Abstract
By analyzing the genomic sequences of 12 Bacteroidales species, we found that all intestinal species have numerous polysaccharide biosynthesis loci, many with promoters that we demonstrate undergo DNA inversion. This feature is not conserved in the Bacteroidales order as a whole, as oral species do not share these genetic features.
The alimentary tract is home to the densest populations of organisms in the human body. Within the digestive tract, the colonic microbiota far exceeds the density of organisms in other sites and is comprised of hundreds of species of microorganisms that coexist in mutualistic relationships with each other and their host. The oral cavity also contains a robust consortium of organisms that accumulate over time and, although they have cooperative relationships with each other, ultimately provide no benefit to their host and are often pathogenic. Members of the phylum Bacteroidetes are key players in both of these ecosystems. This phylum contains many diverse organisms, including human pathogens, mammalian symbionts, nonmammalian endosymbionts, and marine organisms. Bacteroidales, an order within the phylum Bacteroidetes, contains at least four distinct families whose members reside in a mammalian host: the Bacteroidaceae, the Prevotellaceae, the Porphyromonaceae, and the Rikenellaceae. Members of the Bacteroidales order are the most abundant of the cultured gram-negative organisms in the human colonic microbiota, where they provide beneficial properties to their host (13, 17, 22). Members of the Bacteroidales are also found within the human oral cavity and are among the most important of the periodontal pathogens, contributing to periodontitis and other local sequelae as well as systemic diseases (reviewed in reference 9). Bacteroidales species occupy very strict niches within the digestive tract, being present in either the oral cavity or intestine but not in both ecosystems. For example, Bacteroides fragilis, Bacteroides thetaiotaomicron, and Parabacteoides distasonis are members of the normal colonic microbiota, whereas Porphyromonas gingivalis, Prevotella intermedia, and Tannerella forsythensis are oral pathogens.
The organisms of the normal intestinal microbiota establish long-term relationships with the host, where they must peacefully coexist and successfully compete with other members of the microbiota for nutrients and space. They must also withstand attacks from a variety of sources, including phages, antibacterial products from the host, and harmful products of other bacteria. Similar to the case for pathogens, the ability of these organisms to alter their surfaces would seem an obvious survival advantage in this ecosystem. B. fragilis was the first bacterium shown to produce an extensive number of surface capsular polysaccharides: eight phenotypically confirmed molecules from a single strain (15). The expression of these polysaccharides is phase variable, dictated by DNA inversion of the promoter upstream of each polysaccharide biosynthesis locus (15). A second intestinal Bacteroides species, B. thetaiotaomicron, contains the genetic material for the synthesis of seven capsular polysaccharides, four of which have invertible promoter regions (24). The inversion of each of the B. fragilis polysaccharide promoters is mediated by a single global DNA invertase, designated Mpi (6), which is a member of the serine family of site-specific recombinases (Ssrs). In contrast, inversion of the B. thetaiotaomicron polysaccharide promoters is likely mediated by individual tyrosine family site-specific recombinases (Tsrs) encoded by separate genes present just upstream of each of the invertible polysaccharide promoters (24). Therefore, although the mechanisms of inversion differ between the two species, the phenotypic outcome is the same: extensive and rapid surface diversity.
The synthesis of such a large number of capsular polysaccharides has not been demonstrated in any bacterial species outside the Bacteroidales order, and no other bacterial capsular polysaccharides are known to undergo phase variation by inversion of promoter regions. This study was undertaken to determine if these important genetic characteristics that allow such drastic surface variability are conserved among diverse Bacteroidales species or if the synthesis of multiple phase-variable polysaccharides is limited to Bacteroidales of a particular niche or possibly to only a few Bacteroidales species.
Currently, there are a significant number of Bacteroidales species for which there is a complete or partial genomic sequence available. We began these analyses by compiling a list of all species taxonomically categorized by NCBI within the Bacteroidetes phylum and for which there is a publicly available genomic sequence. Other Bacteroidetes species, not listed by NCBI but for which there is a genomic sequence publicly available, were subsequently added to the list.
We retrieved the 16S rRNA gene sequence for each member from those available at the Ribosomal Database Project II (RDP) (release 9.49, 3 April 2007) maintained by the Center for Microbial Ecology at Michigan State University (http://rdp.cme.msu.edu/). Many of these sequences contained a significant number of uncalled or ambiguous bases and were thus replaced where possible with cleaner sequence from the same species by using the RDP sequences as queries in nucleotide-nucleotide BLAST searches against either the GenBank database or a custom database prepared locally from the appropriate genomic sequence. The Tree Builder program provided by the RDP site was then used to create a phylogenetic tree using the Eubacteria alignment model and including the sequence of the Escherichia coli K-12 MG1655 16S rRNA gene (GenBank accession number U00096, bases 4164682 through 4166223) as an outgroup. Species which were phylogenetically misplaced, such as Bacteroides capsillosus (14), were removed from the list.
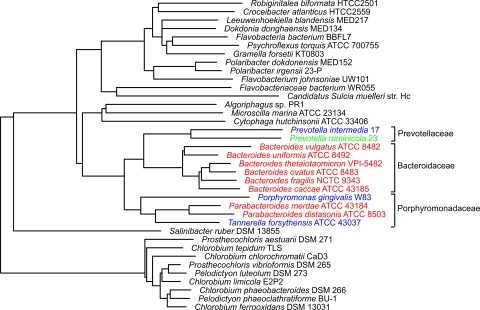
Inspection of the Bacteroidetes 16S rRNA gene cladogram reveals that members of the order Bacteroidales segregate to a distinct branch (Fig. 1). We obtained complete or partial genome sequence for three families of Bacteroidales: the Bacteroidaceae (six species), the Prevotellaceae (two species), and the Porphyromonadaceae (four species). These sequenced members include three oral species (P. gingivalis, T. forsythensis, and P. intermedia), one ruminal species (Prevotella ruminicola), and eight intestinal species (B. fragilis, B. thetaiotaomicron, Bacteroides vulgatus, Bacteroides caccae, Bacteroides ovatus, Bacteroides uniformis, P. distasonis, and Parabacteoides merdae). As shown in Fig. 1, the habitats of these organisms do not strictly segregate by phylogeny, as some members of the Porphyromonadacae reside in the intestine while others are found in the oral cavity.
FIG. 1.
16S rRNA gene cladogram of members of the Bacteroidetes phylum for which there is a partial or complete genome sequence. Members of the Bacteroidales order are shown in color. The three families of Bacteroidales present in this cladogram are indicated. Within the Bacteroidales, oral species are in blue, the ruminal species is in green, and the intestinal species are in red.
In order to locate and analyze putative polysaccharide biosynthesis loci from the genomes of these 12 species, all six frames of their genome sequences were translated into amino acid open reading frames (ORFs) using custom Perl scripts. To be included in the output, the ORF had to begin with an ATG start codon, occupy a stretch of at least 300 bases, end with any of the three stop codons, and not be wholly contained within a larger ORF on either strand. For completed and published genomic sequences, the sequencing center's ORF analyses and annotations were used. Local BLAST databases were then prepared from these ORF lists and used in our initial search for putative polysaccharide loci.
The synthesis of diverse bacterial polysaccharides of all phyla of bacteria is mediated by conserved families of enzymes (for example, those involved in the synthesis of nucleotide-linked sugars, in the transfer of the initial monosaccharide of the polysaccharide repeating unit to a lipid carrier, and in the assembly of the polysaccharide and its transport to the cell surface). In addition, the genes involved in polysaccharide production are usually clustered in the genome, often in one operon or a few adjacent operons. We took advantage of these characteristics to locate putative polysaccharide biosynthesis loci in the genomes of the 12 Bacteroidales species.
We used the protein sequences of the B. fragilis 9343 polysaccharide biosynthesis proteins WcgS (BF1909), WcfY (BF1904), WcfS (BF1377), WcgX (BF1914), and UpaY (BF1367) as queries in protein-protein BLAST searches of our locally prepared databases. WcgS and WcfY are a putative nucleotide sugar dehydratase and a UDP-glucose dehydrogenase (7), respectively, which are involved in the formation of nucleotide-activated sugars from less complex precursors. WcfS and WcgX are representatives of two distinct families of initiating glycosyltransferases (7, 8), which link the first monosaccharide of a repeat unit to undecaprenyl phosphate as the first step in the assembly of the repeat unit. UpaY is a member of the UpxY family of putative polysaccharide regulatory proteins (8). Genes encoding UpxY products are the first gene of each of the eight characterized polysaccharide biosynthesis loci of B. fragilis (15) and of each of the seven polysaccharide biosynthesis loci of B. thetaiotaomicron (24).
For each species under investigation, the appropriate local database was searched using the indicated B. fragilis 9343 protein sequences as queries. Matches to any of the query proteins with a reasonably low E value (≤10−4) were collected and sorted by genome position. These similarity searches detected a minimum of 6 and up to 16 distinct genetic regions, depending on the species (Table 1). DNA encompassing ≤10,000 bp on both sides of the match or cluster of matches was extracted from the genome sequence and translated to amino acid ORFs as before. These ORFs were then used in protein-protein BLAST searches against GenBank and were assigned a putative function based on the annotation of similar sequences in the database. Each region was further analyzed to determine if it exhibited the genetic characteristics of a polysaccharide biosynthesis locus, and the extent of DNA sequence analyzed was adjusted to ensure coverage of the locus. The genetic characteristics we used to define a locus included the presence of at least two putative glycosyltransferases and a putative polysaccharide flippase and polymerase or, alternatively, an ABC-type transporter involved in the export and assembly of polysaccharide. Regions containing these features, coupled with the presence of at least one ortholog of the proteins used to initially detect the region, were given a polysaccharide biosynthesis locus designation (Table 1).
TABLE 1.
Relevant genomic sequence information and polysaccharide locus detection results
| Bacteroidales species | Sequence status at time of analysis | No. of:
|
||
|---|---|---|---|---|
| Orthologous regions detected | Regions designated polysaccharide loci | Polysaccharide loci with IR-flanked promoters | ||
| T. forysthensis | Complete | 9 | 2 | 0 |
| P. gingivalis | Complete | 7 | 2 | 0 |
| P. intermedia | Complete | 7 | 2 | 1 |
| P. ruminicola | 7 contigs | 6 | 4 | 0 |
| B. caccae | 21 contigs | 8 | 5 | 4 |
| B. ovatus | 105 contigs | 16 | 7 | 2 |
| B. thetaiotaomicron | Complete | 13 | 8 | 4 |
| B. vulgatus | 585 contigs | 14 | 6 | 2 |
| B. fragilis | Complete | 17 | 10 | 7 |
| B. uniformis | 67 contigs | 16 | 9 | 6 |
| P. merdae | 61 contigs | 12 | 6 | 3 |
| P. distasonis | 13 contigs | 16 | 13 | 7a |
Two of these seven loci each have two IR-flanked promoter regions upstream.
This detection method proved relatively robust, in that all previously identified polysaccharide loci were accurately detected by this procedure. All eight of the characterized polysaccharide biosynthesis loci of B. fragilis (15) were located, as well as two other loci revealed by genome sequencing (4) but not yet confirmed by phenotype. These analyses also detected each of the seven polysaccharide biosynthesis loci of B. thetaiotaomicron (24) and one additional locus. Also, the capsular polysaccharide biosynthesis locus (1, 10) and the porR-porS polysaccharide region (21) of P. gingivalis were identified.
The number of polysaccharide biosynthesis loci detected for each species correlates with the specific niche of each of the organisms in the mammalian alimentary tract. For example, we detected only two polysaccharide loci in the genomes of each of the three oral species. This is in contrast to the genomes of the intestinal species, which contain at least 5 (B. caccae) and up to 13 (P. distasonis) distinct polysaccharide biosynthesis loci. In contrast to those of the oral species, many of the genome sequences of the intestinal species are incomplete, so the actual number of polysaccharide loci in these species may be even greater. For the one ruminal species, P. ruminicola, we detected four polysaccharide biosynthesis loci, a number between those detected for the oral and the intestinal species. The rumen is an intermediate alimentary tract location, distal to the oral cavity but proximal to the intestine. These data reveal that the number of polysaccharide biosynthesis loci contained in the Bacteroidales species analyzed correlates well with their habitat: without exception, those species residing in the intestine contain a greater number of polysaccharide biosynthesis loci than those in the more proximal alimentary sites. P. distasonis and P. merdae are phylogenetically closer to the oral organisms T. forsythensis and P. gingivalis than to the intestinal Bacteroides spp., yet, like the Bacteroides spp., they also have an extensive number of polysaccharide biosynthesis loci. Therefore, the synthesis of multiple polysaccharides is a general characteristic of the intestinal Bacteroidales.
This niche-specific characteristic does not hold for another glycan-containing class of molecules. The closely related organisms T. forsythensis and P. distasonis each synthesize a family of surface layer glycoproteins (12, 16). The genome of T. forsythensis contains two genes whose products belong to the same family as at least nine glycoproteins of P. distasonis. We analyzed the genomes of each of the other 10 Bacteroidales species and found that only P. merdae contains genes for the synthesis of similar S-layer glycoproteins (data not shown). P. merdae is most closely related to P. distasonis and T. forsythensis (Fig. 1); thus, the presence of genes encoding this family of glycoproteins correlates with phylogenetic relatedness rather than niche. Of note, however, is the fact that the oral organism T. forsythensis contains only 2 glycoprotein orthologs, whereas P. distasonis and P. merdae contain at least 9 and 10 orthologs, respectively. Therefore, within these three related species, the two intestinal members contain more orthologs than the oral member, paralleling the situation found with the polysaccharide biosynthesis loci.
The large number of polysaccharide biosynthesis loci present in the genomes of the intestinal species suggests extensive transfer of DNA between these species, with a selection for strains encoding multiple polysaccharides. Bacteroides spp. have a large arsenal of mobile genetic elements involved in DNA transfer (20). Oral Bacteroidales species also have genetic material involved in DNA transfer (11); however, because their association with the host is pathogenic and often intermittent rather than mutualistic and lifelong like that with the intestinal species, their environment may not be optimal for DNA transfer. Alternatively, the synthesis of multiple surface polysaccharides may not confer a selective advantage to organisms in the oral cavity.
We next set out to determine if the ability of Bacteroidales species to phase vary synthesis of their polysaccharides by DNA inversion of promoters is also a niche-specific characteristic. We previously showed that seven of the eight characterized polysaccharide biosynthesis loci of B. fragilis have promoters that undergo DNA inversion (15). These promoters are contained in 168- to 193 bp-regions that are flanked by inverted repeats (IRs) of 19 to 25 bp. To find IR regions in the 74 different Bacteroidales polysaccharide loci listed in Table 1, the EMBOSS program (18) Einverted was used to retrieve all IRs in the selected regions that reached a program threshold of 35 and were separated by no more than 500 bp. IRs detected by this analysis were further considered only if they were contained within an intergenic region, were at least 15 bp long, and contained sufficient DNA between them to contain a promoter. IR-flanked regions meeting these criteria were analyzed for the presence of a B. fragilis-like consensus promoter sequence (3) using MEME (2), ClustalW (5), and custom Perl scripts. For the 74 different polysaccharide biosynthesis loci analyzed, we found 36 that contained IR regions that met our criteria (Table 2). Thirty-five of these IR regions were present just upstream of the polysaccharide biosynthesis loci of intestinal Bacteroidales, and each intestinal species contained at least two polysaccharide loci with such an IR region. The only oral species with an IR region meeting each of the criteria was P. intermedia. The position of this IR region differs from those of the intestinal species, as it is in the middle rather than at the start of the polysaccharide locus and is present just upstream of a gene encoding a putative transposase. For two of the P. distasonis polysaccharide regions, PS9 and PS12, two sets of adjacent but nonoverlapping IR regions were detected. The IR pairs that were in closer proximity to the first gene of the polysaccharide regions had a better match with the consensus promoter sequence and were further analyzed. Although there was some variability in the amount of DNA between each of the 36 IR pairs and between the downstream IR and the first gene of the locus, these distances were never more than 321 bp and 307 bp, respectively (Table 2).
TABLE 2.
Clustering and positional information for Bacteroidales polysaccharide loci IRs
| Group and locus | Invertible region (bp) | Downstream IR | Distance to first gene (bp) |
|---|---|---|---|
| 1 | |||
| Tsr15a | 248 | TCCCGTTACCTACGAAGTAACGG | 73 |
| Bt_PS6 | 242 | CGTTACCTAAGAAGTAACG | 23 |
| Bt_PS1 | 243 | ACTTCCGTTACCTAAGAAGTAACGGAAACATAT | 14 |
| Bo_PS3 | 250 | TCCGTTACCTAAGAAGTAAC | 24 |
| Bt_PS3 | 248 | TCCGTTACCTAAGAAGTAAC | 24 |
| Bo_PS6 | 249 | TCCGTTACCTAAGAAGTAAC | 24 |
| Bc_PS1 | 249 | TCCGTTACTTAAGAAGTAACAG | 22 |
| Bc_PS3 | 251 | CTGTTACCTAAGAAGTAACAGATATAC | 16 |
| Bc_PS2 | 248 | TCCGTTACCTTTAAAGTAAC | 24 |
| Tsr25a | 266 | TGCCGTTACCTACAAAGTAAC | 31 |
| Tsr26a | 287 | GTTACCTACAAAGTAACT | 106 |
| Tsr19a | 315 | TGTTCTATTACCTAATAAGTAAG | 84 |
| Bu_PS2 | 284 | CTCTTACCTTGTAAGTAAGAGAGTA | 112 |
| 2 | |||
| Pd_PS2 | 317 | GCTACTCCCCAAGTAGCCT | 157 |
| Pd_PS7 | 321 | TAGGCTACTCCCCAAGTAGCCTA | 156 |
| Pd_PS13 | 312 | CGCTACTCACTAAGTAGCCTAAT | 66 |
| Pd_PS1 | 314 | TACGCTACTTCCCGAGTAGCTAA | 151 |
| 3 | |||
| Bv_PS3 | 284 | TACCCTCTCTTACATAGAAACGGTA | 306 |
| Pm_PS2 | 283 | TACCCTCTCTTACATAGAAACGGTA | 306 |
| Bu_PS6 | 286 | TACCCTCTCTTACATAGAAACGGTA | 306 |
| Pd_PS10 | 272 | TATTTGTACTGTTTCTTACCAAGAAACGGTAGAA | 307 |
| Pd_PS9_a | 172 | ATATTTCTCTTGCCAAGAGAAGTAGC | 161 |
| Pm_PS1 | 169 | TAATATTTCTCTTGCCAAGAGAA | 152 |
| Pd_PS12_a | 172 | ATATTTCTCTTTCCAAGAGAAGTAGC | 161 |
| Bc_PS5 | 175 | TATTTATCTTATAGAGAGAAATA | 168 |
| Pd_PS9_b | 160 | ATATTTCTCTTAAAAAGAAAAG | 524 |
| Pd_PS12_b | 160 | ATATTTCTCTTAAAAAGAAAAG | 525 |
| Bt_PS8 | 166 | ATTATACATCTCTTATGAAGAGA | 179 |
| Pm_PS6b | 171 | ATAGATGTACTTCTCTTAAGAAGAGTAGTATGTTTT | 159 |
| Bv_PS2 | 150 | GGGGATCTCTTACTAAGAGAACCCCA | 62 |
| 4 | |||
| Bf_PSA | 193 | ACGAACGTTTTTTGAAACA | 127 |
| Bf_PSB | 181 | ACGAACGTTTTTTGAAACA | 128 |
| Bf_PSE | 168 | ACGAACGTTTTTTGAAACA | 137 |
| Bf_PSH | 192 | ACGAACGTTTTTTGAAACA | 225 |
| Bf_PSD | 189 | TAGACGATCGTCTATTGAAACA | 214 |
| Bf_PSF | 187 | TTAAACGAACGTCTATTGAAACACT | 186 |
| Bu_PS1 | 173 | CGTTCAT-TAAACGAACGT | 84 |
| Bu_PS4 | 173 | CGTTCAT-TAAACGAACGTCTATTGAACGCTTTTT | 188 |
| Bf_PSG | 174 | GTTCAAATAGACGAACGTTT | 198 |
| Bu_PS7 | 173 | GTTCAAATAGACGAACGTTT | 218 |
| Bu_PS3 | 176 | GTTTTT-TAAACGTACGATTAAAAAAC | 202 |
| Pi_PS1 | 189 | AATTCTTATATCGAACTCACGTT | 125 |
Previously identified inverted repeat known to be inverted by Tsrs, used for comparative purposes.
This IR is 91 bp long; only the final 36 bp is shown.
The −7 promoter region (TANNTTTGY) is remarkably well conserved among all nine species (Table 3). The −33 region (TtTG) is less stringently conserved, with some regions having several overlapping or adjacent sequences similar to the −33 consensus sequence, making identification of the true −33 site difficult. The positions occupied by these putative promoters within the IR regions are of note: there are four promoters which use the first base of the downstream IR (closest to the locus) as the final base of the −7 region; for the rest, this base is separated from the first base of the downstream IR by between 0 and 20 bp (mean, 9.7; mode, 8).
TABLE 3.
Sequences of polysaccharide biosynthesis loci promoters that are flanked by IRs
| Species_locusa | Putative promoter sequenceb |
|---|---|
| Consensus | TtTG TAnnTTTGY |
| Bc_PS1 | aagaagtttttacttgtatgctattagtatttttacctatctttgcaccgaataaattTCCGT |
| Bc_PS2 | aaggaaagtttacttgtatgctattaatatttttacctatatttgcatcgaaatattTCCGTT |
| Bc_PS3 | aatgggaaaatatttgtttactatcttaaaatatgtctatctttgcgtcaaatttaattCTGT |
| Bc_PS5 | tacatctgttttatgctgttatttccaataaattacgtacctttgtgcccgttagTATTTATC |
| Bo_PS3 | agtggaggaatatttgttttctatttgtgtttttgtctatctttgcgtcaaatttaatTCCGT |
| Bo_PS6 | agtggaagaatatttgcttactatttatgtttttgtctatctttgcgtcaaatttaagtTCCG |
| Bt_PS8 | atttgttcatttttgtggttattctcaatattttgtgtatttttgcattcaATTATACATCTC |
| Bt_PS1 | agtgggctttaatttgtttcttatttgtgtttttgtctatctttgcatcaaatttACTTCCGT |
| Bt_PS3 | agtaggtttttatttgtttaatacatgtgtttttgtctatctttgcgtcaaatttaaaTCCGT |
| Bt_PS6 | aatttcatttttgtttgcgtattttcataaatatgtccatctttgcgccatgaatcattcCGT |
| Bu_PS1 | ttttattaaacattttgttagttcaaaaaatacggcttatctttgCGTTCATTAAACGAACGT |
| Bu_PS2 | taaatctatttttttgcgattgtagtaatctttttcatatctttgcagccacaaaacatattC |
| Bu_PS3 | tttctatcaaacattttgccattacaaaaaacgtctctatctttgcGTTTTTTAAACGTACGA |
| Bu_PS4 | ttttattaaacattttgcgagattcaaaaatacacgttatctttgCGTTCATTAAACGAACGT |
| Bu_PS6 | aacgtttattttacttttcagttttactgagaatcattacctttgcagtgtgttttTACCCTC |
| Bu_PS7 | ttttattaaacattttgttagttcaaaaaatacagcttatctttgCGTTCATTAAACGAACGT |
| Bv_PS2 | aagcaacaaaagatttccatttcttgggaattatagctatttttgcagccaaatGGGGATCTC |
| Bv_PS3 | aacgtttattttacttttcagttttactgagaatcattacctttgcagtgtgttttTACCCTC |
| Pd_PS1 | aagcggatctattcgcttgtttttagttattttttactatattcgcggcatgaaagaaTACGC |
| Pd_PS2 | taaaactaatgattgcttgttatttgctattttttgctacatttgcaccgtgaaatgaacGCT |
| Pd_PS7 | aagcgcatttgtttgcttgtttttagttattttttactacatttgcgccgtgaaagaaTAGGC |
| Pd_PS9 | tattacccgttttctattgttttttcaatattttacgtaactttgcgcattgtaATATTTCTC |
| Pd_PS10 | aatcccccctttatttttttgttttgcagattataattacatttgcgacgTATTTGTACTGTT |
| Pd_PS12 | tatcacccgttttctattgttttttcaatattttacgtaactttgcacattgtaATATTTCTC |
| Pd_PS13 | aaaccctaatgattgcttgttttttgttattttttactacatttgcggcgtgaaacaaaCGCT |
| Pi_PS1 | aataatgacttgatatgcttccaataaaaaaacggaatatctctgtctAATTCTTATATCGAA |
| Pm_PS1 | tatcacctgttttctatcgtttcttcaatattttacataactttgcacattgTAATATTTCTC |
| Pm_PS2 | aacgtttattttacttttcagttttactgagaatcattacctttgcagtgtgttttTACCCTC |
| Pm_PS6 | aagttcactttatgccttgatttttagatttttttcgtacttttgcaaacgaaATGTACTTCT |
| Bf_PSA | atttataaaaatattttgttgttataaaaatgtgccttacctttgtgtttataaACGAACGTT |
| Bf_PSB | tatccactaaatattttgtacattaaaacagactccttacctttgttcaatcaaACGAACGTT |
| Bf_PSD | tttttattaaacattttgcagttatgaaaataccctctatctttgcgTTCAATAGACGATCGT |
| Bf_PSE | tcatctaaaaatattttgcacatcaaaatttagtttatacctttgtgctattaaACGAACGTT |
| Bf_PSF | tttttattaaacattttgtagttctaaaaataccccctatctttgcgttcaaTTAAACGAACG |
| Bf_PSG | ttttaattaaacattttgctattatgaaaataccccttatctttgGTTCAAATAGACGAACGT |
| Bf_PSH | atttataaaaatattttgctatagtaaaaatactgtttacctttgttccattgaACGAACGTT |
Bc, B. caccae; Bo, B. ovatus; Bt, B. thetaiotaomicron; Bu, B. uniformis; Bv, B. vulgatus; Pd, P. distasonis; Pi, P. intermedia; Pm, P. merdae; Bf, B. fragilis.
The putative promoter sequence for each polysaccharide locus is underlined and compared to the consensus B. fragilis promoter sequence. The 5′ sequences of the downstream IRs are shown in bold capital letters.
The downstream IRs of these 36 different pairs were aligned using ClustalW (5), which allowed us to cluster them into four distinct groups, with the P. intermedia IR as an outlier. This analysis revealed extensive DNA relatedness between IRs of different species (Table 2). In fact, the 25-bp B. vulgatus PS3, P. merdae PS2, and B. uniformis PS6 IRs are exactly the same. Moreover, the DNA between these IRs is nearly identical: the B. vulgatus and P. merdae regions vary by one base, and the B. uniformis region varies from the other two by just 15 bp over this 283-bp stretch. Groups 1 to 3 are similar in that each IR region is immediately preceded by a gene encoding a Tsr. This genetic architecture is similar to that of the genetic regions of four previously characterized B. fragilis Tsrs with DNA invertase activity: Tsr15, Tsr19, Tsr25, and Tsr26 (19, 23). These DNA invertases act locally to invert promoters contained between IRs in their immediate downstream region, affecting the expression of outer surface proteins and uncharacterized products. The IR regions of these four locally acting DNA invertases have significant DNA identity with group 1 Bacteroidales polysaccharide IRs (Table 3), suggesting that their inversions are also mediated by the Tsrs encoded in their upstream regions.
The genetic architecture of the group 4 IRs, which include the seven B. fragilis polysaccharide loci IRs and four of the six B. uniformis IRs, differs from that of the other four groups in that these IRs are not present downstream of Tsr genes. Inversion of the seven B. fragilis capsule polysaccharide promoters is mediated by a single global DNA invertase, Mpi, which is an Ssr (6). The similarity of the four B. uniformis IRs to those of B. fragilis and the lack of an upstream Tsr gene suggest that inversion of these IR regions is also mediated by a global Ssr. We found that of the 12 Bacteroidales species analyzed, B. uniformis encodes a product that is the most similar to Mpi, demonstrating 90% similarity along the lengths of the products. The next most similar product to Mpi is Gfi, encoded by P. distasonis. Gfi is another Ssr that acts as a global DNA invertase, mediating promoter inversions of glycoprotein genes (12).
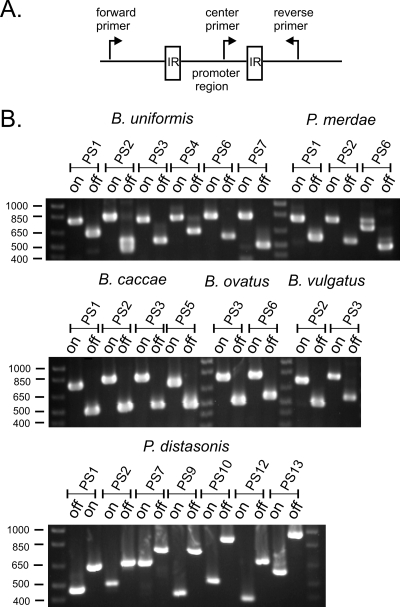
Inversion of the seven B. fragilis and four B. thetaiotaomicron polysaccharide promoters has been previously described (15, 24). To verify our predictions that the other 24 promoters between each of the IR pairs upstream of the intestinal Bacteroidales polysaccharide biosynthesis loci indeed invert, we performed PCR analysis as shown in Fig. 2. Two PCRs were performed for each region, one using a forward primer upstream of the IR region with a central primer between the IRs and a second using the same central primer with a downstream reverse primer outside of the IR region (for primer sequences, see Table S1 in the supplemental material). PCR products would result from both PCRs only if the DNA between the IRs inverted. As shown in Fig. 2, PCR products were generated for both PCRs for each locus. The sizes of the corresponding products correlate exactly with the sizes predicted if the regions inverted exactly between the IRs (Table S1 in the supplemental material). Therefore, each of the polysaccharide promoters that is predicted to undergo inversion does indeed invert.
FIG. 2.
DNA regions between each of the IRs undergo inversion. (A) Diagrammatic representation of the PCR method used to detect DNA inversions of each of the invertible promoter regions. Each of the primers contained within the IR (center primer) was used in a PCR with both the forward and reverse primers individually. (B) Ethidium bromide-stained agarose gels of PCR products, demonstrating inversion of the DNA between each of the IRs for each of the designated regions.
The combined data from these analyses suggest that of the 35 invertible polysaccharide promoter regions identified in the intestinal species, the inversions of 24 of them are mediated by local Tsrs, while those of 11 are mediated by global Ssrs. B. uniformis is the only species besides B. fragilis that is predicted to have polysaccharide promoters whose inversion is mediated by an Ssr. Unlike B. fragilis, however, and unique among the species analyzed here, B. uniformis also has polysaccharide promoters predicted to be inverted by local Tsrs.
This study has revealed that the ability of intestinal Bacteroidales species to synthesize multiple polysaccharides and to phase vary their expression by promoter inversion is a remarkably conserved characteristic. This conservation likely highlights the importance of this process to the survival of these species in the competitive human intestinal ecosystem. The mechanism of inversion, whether mediated by local Tsrs or by a global Ssr, seems less important than the result: extensive and rapid surface variability. This feature ensures that, for a single strain, there will always be a heterogeneous population of organisms, some of which may be more resistant to an attack, whether from a phage, from other bacterial members, or from the host.
Supplementary Material
Acknowledgments
We thank C. M. Fletcher and M. Chatzidaki-Livanis for helpful discussions. Preliminary sequence data for P. intermedia, P. ruminicola, and T. forsythensis were obtained from The Institute for Genomic Research through the website at http://www.tigr.org. Sequencing of P. intermedia and T. forsythensis was accomplished by The Institute for Genomic Research with support from NIDCR, and support for sequencing of P. ruminicola was provided by the USDA. Sequence data for B. caccae, B. ovatus, B. thetaiotaomicron, B. uniformis, B. vulgatus, P. distasonis, and P. merdae were produced by the Genome Sequencing Center at Washington University School of Medicine in St. Louis, MO, and were downloaded from the NCBI Genome Project at http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi.
This work was supported by grant AI44193 from the National Institutes of Health (NIAID).
Footnotes
Published ahead of print on 9 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aduse-Opoku, J., J. M. Slaney, A. Hashim, A. Gallagher, R. P. Gallagher, M. Rangarajan, K. Boutaga, M. L. Laine, A. J. Van Winkelhoff, and M. A. Curtis. 2006. Identification and characterization of the capsuylar polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect. Immun. 74449-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28-36. In R. Altman, D. Brutlag, P. Karp, R. Lathrop, and D. Searls (ed.), Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA. [PubMed]
- 3.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193149-154. [DOI] [PubMed] [Google Scholar]
- 4.Cerdeno-Tarraga, A., S. Patrick, L. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 3071463-1465. [DOI] [PubMed] [Google Scholar]
- 5.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 313497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coyne, M., K. Weinacht, C. Krinos, and L. Comstock. 2003. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc. Natl. Acad. Sci. USA 10010446-10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyne, M. J., W. Kalka-Moll, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2000. Bacteroides fragilis NCTC9343 produces at least three distinct capsular polysaccharides: cloning, characterization, and reassignment of the PS B and PS C biosynthesis loci. Infect. Immun. 686176-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne, M. J., A. O. Tzianabos, B. Mallory, D. L. Kasper, and L. E. Comstock. 2001. A polysaccharide biosynthesis locus required for virulence of Bacteroides fragilis. Infect. Immun. 694342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Aiuto, F., F. Graziani, S. Tete', M. Gabriele, and M. S. Tonetti. 2005. Periodontitis: from local infection to systemic diseases. Int. J. Immunopathol. Pharmacol. 181-12. [PubMed] [Google Scholar]
- 10.Davey, M. E., and M. J. Duncan. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J. Bacteriol. 1885510-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan, M. J. 2003. Genomics of oral bacteria. Crit. Rev. Oral Biol. Med. 14175-187. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher, C., M. Coyne, D. Bentley, O. Villa, and L. Comstock. 2007. Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc. Natl. Acad. Sci. USA 1042413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper, L., T. Stappenbeck, C. Hong, and J. Gordon. 2003. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4269-273. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins, S. A., D. B. Drucker, V. F. Hillier, and L. A. Ganguli. 1992. Numerical taxonomy of Bacteroides and other genera of Gram-negative anaerobic rods. Microbios 69139-154. [PubMed] [Google Scholar]
- 15.Krinos, C. M., M. J. Coyne, K. G. Weinacht, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414555-558. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S. W., M. Sabet, H. S. Um, J. Yang, H. C. Kim, and W. Zhu. 2006. Identification and characterization of the genes encoding a unique surface (S−) layer of Tannerella forsythia. Gene 371102-111. [DOI] [PubMed] [Google Scholar]
- 17.Mazmanian, S., C. Liu, A. Tzianabos, and D. Kasper. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122107-118. [DOI] [PubMed] [Google Scholar]
- 18.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology open software suite. Trends Genet. 16276-277. [DOI] [PubMed] [Google Scholar]
- 19.Roche-Hakansson, H., M. Chatzidaki-Livanis, M. Coyne, and L. Comstock. 2007. Bacteroides fragilis synthesizes a DNA invertase affecting both a local and distant region. J. Bacteriol. 1892119-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12412-416. [DOI] [PubMed] [Google Scholar]
- 21.Shoji, M., D. B. Ratnayake, Y. Shi, T. Kadowaki, K. Yamamoto, F. Yoshimura, A. Akamine, M. A. Curtis, and K. Nakayama. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 1481183-1191. [DOI] [PubMed] [Google Scholar]
- 22.Stappenbeck, T., L. Hooper, and J. Gordon. 2002. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Nat Acad. Sci. USA 9915451-15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinacht, K. G., H. Roche, C. M. Krinos, M. J. Coyne, J. Parkhill, and L. E. Comstock. 2004. Tyrosine site-specific recombinases mediate DNA inversions affecting the expression of outer surface proteins of Bacteroides fragilis. Mol. Microbiol. 531319-1330. [DOI] [PubMed] [Google Scholar]
- 24.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 2992074-2076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.