Abstract
To identify components of the copper homeostatic mechanism of Lactococcus lactis, we employed two-dimensional gel electrophoresis to detect changes in the proteome in response to copper. Three proteins upregulated by copper were identified: glyoxylase I (YaiA), a nitroreductase (YtjD), and lactate oxidase (LctO). The promoter regions of these genes feature cop boxes of consensus TACAnnTGTA, which are the binding site of CopY-type copper-responsive repressors. A genome-wide search for cop boxes revealed 28 such sequence motifs. They were tested by electrophoretic mobility shift assays for the interaction with purified CopR, the CopY-type repressor of L. lactis. Seven of the cop boxes interacted with CopR in a copper-sensitive manner. They were present in the promoter region of five genes, lctO, ytjD, copB, ydiD, and yahC; and two polycistronic operons, yahCD-yaiAB and copRZA. Induction of these genes by copper was confirmed by real-time quantitative PCR. The copRZA operon encodes the CopR repressor of the regulon; a copper chaperone, CopZ; and a putative copper ATPase, CopA. When expressed in Escherichia coli, the copRZA operon conferred copper resistance, suggesting that it functions in copper export from the cytoplasm. Other member genes of the CopR regulon may similarly be involved in copper metabolism.
Lactic acid bacteria constitute a heterogeneous group of gram-positive bacteria that are traditionally used to produce fermented food (29). They usually produce lactic acid as the major end product of their fermentative metabolism. Lactococcus lactis is one of the most extensively studied species among the lactic bacteria. Through the complete sequencing of the L. lactis IL1403 genome, this organism became amenable to proteomics (4). We here combined proteomics, bioinformatics, and molecular biology to obtain information on how L. lactis deals with copper.
Copper is an essential micronutrient for both prokaryotes and eukaryotes. It is used as a cofactor in many enzymes such as superoxide dismutase, cytochrome c oxidase, and lysyl oxidase. The two oxidation states of copper, Cu+ and Cu2+, not only allow its participation in essential redox reactions but also can form reactive oxygen species that cause cellular damage (30). Thus, copper homeostasis has to be tightly regulated to preclude toxic effects (12).
Enterococcus hirae is the closest relative of L. lactis in which copper metabolism has extensively been studied (25). The major copper homeostatic system of E. hirae is the cop operon. It encodes four proteins: two ATPases, CopA and CopB; a copper-responsive repressor, CopY; and the copper chaperone, CopZ (16, 17). CopA and CopB are transmembranous ATPases which appear to transport copper ions across the cytoplasmic membrane (18, 24). They belong to the family of heavy-metal CPx-type ATPases that includes enzymes transporting Cu+, Ag+, Co2+, Zn2+, Cd2+, Hg2+, and Pb2+ (26). It is apparent from the genome sequence that L. lactis IL1403 possesses a similar copRZA operon, encoding a CopY-type repressor, CopR; a CopZ copper chaperone; and a CopA copper ATPase. Interestingly, a protein homologous to CopB of E. hirae is absent from the copRZA operon but is encoded by an unlinked gene. The roles of the L. lactis copRZA and copB genes in copper homeostasis have not yet been established.
To identify genes which could be involved in copper homeostasis in L. lactis, the response of the proteome to copper exposure was analyzed by two-dimensional (2D) gel electrophoresis. Mass spectrometry was used to identify proteins which responded to copper in terms of their expression level. Three proteins upregulated by copper were identified, namely glyoxylase I, YaiA; a nitroreductase, YtjD; and lactate oxidase, LctO. None of these proteins has previously been implicated in copper homeostasis.
The lctO and ytjD genes contained cop box promoter elements near the translational start site. cop boxes are short inverted repeats of consensus TACAnnTGTA and are the binding sites of CopY-type repressors, which regulate the expression of copper ATPases in firmicutes (20). The surprising finding of cop box elements in the promoters of two genes with no obvious connection to copper prompted us to perform a genome-wide analysis of genes which could be under the control of cop box promoters and thus CopR. Seven cop boxes in the promoter regions of six genes and two operons were found to interact with CopR in vitro, and all but two of these promoters were shown to be induced by copper in vivo. This constitutes a CopR regulon, encompassing 11 copper-inducible genes. The cloned copRZA operon conferred copper resistance to a copper-sensitive Escherichia coli strain, supporting a function of CopA in copper export. The function of LctO has been addressed in a separate report (3), while the elucidation of the function of the remaining genes of the CopR regulon requires further investigation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. lactis IL1403 was obtained from Emmanuelle Maguin (INRA, Jouy-en-Josas, France) and was grown semianaerobically (air-saturated medium in sealed bottles) at 30°C in M17 medium. Medium components were from Axon Lab AG, Dättwil, Switzerland.
Sample preparation for proteomics.
From overnight cultures of L. lactis IL1403 in M17 medium, inocula of 1% (vol/vol) were added to 500 ml of fresh M17 medium (28). At an optical density at 548 nm (OD548) of 0.4 to 0.5, cultures were evenly split and 200 μM CuSO4 was added to one of the cultures. Growth was continued for 45 min before the cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C. All further steps were conducted at 4°C. The cells were washed twice with 20 mM Tris-Cl (pH 8) and 1 mM EDTA. The final pellet was resuspended in 10 ml of the same buffer. Cells were broken by three passages through a French press at 70 MPa. Cell debris was removed by centrifugation at 4,000 × g for 20 min. The supernatant was subjected to ultracentrifugation at 100,000 × g for 45 min, and the supernatant was used for 2D gel electrophoresis. Protein concentrations were determined with the Bio-Rad protein assay, using bovine serum albumin as a standard.
2D gel electrophoresis.
The appropriate volume of sample containing 300 μg of protein was precipitated with methanol-chloroform as described previously (31). The pellet was air dried and dissolved in 300 μl of isoelectric focusing buffer (9). Isoelectric focusing strips (Bio-Rad Ready Strips IPG; pH 4 to 7, 17 cm) were passively rehydrated overnight with the sample to be analyzed. Isoelectric focusing was performed with a Bio-Rad Protean isoelectric focusing cell. Following rapid voltage ramping from 0 to 250 V in 20 min, the tension was raised linearly to 4,000 V in 2 h, followed by ramping to 10,000 V in 10 h. Following electrofocusing, the strips were equilibrated for 20 min with 10 ml of NuPAGE solution (Invitrogen) containing 155 mg of dithiothreitol, followed by a 20-min incubation with 10 ml of the same solution containing 250 mg of iodoacetamide. The equilibrated strips were loaded on a standard 12% sodium dodecyl sulfate-polyacrylamide gel (11) and run for 4 h at 180 V. The gels were washed three times for 5 min with distilled water, followed by staining with homemade colloidal Coomassie blue and destaining with distilled water. The protein content of spots was quantified by densitometry of gels from four independent experiments.
In-gel protein digestion.
Coomassie blue-stained gel spots of interest were excised from the gels and washed with 50 mM NH4HCO3 in 50% (vol/vol) acetonitrile for 30 min. The gel pieces were dried in a SpeedVac vacuum drier and rehydrated at 4°C for 60 min in 15 μl of digestion solution (20 mM NH4HCO3 [pH 7.8], 2 ng/μl of sequencing-grade trypsin [Promega]). Digestion was carried out overnight at 37°C under shaking. The gel slices were then extracted once with 30 μl of 1% formic acid and once with acetonitrile for 30 min at 37°C with shaking. The combined supernatants were dried in a SpeedVac, and the pellets were dissolved in 10 μl of 0.1% formic acid for matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS).
MALDI-TOF MS.
Analysis was performed using an Ultraflex MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). Samples were spotted on an AnchorChip target (Bruker Daltonics) with 1 μl of freshly prepared α-cyano-4-hydroxicinnamic acid matrix solution (1 mg/ml). External calibration was performed for each run using peptide calibrants (Bruker Daltonics; part no. 202570). Signals from 100 to 200 laser shots were summed up per analysis. Mass accuracy was estimated at ±0.2 Da.
Database search parameters.
Searches were performed against the NCBInr and Swiss-Prot databases using the MASCOT Peptide Mass Fingerprint database search software (www.matrixscience.com). The L. lactis IL1403 database (MOLOKO) was also searched using the PeptOko software (http://spock.jouy.inra.fr/). Methionine oxidation was included as a possible modification in all searches, and alkylation of cysteines was included where appropriate. One missed tryptic cleavage was considered, and the mass tolerance for the monoisotopic peptide masses was set to ±0.2 Da.
EMSA.
The primers used for electrophoretic mobility shift assay (EMSA) are listed in Table 1. Primers were heated to 95°C for 10 min, slowly cooled to room temperature, and used for EMSA according to Parkhill et al. (19). The incubation buffer was prepared as 2× stock solution [40 mM Tris-acetate (pH 8), 10 mM Mg-acetate, 100 mM Na-acetate, 24% glycerol, 2 mM Ca(OH)2, 5 mM Na-ascorbate, 50 μg/ml bovine serum albumin], and aliquots were stored at −20°C. CopR, prepared as described previously (21), was mixed with DNA in incubation buffer plus 5 mM dithiothreitol, and the reaction mixtures were gently mixed and incubated at room temperature for 30 min, followed by separation on 15% TAE (40 mM Tris-acetate [pH 8], 1 mM EDTA)-polyacrylamide gels, prerun at 165 V for 90 min. Gels were stained with 10 μg/ml of ethidium bromide and photographed under UV light.
TABLE 1.
List of primers used in this study
| Primer | Sequence (5′ to 3′)a | cop box/geneb | Annotation |
|---|---|---|---|
| dm1 | CCCAAGCTTGGGCGGTTGCCGTCAATGAAGTG | copRZA | Intergenic |
| dm2 | GCCTAGGGCTTGCTGTCAGCATCCCTGTT | copRZA | Intergenic |
| dm9 | CGGAATCGACTGATTTAGCG | copA | AAK04932.1 |
| dm10 | AACAGGTAAGGCTTCACCTG | copA | AAK04932.1 |
| dm7 | GTGGCTCAACCATTGTATGC | 16S RNA | L200159 |
| dm8 | AGCCTCAGTGTCAGTTACAG | 16S RNA | L200159 |
| dm34 | CATCATCATGGCTGGCTTAC | copB | AAK04949.1 |
| dm35 | TCACCAATCGCTGGCTTTAC | copB | AAK04949.1 |
| LactO1 | GAGGCTGCGGGAAATAAAGG | lctO | AAK05350.1 |
| LactO2 | TACCGCAACAACATCTGCTC | lctO | AAK05350.1 |
| ob11 | TTATAAAGTTTACATCTGTAGATGATAGAT | lctO* | AAK05350.1 |
| ob12 | ATCTATCATCTACAGATGTAAACTTTATAA | lctO* | AAK05350.1 |
| ob14 | TTTTTATTGTTTACATATGTAATCTTAT | ytjD-1 | AAK06011.1 |
| ob15 | ATAAGATTACATATGTAAACAATAAAAA | ytjD-1 | AAK06011.1 |
| ob16 | CAAATAAATTTACACTTGTAAACTATTTTG | ytjD-2 | AAK06011.1 |
| ob17 | CAAAATAGTTTACAAGTGTAAATTTATTTG | ytjD-2 | AAK06011.1 |
| ob18 | GTAAAATGTTTACATGTGTAAATTTTCACG | acpD | AAK04479.1 |
| ob19 | CGTGAAAATTTACACATGTAAACATTTTAC | acpD | AAK04479.1 |
| ob20 | TTTTTGCTATTACACTTGTATCACATAAAT | yxdE | AAK06348.1 |
| ob21 | ATTTATGTGATACAAGTGTAATAGCAAAAA | yxdE | AAK06348.1 |
| ob22 | TATAAGTATATACATCTGTAAAAGTGAAAG | yfhF | AAK04678.1 |
| ob23 | CTTTCACTTTTACAGATGTATATACTTATA | yfhF | AAK04678.1 |
| ob24 | TTTTAGTGTTTACACGTGTAAACTTATCTT | copR | AAK04930.1 |
| ob25 | AAGATAAGTTTACACGTGTAAACACTAAAA | copR | AAK04930.1 |
| ob26 | TCTTTTCGTTTACAATTGTAAACATAGAAA | yahC | NP_835288.1 |
| ob27 | TTTCTATGTTTACAATTGTAAACGAAAAGA | yahC | NP_835288.1 |
| ob28 | TAGAGCAATTTACACTTGTAAATTAGATCA | rgpC* | AAK04300.1 |
| ob29 | TGATCTAATTTACAAGTGTAAATTGCTCTA | rgpC* | AAK04300.1 |
| ob30 | GTTATAAAGATACAGATGTAATGGGAGAAG | yleE* | AAK05230.1 |
| ob31 | CTTCTCCCATTACATCTGTATCTTTATAAC | yleE* | AAK05230.1 |
| ob32 | GTAAAATGTTTACATGTGTAAATTTTCACG | ydiD | AAK04479.1 |
| ob33 | CGTGAAAATTTACACATGTAAACATTTTAC | ydiD | AAK04479.1 |
| ob34 | TGCCTAGAAATACATTTGTAATTGGTCTTA | yfeA* | AAK04652.1 |
| ob35 | TAAGACCAATTACAAATGTATTTCTAGGCA | yfeA* | AAK04652.1 |
| ob36 | TTAGTTTGAGTACAATTGTATTTGTTGTTA | ygbE* | AAK04726.1 |
| ob37 | TAACAACAAATACAATTGTACTCAAACTAA | ygbE* | AAK04726.1 |
| ob38bis | CCACATTTGATACAAGTGTATTGTTGTTGA | yhcC* | AAK04813.1 |
| ob39bis | TCAACAACAATACACTTGTATCAAATGTGG | yhcC* | AAK04813.1 |
| ob40 | ATCATGGAATTACAGCTGTAGCACAAGATC | noxA* | AAK04926.1 |
| ob41 | GATCTTGTGCTACAGCTGTAATTCCATGAT | noxA* | AAK04926.1 |
| ob42 | CATTTGCAAATACAAATGTAGAGGCCTATT | yihF* | AAK04954.1 |
| ob43 | AATAGGCCTCTACATTTGTATTTGCAAATG | yihF* | AAK04954.1 |
| ob44bis | CACCAGATTCTACAGCTGTATCAACAGCTT | trpF* | AAK05564.1 |
| ob45bis | AAGCTGTTGATACAGCTGTAGAATCTGGTG | trpF* | AAK05564.1 |
| ob47bis | TTGCATCGCGTACATTTGTAGCGAGCTCTT | dxsA* | AAK05579.1 |
| ob48bis | AAGAGCTCGCTACAAATGTACGCGATGCAA | dxsA* | AAK05579.1 |
| ob49 | ACCAATCTGGTACATGTGTAAAGTTGTTTT | ypcG* | AAK05593.1 |
| ob50 | AAAACAACTTTACACATGTACCAGATTGGT | ypcG* | AAK05593.1 |
| ob51 | ATCCTGCATTTACAATTGTAAAATATTTTA | ysdE* | AAK05876.1 |
| ob52 | TAAAATATTTTACAATTGTAAATGCAGGAT | ysdE* | AAK05876.1 |
| ob53bis | TAGGTAATTCTACATCTGTACGGCCGAAGT | metK* | AAK06000.1 |
| ob54bis | ACTTCGGCCGTACAGATGTAGAATTACCTA | metK* | AAK06000.1 |
| ob57 | CACGATGTCCTACACCTGTAATTTCAGTAA | ackA1* | AAK06111.1 |
| ob58 | TTACTGAAATTACAGGTGTAGGACATCGTG | ackA1* | AAK06111.1 |
| ob59 | CAGCCGCAATTACAACTGTAATTAAAGTTG | zitP* | AAK06211.1 |
| ob60 | CAACTTTAATTACAGTTGTAATTGCGGCTG | zitP* | AAK06211.1 |
| ob61 | GTGCCCCTACTACACTTGTATCACCAGGAT | ywfH* | AAK06279.1 |
| ob62 | ATCCTGGTGATACAAGTGTAGTAGGGGCAC | ywfH* | AAK06279.1 |
| PcopB3 | TTTGATAGTTTACAATTGTAAACTATATAT | copB | AAK04949.1 |
| PcopB4 | ATATATAGTTTACAATTGTAAACTATCAAA | copB | AAK04949.1 |
The consensus sequences of the cop boxes are underlined.
For primers binding to cop boxes, the gene name is underlined. An asterisk indicates that the cop box lies within the annotated reading frame of the gene.
Real-time quantitative PCR.
For RNA isolation, fresh mid-log cultures of L. lactis IL1403 in M17 medium were, where required, induced for 45 min with CuSO4. Cells from 1 ml were then harvested by centrifugation, and the pellet was used immediately for RNA isolation with the RNeasy kit from Qiagen, according to the manufacturer's instructions. The optional treatment with DNase I (RNase-free) for 30 min at room temperature was included. RNA concentrations were measured with a NanoDrop ND-1000 spectrophotometer. RNA quality was assessed on 1.2% agarose-formaldehyde gels stained with ethidium bromide. RNA was stored at −80°C. cDNA was reverse transcribed from 1 μg of total RNA per reaction using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's protocol. Real-time quantitative PCR was performed on the LightCycler (Roche), using SYBR Premix Ex Taq (TaKaRa BIO Inc., Otsu, Japan) according to the manufacturer's instructions. Quantitative PCR conditions were as follows: one cycle at 95°C for 10 s, plus 40 cycles of annealing at 61°C for 20 s, extension at 72°C for 45 s, and denaturation at 95°C for 6 s. After the LightCycler PCRs, the products were analyzed for specificity and homogeneity by a melting curve analysis and additionally by electrophoresis on 1% agarose gels. 16S RNA was determined as an internal reference with primers dm7 and dm8, and all real-time quantitative PCR results were expressed relative to 16S RNA.
Cloning of the copRZA operon of L. lactis IL1403.
Primers dm1 and dm2 were used to amplify the cop operon of L. lactis IL1403 with its promoter, using LA Taq DNA polymerase (TaKaRa). The PCR product was purified by agarose gel electrophoresis, followed by extraction with the QIAquick kit (Qiagen). The purified PCR product was cloned into the pCRBluntII Topo vector (Invitrogen), resulting in pCRZA27. The cop operon was then subcloned by cutting pCRZA27 with BamHI and HindIII and ligating the fragment containing the copRZA operon with pSU18, cut with the same enzymes, resulting in pSURZA. Standard molecular biology procedures were conducted as described previously (2).
Biosensor construction.
A biosensor to measure copRZA promoter activity in E. coli, pCRZL, was constructed by digesting pCRZA27 with MunI and BamHI, making it blunt ended with Klenow polymerase, and ligating it with the lux gene cluster, which had been excised from pUWL500 (Photodyne Technologies, Los Angeles, CA) with XbaI and NcoI, followed by filling of the sticky ends with Klenow DNA polymerase. Ligation was performed overnight at 16°C using T4 ligase (Roche).
Luminescence assays.
Cultures of 20 ml of LB medium of E. coli DH5α containing pCRZL were inoculated with 5% of an overnight culture, grown at 37°C with shaking to an OD0.5 at 595 nm, and 1-ml aliquots were induced with CuSO4, as indicated in the experiments, for 1 h at 37°C. Luminescence of 300-μl aliquots was recorded in microtiter plates with a charge-coupled device camera (LAS-1000; Fuji), and the signals were quantified with AIDA software (Raytest).
Complementation assays.
The cop operon was excised from pCRZA27 with BamHI and HindIII and ligated into pSU18, cut with the same enzymes, resulting in pSURZA. For growth measurements, overnight cultures of E. coli W3110ΔcopA containing pSURZA or the control plasmid pSU18 were diluted 100-fold and grown aerobically for 1 h at 37°C before induction with CuSO4.
Growth measurements.
Growth measurements were conducted in sealed 1-ml cuvettes at 30°C. M17 medium (1 ml), containing the additives detailed in Results, was inoculated with 10 μl of an overnight culture. The OD546 was measured directly in the cuvettes. For competition assays, 25-ml cultures with M17 medium, without or with 1 mM copper, were inoculated with wild-type and ΔcopA cells in a ratio of 1:100 and grown for 28 generations by serial 100-fold dilutions into fresh medium every 24 h. After four transfers, the CFU of wild-type and ΔcopA cells were determined by plating serial dilutions on M17 plates with or without erythromycin.
RESULTS
Identification of copper-regulated proteins by proteomics.
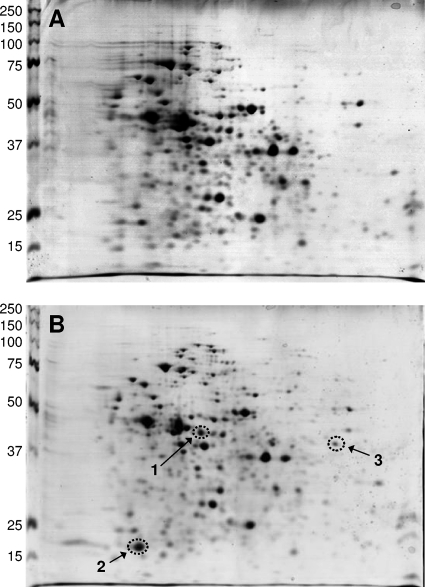
The response of protein expression to copper was investigated by comparing the cytosolic proteome of L. lactis cells grown in the absence of copper to that of cells challenged with 200 μM copper for 45 min. The rationale for these conditions was the previous observation that copper-regulated promoters are significantly induced under these conditions, while cell growth is not affected. Twelve gels of four independent experiments were analyzed visually as well as with the PDQuest software (GE Health Care, Fairfield, CT). Figure 1 shows a set of 2D gels of the cytosolic fraction of L. lactis with and without copper induction. Three spots were reproducibly induced by copper; no down-regulated proteins could be detected. The spots corresponding to the copper-induced proteins were excised from the gels and digested with trypsin, and the peptides were analyzed by MALDI-TOF MS analysis. Database searching allowed the unambiguous identification of these proteins (Table 2).
FIG. 1.
Examples of 2D gels of the cytosolic proteins of L. lactis IL1403 not induced (A) or induced with 200 μM copper for 45 min (B). Isoelectric focusing was in the horizontal direction with a pH gradient (from left to right) of 4 to 7, followed by sodium dodecyl sulfate gel electrophoresis on 12% polyacrylamide gels in the vertical direction. The scales on the left indicate the sizes of molecular mass markers in kDa. The gels were stained with colloidal Coomassie blue. Spot 1, glyoxylase I (YaiA); spot 2, nitroreductase (YtjD); spot 3, l-lactate oxidase (LctO).
TABLE 2.
Cytoplasmic proteins affected by copper, identified by 2D gel electrophoresis
| Spot | Protein | No. of peptides identified | % Sequence coverage | Mascot score | Mol mass (kDa) | Accession no. | Function | Fold induction by coppera |
|---|---|---|---|---|---|---|---|---|
| 1 | YaiA | 19 | 61 | 83 | 43,314 | Q9CJC0 | Glyoxylase I | 30.3 ± 5.4 |
| 2 | YtjD | 12 | 48 | 95 | 22,528 | Q9CED0 | Nitroreductase | 34.7 ± 3.3 |
| 3 | LctO | 18 | 46 | 139 | 40,946 | Q9CG58 | Lactate oxidase | 17.1 ± 3.5 |
Induction by 200 μM extracellular CuSO4 ± standard deviation from four independent experiments.
The protein of spot 1 was upregulated 30-fold by copper and was identified as YaiA. The predicted protein of 389 amino acids and a pI of 4.84 exhibits sequence similarity to glyoxylase I (lactoylglutathione lyase). Spot 2 corresponded to YtjD, a protein of 202 amino acids and a pI of 4.47 which exhibits sequence similarity to nitroreductases. It was upregulated 35-fold by copper. Nitroreductases are a group of proteins that catalyze pyridine nucleotide-dependent reduction of nitro compounds. The in vivo function of most members of this family of enzymes, including YtjD, remains unknown. Spot 3, which was upregulated 17-fold, had been identified previously as LctO, an NAD-independent lactate oxidase. An lctO-knockout mutant did not display enhanced copper resistance, and it was hypothesized that LctO serves in the consumption of molecular oxygen under copper stress (3).
Identification of cop box promoters.
Inspection of the genomic context of these three copper-induced genes revealed some surprises. The yaiA gene is part of a predicted operon (7) consisting of yahD, yaiA, and yaiB. Upstream of the yahD gene, there is an open reading frame which was not annotated in the genome sequence. It encodes a hypothetical protein (L200005; accession no. NP_835288) of 65 amino acids and a pI of 4.5. This gene was here named “yahC” as it lies between yahB and yahD. The second gene of the yahCD-yaiAB operon, yahD, encodes a predicted protein of 206 amino acids and a pI of 4.58. It shares sequence similarity with a family of phospholipases/carboxyesterases with broad substrate specificity. The gene following the putative YaiA glyoxylase gene, yaiB, encodes a putative protein of 196 amino acids and a pI of 4.96. It belongs to a family of poorly characterized proteins containing flavoprotein oxygenase domains. Interestingly, the putative promoter region of the yahCD-yaiAB operon features a cop box (Fig. 2).
FIG. 2.
cop boxes in L. lactis promoters. The cop boxes of consensus TACAnnTGTA present in the promoter regions of L. lactis genes are boxed, and the presumed ribosome binding sites and ATG start codons are underlined. yahCD-yaiAB, promoter of the operon encoding, among others, glyoxylase I (accession no. AE006246); ytjD, nitroreductase promoter (accession no. AE006421); lctO, l-lactate oxidase promoter (accession no. AE006357); copB, promoter of a putative copper ATPase (accession no. NC_009004); copRZA, promoter of the copRZA operon encoding a repressor, a copper chaperone, and a copper ATPase (accession no. AE006316); ydiD, promoter of a hypothetical azoreductase (accession no. AE006275); yfhF, predicted membrane protein of unknown function (accession no. AE006291); and yxdE, predicted short-chain alcohol dehydrogenase (accession no. AE006454).
cop boxes are sequence elements of consensus TACAnnTGTA and have been identified in E. hirae and other firmicutes as the motif interacting with CopY-type copper-responsive repressors to control gene expression. cop boxes occur in one or two copies in the promoter regions of cop operons, which usually consist of the repressor gene, one or two putative copper ATPases, and a copper chaperone in various gene orders (10). The interaction of CopY of E. hirae with the two cop boxes of the cop operon has been studied in detail. CopY represses transcription of the copYZAB operon of E. hirae under low-copper conditions by binding to the cop boxes; excess copper releases CopY from the DNA, allowing transcription to ensue (5, 6, 27). In the L. lactis genome, there is a single CopY homologue, CopR, which has previously been shown to also interact with cop boxes (20). The presence of a cop box in the promoter region of the yahCD-yaiAB operon suggests that this operon is under the control of copper via the CopR repressor and that the gene products function in copper metabolism. YtjD, the nitroreductase identified by proteomics, is encoded by a monocistronic gene. The promoter region of the ytjD gene features two cop boxes, again suggesting that this gene is under the control of CopR (Fig. 2). Similarly, the monocistronic lctO gene features a cop box in the promoter region and is thus under the control of CopR (3).
Testing of the interaction of CopR with cop box promoters—defining the CopR regulon.
The occurrence of cop boxes in the promoter regions of all three genes whose gene products were identified by proteomics raised the question of to what extent cop boxes are involved in the response of L. lactis to copper. Searching the L. lactis IL1403 genome for the cop box consensus sequence TACAnnTGTA revealed 28 occurrences of this motif. Of these, 17 appeared to be within coding regions of chromosomal genes and 2 were in the coding regions of prophage genes. Given the possibility that some of the annotated genes could be in error, as appears to be the case for lctO (3), we tested the interaction of all the genomic cop boxes with the purified L. lactis CopR repressor by EMSA. When 30-mer double-stranded oligonucleotides containing the different cop boxes were incubated with purified CopR, seven exhibited an electrophoretic mobility shift (Fig. 3). These seven cop boxes are in the promoter regions of four monocistronic genes, namely lctO, ytjD (two cop boxes), ydiD, and copB; and two operons, yahCD-yaiAB and copRZA. No interaction was observed with the cop boxes in the promoter regions of yfhF and yxdE, although this would have been expected on an empirical basis (cf. Fig. 2). In all cases in which CopR interacted with the cop box, copper led to the release of the repressor from the DNA. There are thus a total of six promoters which are apparently copper regulated by the CopR repressor, controlling the expression of a total of 11 genes. This defines a CopR regulon in L. lactis.
FIG. 3.
EMSA of the interaction of CopR with different cop boxes. (A to C) Thirty-base-pair double-stranded oligonucleotides containing the cop box motif flanked by 10 bp on either side were incubated with purified CopR of L. lactis. Samples were electrophoresed on nondenaturing, 15% polyacrylamide gels and stained with ethidium bromide. The arrows indicate the migration positions of free DNA, and the double arrows indicate those of the DNA-CopR complexes. Each lane contains 18 pmol of DNA and 175 pmol of CopR. The − and + signs indicate the absence and presence, respectively, of 5 μM copper.
In vivo induction of the CopR regulon by copper.
To verify copper regulation of the genes and operons of the CopR regulon in vivo, expression was analyzed by real-time quantitative PCR. Copper (200 μM CuSO4) induced all six promoters for which interaction of the cop boxes with CopR had been shown (Fig. 4). Induction was between 3- and 12-fold, but no attempt was made to determine the optimal copper concentration. No induction was observed for the yfhF and yxdE genes, in line with the failure of their cop boxes to interact with CopR. There is thus agreement between in vitro DNA-repressor interaction and in vivo gene expression analysis.
FIG. 4.
Real-time quantitative PCR analysis of mRNA expression from genes with cop box promoters. RNA was isolated from cells which were not induced (open bars) or from cells induced with 200 μM of copper for 45 min (filled bars). Gene names are given on the abscissa.
To find a reason for the failure of CopR to interact with the cop boxes of the yfhF and yxdE genes and the failure of copper to induce expression of these genes, we analyzed the sequences surrounding the consensus cop box motif TACAnnTGTA. Figure 5 shows a ClustalW alignment of the nine cop boxes which were found in promoter regions in L. lactis. There appears to be an extended consensus binding motif for cop boxes which interact with CopR of sequence ttTACAnnTGTAAA, with A or C allowed in the first and second positions. The cop box of the yfhF gene would conform to this rule but did not interact with CopR. The reason for this is not clear. The cop box of the yxdE gene, which also failed to interact with CopR, does not match the extended cop box consensus sequence. When sequencing the lctO gene from our L. lactis IL1403 strain, we found approximately 1 “mutation” per 30 bases compared to the genome sequence in the database (3). We do not know the reason for this discrepancy: we received our strain from the lab where it was sequenced. Conceivably, there are also polymorphisms in the cop boxes, which could account for the inconsistencies.
FIG. 5.
ClustalW alignment of cop boxes present in the promoter regions of L. lactis genes. Sequences are ordered by decreasing pairwise similarity and labeled with the first downstream gene. The most conserved residues are depicted in inverse colors. The extended cop box consensus sequence is indicated below the alignment.
Function of genes of the CopR regulon.
The CopR regulon thus encompasses 11 genes, organized into 4 monocistronic genes, ytjD, lctO, copB, and ydiD; and two operons, yahCD-yaiAB and copRZA. Only YaiA, YtjD, and LctO were detected by proteomics, as mentioned above. Some of the additional genes of the CopR regulon, identified on the basis of the cop box, appear to have a direct role in copper homeostasis. The copR gene is the first gene of the polycistronic copRZA operon (Fig. 6). It encodes the CopR repressor, which regulates the CopR regulon, a CopZ-like copper chaperone which probably functions in intracellular copper routing (1, 6), and the CopA ATPase. The latter is a member of the CPx-type ATPase family (26). It features a CxxC motif in the N terminus and thus possesses a heavy-metal-associated domain typical of heavy-metal ATPases. The protein most closely related to CopA of L. lactis is CopA of E. hirae, which exhibits 45% sequence identity. CopA of E. hirae has been postulated to have a role in copper uptake, but the evidence for this is not conclusive (15, 18).
FIG. 6.
Copper homeostatic genes of L. lactis. (A) Schematic representation of the copRZA operon and the copB gene. PcopR and PcopB indicate the promoter regions containing the cop boxes, and the arrows indicate the conjectured start of transcription. The genes are not drawn to scale. (B and C) Alignment of the N termini of CopA and CopB, respectively, of L. lactis (Ll) with the corresponding enzymes of E. hirae (Eh). Identical or similar amino acid residues are indicated in reverse colors; dissimilar residues are emphasized in gray.
A second CPx-type ATPase is encoded by the unlinked, monocistronic copB gene, which is also a member of the CopR regulon (Fig. 6). CopB features a histidine-rich N-terminal heavy-metal-associated domain similar to that of CopB of E. hirae. Of the proteins in the database, CopB of L. lactis is most closely related to CopB of E. hirae (55% sequence identity). CopB of E. hirae has been shown to serve in the extrusion of cytoplasmic copper (18, 24). It is notable that in E. hirae, copA and copB are organized in the same operon and coregulated and cotranscribed, while in L. lactis, the copB gene is unlinked to the copRZA operon. Finally, the monocistronic ydiD gene of the CopR regulon encodes a predicted protein of 201 amino acids and a pI of 4.6. Based on sequence similarity, YdiD is predicted to be a flavin mononucleotide (FMN)-dependent NADH-azoreductase. The same conserved domain is, however, also present in acyl carrier protein phosphodiesterases, and the elucidation of the function of YdiD will require detailed biochemical studies.
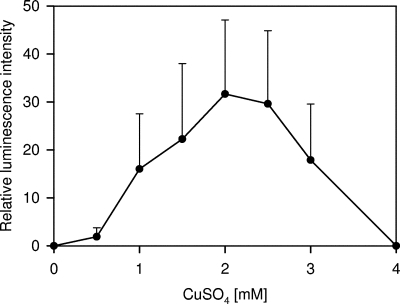
To assess if the regulation by CopR was solely governed by the cop box and the CopR repressor (possibly aided by the CopZ copper chaperone), a lux-based biosensor for E. coli was constructed. It contained the promoter of the L. lactis copRZA operon and the copR and copZ genes upstream of a promoter-less luxCDABE gene cluster. This biosensor, pCRZL, displayed copper-induced luminescence in E. coli (Fig. 7). Maximal stimulation of light emission was observed at a copper concentration of 2 mM in medium. The copper-dependent regulation of the copRZA promoter observed in E. coli suggests that the regulation of this promoter, and most likely all of the cop box promoters, is accomplished by CopR (and CopZ), without the contribution of other cellular factors.
FIG. 7.
Biosensor assay of CopR regulation by copper. E. coli DH5α containing the plasmid pCRZL with the lux genes under the control of the copRZA promoter and the CopR repressor was induced with different CuSO4 concentrations, and luminescence was measured. The error bars indicate standard deviations.
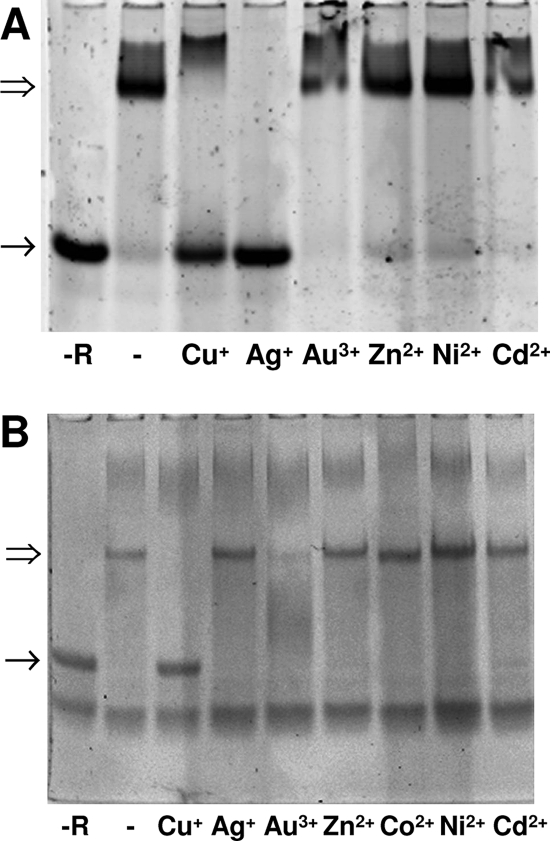
Metal specificity of CopR-cop box interactions.
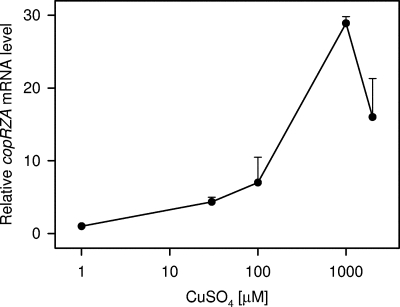
Since the copRZA operon and the copB gene are most likely involved in copper homeostasis in a direct way, the regulation of these genes was analyzed in more detail. Figure 8 shows the metal specificity of the interaction of CopR with the promoters of the copRZA operon and the copB gene. Cu+ and Ag+ released CopR from the copRZA promoter, while Au3+, Zn2+, Ni2+, and Cd2+ did not. This suggests that the copRZA operon is induced by copper and silver, but not by the other metal ions tested. In the case of the copB promoter, Cu+ also released the repressor, while Zn2+, Co2+, Ni2+, and Cd2+ did not. Au3+ generated a gel artifact and could not be assessed. Since silver and gold are not biological ions, their effects were not further investigated. The induction of the copRZA operon by copper was also assessed in vivo by real-time quantitative PCR. As can be seen in Fig. 9, a maximal induction of 30-fold, compared to uninduced cells, was observed with 1 mM externally added CuSO4 in complex growth medium. This copper induction again suggests a function for CopA in copper resistance.
FIG. 8.
Metal specificity of CopR-promoter interactions. The release of CopR from the copRZA and the copB cop boxes was assessed by EMSA. Oligonucleotides (2.5 pmol) were incubated with a 20-fold molar excess of purified CopR in presence of 50 μM of the metal ions indicated in the figure and resolved on polyacrylamide gels as described in Materials and Methods. (A) cop box of the copRZA operon. (B) cop box of the copB gene. −R, control without CopR.
FIG. 9.
Real-time quantitative PCR measurement of copRZA mRNA expression in response to copper. Cultures of wild-type L. lactis in complex media were induced in mid-log phase with the indicated concentrations of CuSO4 for 45 min, RNA was extracted and reverse transcribed, and cop mRNA levels were determined by real-time quantitative PCR with primers dm9 and dm10, directed against the copA gene. The error bars indicate standard deviations.
Analysis of knockout mutants in putative copper ATPase genes.
To further assess the roles of CopA and CopB in copper homeostasis, knockout mutants were constructed by homologous recombination as described previously (8) and tested for growth in the presence of copper. Compared to the wild type, the ΔcopA mutant, ΔcopB mutant, and ΔcopA ΔcopB double mutant all displayed only marginally increased copper sensitivity (not shown). To provide additional evidence for a role of the copRZA operon in copper resistance, we cloned the operon in an E. coli strain deficient in its endogenous cytoplasmic copper export pump (W3110ΔcopA) and thus much more copper sensitive than the wild type (22). Transformation of this strain with a plasmid containing the L. lactis copRZA operon significantly increased copper resistance (Fig. 10). To obtain further support for a role of CopA in copper resistance, growth competition experiments were performed. Mixed cultures of L. lactis wild-type and ΔcopA cells were inoculated at a ratio of 1:100. Following growth for 28 generations in the presence of 1 mM copper, wild-type and ΔcopA cells were recovered at a ratio of 31:1, while the ratio was 1:14 when grown in the absence of copper. This documents a clear growth advantage of the wild type over the ΔcopA mutant in the presence of copper. Taken together, these observations strongly support a role of the copRZA operon of L. lactis in copper resistance, presumably by CopA acting as a copper efflux pump.
FIG. 10.
Copper resistance in E. coli conferred by the L. lactis copRZA operon. (A) Growth response of E. coli W3110ΔcopA containing the control vector pSU18 (open circles) or the vector pSURZA containing the L. lactis copRZA operon (solid circles). The indicated copper concentrations were added to the media, and the OD was measured after 8 h of growth. (B) Replica plating of strains on a plate without added copper (left) or with 2 mM CuSO4 (right). Section 1, E. coli W3110 wild type; section 2, E. coli W3110ΔcopA with pSURZA containing the copRZA operon; section 3, E. coli W3110ΔcopA containing the pSU18 control vector.
DISCUSSION
We here describe the identification of the CopR regulon of L. lactis. Copper-induced expression of three proteins, namely YtjD, LctO, and YaiA, was initially demonstrated by proteomics. Based on a common cop box signature in the promoters of these genes, an extended CopR regulon could be identified by a genome-wide search for this signature, followed by in vitro analysis of promoter-repressor interactions and measurement of gene induction by copper. The genes of the CopR regulon are organized into four monocistronic genes, ytjD, lctO, ydiD, and copB; and two operons, copRZA and yahCD-yaiAB. Why were not all gene products of the CopR regulon detected by proteomics? CopZ and YahC have predicted molecular masses of 7 kDa and are too small to be detected on our 2D gels. CopA and CopB, on the other hand, are membrane proteins, which were not included in the proteomics analysis. The remaining four proteins, CopR, YahD, YaiB, and YdiD, could in principle have been detected on 2D gels but may be of low solubility or low in abundance. This remains an issue for speculation. It has to be kept in mind that proteomics does not provide a complete picture of the proteome—it is just a window of limited coverage.
What are the functions of the 11 genes of the CopR regulon? We started to address this question by analyzing some of the gene products in detail. YtjD, the protein most strongly induced by copper, belongs to a family of nitroreductases which have a broad spectrum of substrate specificities and their in vivo function is generally unknown. Purified YtjD contained an FMN cofactor and exhibited a specific activity of 0.1 U/mg for the reduction of nitrosoglutathione, using NADPH as reductant (F. Mourlane, unpublished observation). The in vivo function of YtjD remains unknown, but the induction of a nitroreductase by copper could hint at a connection of copper stress to nitrosative stress. Further analysis of the function of YtjD is currently in progress in our laboratory.
LctO was previously shown to be an NAD-independent lactate oxidase which catalyzes the conversion of lactate to pyruvate with use of molecular oxygen and generation of hydrogen peroxide (3). A Fenton-type reaction with copper and hydrogen peroxide could, of course, generate toxic hydroxyl radicals, but this may be less of a problem than has been surmised. It was recently shown that copper-loaded E. coli cells were less sensitive to killing by hydrogen peroxide than cells grown without copper supplementation (13). Also, “paradoxical” increase in H2O2 is common in lactic acid bacteria (14, 23). In a nonrespiring organism like L. lactis, it may be more important to keep oxygen low, rather than hydrogen peroxide. In Lactobacillus delbrueckii subsp. bulgaricus, an oxygen-consuming NADH oxidase serves in eliminating oxygen (14). In a separate report, we recently proposed a similar function for LctO in oxygen removal (3).
The monocistronic copB gene encodes a CPx-type ATPase similar to CopB of E. hirae, which has been shown to extrude excess cytoplasmic copper (18, 24). The similarity of L. lactis CopB to E. hirae CopB and the induction by copper suggest that these enzymes have similar functions in copper export. Further studies to firmly establish this are in progress.
For several genes of the CopR regulon, the in vivo functions and their connections to copper metabolism remain obscure and will require extensive additional work. The predicted protein encoded by the ydiD gene exhibits sequence similarity to FMN-dependent NADH-azoreductase, but its function remains unknown. Also, the possible function of the yahCD-yaiAB operon in copper metabolism is unclear. The operon encodes, in order, a predicted protein of unknown function, hypothetical phospholipases/carboxyesterases, a glyoxylase I, and putative flavoprotein oxygenases. Clearly, the assignment of a function to the yahCD-yaiAB operon will have to await further data.
The second operon of the CopR regulon, copRZA, encodes the CopR repressor, the CopZ copper chaperone, and the CopA copper ATPase. The latter exhibits 45% sequence identity to CopA of E. hirae, which has been proposed to be a copper importer (25), while similar enzymes of other microorganisms clearly serve in copper extrusion (22). Recent findings cast doubt on the function of E. hirae CopA in copper import (M. Solioz, unpublished), and three experimental findings support a role for L. lactis CopA in copper export. First, CopA is under the control of CopR and is induced by copper. Second, the copper-sensitive phenotype of an E. coli strain deficient in cytoplasmic copper export could be complemented by CopA. Third, the wild-type strain exhibited a strong growth advantage over a ΔcopA strain when grown in mixed cultures in the presence of 1 mM copper. These findings strongly support a function of L. lactis CopA in copper resistance.
Taken together, we here identified a CopR regulon in L. lactis which controls the copper-dependent expression of 11 genes with a potential role in copper metabolism. We also showed that the copRZA operon is involved in copper resistance. The functions of other genes of the CopR regulon in copper homeostasis remain unclear, and further investigations are under way in our laboratory. The trans-regulation of a group of genes by the copper-responsive CopR repressor moves a number of new, unexpected genes into the spotlight of copper metabolism. The current work thus sets the stage for the investigation of novel proteins with putative involvement in copper metabolism.
Acknowledgments
We thank Jivko Stoyanov and Thomas Weber for technical support, André Schaller and Daniel Molina for help with mass spectrometry, and Bernhard Erni for valuable suggestions.
This work was supported by grant 3100A0-109703 from the Swiss National Foundation and by a grant from the International Copper Association.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Arnesano, F., L. Banci, I. Bertini, S. Ciofi-Baffoni, E. Molteni, D. L. Huffman, and T. V. O'Halloran. 2002. Metallochaperones and metal-transporting ATPases: a comparative analysis of sequences and structures. Genome Res. 12255-271. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, R. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 3.Barré, O., F. Mourlane, and M. Solioz. 2007. Copper induction of lactate oxidase of Lactococcus lactis: a novel metal stress response. J. Bacteriol. 1895947-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobine, P., W. A. Wickramasinghe, M. D. Harrison, T. Weber, M. Solioz, and C. T. Dameron. 1999. The Enterococcus hirae copper chaperone CopZ delivers copper(I) to the CopY repressor. FEBS Lett. 44527-30. [DOI] [PubMed] [Google Scholar]
- 6.Cobine, P. A., G. N. George, C. E. Jones, W. A. Wickramasinghe, M. Solioz, and C. T. Dameron. 2002. Copper transfer from the Cu(I) chaperone, CopZ, to the repressor, Zn(II)CopY: metal coordination environments and protein interactions. Biochemistry 415822-5829. [DOI] [PubMed] [Google Scholar]
- 7.Ermolaeva, M. D., O. White, and S. L. Salzberg. 2001. Prediction of operons in microbial genomes. Nucleic Acids Res. 291216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber, S. D., and M. Solioz. 2007. Efficient transformation of Lactococcus lactis IL1403 and generation of knock-out mutants by homologous recombination. J. Basic Microbiol. 47281-286. [DOI] [PubMed] [Google Scholar]
- 9.Guillot, A., C. Gitton, P. Anglade, and M. Y. Mistou. 2003. Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics 3337-354. [DOI] [PubMed] [Google Scholar]
- 10.Jordan, I. K., D. A. Natale, and M. Y. Galperin. 2000. Copper chaperones in bacteria: association with copper-transporting ATPases. Trends Biochem. Sci. 25480-481. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. J. Biol. Chem. 80575-599. [DOI] [PubMed] [Google Scholar]
- 12.Linder, M. C., and M. Hazegh Azam.1996. Copper biochemistry and molecular biology. Am. J. Clin. Nutr. 63797S-811S. [DOI] [PubMed] [Google Scholar]
- 13.Macomber, L., C. Rensing, and J. A. Imlay. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 1891616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marty-Teysset, C., F. de la Torre, and J.-R. Garel. 2000. Increased production of hydrogen peroxide by Lactobacillus delbrueckii subsp. bulgaricus upon aeration: involvement of an NADH oxidase in oxidative stress. Appl. Environ. Microbiol. 66262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odermatt, A., R. Krapf, and M. Solioz. 1994. Induction of the putative copper ATPases, CopA and CopB, of Enterococcus hirae by Ag+ and Cu2+, and Ag+ extrusion by CopB. Biochem. Biophys. Res. Commun. 20244-48. [DOI] [PubMed] [Google Scholar]
- 16.Odermatt, A., and M. Solioz. 1995. Two trans-acting metalloregulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. J. Biol. Chem. 2704349-4354. [DOI] [PubMed] [Google Scholar]
- 17.Odermatt, A., H. Suter, R. Krapf, and M. Solioz. 1992. An ATPase operon involved in copper resistance by Enterococcus hirae. Ann. N. Y. Acad. Sci. 671484-486. [DOI] [PubMed] [Google Scholar]
- 18.Odermatt, A., H. Suter, R. Krapf, and M. Solioz. 1993. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J. Biol. Chem. 26812775-12779. [PubMed] [Google Scholar]
- 19.Parkhill, J., A. Z. Ansari, J. G. Wright, N. L. Brown, and T. V. O'Halloran. 1993. Construction and characterization of a mercury-independent MerR activator (MerRAC): transcriptional activation in the absence of Hg(II) is accompanied by DNA distortion. EMBO J. 12413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Portmann, R., D. Magnani, J. V. Stoyanov, A. Schmechel, G. Multhaup, and M. Solioz. 2004. Interaction kinetics of the copper-responsive CopY repressor with the cop promoter of Enterococcus hirae. J. Biol. Inorg. Chem. 9396-402. [DOI] [PubMed] [Google Scholar]
- 21.Portmann, R., K. R. Poulsen, R. Wimmer, and M. Solioz. 2006. CopY-like copper inducible repressors are putative ‘winged helix’ proteins. Biometals 1961-70. [DOI] [PubMed] [Google Scholar]
- 22.Rensing, C., B. Fan, R. Sharma, B. Mitra, and B. P. Rosen. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 97652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rochat, T., J.-J. Gratadoux, A. Gruss, G. Corthier, E. Maguin, P. Langella, and M. van de Guchte. 2006. Production of a heterologous nonheme catalase by Lactobacillus casei: an efficient tool for removal of H2O2 and protection of Lactobacillus bulgaricus from oxidative stress in milk. Appl. Environ. Microbiol. 725143-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solioz, M., and A. Odermatt. 1995. Copper and silver transport by CopB-ATPase in membrane vesicles of Enterococcus hirae. J. Biol. Chem. 2709217-9221. [DOI] [PubMed] [Google Scholar]
- 25.Solioz, M., and J. V. Stoyanov. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27183-196. [DOI] [PubMed] [Google Scholar]
- 26.Solioz, M., and C. Vulpe. 1996. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 21237-241. [PubMed] [Google Scholar]
- 27.Strausak, D., and M. Solioz. 1997. CopY is a copper-inducible repressor of the Enterococcus hirae copper ATPases. J. Biol. Chem. 2728932-8936. [DOI] [PubMed] [Google Scholar]
- 28.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urbach, G. 1995. Contribution of lactic acid bacteria to flavor compound formation in dairy products. Int. Dairy J. 5877-903. [Google Scholar]
- 30.Urbanski, N. K., and A. Beresewicz. 2000. Generation of *OH initiated by interaction of Fe2+ and Cu+ with dioxygen: comparison with the Fenton chemistry. Acta Biochim. Pol. 47951-962. [PubMed] [Google Scholar]
- 31.Wessel, D., and U. I. Flügge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138141-143. [DOI] [PubMed] [Google Scholar]