet the cells appeared to be nonmotile (18). How the c-di-GMP generated by DgcA and that bound by DgrA and DgrB influence flagellar function remains to be determined; however, several intriguing clues exist. First, overexpression of DgrA, but not of DgrB, reduced the levels of a single flagellum-associated protein, FliL (18). Second, in C. crescentus, disruption of fliL was shown to result in paralyzed flagella (42). Third, in C. crescentus, FliL is present throughout the inner membrane rather than localized to the vicinity of the flagellum, a position that could facilitate its ability to interact with c-di-GMP wherever it exists. Fourth, FliL is the product of the first gene in a large operon that encodes motor/switch and export apparatus proteins, but it has not yet been assigned a specific function (91). Recently, however, it was shown to be involved in the transition of Proteus mirabilis from a vegetative swimmer cell to an elongated swarmer cell (6): polar insertions into fliL mutants resulted in a nonmotile, hyperelongated phenotype, while cells (including wild type) that overexpressed FliL alone did not elongate but remained nonmotile. Furthermore, expression of P. mirabilis FliL in wild-type E. coli resulted in reduced motility. Whether the P. mirabilis FliL functions in a c-di-GMP-dependent pathway and, if so, what level of regulation is impacted remain to be determined.
How might c-di-GMP influence C. crescentus flagellar function? The strong similarities to the YhjH/YcgR story, coupled with the observations that fliL mutants of C. crescentus assemble paralyzed flagella and that overexpression of P. mirabilis FliL inhibits E. coli motility, led Christen and coworkers to argue that FliL plays a key role in a universal mechanism that controls the transition between the motile planktonic and sessile biofilm lifestyle (18). In this model (Fig. 6B), the c-di-GMP produced by DgcA binds to DgrA and DgrB. The c-di-GMP-DgrA complex inhibits FliL, which is required for flagellar rotation. The c-di-GMP-DgrB complex also inhibits rotation, but the mechanism remains uncertain. Whether it involves the interaction between the MotAB energy transduction apparatus and FliG of the switching device, as proposed by Ko and Park (47), remains to be determined.
How does the c-di-GMP-DgrA complex regulate FliL? While the mechanism is unclear, the evidence suggests that it works posttranscriptionally. First, despite its position as the first gene in its operon, only FliL is regulated by DgrA. Furthermore, a search for extragenic suppressors of DgrA overexpression revealed rpsA, which encodes S1, a ribosomal protein that enhances translation initiation (18). An additional link between FliL, motility, and c-di-GMP is described later in the review.
SadC/BifA.
In four recent manuscripts, O'Toole and colleagues have sketched out a c-di-GMP-dependent pathway that appears to control the initiation of biofilm development in P. aeruginosa in part by regulating the flagellar reversal rate of swarming cells (11, 49, 66, 102).
Biochemical assays demonstrated that SadC, a GGDEF domain protein, functions as a DGC (66), while BifA, a GGDEF-EAL domain hybrid, functions as a PDE (49). Epistasis experiments provided evidence that, although other DGCs might be involved, these two integral cytoplasmic membrane proteins primarily set the steady-state level of a c-di-GMP pool responsible for controlling biofilm formation, wrinkled colony morphology, extracellular polysaccharide (EPS) synthesis, and swarming (Fig. 6C). The cumulative data support the hypothesis that c-di-GMP accumulation enhances biofilm formation by increasing pel-dependent EPS synthesis and inhibiting swarming. This inhibition, however, does not involve EPS: pelA bifA mutants are no more motile than bifA mutants (49, 66).
Instead, c-di-GMP-dependent inhibition of swarming correlates with a decrease in the rate at which flagellar rotation switches between the clockwise and counterclockwise states. Flagella are intimately involved in reversible attachment, the initial step of biofilm development (11, 71, 102). Furthermore, cells that switch rapidly (e.g., sadC mutants) are less likely to become irreversibly attached. Thus, O'Toole and coworkers suggested that the reduced switching rate provides the cells with a longer window of opportunity to interact lengthwise with the surface and increases the likelihood of irreversible attachment, the committed step toward biofilm formation (11). They argue that this behavior is analogous to that of switching-deficient E. coli mutants, which tend to become trapped in blind alleys within the liquid-filled maze of channels that comprises semisolid (soft) agar (108).
How does this pool of c-di-GMP influence motor reversals and, hence, the transition between biofilm formation and swarming? The details of the mechanism remain unsolved; however, the authors have used epistasis analyses to conclude that SadB is a required downstream pathway component that regulates both EPS synthesis and motor reversals (11, 49, 66). Downstream of SadB is the CheIV chemotaxis-like cluster, which includes the chemoreceptor PilJ and the methylesterase ChpB (Fig. 6C). It appears that the methylation state of PilJ determines the reversal rate (11), a process that involves the energy transduction complexes MotAB and MotCD (102).
Flagellar ejection.
In C. crescentus, motility is controlled at specific times in the cell cycle by flagellar assembly and subsequent ejection of a fully functional flagellum. Release of the Caulobacter flagellum occurs concomitantly with the proteolysis of FliF (41). FliF is the first flagellar protein assembled and comprises the MS ring that anchors the flagellum into the cytoplasmic membrane (Fig. 1) (41, 103). Given its importance in assembly, it would thus not be surprising if it also served as the target for the control of flagellar ejection (41). Indeed, upon proteolysis of FliF, the flagellar filament along with the hook and the distal portion of the rod is released into the extracellular medium (41, 43). However, recent data have failed to support this model. For example, proteolysis-resistant derivatives of FliF retained normal flagellar function and ejection (30). Similarly, a mutant that lacked ClpA, a protein that together with ClpP forms the ClpAP protease (75), failed to degrade FliF yet ejected flagella normally (30). Together, these data argue that ejection does not require cleavage of FliF but remain consistent with a model that involves the loss of one protein or a small complex of proteins located near FliF at the cytoplasmic-proximal portion of the rod (43). Other FliF-proximal flagellar proteins, the switching proteins FliG and FliM, also appear to undergo cell cycle-dependent degradation (41, 42) and thus could also participate in flagellar ejection.
Although the cause of flagellar ejection cannot be ascribed to proteolysis of FliF, this developmental event remains proximal to ejection and thus important as a marker for development. One regulator known to control FliF stability and flagellar ejection is PleD, the best-characterized regulator of c-di-GMP levels and motility in C. crescentus (2, 4, 33, 93). Examination of PleD provided biochemical proof that the GGDEF domain was responsible for DGC activity (73). Furthermore, PleD is the first DGC for which a crystal structure has been obtained. Most importantly, this structural work revealed the active site for c-di-GMP synthesis and identified a site for allosteric inhibition (13, 17).
Like many other regulators of c-di-GMP, PleD controls the motile-sessile transition: the evidence for its role in ejection is substantial, as are the data that this protein participates in the subsequent development of the stalk (2, 33, 52, 93). In a pleD mutant, the flagellum is not ejected. This effect is independent of fliF transcription; instead, the FliF protein is not efficiently processed, i.e., the stability of the FliF protein increases from 67 min to 94 min, a change probably sufficient to account for the retention of FliF by the pleD mutant (2). The C-terminal GGDEF domain of PleD is required for appropriate degradation of FliF, as mutations that deleted GGDEF or changed the GG to DE failed to induce normal proteolysis (2). These data suggest that the DGC activity of PleD is important for flagellar ejection (Fig. 7).
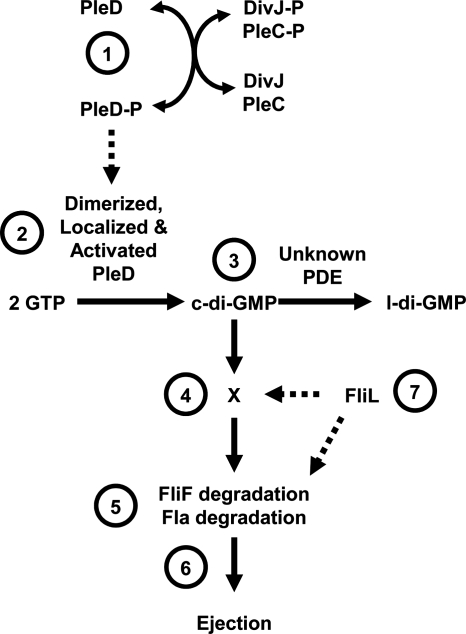
FIG. 7.
Working model for how c-di-GMP influences flagellar ejection. The DGC PleD controls flagellar ejection. (Label 1) Activation of PleD depends upon phosphorylation, which is controlled by two sensor kinases, DivJ and PleC. (Label 2) Phosphorylated PleD (PleD-P) dimerizes, and these dimers can now be polarly localized. (Label 3) The dimer form of PleD is now active as a DGC, and it and an unidentified PDE set the steady-state levels of c-di-GMP. (Label 4) c-di-GMP binds to an unknown c-di-GMP-binding protein, which (label 5) helps to destabilize FliF and possibly another flagellar protein(s) in the proximity of FliF. (Label 6) Destabilization of this protein(s) leads to ejection. (Label 7) FliL appears to be a component of the PleD pathway, but its position in that pathway remains uncertain. l-di-GMP, linear diguanylate.
PleD does not impact generalized cell cycle-dependent degradation, as indicated by the normal degradation in a pleD mutant of the cell cycle-dependent McpA chemoreceptor (2). Thus, ejection of the flagellum and degradation of the chemotaxis machinery appear to be independent events. Indeed, proteolysis of FliF is independent of a number of other flagellar proteins, including rod, hook, and switch proteins (2). Intriguingly, however, proteolysis is decreased in cells lacking FliL, a protein that plays a role in flagellar rotation (2, 42) and whose levels are decreased in cells that overexpress the PilZ domain protein DgrA (18). Thus, both PleD and FliL appear to destabilize FliF. Furthermore, the stability of FliF in a pleD fliL double mutant was similar to that of the pleD single mutant, a result that suggests they may function in the same pathway (2). Whether DgrA is also part of this pathway remains to be determined. A dissection of FliL revealed that its ability to induce normal FliF proteolysis depends upon a highly charged internal domain (ΔfliL1), a region not required for motility (42). These data support the hypothesis that FliL plays distinct roles in both motility and FliF degradation.
The connection between PleD and FliL indicates that PleD may contribute to control of flagellar rotation. Indeed, this possibility was first suggested by the isolation of pleD as a motile suppressor of the paralyzed pleC mutant (93). Furthermore, flagellar biogenesis but not rotation could be restored to the pleD mutant by a constitutively active allele of the P. fluorescens WspR DGC (4), further establishing this link. However, the specific role of PleD, if any, in promoting flagellar rotation remains to be determined.
How is the activity of PleD controlled? PleD is composed of three domains: the GGDEF domain and two copies of the two-component response regulator domain. This architecture is unusual on two counts: it appears to have two input domains, and those input domains are attached to a GGDEF domain instead of a DNA-binding domain (2, 33). This structure suggests that phosphorylation modulates DGC activity, and the evidence supports this hypothesis (72, 73). Furthermore, this phosphorylation controls localization. PleD localizes to the stalked pole at the swarmer cell-to-stalked cell transition (73). This localization depends upon activation of PleD: PleD fails to localize to the poles when it is unable to be phosphorylated, either through mutation of the site of phosphorylation or by disruption of the two kinases (DivJ and PleC) known to control the phosphorylation status of PleD (4, 73). In contrast, localization is independent of the GGDEF domain. A dominant negative allele of pleD, which produces a protein (PleD*) with four substitutions, exhibited substantially reduced motility (4). PleD* functions independently of phosphorylation signals, as disruption of the site of phosphorylation (Asp53) did not impact its activity. In contrast, the addition of a mutation that prevents PleD dimerization (Y26A) disrupted localization of PleD* (72), thus supporting the importance of dimerization for localization of PleD. In summary, the activity of PleD is controlled by phosphorylation, which promotes formation of a dimer, the form of PleD associated with DGC activity (Fig. 7) (72, 73). Thus, it is clear from the investigation of this one DGC that cells can tightly control the activity of a DGC to ultimately regulate motility. In this case, the posttranscriptional mechanisms are phosphorylation and localization, which are further controlled in Caulobacter by links to the cell cycle (59). In other organisms under different environmental and developmental scenarios, the mechanisms will likely differ.
Concluding remarks.
Many studies link c-di-GMP and motility. However, to understand the specific roles of c-di-GMP in controlling motility, it is necessary to look beyond studies that rely on DGC or PDE overexpression, as the observed impacts on motility may, in many cases, turn out to be nonspecific, artifactual consequences of high c-di-GMP levels on cellular physiology. Rigorous proof that a particular pathway exerts a specific effect on motility requires the construction of nonpolar chromosomal mutations, verification by single-copy complementation, and, where appropriate, epistasis tests to evaluate relationships within and between pathways.
Despite the limited number of studies in which such experiments have been completed, there is nonetheless clear evidence that c-di-GMP exerts specific effects on flagellar biogenesis. The mechanisms by which c-di-GMP appear to impact motility are as diverse as the levels of control that are already known to exist. A better understanding of the role of c-di-GMP in controlling motility, however, will be advanced substantially through additional efforts to fence in the level at which the c-di-GMP-dependent effect occurs. While it is relatively simple to investigate motility by using soft agar assays (108), too many studies appear to rely solely upon evaluating single time point data. This is particularly problematic because subtle phenotypes, such as the reduced motility reported in many investigations of c-di-GMP-dependent pathways, can be caused by a number of reasons. These include delayed migration due to a failure to express flagella at the time of inoculation, a subtle chemotactic defect, reduced numbers of flagella, and mildly defective flagella. Detailed knowledge of migration history can help distinguish among these diverse possibilities (for examples, see references 69 and 70). To complete the evaluation, other assays must be employed, including light microscopy to examine swimming behavior and motor rotation, electron microscopy to observe flagella and/or incomplete flagellar structures, reporters to test gene and protein expression, and biochemical studies (e.g., immunoblot analysis) to verify the conclusions.
Once it becomes clear that c-di-GMP is involved in controlling motility, the next goal should be to identify the effector. The discovery of the PilZ domain has provided insights and promise for identifying these effectors and elucidating their pathways. For the few PilZ proteins whose roles have been ascertained, however, their functions have been revealed only by their overexpression or when either a DGC has been overexpressed or a PDE disrupted. Are there circumstances in which these proteins play a role in the absence of high levels of c-di-GMP? One possibility is that the PilZ domain proteins do not become important except under conditions not typically assayed in the laboratory (e.g., swarming across surfaces or survival in a host). Alternatively, or in addition, the activity of certain DGCs and PDEs may be limited to specific locations and times. This certainly is the case for Caulobacter PleD, whose impact on flagellar ejection depends upon a number of factors, including the receipt of specific cell cycle-dependent signals (59), its status relative to phosphorylation and the monomer-dimer transition, and its localization. Other signals in other organisms are known or suspected to act upon the levels of c-di-GMP or its downstream output. These include Mg2+ (V. fischeri) (69, 70), norspermidine (V. cholerae) (44), phosphate (P. fluorescens) (67), and aminoglycosides (P. aeruginosa) (35). Even with evidence of specific signals that induce various c-di-GMP-dependent pathways, it would seem surprising if the very limited numbers of PilZ domain proteins identified by bioinformatics (5) alone can carry out all of the downstream consequences of c-di-GMP signaling. Perhaps some PilZ domain proteins are not highly conserved and, thus, have not been identified as such. Another possibility is that some of the proteins that carry less well-conserved EAL/GGDEF signature motifs will turn out to be effectors of the target rather than controllers of the signal. Moreover, other c-di-GMP-binding domains may yet exist. Alternatively, or additionally, c-di-GMP might bind directly to specific flagellar components.
In writing this review, two common themes we noticed were the connection between c-di-GMP and FliL in controlling motility and the importance of c-di-GMP in swarming motility of multiple organisms. FliL, although part of the flagellar regulon, does not seem to be an integral component of the flagellum in the enterics. In Caulobacter, FliL also is not required to make a flagellum, but it is required for flagellar function, e.g., rotation. Furthermore, the levels of FliL are decreased by overexpression of the c-di-GMP receptor protein DgrA, which also causes the production of paralyzed flagella. It would be interesting to know whether the dual overexpression of FliL can restore some motility to cells that overexpress DgrA. Intriguingly, the loss of either FliL or PleD increases the stability of FliF, a phenotype that occurs coincident with flagellar ejection. Together, these data lead us to wonder about the role of FliL. Perhaps it is a sensor that recognizes that a cell is touching a surface and thereby needs to induce lateral flagella for swarming or that a flagellum is no longer required and thereby signals its ejection. That a protein such as FliL might be involved in relaying information about surface recognition is not too far-fetched. Indeed, for Salmonella, the flagellum has been shown to sense surface wetness and, in response, control filament length of swarming cells (106). Interestingly, the ability to reverse flagellar rotation seems to be all that is required to enhance surface wetness and restore filaments to their maximal length (56). The latter phenomenon could provide an alternative explanation for the swarming and biofilm phenotypes observed by O'Toole and colleagues. P. aeruginosa sadC mutants switch rapidly, potentially increasing surface hydration and promoting swarming. In contrast, bifA mutants reverse less frequently and would not hydrate their surfaces. The result would be reduced swarming and enhanced biofilm formation.
Finally, the variety of levels at which c-di-GMP can impact motility in the relatively few organisms studied begs the following question: how universal are these mechanisms? Given the evolutionary relationship between flagellar and nonflagellar TTS systems (organelles through which pathogens inject their virulence factors), we might expect to shortly see evidence that c-di-GMP controls virulence through impacting the transcription, translation, assembly, and function of such organelles. The groundwork that we lay down in understanding the impact of c-di-GMP on flagellar biogenesis and function, in that case, will be far-reaching.
ADDENDUM IN PROOF
Recent work studying ScrC suggests that this protein can function as both a PDE and a DGC (R. B. R. Ferreira, L. C. M. Antunes, E. P. Greenberg, and L. L. McCarter, J. Bacteriol., in press). ScrC may thus be the “unknown DGC” shown in Fig. 4A, and ScrB may modulate these two activities.
Acknowledgments
There are many stories of c-di-GMP control of flagellar biogenesis that are developing. We apologize to those whose research we were unable to include here. We thank Christine Anderson for her helpful comments.
This work was supported by the estate of William G. Potts in support of medical research at the Stritch School of Medicine at Loyola Univeristy Chicago, by NIH grant GM59690 awarded to K.L.V., and by NIH grant GM066130 awarded to A.J.W.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5160-165. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32379-391. [DOI] [PubMed] [Google Scholar]
- 3.Aldridge, P., J. E. Karlinsey, and K. T. Hughes. 2003. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol. Microbiol. 491333-1345. [DOI] [PubMed] [Google Scholar]
- 4.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 471695-1708. [DOI] [PubMed] [Google Scholar]
- 5.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 223-6. [DOI] [PubMed] [Google Scholar]
- 6.Belas, R., and R. Suvanasuthi. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J. Bacteriol. 1876789-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertin, P., E. Terao, E. H. Lee, P. Lejeune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 1765537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 1883600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247123-130. [DOI] [PubMed] [Google Scholar]
- 10.Boles, B. R., and L. L. McCarter. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 1845946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caiazza, N. C., J. H. Merritt, K. M. Brothers, and G. A. O'Toole. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1893603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 3111113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, C., R. Paul, D. Samoray, N. C. Amiot, B. Giese, U. Jenal, and T. Schirmer. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 10117084-17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, J. C., and C. Stephens. 2007. Bacterial cell cycle: completing the circuit. Curr. Biol. 17R203-R206. [DOI] [PubMed] [Google Scholar]
- 15.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choy, W. K., L. Zhou, C. K. Syn, L. H. Zhang, and S. Swarup. 2004. MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J. Bacteriol. 1867221-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christen, B., M. Christen, R. Paul, F. Schmid, M. Folcher, P. Jenoe, M. Meuwly, and U. Jenal. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 28132015-32024. [DOI] [PubMed] [Google Scholar]
- 18.Christen, M., B. Christen, M. G. Allan, M. Folcher, P. Jeno, S. Grzesiek, and U. Jenal. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 1044112-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 28030829-30837. [DOI] [PubMed] [Google Scholar]
- 20.Cotter, P. A., and S. Stibitz. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 1017-23. [DOI] [PubMed] [Google Scholar]
- 21.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 1502497-2502. [DOI] [PubMed] [Google Scholar]
- 22.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50809-824. [DOI] [PubMed] [Google Scholar]
- 23.Donato, G. M., and T. H. Kawula. 1998. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J. Biol. Chem. 27324030-24036. [DOI] [PubMed] [Google Scholar]
- 24.Dutton, R. J., Z. Xu, and J. W. Gober. 2005. Linking structural assembly to gene expression: a novel mechanism for regulating the activity of a σ54 transcription factor. Mol. Microbiol. 58743-757. [DOI] [PubMed] [Google Scholar]
- 25.Ferris, H. U., and T. Minamino. 2006. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol. 14519-526. [DOI] [PubMed] [Google Scholar]
- 26.Fouhy, Y., J. F. Lucey, R. P. Ryan, and J. M. Dow. 2006. Cell-cell signaling, cyclic di-GMP turnover and regulation of virulence in Xanthomonas campestris. Res. Microbiol. 157899-904. [DOI] [PubMed] [Google Scholar]
- 27.Frye, J., J. E. Karlinsey, H. R. Felise, B. Marzolf, N. Dowidar, M. McClelland, and K. T. Hughes. 2006. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1882233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 20311-21. [DOI] [PubMed] [Google Scholar]
- 30.Grünenfelder, B., S. Tawfilis, S. Gehrig, M. Østerås, D. Eglin, and U. Jenal. 2004. Identification of the protease and the turnover signal responsible for cell cycle-dependent degradation of the Caulobacter FliF motor protein. J. Bacteriol. 1864960-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Güvener, Z. T., and L. McCarter. 2003. Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J. Bacteriol. 1855431-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harshey, R. M. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57249-273. [DOI] [PubMed] [Google Scholar]
- 33.Hecht, G., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer- to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 1776223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 10214422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman, L. R., D. A. D'Argenio, M. J. MacCoss, Z. Zhang, R. A. Jones, and S. I. Miller. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 4361171-1175. [DOI] [PubMed] [Google Scholar]
- 36.Huitema, E., S. Pritchard, D. Matteson, S. K. Radhakrishnan, and P. H. Viollier. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 1241025-1037. [DOI] [PubMed] [Google Scholar]
- 37.Jaques, S., and L. L. McCarter. 2006. Three new regulators of swarming in Vibrio parahaemolyticus. J. Bacteriol. 1882625-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7185-191. [DOI] [PubMed] [Google Scholar]
- 39.Jenal, U. 2000. Signal transduction mechanisms in Caulobacter crescentus development and cell cycle control. FEMS Microbiol. Rev. 24177-191. [DOI] [PubMed] [Google Scholar]
- 40.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40385-407. [DOI] [PubMed] [Google Scholar]
- 41.Jenal, U., and L. Shapiro. 1996. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 152393-2406. [PMC free article] [PubMed] [Google Scholar]
- 42.Jenal, U., J. White, and L. Shapiro. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J. Mol. Biol. 243227-244. [DOI] [PubMed] [Google Scholar]
- 43.Kanbe, M., S. Shibata, Y. Umino, U. Jenal, and S.-I. Aizawa. 2005. Protease susceptibility of the Caulobacter crescentus flagellar hook-basal body: a possible mechanism of flagellar ejection during cell differentiation. Microbiology 151433-438. [DOI] [PubMed] [Google Scholar]
- 44.Karatan, E., T. R. Duncan, and P. I. Watnick. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 1877434-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kazmierczak, B. I., M. B. Lebron, and T. S. Murray. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 601026-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim, Y. K., and L. L. McCarter. 2007. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 1894094-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko, M., and C. Park. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 303371-382. [DOI] [PubMed] [Google Scholar]
- 48.Kovacikova, G., W. Lin, and K. Skorupski. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol. Microbiol. 57420-433. [DOI] [PubMed] [Google Scholar]
- 49.Kuchma, S. L., K. M. Brothers, J. H. Merritt, N. T. Liberati, F. M. Ausubel, and G. A. O'Toole. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1898165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulasakara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 1032839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee, V. T., J. M. Matewish, J. L. Kessler, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 651474-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levi, A., and U. Jenal. 2006. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J. Bacteriol. 1885315-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li, R., and S. C. Wai. 2004. Bacterial cell polarity: a “swarmer-stalked” tale of actin. Trends Cell Biol. 14532-536. [DOI] [PubMed] [Google Scholar]
- 54.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60331-348. [DOI] [PubMed] [Google Scholar]
- 55.Macnab, R. M. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694207-217. [DOI] [PubMed] [Google Scholar]
- 56.Mariconda, S., Q. Wang, and R. M. Harshey. 2006. A mechanical role for the chemotaxis system in swarming motility. Mol. Microbiol. 601590-1602. [DOI] [PubMed] [Google Scholar]
- 57.Martin, M. E., and Y. V. Brun. 2000. Coordinating development with the cell cycle in Caulobacter. Curr. Opin. Microbiol. 3589-595. [DOI] [PubMed] [Google Scholar]
- 58.Marykwas, D. L., and H. C. Berg. 1996. A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J. Bacteriol. 1781289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matroule, J. Y., H. Lam, D. T. Burnette, and C. Jacobs-Wagner. 2004. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118579-590. [DOI] [PubMed] [Google Scholar]
- 60.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56289-314. [DOI] [PubMed] [Google Scholar]
- 61.McCarter, L. 1999. The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 151-57. [PubMed] [Google Scholar]
- 62.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mendez-Ortiz, M. M., M. Hyodo, Y. Hayakawa, and J. Membrillo-Hernandez. 2006. Genome wide transcriptional profile of Escherichia coli in response to high levels of the second messenger c-di-GMP. J. Biol. Chem. 2818090-8099. [DOI] [PubMed] [Google Scholar]
- 64.Merighi, M., V. T. Lee, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65876-895. [DOI] [PubMed] [Google Scholar]
- 65.Merino, S., J. G. Shaw, and J. M. Tomás. 2006. Bacterial lateral flagella: an inducible flagella system. FEMS Microbiol. Lett. 263127-135. [DOI] [PubMed] [Google Scholar]
- 66.Merritt, J. H., K. M. Brothers, S. L. Kuchma, and G. A. O'Toole. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 1898154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monds, R. D., P. D. Newell, R. H. Gross, and G. A. O'Toole. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63656-679. [DOI] [PubMed] [Google Scholar]
- 68.Muir, R. E., and J. W. Gober. 2004. Regulation of FlbD activity by flagellum assembly is accomplished through direct interaction with the trans-acting factor, FliX. Mol. Microbiol. 54715-730. [DOI] [PubMed] [Google Scholar]
- 69.O'Shea, T. M., C. R. DeLoney-Marino, S. Shibata, S.-I. Aizawa, A. J. Wolfe, and K. L. Visick. 2005. Magnesium promotes flagellation of Vibrio fischeri. J. Bacteriol. 1872058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Shea, T. M., A. H. Klein, K. Geszvain, A. J. Wolfe, and K. L. Visick. 2006. Diguanylate cyclases control magnesium-dependent motility of Vibrio fischeri. J. Bacteriol. 1888196-8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30295-304. [DOI] [PubMed] [Google Scholar]
- 72.Paul, R., S. Abel, P. Wassmann, A. Beck, H. Heerklotz, and U. Jenal. 2007. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J. Biol. Chem. 28229170-29177. [DOI] [PubMed] [Google Scholar]
- 73.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pei, J., and N. V. Grishin. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42210-216. [DOI] [PubMed] [Google Scholar]
- 75.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32449-458. [DOI] [PubMed] [Google Scholar]
- 76.Pratt, J. T., R. Tamayo, A. D. Tischler, and A. Camilli. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 28212860-12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 391595-1609. [DOI] [PubMed] [Google Scholar]
- 78.Prüβ, B. M., D.-J. Kim, S. Forst, R. T. Fleming, K. L. Visick, and A. J. Wolfe. 2005. Genomics of flagella, p. 1-12. In B. M. Prüβ (ed.), Global regulatory networks in enteric bacteria. Research Signpost, Kerala, India.
- 79.Rahman, M., R. Simm, A. Kader, E. Basseres, U. Romling, and R. Mollby. 2007. The role of c-di-GMP signaling in an Aeromonas veronii biovar sobria strain. FEMS Microbiol. Lett. 273172-179. [DOI] [PubMed] [Google Scholar]
- 80.Ramelot, T. A., A. Yee, J. R. Cort, A. Semesi, C. H. Arrowsmith, and M. A. Kennedy. 2007. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins 66266-271. [DOI] [PubMed] [Google Scholar]
- 81.Romling, U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 621234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Romling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9218-228. [DOI] [PubMed] [Google Scholar]
- 83.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. c-di-GMP: the dawning of a novel bacterial signaling system. Mol. Microbiol. 57629-639. [DOI] [PubMed] [Google Scholar]
- 84.Ross, P., R. Mayer, and M. Benziman. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 5535-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 1036712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Ryan, R. P., Y. Fouhy, J. F. Lucey, and J. M. Dow. 2006. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 1888327-8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rychlik, I., G. Martin, U. Methner, M. Lovell, L. Cardova, A. Sebkova, M. Sevcik, J. Damborsky, and P. A. Barrow. 2002. Identification of Salmonella enterica serovar Typhimurium genes associated with growth suppression in stationary-phase nutrient broth cultures and in the chicken intestine. Arch. Microbiol. 178411-420. [DOI] [PubMed] [Google Scholar]
- 88.Ryjenkov, D. A., R. Simm, U. Romling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP. The PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 28130310-30314. [DOI] [PubMed] [Google Scholar]
- 89.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 1871792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 1874774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schoenhals, G. J., and R. M. Macnab. 1999. FliL is a membrane-associated component of the flagellar basal body of Salmonella. Microbiology 1451769-1775. [DOI] [PubMed] [Google Scholar]
- 92.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 531123-1134. [DOI] [PubMed] [Google Scholar]
- 93.Sommer, J. M., and A. Newton. 1989. Turning off flagellum rotation requires the pleiotropic gene pleD: pleA, pleC, and pleD define two morphogenic pathways in Caulobacter crescentus. J. Bacteriol. 171392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soutourina, O. A., and P. N. Bertin. 2003. Regulation cascade of flagellar expression in gram-negative bacteria. FEMS Microbiol. Rev. 27505-523. [DOI] [PubMed] [Google Scholar]
- 95.Stephens, C. 2004. Prokaryotic development: a new player on the cell cycle circuit. Curr. Biol. 14R505-R507. [DOI] [PubMed] [Google Scholar]
- 96.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 1804416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tamayo, R., J. T. Pratt, and A. Camilli. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61131-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 28033324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomas, J., G. P. Stafford, and C. Hughes. 2004. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. USA 1013945-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thormann, K. M., S. Duttler, R. M. Saville, M. Hyodo, S. Shukla, Y. Hayakawa, and A. M. Spormann. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 1882681-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Toutain, C. M., N. C. Caizza, M. E. Zegans, and G. A. O'Toole. 2007. Roles for flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res. Microbiol. 158471-477. [DOI] [PubMed] [Google Scholar]
- 103.Ueno, T., K. Oosawa, and S. Aizawa. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227672-677. [DOI] [PubMed] [Google Scholar]
- 104.Van Way, S. M., E. R. Hosking, T. F. Braun, and M. D. Manson. 2000. Mot protein assembly into the bacterial flagellum: a model based on mutational analysis of the motB gene. J. Mol. Biol. 2977-24. [DOI] [PubMed] [Google Scholar]
- 105.Wang, Q., J. G. Frye, M. McClelland, and R. M. Harshey. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52169-187. [DOI] [PubMed] [Google Scholar]
- 106.Wang, Q., A. Suzuki, S. Mariconda, S. Porwollik, and R. M. Harshey. 2005. Sensing wetness: a new role for the bacterial flagellum. EMBO J. 242034-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weber, H., C. Pesavento, A. Possling, G. Tischendorf, and R. Hengge. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 621014-1034. [DOI] [PubMed] [Google Scholar]
- 108.Wolfe, A. J., and H. C. Berg. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 866973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu, J., and A. Newton. 1997. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol. Microbiol. 24233-239. [DOI] [PubMed] [Google Scholar]