Abstract
YsxC is a small GTPase of Bacillus subtilis with essential but still unknown function, although recent works have suggested that it might be involved in ribosome biogenesis. Here, purified YsxC overexpressed in Escherichia coli was found to be partly associated with high-molecular-weight material, most likely rRNA, and thus eluted from gel filtration as a large complex. In addition, purification of ribosomes from an E. coli strain overexpressing YsxC allowed the copurification of the YsxC protein. Purified YsxC was shown to bind preferentially to the 50S subunit of B. subtilis ribosomes; this interaction was modulated by nucleotides and was stronger in the presence of a nonhydrolyzable GTP analogue than with GTP. Far-Western blotting analysis performed with His6-YsxC and ribosomal proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed that YsxC interacted with at least four ribosomal proteins from the 50S subunit. Two of these putative protein partners were identified by mass spectrometry as L1 and L3, while the third reactive band in the one-dimensional gel contained L6 and L10. The fourth band that reacted with YsxC contained a mixture of three proteins, L7/L12, L23, and L27, suggesting that at least one of them binds to YsxC. Coimmobilization assays confirmed that L1, L6, and L7/L12 interact with YsxC. Together, these results suggest that YsxC plays a role in ribosome assembly.
GTPases are found in all three kingdoms of life (9, 13, 50, 63) and can be broadly classified into four large subfamilies according to their cellular roles and molecular weights: small GTP-binding proteins involved in cell proliferation, translational GTPases, α-subunits of heterotrimeric G proteins involved in cell signaling, and large GTP-binding proteins (50). In eukaryotes, all these families are present, and the last two decades have witnessed a tremendous increase in our understanding of the structures and functions of many of their members (9, 63, 67, 70, 71). In contrast, GTPases are rather scarce in prokaryotes, as some families, such as the α-subunits of heterotrimeric G proteins or large GTP-binding proteins, appear to be missing, and small GTPases are largely underrepresented (13, 50). Besides, apart from the well-characterized translation factors, including EF-G, EF-Tu, and IF2, there is still a paucity of information concerning most of the remaining bacterial GTPases. At first glance, this might seem paradoxical, as many putative GTPases inferred from the genome-sequencing programs have been shown to be essential for bacterial growth, but their discovery is still in an early stage (11, 13, 45). This emphasizes the need to urgently address the fundamental question regarding the cellular roles of these uncharacterized enzymes (56).
Many reports have suggested that the functions of most of the unknown conserved putative bacterial GTPases are somehow linked with the ribosomes and nucleic acid binding (11, 12, 15). Over the past few years, biochemical evidence has accumulated to support this view for several GTPases, including YlqF/RbgA (40, 66), YloQ (14, 16), Era (32, 41, 60), Obg/CgtA (59, 72, 75), and EngA/Der (7, 28, 61). Bacillus subtilis YsxC (termed YihA in Escherichia coli) is part of another family of GTPases broadly conserved in bacteria that has been recently selected as a priority target for functional characterization (22). Genetic studies have shown that this protein is essential to the growth of B. subtilis and E. coli (2, 53, 69). Depletion of YsxC resulted in cell elongation, abnormal cell curvature, and nucleoid condensation in B. subtilis (45). Depletion of YihA in E. coli results in impaired cell division (17). The ysxC gene was shown to be transcribed together with the lon gene (55), and both gene products are considered heat shock proteins, since they could be induced by heat and other stresses (3, 52, 55). Both YihA and YsxC have been purified (35, 57), and determination of the three-dimensional structure of YsxC (57) confirmed that the protein belongs to the large superfamily of translation factor-related (TRAFAC) GTPases, as defined by Leipe and coworkers (36). Regarding the function of YsxC, a recent study suggested that it is required for large-ribosomal-subunit biogenesis in B. subtilis, since YsxC-depleted cells accumulate immature ribosomal-subunit intermediates (61). These immature subunits lack three ribosomal proteins, L16, L27, and L36 (61).
In the current work, we show that during purification of YsxC overexpressed in E. coli, a high-molecular-weight RNA species, possibly rRNA, copurified with the protein. We have further investigated the association between YsxC and B. subtilis ribosomes using pelleting assays and velocity gradient centrifugation techniques. YsxC was shown to preferentially bind to the 50S ribosomal subunit, and this binding was strengthened in the presence of a nonhydrolyzable GTP analogue. A far-Western approach was then used to pinpoint the putative ribosomal-protein partners of YsxC, revealing four reactive bands, all from the large ribosomal subunit. Two of these bands were assigned to L1 and L3 by mass spectrum analysis, while the two additional bands contained a mixture of two (L6 and L10) or three (L7/L12, L23, and L27) subunits, suggesting that at least one of them in each band was an interacting partner. Of these proteins, six could be overexpressed under a soluble form: L1, L6, L10, L7/L12, L23, and L27. Pull-down experiments were then performed with these proteins and confirmed that YsxC could interact with at least L1, L6, and L7/L12. Overall, our results strongly support a direct role of YsxC during the biogenesis of the large ribosomal subunit.
MATERIALS AND METHODS
Plasmids, bacterial strains, and culture conditions.
To clone the ysxC gene from B. subtilis, the genomic DNA of B. subtilis strain 168 was prepared according to the method of te Riele et al. (64). The ysxC gene was amplified from B. subtilis genomic DNA by PCR using Pfu Turbo DNA polymerase from Stratagene and oligonucleotides a (CGGGATCCATGAAAGTCACAAAGTCAGAAATCGTGATC; a BamHI site is underlined) and b (GGGCTCGAGCGACCGGTTTATCATTTTTTTGATCG; an XhoI site is underlined), cut with BamHI and XhoI, and cloned into the BamHI-XhoI sites of pGEX-4T1 (Amersham Biosciences). The His6-ysxC fusion plasmid has already been described (61). Oligonucleotides b and c (GGCATATGAAAGTCACAAAGTCAGAAATC; an NdeI site is underlined) were used to perform another PCR, and the PCR product was digested with NdeI and XhoI and ligated into the NdeI-XhoI sites of pET-21b (Novagen) (to express YsxC without any tag). After transformation of the ligation mixtures into E. coli DH5α cells, positive clones were confirmed by DNA sequencing (Genome Express).
B. subtilis ribosomal-protein genes coding for L1, L3, L6, L7, L10, L23, and L27 were amplified from B. subtilis genomic DNA by PCR using Pfu Turbo DNA polymerase and the oligonucleotides listed in Table S1 in the supplemental material. The resulting PCR products were digested either by NdeI and BamHI and ligated into the NdeI-BamHI sites of pET-15b, by BamHI and XhoI and ligated into the BamHI-XhoI sites of pET-21b, or by BamHI and XhoI and ligated into the BamHI-XhoI sites of pGEX-4T-1 (see Table S2 in the supplemental material). After transformation of the ligation mixtures into E. coli DH5α cells, positive clones were confirmed by DNA sequencing (Genome Express).
E. coli cells, strain BL21(DE3), were transformed with the YsxC plasmids or the plasmids encoding the ribosomal proteins. All E. coli strains that express the ysxC gene or the ribosomal genes were first grown overnight at 37°C with vigorous shaking in LB medium (Sigma) supplemented with 100 μg of ampicillin per ml. The overnight cultures were inoculated into LB medium containing ampicillin and then induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (Euromedex) at an optical density at 600 nm (OD600) of 0.6 for 3 h at 37°C for YsxC proteins or overnight at 15°C for ribosomal proteins when necessary (see Table S2 in the supplemental material). Cells were harvested by centrifugation at 4,000 × g for 10 min. The pellets were frozen at −80°C until they were used.
Purification of YsxC proteins.
The bacterial pellets were thawed on ice and resuspended in 25 ml of cold lysis buffer (50 mM NaPO4, 1 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride) (Fluka), 10 μΜ leupeptin (Euromedex), and 6 μM pepstatin (Euromedex) at pH 8 for YsxC and 8.5 for glutathione S-transferase (GST)-YsxC and His6-YsxC. The cells were disrupted by ultrasonication three times for 20 s each time and spun at 39,000 × g for 20 min at 4°C. The supernatant fraction was saved.
The GST-YsxC protein was purified by glutathione-Sepharose affinity column chromatography by loading the supernatant onto a 4-ml glutathione-Sepharose column (Sigma). The column was washed with 100 ml of buffer A (50 mM NaPO4, 1 mM MgCl2, 0.1 M NaCl, pH 8). At this point, either the GST-tagged protein was eluted with about 15 ml of freshly prepared 20 mM glutathione in buffer A or the GST tag was cleaved by proteolysis while bound to glutathione-Sepharose. In the latter case, the resin was suspended in the same volume of buffer A, and thrombin (Calbiochem; 25 U/ml of resin) was added. The digestion proceeded for 2 h at 4°C under gentle rotation to keep the beads in suspension. The flowthrough was then recovered, and the column was rinsed with 1 resin volume of buffer A. Phenylmethylsulfonyl fluoride (1 mM) was added to inactivate the thrombin.
His6-YsxC was purified as described previously (61).
The native (nontagged) YsxC protein was purified in two steps. The supernatant fraction was loaded onto a 10-ml DEAE-cellulose (Sigma) column equilibrated with buffer C (50 mM NaPO4, 1 mM MgCl2, pH 8). The YsxC protein was recovered in the flowthrough, and the column was rinsed with 1 resin volume of buffer C. The protein solution was then loaded onto a 5-ml SP-Sepharose column (Sigma) equilibrated with buffer C, and the column was washed with 100 ml buffer C containing 0.3 M NaCl and eluted with 10 ml 0.6 M NaCl in buffer C.
GST was purified from E. coli cells, strain BL21(DE3), transformed with pGEX-4T1 vector (Amersham) by using a glutathione-Sepharose affinity column as described above for the GST-YsxC protein.
The eluted fractions containing nontagged or tagged YsxC were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (34) and stained with Coomassie blue (protein molecular weight marker from Fermentas). They were then concentrated in centrifugal filter devices (Centricon from Amicon) up to a final OD280 of 1 to 2. Glycerol was added to the YsxC preparations at a final concentration of 15% (vol/vol), and after a freezing step in liquid nitrogen, aliquots were stored at −80°C. The protein concentrations were determined using a Lowry Protein Assay Reagent (Pierce), with bovine serum albumin as a standard.
In a few cases, protein-concentrated aliquots were gel filtered on a Superdex 200 10/300GL (Amersham) equilibrated in buffer A (50 mM NaPO4, pH 8, 0.1 M NaCl). The resulting fractions (0.5 ml each) were subjected to SDS-PAGE and stained with Coomassie blue or electroblotted onto Immobilon-P transfer membranes (Millipore) for Western blotting analysis. The blots were blocked with 25 mg albumin bovine fraction V (Sigma) per ml Tris-buffered saline (10 mM Tris, 0.137 M NaCl, pH 7.6)-Tween 0.05% for 1 h. Primary antibodies against GST (anti-GST-horseradish peroxidase [HPR] conjugate; Euromedex) or His tag (HisProbe-HPR; Pierce) were incubated with membranes in the presence of albumin in Tris-buffered saline-Tween for 1 h at room temperature at dilutions of 1:5,000. The antibodies were detected by fluorography with an enhanced chemiluminescent substrate (Super-Signal Pico chemiluminescent substrate; Pierce) as recommended by the manufacturer.
Purification of recombinant ribosomal proteins.
His6-ribosomal proteins were purified following the same protocol as described for His6-YsxC (61). The GST-ribosomal proteins were purified by glutathione-Sepharose affinity column chromatography as described above for the GST-YsxC protein.
Analysis of the high-molecular-weight material associated with YsxC.
To analyze the material associated with YsxC, GST-YsxC was treated with either 1 mg/ml RNase A (Sigma) or 0.2 mg/ml DNase I (Euromedex) for 15 min at room temperature and loaded onto a Superdex 200 10/300GL (equilibrated with buffer A), as described above, that had been calibrated with the following protein standards (Amersham): RNase (13.7 kDa), ovalbumin (43 kDa), bovine serum albumin (67 kDa), and Blue Dextran (2,000 kDa). Each peak resolved was subjected to SDS-PAGE or Western blotting analysis as described above.
To check for the presence of RNA in the high-molecular-weight material associated with YsxC, purified GST-YsxC was extracted with an equal volume of phenol-chloroform-isoamyl alcohol, and RNA was precipitated with ethanol (58). The pellet was resuspended in water and analyzed with 1.5% agarose gels containing formaldehyde (58), with E. coli rRNAs as molecular weight markers. E. coli ribosome preparation from strain BL21(DE3) is described below, and rRNA extraction was performed as described above (phenol-chloroform-isoamyl alcohol extraction).
Ribosome purification and preparation of 30S and 50S subunits.
Highly purified ribosomes were prepared from B. subtilis (strain 168) and from E. coli cells [strain BL21(DE3), wild type or transformed with YsxC or GST plasmids and induced with IPTG for 3 h, thus expressing YsxC proteins or GST, respectively], essentially as described by Daigle and Brown (16). For all strains, 2 liters of LB was inoculated with 20 ml of an overnight culture of B. subtilis or E. coli. The cells were grown in LB medium at 37°C and collected at an OD600 of 0.8. Chloramphenicol was added to a final concentration of 100 μg/ml 3 min prior to harvesting (37). The ribosomal pellet was resuspended in a small volume of buffer (0.1 mM magnesium acetate, 0.005 mM EDTA, 10 mM Tris-HCl, pH 7.5, 30 mM KCl, 6 mM mercaptoethanol, 1.25 mM dithiothreitol) and stored at −80°C until they were used.
Separation of the 70S ribosome into 30S and 50S subunits.
Chloramphenicol (100 μg/ml) was added to the ribosome preparation, and 5 OD260 units of the suspension was layered onto 5 to 20% (wt/vol) sucrose RNase- and DNase-free-gradients made up in 10 mM Tris-HCl, pH 7.5, 0.1 mM magnesium acetate, 30 mM KCl, followed by centrifugation at 200,000 × g at 4°C for 190 min in a SW41Ti rotor (Beckman). Fractions (0.5 ml) were collected, the pump flow rate was set to 0.75 ml/min, and the UV absorbance (OD260) of the sample was monitored. Each fraction was precipitated by the addition of 10% trichloroacetic acid (TCA) and 0.02% deoxycholate, resuspended in SDS loading buffer, analyzed by SDS-12% PAGE, and immunoblotted with anti-His antibodies.
When 50S and 30S subunits were needed, fractions containing the 50S and 30S subunits were collected, combined, and precipitated in the presence of 20 mM magnesium acetate by mixing the solution with an equal volume of ice-cold ethanol. After incubation at −20°C for 1 h, subunits were pelleted by centrifugation at 10,000 × g for 30 min at 4°C and resuspended in a small volume of buffer (0.1 mM magnesium acetate, 5 μM EDTA, 10 mM Tris-HCl, pH 7.5, 30 mM KCl, 6 mM β-mercaptoethanol, 1.25 mM dithiothreitol). They were fractionated again on a 5 to 20% sucrose gradient, collected, and stored at −80°C. Quantitation of subunits was done by absorbance at 260 nm (1 A260 unit is equivalent to 69 or 34.5 pmol of 30S or 50S ribosomes, respectively) (16).
Binding of YsxC to 70S ribosomes. (i) Pelleting assays.
Pelleting assays were performed with a mixture of B. subtilis ribosomes (total A260 = 10, i.e., 114 picomoles) and 1 μg His6-YsxC (180 picomoles), as described previously (61).
(ii) Effects of excess guanine nucleotides on YsxC association with ribosome 30S and 50S subunits.
A mixture of B. subtilis ribosomes (OD260 = 10) preincubated with 1 μg His6-YsxC in the presence of 1 mM GDP, GTP, GMPPNP, or no added nucleotides was sedimented through a 5 to 20% (wt/vol) sucrose gradient in standard buffer (10 mM Tris-HCl, pH 7.5, 0.1 mM magnesium acetate, 30 mM KCl), with an additional 100 μM GDP, GTP, GMPPNP (guanylyl-imidodiphosphate), or nothing, respectively, at 200,000 × g at 4°C for 190 min in a SW41Ti rotor (Beckman), and the resulting fractions were analyzed as described above (absorbance at 260 nm, precipitation by 10% TCA and 0.02% deoxycholate, and immunoblotting with anti-His antibodies).
(iii) Far-Western blot analysis.
Ribosomal proteins from B. subtilis were separated by SDS-PAGE, using the electrophoretic system Protean II xi Cell device from Bio-Rad and a 14 to 18% or 14 to 20% acrylamide gradient. A prestained protein molecular weight marker (Fermentas), Seeblue Plus 2-stained standard (Invitrogen), or Mark 12 MW Standard (Invitrogen), was used. Proteins from the gel were then transferred to Immobilon-P transfer membranes (Millipore), washed, and incubated with histidine-tagged YsxC following precisely the protocol described by Bouvet et al. (10). Histidine-tagged YsxC was revealed by immunoblotting with anti-His antibodies, as described above.
Association of YsxC and recombinant ribosomal proteins. (i) Pull-down assay.
About 50 μg GST-tagged bait protein (GST-YsxC, GST-L10, GST-L27, or GST protein) was bound to about 50 μl of an immobilized glutathione support (glutathione-Sepharose) for 35 min in a minicolumn in buffer A (50 mM NaPO4, 0.3 M NaCl, pH 8). The column was then washed with 3 ml buffer A; loaded with L1, L6, L7, or L23 (when the bait protein was GST-YsxC or GST protein) or His6-YsxC (when the bait was GST-L10, GST-L27, or GST); incubated for 45 min; and then washed again with 3 ml buffer A, and the protein complex was eluted from the affinity support with 100 μl elution buffer (buffer A plus 100 mM glutathione). Protein complexes contained in eluted samples were then visualized by SDS-PAGE, and when necessary, histidine-tagged prey proteins were revealed by immunoblotting with anti-His antibodies, as described before.
(ii) Protein digestion and mass spectrometry analysis.
Protein bands were manually excised from Coomassie-blue-stained one-dimensional gels and washed several times with destaining solutions (25 mM NH4HCO3 for 15 min and then 50% [vol/vol] acetonitrile containing 25 mM NH4HCO3 for 15 min). Gel pieces were then dehydrated with 100% acetonitrile and dried under vacuum on a centrifugal evaporator. Samples were incubated with 7% H2O2 for 15 min for cysteine bond reduction before being washed again several times with the destaining solutions described above. After dehydration with 100% acetonitrile, the gel pieces were dried under vacuum. Depending on the protein amount, 0.2 μg to 0.5 μg of modified trypsin (Promega; sequencing grade) in 25 mM NH4HCO3 was added to the dehydrated gel bands. After 30 min of incubation at room temperature, 10 μl to 20 μl of 25 mM NH4HCO3 was added to submerge the gel pieces before incubation overnight at 37°C (29).
(iii) Nano-LC-MS/MS analysis of samples.
For nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) analysis, peptides were extracted from gel pieces by incubation for 15 min first in 50% acetonitrile, followed by 5% formic acid and finally 100% acetonitrile. The pooled supernatants were then transferred into microcentrifuge tubes and dried under vacuum. The dried extracted peptides were solubilized in water containing 2% acetonitrile and 7% trifluoroacetic acid before being transferred in vials compatible with nano-LC-MS/MS analysis (CapLC [Waters] and Q-TOF Ultima [Micromass], United Kingdom). The method consisted of a 60-min run at a flow rate of 200 nl/min using a gradient from two solvents, A (5% acetonitrile and 0.1% formic acid in water) and B (80% acetonitrile and 0.08% formic acid in water). The system included a 300-μm by 5-mm PepMap C18 precolumn to preconcentrate peptides and a 75-μm by 150-mm C18 column (LC Parkings Dionex) used for peptide elution. For automatic LC-MS/MS analysis, the QTOF Ultima instrument was run in data-dependent mode. In MS/MS mode, spectra were calibrated thanks to the fragmentation of the glufibrino-peptide. MS and MS/MS data were acquired and processed automatically using MassLynx 4.0 software (Waters). Each sample was submitted to searches against the Swiss Prot Trembl database (4,967 sequences for the B. subtilis taxonomy) using an in-house Mascot server (version 2.1; Matrix Sciences, London, United Kingdom). The Mascot search parameters used were as follows: enzyme, trypsin/P; one missed cleavage allowed; peptide tolerance, 0.4 Da; MS/MS tolerance, 0.4 Da; monoisotopic peptide charges, 2+ and 3+; and variable modifications, acetyl (N-ter)/oxidized methionine under sulfone and sulfoxide form/FMA + 1/FMA − 1/cysteic acid. Proteins that were identified with at least two peptides, both showing Mascot scores higher than 40, were validated without any manual validation. For proteins identified by only one peptide having a score higher than 40, the peptide sequence was checked manually. Peptides, with scores higher than 20 and lower than 40 were systematically checked and/or interpreted manually to confirm or cancel the Mascot suggestion.
RESULTS
Purification of GST-YsxC overexpressed in E. coli pulls down high-molecular-weight compounds.
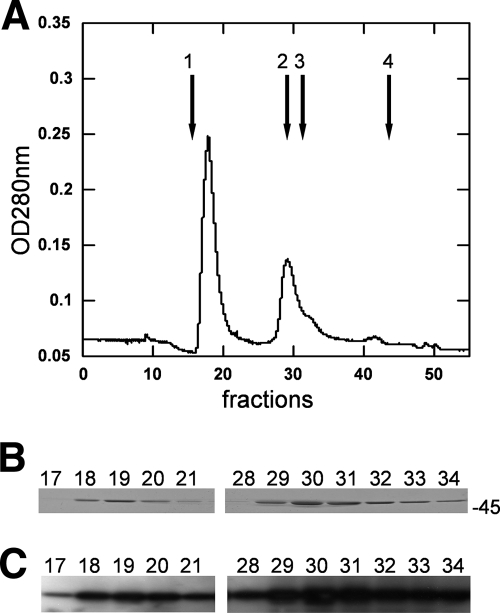
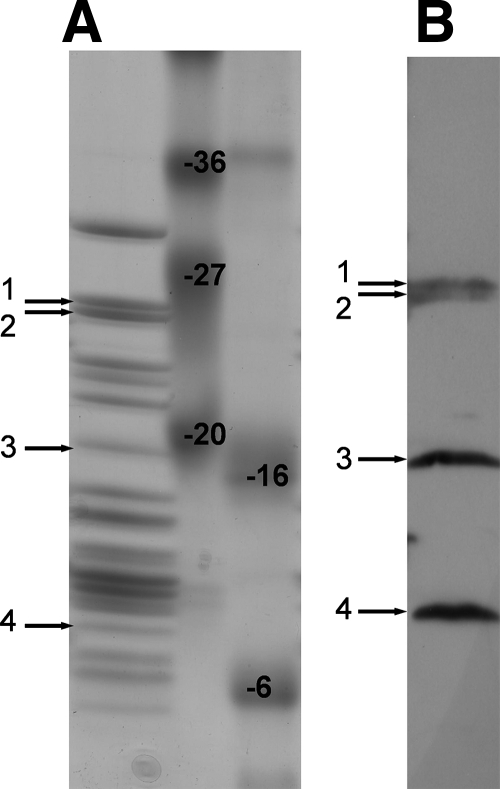
After overexpression of the GST-YsxC fusion protein in E. coli, its purification by a standard procedure using glutathione affinity chromatography gave essentially a single “pure” band when analyzed by SDS-PAGE, with an apparent molecular mass of ∼45 kDa (data not shown). Unexpectedly, however, analysis of the “purified” GST-YsxC fusion protein by size exclusion chromatography revealed the presence of two main peaks, eluting in fractions 18 to 20 and 29 to 31 (Fig. 1A). The second peak (fractions 29 to 31) eluted with an apparent molecular mass of ∼45 kDa and, as expected, contained the GST-YsxC protein, as shown in Fig. 1B. This was confirmed by Western blot analysis using anti-GST antibodies (Fig. 1C). The first peak (fractions 18 to 20) eluted in the void volume of the column and therefore had an estimated molecular mass of over 400 kDa. SDS-PAGE showed that these fractions contained only protein(s) with an apparent molecular mass of ∼45 kDa (Fig. 1B), suggesting that GST-YsxC also eluted in these fractions. No additional protein could be visualized on the stained gel. The presence of the GST moiety was confirmed in these fractions using anti-GST antibodies (Fig. 1C), thus demonstrating that GST-YsxC was indeed the protein present in this peak. The amount of GST-YsxC in the first peak was estimated to be ∼50% of that in the second peak (Fig. 1B and C), whereas the absorbance at 280 nm of the first peak reached a value almost twice as high as that in the second peak (Fig. 1A). A further analysis of this peak of high molecular mass revealed that it absorbed strongly at 260 nm (OD260/OD280 > 1.6) suggesting that it likely contained nucleic acid associated with the GST-YsxC protein.
FIG. 1.
Analysis of GST-YsxC by gel filtration chromatography. (A) Purified GST-YsxC was subjected to gel filtration chromatography through a Superdex-200 column, as described in Materials and Methods, and 0.5-ml fractions were collected and analyzed. The absorbance at 280 nm (OD280) is indicated. The elution positions of molecular mass markers (Amersham) are indicated by arrows 1 to 4, respectively, as follows: Blue Dextran 2000 (mass, 2,000 kDa), albumin (mass, 66 kDa), ovalbumin (mass, 45 kDa), and RNase A (mass, 13.7 kDa). (B) Fractions were analyzed by 12% acrylamide SDS-PAGE and stained with Coomassie blue. The position of the 45-kDa molecular mass marker is indicated. (C) Analysis by Western blotting of the fractions eluted from gel filtration. The fractions were transferred onto an Immobilon-P transfer membrane and probed with an anti-GST antibody, as described in Materials and Methods.
The B. subtilis YsxC protein strongly interacts with ribosomal material from E. coli.
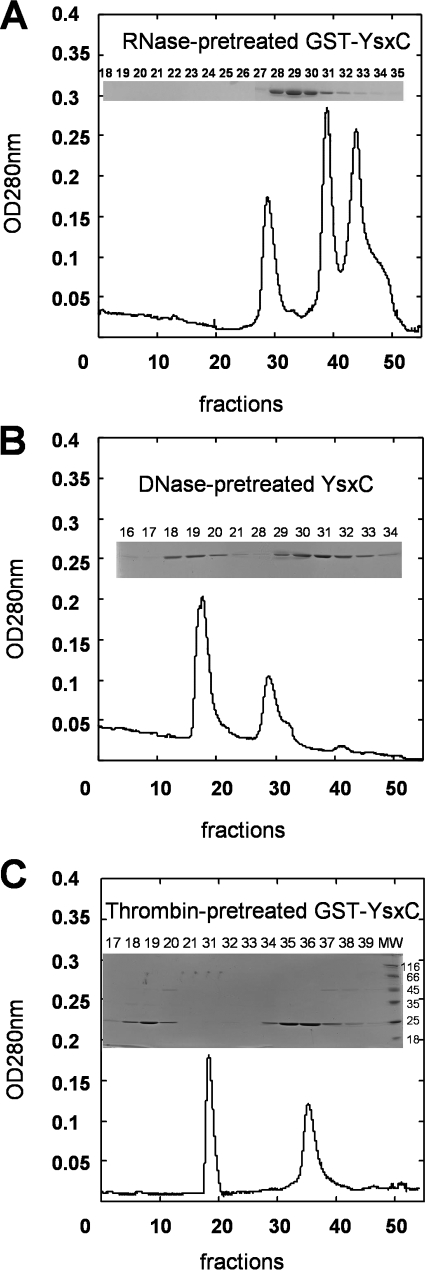
In order to analyze the nature of the material associated with the GST-YsxC fusion protein, samples were first treated with RNase or DNase prior to being subjected to size exclusion chromatography, as previously reported for the Era protein (41). When the GST-YsxC preparation was first treated with 1 mg/ml of RNase A (previously boiled for 15 min at 100°C to inactivate the possibly contaminating DNases), the peak of high molecular mass totally disappeared from the elution profile (Fig. 2A). Concomitantly, the peak with an apparent molecular mass of about 45 kDa, corresponding to the GST-YsxC protein alone, increased slightly but significantly. Two new peaks also appeared: one, containing fractions 39 to 40, corresponded to RNase (apparent molecular mass, ∼14 kDa), and the last peak (around fraction 45) corresponded to very low-molecular-mass material (probably nucleotides and salts). These results strongly suggested that the GST-YsxC fusion protein formed a complex with some E. coli RNA. By contrast, prior treatment of GST-YsxC with 0.2 mg/ml of DNase I (RNase free) did not modify the elution profile of GST-YsxC (Fig. 2B).
FIG. 2.
Gel filtration chromatography profiles of GST-YsxC preparations from a Superdex-200 column. The experiments were conducted exactly as described in the legend to Fig. 1. The insets show SDS-PAGE gels, stained with Coomassie blue, of the different fractions eluted from the gel filtration column. (A) Elution profile of RNase-treated GST-YsxC. Elution of 45-kDa material occurs at fractions 29 and 30. (B) Elution profile of DNase-treated GST-YsxC. (C) Elution profile of GST-YsxC, where the GST moiety was digested by thrombin while bound to glutathione-Sepharose beads prior to the gel filtration.
To assess which moiety of the GST-YsxC fusion interacted with E. coli RNA, several controls were made. First, purification of the His6-YsxC tagged protein led to a column profile similar to that seen with the GST-YsxC fusion protein when analyzed by size exclusion chromatography (data not shown). Conversely, purification of the GST tag alone run on size exclusion chromatography did not allow the detection of high-molecular-mass species (data not shown). Also, a thrombin digestion was performed to remove the GST moiety from the GST-YsxC fusion protein, thereby allowing the purification of the YsxC moiety alone (apparent molecular mass, ∼22 kDa). This sample was then submitted to size exclusion chromatography as before, and again, a peak of high molecular mass was obtained, which contained the YsxC protein alone (fractions 18 to 20), while free YsxC eluted from the column with a higher retention time (fractions 35 to 37), due, as expected, to its lower molecular mass (Fig. 2C). Collectively, these results show that high-molecular-mass material, most likely the ribosome, is associated with the YsxC moiety and not with the GST moiety.
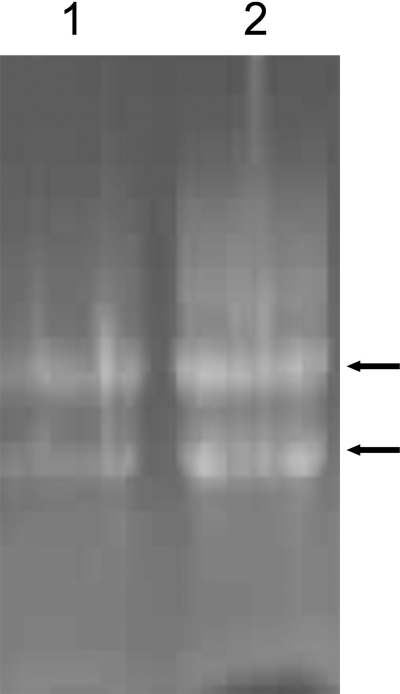
To further analyze the nature of the high-molecular-mass compounds copurified with the GST-YsxC fusion, the mobility of phenol-chloroform-extracted material from a GST-YsxC preparation was checked using denaturing agarose gel electrophoresis and staining with ethidium bromide. Two bands with apparent mobilities of ∼1.5 and ∼3 kb were visualized for GST-YsxC-associated material, and two bands with similar apparent mobilities were also observed for the E. coli RNA extracted from the ribosomes (Fig. 3, lanes 1 and 2). Together, these experiments show that rRNA is part of the high-molecular-mass material copurified with GST-YsxC.
FIG. 3.
Analysis of the material associated with YsxC. Analysis was performed on a 1.5% agarose gel containing formaldehyde that was stained with ethidium bromide. Lane 1, phenol-chloroform-isoamyl alcohol-extracted material from a GST-YsxC preparation; lane 2, phenol-chloroform-isoamyl alcohol-extracted RNA from E. coli ribosomes. The arrows indicate the migrations of 1.5-kb and 3-kb DNA fragments.
YsxC overexpressed in E. coli can be copurified with the ribosomal fraction.
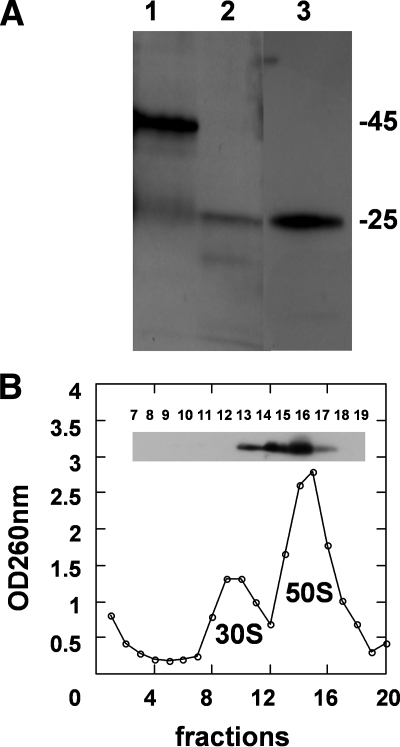
To further show that overexpressed YsxC was bound to the ribosome, we purified ribosomes from E. coli bacteria following overexpression of GST alone, GST-YsxC, or His6-YsxC, as checked by an analytical SDS-PAGE (data not shown). The ribosomal fractions were then loaded onto a 12% SDS-PAGE gel, and the overexpressed proteins putatively copurified with the ribosomes were visualized by Western blotting. The presence of the GST-YsxC fusion protein was detected, with an expected molecular mass of ∼45 kDa, and a faint band with an apparent molecular mass of ∼25 kDa was also present (Fig. 4A, lane 1). The latter band most likely corresponded to the GST moiety, probably resulting from some proteolytic degradation of the fusion protein. Ribosomes prepared from bacteria overexpressing GST alone also gave a similar faint signal (Fig. 4A, lane 2). This reflected some nonspecific binding of the GST protein alone to the ribosomes. When a similar experiment was carried out with ribosomes prepared from E. coli overexpressing His6-YsxC, the presence of this tagged-protein was detected as well, using a HisProbe-HPR. It must be noted that control experiments performed from ribosomes obtained from “native,” nontransformed E. coli cells showed no detectable bands at the corresponding molecular weights when probed with either anti-GST or HisProbe-HPR (data not shown).
FIG. 4.
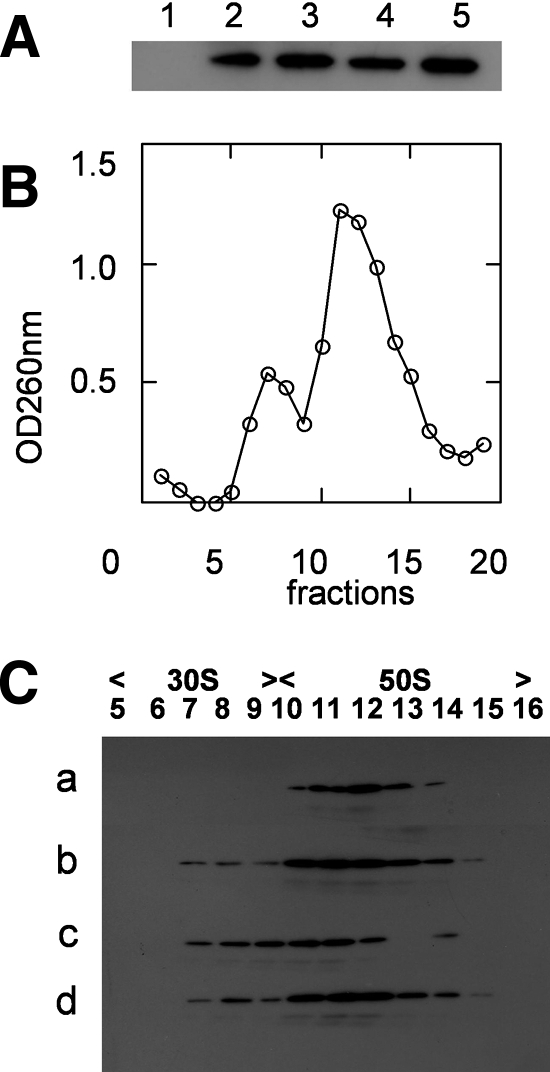
Analysis of ribosomes from E. coli cells overexpressing YsxC. (A) Western blot of purified ribosomes from E. coli cells, where GST-YsxC (lane 1), GST (lane 2), or His6-YsxC (lane 3) was overexpressed. Ribosomes (about 50 pmol) were loaded onto a 14% SDS-PAGE gel, and proteins were transferred onto an Immobilon-P transfer membrane and probed with an anti-GST antibody (lanes 1 and 2) or with an anti-His antibody (lane 3), as described in Materials and Methods. The positions of the 25-kDa (REase Bsp981) and 45-kDa (ovalbumin) molecular mass markers are indicated. (B) YsxC cofractionates with the 50S ribosomal subunit by sucrose density centrifugation. E. coli ribosomes were purified from His6-YsxC-induced cells. Shown is the UV profile at 260 nm of the different fractions of the 5 to 20% sucrose gradient, with the top of the gradient on the left. Fractions (0.5 ml) were precipitated with TCA and separated by SDS-12% PAGE, and after transfer onto Immobilon-P, the presence of His6-YsxC was revealed with anti-His antibodies (shown as an inset).
In order to pinpoint which ribosomal subunit YsxC was bound to, ribosomes prepared from His6-YsxC-overexpressing E. coli were dissociated under a low Mg2+ concentration and high-salt conditions in the presence of chloramphenicol. The mixture of 30S and 50S ribosomal subunits was layered onto a 5 to 20% sucrose gradient and separated by ultracentrifugation. Each fraction of the gradient was analyzed by OD260, and the result is shown in Fig. 4B. To detect the presence of YsxC, fractions of the gradient were TCA precipitated and loaded onto an SDS-PAGE gel, and the presence of His6-YsxC was revealed by Western blot analysis using a HisProbe-HPR. Clearly, His6-YsxC was found associated with the 50S ribosomal subunit only (fractions 13 to 17) (Fig. 4B, inset). These observations corroborate the conclusion that YsxC can be found in vivo associated with ribosomes.
Purified YsxC interacts mainly with the 50S ribosomal subunit of B. subtilis.
Because the previous experiments reflected “nonnative” interaction between E. coli ribosomes and overexpressed YsxC, we also investigated the interaction of YsxC with its cognate ribosomes purified from B. subtilis. In a previous report, we had shown that YsxC was able to interact with the 70S B. subtilis ribosomes, and this interaction seemed to be increased by the presence of either GDP or GTP (61). Figure 5A confirms and extends these results, since both nucleotides favored the binding of YsxC to ribosomes. In addition, it is shown here that the nonhydrolyzable GTP analogue GMPPNP strongly stimulated the binding of YsxC to ribosomes. To identify the ribosome subunit to which YsxC was bound, 30S and 50S subunits were separated by sucrose gradient, and the fraction containing His6-YsxC was revealed as shown in Fig. 4. It is important to note here that the addition of nucleotides did not change the overall profiles of ribosomes dissociated into 30S and 50S subunits (only the absorbance profile obtained in the absence of nucleotide is shown in Fig. 5B). In the absence of any nucleotide, YsxC was found to be associated with the 50S subunit only (Fig. 5C, line a). Addition of the nucleotide GDP, GTP, or GMPPNP (Fig. 5C, lines b, c, and d, respectively) still allowed binding to the 50S subunit, but marginal binding to the 30S subunit was also detected for each nucleotide. GDP or GMPPNP was preferred over GTP for binding to the 50S subunit, suggesting either that during GTP hydrolysis YsxC goes through a cycle of interaction/dissociation with the 50S subunit or that in its GTP-bound state, the affinity of YsxC for this subunit is somehow reduced. The latter explanation, however, seems unlikely since in the presence of GMPPNP, which mimics a GTP-bound state, YsxC appeared quite able to interact with the 50S subunit.
FIG. 5.
Binding of YsxC to 70S ribosomes and ribosomal subunits. (A) His6-YsxC was tested for the ability to interact with 70S ribosomes following a 15-min incubation at room temperature in the presence or absence of GDP, GTP, or GMPPNP (1 mM each). Pelleting assays were conducted as described in Materials and Methods. The pellets were analyzed by SDS-PAGE and Western blotting, using anti-His antibody, as described in Materials and Methods. Lane 1, His6-YsxC; lanes 2 to 5, His6-YsxC incubated with 70S ribosomes in the absence (lane 2) or in the presence of GDP (lane 3), GTP (lane 4), or GMPPNP (lane 5). (B) Typical absorbance profile of B. subtilis ribosomes sedimented through 5 to 20% sucrose gradients, with the top of the gradient on the left. (C) Effects of nucleotides on YsxC sedimentation. Shown is Western blot analysis of fractions obtained by velocity centrifugation of B. subtilis ribosomes through 5 to 20% sucrose gradients containing no nucleotide (line a), 100 μM GDP (line b), 100 μM GTP (line c), or 100 μM GMPPNP (line d) after a prior incubation with His6-YsxC with no added nucleotide (line a), 1 mM GDP (line b), 1 mM GTP (line c), or 1 mM GMPPNP (line d).
In order to address the question of which ribosomal proteins YsxC was able to interact with, a far-Western blotting approach was used. In the first step, 30S or 50S ribosomal proteins from B. subtilis were separated by SDS-PAGE, transferred to Immobilon-P membranes, renatured on the membrane as previously described (10), and incubated with His6-YsxC. After extensive washing of the membrane, the presence of the His6-YsxC protein bound to ribosomal proteins was detected using the HisProbe-HPR. Under these conditions, no ribosomal proteins that belonged to the 30S subunit were found to interact with YsxC (not shown). In contrast, four reactive bands from the 50S subunit were revealed as putative partners of His6-YsxC (Fig. 6B). When far-Western blotting was performed using total ribosomal proteins instead of 50S ribosomal proteins, similar bands were found to react with the His probe (data not shown). A control experiment was performed in which the His probe was directly incubated with ribosomal proteins previously separated by SDS-PAGE and transferred to an Immobilon-P membrane. Under these conditions, where no YsxC was added, no signal was detected, showing that no nonspecific recognition of ribosomal proteins occurred using the His probe (data not shown). To assign the ribosomal proteins to which the His6-YsxC protein was bound, superimposition of the detected bands with those on the membrane previously stained with Ponceau red was carried out and compared to an SDS-PAGE of the 50S ribosomal proteins migrating under similar conditions. The corresponding bands, shown in Fig. 6A, were excised, and the protein content of each band was analyzed by nano-LC-MS/MS. Bands 1 and 2 were in some cases well separated on the gradient gel, and both were reactive toward His6-YsxC. The identified proteins are listed in Table 1. The proteins present in the doublet of the two upper bands in Fig. 6B were L1 and L3 in both samples (Table 1, no. 1 and 2) but in different ratios; the upper band contained essentially L1 (11 peptides compared to 1), whereas the lower band contained a little bit more L3 (11 peptides out of 20). In the third band, the most abundant protein was L10 (with nine peptides and a coverage of the sequence of 38.2%), and one peptide corresponded to L6. In the fourth band, the most abundant protein was L27 (7 peptides out of 11), but L7/L12 was also present (3 peptides), as well as L23 (1 peptide).
FIG. 6.
Interaction of YsxC with a subset of ribosomal proteins. Proteins from B. subtilis 50S ribosomal subunits were analyzed by SDS-PAGE. After transfer onto an Immobilon-P membrane, the proteins were stained with Ponceau red dye. Ribosomal proteins bound to the membrane were renatured and incubated with purified His6-YsxC. After extensive washing, YsxC was detected by immunoblotting using an anti-His antibody and revealed on a film using enhanced chemiluminescence. (A) Separation of 50S ribosomal proteins through a 14 to 18% SDS-PAGE gel. The 20-, 27-, and 36-kDa molecular mass markers were the Prestained Protein Molecular Weight Markers from Fermentas, and the 6- and 16-kDa molecular mass markers were the Seeblue Plus 2-stained standard from Invitrogen. (B) Detection of bound YsxC by immunoblotting. The film was superimposed on the corresponding Immobilon-P membrane stained with Ponceau red dye to localize the protein bands (indicated by arrows).
TABLE 1.
Protein identification by MS/MSa
| Band no. | Protein description | Accession no. | Scoreb | Mass (Da) | Coverage (%)c | No. of peptidesd |
|---|---|---|---|---|---|---|
| 1 | L1 (RL1_BACSU) | Q06797 | 762.4 | 24,776 | 47.2 | 11 |
| L3 (RL3_BACSU) | P42920 | 92.4 | 22,669 | 7.3 | 1 | |
| 2 | L3 (RL3_BACSU) | P42920 | 722.9 | 22,669 | 50.7 | 11 |
| L1 (RL1_BACSU) | Q06797 | 542.3 | 24,776 | 36.4 | 9 | |
| 3 | L10 (RL10_BACSU) | P42923 | 401.4 | 17,887 | 38.2 | 9 |
| L6 (RL6_BACSU) | P46898 | 69.6 | 19,366 | 6.8 | 1 | |
| 4 | L27 (RL27_BACSU) | P05657 | 320.5 | 10,366 | 47.9 | 7 |
| L7/L12 (RL7_BACSU) | P02394 | 170.8 | 12,612 | 23.8 | 3 | |
| L23 (RL23_BACSU) | P42924 | 35.2 | 10,922 | 13.1 | 1 |
Proteins were identified by nano-LC-ESI (electrospray ionization) MS/MS, followed by protein database mining.
Score attributed to the identified protein by Mascot software (Matrix Science).
Coverage (%), percentage of the full-length sequence covered by the matching peptides.
No of peptides, number of peptides assigned to the protein.
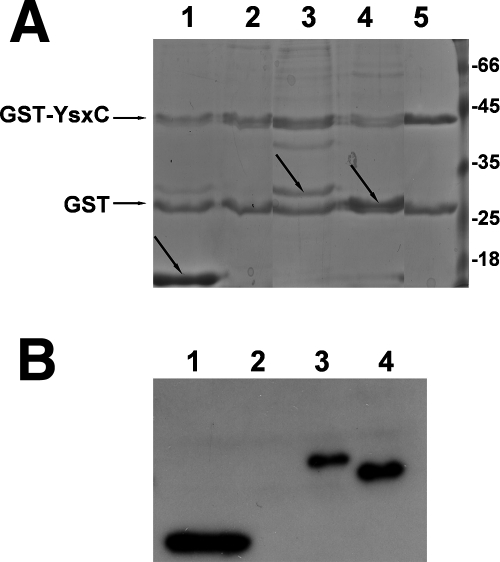
To validate our results, we used a pull-down assay, in which one of the proteins (YsxC or a ribosomal protein) was cloned with a GST tag at one of its extremities and bound to a glutathione-agarose column. If a putative interacting protein (lacking a GST tag) bound to the immobilized target protein and coeluted from the column when the target was eluted with reduced glutathione, then one could infer that the two proteins interacted. All of the putative interacting ribosomal proteins (L1, L3, L6, L7/L12, L10, L23, and L27) were thus cloned with a His6 tag to facilitate their purification and, when soluble (see Table S2 in the supplemental material for production and solubility), were used as “prey” proteins, the immobilized target protein (the “bait” protein) being GST-YsxC (or GST protein as a control). In these cases, L7/L12 (mass, 14.9 kDa), L1 (mass, 29 kDa), and L6 (mass, 22.7 kDa) were eluted from the glutathione-agarose column, together with GST-YsxC (mass, 45 kDa) (but not with GST protein alone, as a control), whereas L23 (mass, 12.6 kDa) did not coelute with GST-YsxC (Fig. 7A). During the pull-down assay, there was some proteolysis of the GST-YsxC fusion protein, resulting in the presence of two protein bands on the gel: GST-YsxC and GST (YsxC was washed away during the washing step). In the case of L6 (mass, 22.7 kDa), its migration was almost identical to that of the GST protein (25.2 kDa); therefore, the presence of His6-L6 was checked and confirmed by immunoblotting, using anti-His antibody (Fig. 7B). His6-L10 and His6-L27 were not soluble. GST-L10 and GST-L27 were therefore produced but were poorly soluble (see Table S2 in the supplemental material). Coimmobilization assays were then performed with GST-L10 and GST-L27 as the prey proteins, using His6-YsxC as bait, but no interaction could be detected between these two ribosomal proteins and YsxC. However, one has to keep in mind that an additional interaction(s) between YsxC and some other putative ribosomal partner(s) might have occurred but that it was too loose to be retained during the washing steps. In conclusion, we were able to show that the L1, L6, and L7/L12 ribosomal proteins interact in vitro with YsxC, with the possibility that an additional interaction(s) between YsxC and some other soluble putative ribosomal partner(s) might have occurred but was too loose to be retained during the washing steps.
FIG. 7.
Pull-down assay of YsxC and His-tagged recombinant ribosomal proteins. (A) SDS-PAGE of coeluted GST-YsxC and ribosomal prey proteins from a glutathione-agarose column. The ribosomal proteins used as prey proteins were L7 (lane 1), L23 (lane 2), L1 (lane 3), and L6 (lane 4). GST-YsxC alone was loaded as a control (lane 5) and gave two bands (GST-YsxC and GST, due to partial cleavage of the YsxC moiety). The values of the molecular mass markers (Fermentas) are indicated on the right of the gel in kDa. (B) Western blot analysis using an anti-His antibody of coeluted ribosomal proteins with GST-YsxC. Lane 1, L7; lane 2, L23; lane 3, L1; lane 4, L6.
DISCUSSION
In the past few years, numerous nonribosomal proteins, including many GTPases, have been found to play key roles in ribosome assembly in eukaryotic cells (19, 31), and a similar trend is now emerging for the prokaryotic ribosome (15). YsxC has recently been suggested to be involved in ribosome biogenesis (61), and we report here that YsxC interacts with several ribosomal proteins of the 50S subunit.
We observed above that heterologous expression of recombinant YsxC from B. subtilis in E. coli cells resulted in the association of YsxC with ribosomes, which involved, at least in part, a direct interaction with rRNA. A possible interaction between YsxC and RNA was initially proposed based on the three-dimensional structure of YsxC due to the presence on its surface of a patch of conserved basic residues that might act as a hook to anchor the protein to RNA (57). Moreover, among the newly discovered bacterial GTPases that interact with ribosome, many have been shown to have the ability to bind to rRNA (5, 25, 27, 40, 42, 43), and this is probably a conserved trait originating from an ancient GTPase traceable to the last universal common ancestor (13, 36).
We were able to show that YsxC preferentially binds the 50S subunit and that guanine nucleotide occupancy of YsxC affects its association with the ribosome. This result is consistent with the different conformations adopted by YsxC in the GTP-bound, GDP-bound, and apo-enzyme states (57). Thus, it is likely that the ribosome-associated GTP-bound YsxC is released from the ribosome as a consequence of the hydrolysis of GTP.
To identify the possible ribosomal-protein partners of YsxC, a far-Western approach was used, leading to a positive interaction with L1, L3, at least one protein of a mixture of L6 and L10, and one protein of a mixture of L7/L12 (L7 and L12 are the same protein, but L7 is acetylated), L23, and L27. It is important to stress here the importance of protein L27 for both the assembly and function of the ribosome, since L27 has been suggested to be lacking in the ribosomes of YsxC-depleted cells (61) and deletion of the gene results in a severe growth defect (38, 73). All of the putative protein partners that we identified belong to the 50S subunit, while the 30S subunit failed to reveal any positive band, a result consistent with the interaction of YsxC primarily with the 50S subunit.
In order to validate our far-Western results, we performed pull-down assays with YsxC and recombinant ribosomal proteins and confirmed a direct interaction between L1, L6, or L7/L12 and YsxC. We were unable to detect any interaction between L10 or L27 and YsxC by this approach. However, for L10, its N-terminal part is free in the ribosome, while its C-terminal part interacts with two L7/L12 dimers in E. coli (20, 23, 24, 51). In contrast, in the recombinant protein, its N-terminal part is linked to the GST moiety, and this could have prevented its interaction with YsxC. Thus, we cannot rule out the possibility that L10 is a physiological partner of YsxC.
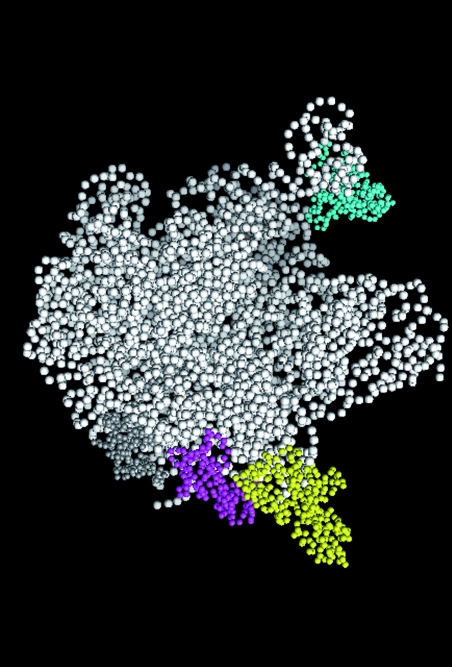
It is remarkable that most of the proteins that potentially interact with YsxC, except L1, are in the same location on the ribosome (Fig. 8 shows the protein localization). The L7/L12 proteins are the only ribosomal proteins that are present in multiple copies in prokaryotic ribosomes. They are organized as two L7/L12 dimers associated with the C-terminal region of protein L10, and at least one of the dimers comprises the stalk protuberance (20, 23, 24, 51). The detailed structure of the pentameric complex has not yet been resolved by X-ray crystallography of ribosomes (4, 6, 26, 33, 62, 74). On the basis of immune electron microscopy, the two L7/L12 dimers have been proposed to bind to different locations and also to adopt different conformations (49): one nonstalk dimer of protein L7/L12 is in a folded conformation on the subunit body, while the second dimer is in an extended conformation in the subunit stalk. Other studies (cross-linking data and cryoelectron microscopic localization) also suggest multiple locations for L7/L12 in the large subunit (18, 44, 65). The ribosomal stalk is highly flexible, and various conformations of the stalk are thought to occur in response to elongation factor binding and GTP hydrolysis (8). As for L1, it has been proposed to be located on the opposite side of the ribosome from the stalk position (39, 68). However, intrinsic flexibility of L1 has been suggested, depending on RNA binding (46-48); it adopts a closed conformation in the absence of RNA (47) but opens upon RNA binding (46, 48). This probably explains the difficulty in precisely determining the structure of the L1 protuberance within the ribosome, due to the high mobility of the region (1, 48). The flexibility of these regions might lead to a possible location of L1 closer to L7/L12 in the three-dimensional structure. Alternatively, one might propose that YsxC is able to interact with L1 and L7/L12 at different locations on the ribosome. The different patterns of interaction of YsxC with the 50S ribosomal proteins, depending on the nature of the nucleotide added (Fig. 5), might support such a hypothesis.
FIG. 8.
Components of the 50S subunit of Thermus thermophilus ribosomes. 23S and 5S rRNAs are shown in white. Only proteins L1 (cyan), L3 (gray), L6 (magenta), and L7/L12 (yellow) are represented. The figure was generated using PYMOL software and the structure of the T. thermophilus 50S ribosome (Protein Data Bank accession number 1GIY) (74). In this structure, L10 and L27 were missing, and only one of the two L7/L12 dimers was represented.
YsxC is required for 50S ribosomal-subunit assembly in vivo, since a 44.5S ribosomal intermediate accumulates in cells depleted of YsxC (61). This intermediate lacks ribosomal proteins L16, L27, and L36 (61), and we also have evidence that YsxC interacts with the 44.5S complex (data not shown). Altogether, these results strongly suggest that YsxC participates in the assembly and/or the processing steps of the preribosomal particle. We propose that YsxC binds to the 44.5S preribosomal complex through interaction with the partner ribosomal proteins identified here and/or possibly through association with rRNA. This association would somehow locally modify the conformation of the presubunit, thereby allowing the incorporation of the other missing proteins.
Supplementary Material
Acknowledgments
We are grateful to Jérôme Garin for preparing Table 1 and for critical reading of the manuscript.
This work was supported by a CNRS “Young Investigator” ATIP Program (J.-M.J.).
Footnotes
Published ahead of print on 2 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agrawal, R. K., R. K. Lata, and J. Frank. 1999. Conformational variability in Escherichia coli 70S ribosome as revealed by 3D cryo-electron microscopy. Int. J. Biochem. Cell Biol. 31243-254. [DOI] [PubMed] [Google Scholar]
- 2.Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16851-856. [DOI] [PubMed] [Google Scholar]
- 3.Arnosti, D. N., V. L. Singer, and M. J. Chamberlin. 1986. Characterization of heat shock in Bacillus subtilis. J. Bacteriol. 1681243-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289905-920. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, J., M. Kallas, and E. Hurt. 2006. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J. Biol. Chem. 28124737-24744. [DOI] [PubMed] [Google Scholar]
- 6.Berk, V., W. Zhang, R. D. Pai, and J. H. Cate. 2006. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. USA 10315830-15834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bharat, A., M. Jiang, S. M. Sullivan, J. R. Maddock, and E. D. Brown. 2006. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 1887992-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bocharov, E. V., A. G. Sobol, K. V. Pavlov, D. M. Korzhnev, V. A. Jaravine, A. T. Gudkov, and A. S. Arseniev. 2004. From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J. Biol. Chem. 27917697-17706. [DOI] [PubMed] [Google Scholar]
- 9.Bourne, H. R., D. A. Sanders, and F. McCormick. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348125-132. [DOI] [PubMed] [Google Scholar]
- 10.Bouvet, P., J. J. Diaz, K. Kindbeiter, J. J. Madjar, and F. Amalric. 1998. Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 27319025-19029. [DOI] [PubMed] [Google Scholar]
- 11.Brown, E. D. 2005. Conserved P-loop GTPases of unknown function in bacteria: an emerging and vital ensemble in bacterial physiology. Biochem. Cell Biol. 83738-746. [DOI] [PubMed] [Google Scholar]
- 12.Caldon, C. E., and P. E. March. 2003. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6135-139. [DOI] [PubMed] [Google Scholar]
- 13.Caldon, C. E., P. Yoong, and P. E. March. 2001. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 41289-297. [DOI] [PubMed] [Google Scholar]
- 14.Campbell, T. L., D. M. Daigle, and E. D. Brown. 2005. Characterization of the Bacillus subtilis GTPase YloQ and its role in ribosome function. Biochem. J. 389843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comartin, D. J., and E. D. Brown. 2006. Non-ribosomal factors in ribosome subunit assembly are emerging targets for new antibacterial drugs. Curr. Opin. Pharmacol. 6453-458. [DOI] [PubMed] [Google Scholar]
- 16.Daigle, D. M., and E. D. Brown. 2004. Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro. J. Bacteriol. 1861381-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dassain, M., A. Leroy, L. Colosetti, S. Carole, and J. P. Bouche. 1999. A new essential gene of the ‘minimal genome’ affecting cell division. Biochimie 81889-895. [DOI] [PubMed] [Google Scholar]
- 18.Dey, D., D. E. Bochkariov, G. G. Jokhadze, and R. R. Traut. 1998. Cross-linking of selected residues in the N- and C-terminal domains of Escherichia coli protein L7/L12 to other ribosomal proteins and the effect of elongation factor Tu. J. Biol. Chem. 2731670-1676. [DOI] [PubMed] [Google Scholar]
- 19.Dez, C., and D. Tollervey. 2004. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7631-637. [DOI] [PubMed] [Google Scholar]
- 20.Diaconu, M., U. Kothe, F. Schlunzen, N. Fischer, J. M. Harms, A. G. Tonevitsky, H. Stark, M. V. Rodnina, and M. C. Wahl. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121991-1004. [DOI] [PubMed] [Google Scholar]
- 21.Falson, P., A. Di Pietro, and D. C. Gautheron. 1986. Chemical modification of thiol groups of mitochondrial F1-ATPase from the yeast Schizosaccharomyces pombe. Involvement of alpha- and gamma-subunits in the enzyme activity. J. Biol. Chem. 2617151-7159. [PubMed] [Google Scholar]
- 22.Galperin, M. Y., and E. V. Koonin. 2004. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 325452-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griaznova, O., and R. R. Traut. 2000. Deletion of C-terminal residues of Escherichia coli ribosomal protein L10 causes the loss of binding of one L7/L12 dimer: ribosomes with one L7/L12 dimer are active. Biochemistry 394075-4081. [DOI] [PubMed] [Google Scholar]
- 24.Gudkov, A. T., L. G. Tumanova, S. Y. Venyaminov, and N. N. Khechinashvilli. 1978. Stoichiometry and properties of the complex between ribosomal proteins L7 and L10 in solution. FEBS Lett. 93215-218. [DOI] [PubMed] [Google Scholar]
- 25.Hang, J. Q., T. I. Meier, and G. Zhao. 2001. Analysis of the interaction of 16S rRNA and cytoplasmic membrane with the C-terminal part of the Streptococcus pneumoniae Era GTPase. Eur. J. Biochem. 2685570-5577. [DOI] [PubMed] [Google Scholar]
- 26.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107679-688. [DOI] [PubMed] [Google Scholar]
- 27.He, W. J., S. Tang, and W. Y. Liu. 2002. In vitro interaction of eukaryotic elongation factor 2 with synthetic oligoribonucleotide that mimics GTPase domain of rat 28S ribosomal RNA. Int. J. Biochem. Cell Biol. 34263-268. [DOI] [PubMed] [Google Scholar]
- 28.Hwang, J., and M. Inouye. 2006. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 611660-1672. [DOI] [PubMed] [Google Scholar]
- 29.Jaquinod, M., F. Villiers, S. Kieffer-Jaquinod, V. Hugouvieux, C. Bruley, J. Garin, and J. Bourguignon. 2007. A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell Proteomics 6394-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffares, D. C., A. M. Poole, and D. Penny. 1998. Relics from the RNA world. J. Mol. Evol 4618-36. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, A. W., E. Lund, and J. Dahlberg. 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27580-585. [DOI] [PubMed] [Google Scholar]
- 32.Johnstone, B. H., A. A. Handler, D. K. Chao, V. Nguyen, M. Smith, S. Y. Ryu, E. L. Simons, P. E. Anderson, and R. W. Simons. 1999. The widely conserved Era G-protein contains an RNA-binding domain required for Era function in vivo. Mol. Microbiol. 331118-1131. [DOI] [PubMed] [Google Scholar]
- 33.Korostelev, A., S. Trakhanov, M. Laurberg, and H. F. Noller. 2006. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 1261065-1077. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 35.Lehoux, I. E., M. J. Mazzulla, A. Baker, and C. M. Petit. 2003. Purification and characterization of YihA, an essential GTP-binding protein from Escherichia coli. Protein Expr. Purif. 30203-209. [DOI] [PubMed] [Google Scholar]
- 36.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 31741-72. [DOI] [PubMed] [Google Scholar]
- 37.Lin, B., D. A. Thayer, and J. R. Maddock. 2004. The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit. J. Bacteriol. 186481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maguire, B. A., A. D. Beniaminov, H. Ramu, A. S. Mankin, and R. A. Zimmermann. 2005. A protein component at the heart of an RNA machine: the importance of protein L27 for the function of the bacterial ribosome. Mol. Cell 20427-435. [DOI] [PubMed] [Google Scholar]
- 39.Malhotra, A., P. Penczek, R. K. Agrawal, I. S. Gabashvili, R. A. Grassucci, R. Junemann, N. Burkhardt, K. H. Nierhaus, and J. Frank. 1998. Escherichia coli 70 S ribosome at 15 Å resolution by cryo-electron microscopy: localization of fMet-tRNAfMet and fitting of L1 protein. J. Mol. Biol. 280103-116. [DOI] [PubMed] [Google Scholar]
- 40.Matsuo, Y., T. Morimoto, M. Kuwano, P. C. Loh, T. Oshima, and N. Ogasawara. 2006. The GTP-binding protein YlqF participates in the late step of 50 S ribosomal subunit assembly in Bacillus subtilis. J. Biol. Chem. 2818110-8117. [DOI] [PubMed] [Google Scholar]
- 41.Meier, T. I., R. B. Peery, S. R. Jaskunas, and G. Zhao. 1999. 16S rRNA is bound to Era of Streptococcus pneumoniae. J. Bacteriol. 1815242-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meier, T. I., R. B. Peery, K. A. McAllister, and G. Zhao. 2000. Era GTPase of Escherichia coli: binding to 16S rRNA and modulation of GTPase activity by RNA and carbohydrates. Microbiology 1461071-1083. [DOI] [PubMed] [Google Scholar]
- 43.Moazed, D., J. M. Robertson, and H. F. Noller. 1988. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334362-364. [DOI] [PubMed] [Google Scholar]
- 44.Montesano-Roditis, L., D. G. Glitz, R. R. Traut, and P. L. Stewart. 2001. Cryo-electron microscopic localization of protein L7/L12 within the Escherichia coli 70 S ribosome by difference mapping and Nanogold labeling. J. Biol. Chem. 27614117-14123. [DOI] [PubMed] [Google Scholar]
- 45.Morimoto, T., P. C. Loh, T. Hirai, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 1483539-3552. [DOI] [PubMed] [Google Scholar]
- 46.Nevskaya, N., S. Tischenko, R. Fedorov, S. Al-Karadaghi, A. Liljas, A. Kraft, W. Piendl, M. Garber, and S. Nikonov. 2000. Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family. Structure 8363-371. [DOI] [PubMed] [Google Scholar]
- 47.Nikonov, S., N. Nevskaya, I. Eliseikina, N. Fomenkova, A. Nikulin, N. Ossina, M. Garber, B. H. Jonsson, C. Briand, S. Al-Karadaghi, A. Svensson, A. Aevarsson, and A. Liljas. 1996. Crystal structure of the RNA binding ribosomal protein L1 from Thermus thermophilus. EMBO J. 151350-1359. [PMC free article] [PubMed] [Google Scholar]
- 48.Nikulin, A., I. Eliseikina, S. Tishchenko, N. Nevskaya, N. Davydova, O. Platonova, W. Piendl, M. Selmer, A. Liljas, D. Drygin, R. Zimmermann, M. Garber, and S. Nikonov. 2003. Structure of the L1 protuberance in the ribosome. Nat. Struct. Biol 10104-108. [DOI] [PubMed] [Google Scholar]
- 49.Olson, H. M., A. Sommer, D. S. Tewari, R. R. Traut, and D. G. Glitz. 1986. Localization of two epitopes of protein L7/L12 to both the body and stalk of the large ribosomal subunit. Immune electron microscopy using monoclonal antibodies. J. Biol. Chem. 2616924-6932. [PubMed] [Google Scholar]
- 50.Pandit, S. B., and N. Srinivasan. 2003. Survey for g-proteins in the prokaryotic genomes: prediction of functional roles based on classification. Proteins 52585-597. [DOI] [PubMed] [Google Scholar]
- 51.Pettersson, I., S. J. Hardy, and A. Liljas. 1976. The ribosomal protein L8 is a complex L7/L12 and L10. FEBS Lett. 64135-138. [DOI] [PubMed] [Google Scholar]
- 52.Phillips, T. A., R. A. VanBogelen, and F. C. Neidhardt. 1984. lon gene product of Escherichia coli is a heat-shock protein. J. Bacteriol. 159283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pragai, Z., and C. R. Harwood. 2000. YsxC, a putative GTP-binding protein essential for growth of Bacillus subtilis 168. J. Bacteriol. 1826819-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pullman, M. E., H. S. Penefsky, A. Datta, and E. Racker. 1960. Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase. J. Biol. Chem. 2353322-3329. [PubMed] [Google Scholar]
- 55.Riethdorf, S., U. Volker, U. Gerth, A. Winkler, S. Engelmann, and M. Hecker. 1994. Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene. J. Bacteriol. 1766518-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts, R. J. 2004. Identifying protein function—a call for community action. PLoS Biol. 2E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruzheinikov, S. N., S. K. Das, S. E. Sedelnikova, P. J. Baker, P. J. Artymiuk, J. Garcia-Lara, S. J. Foster, and D. W. Rice. 2004. Analysis of the open and closed conformations of the GTP-binding protein YsxC from Bacillus subtilis. J. Mol. Biol. 339265-278. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Sato, A., G. Kobayashi, H. Hayashi, H. Yoshida, A. Wada, M. Maeda, S. Hiraga, K. Takeyasu, and C. Wada. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10393-408. [DOI] [PubMed] [Google Scholar]
- 60.Sayed, A., S. Matsuyama, and M. Inouye. 1999. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem. Biophys. Res. Commun. 26451-54. [DOI] [PubMed] [Google Scholar]
- 61.Schaefer, L., W. C. Uicker, C. Wicker-Planquart, A. E. Foucher, J. M. Jault, and R. A. Britton. 2006. Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J. Bacteriol. 1888252-8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Selmer, M., C. M. Dunham, F. V. T. Murphy, A. Weixlbaumer, S. Petry, A. C. Kelley, J. R. Weir, and V. Ramakrishnan. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 3131935-1942. [DOI] [PubMed] [Google Scholar]
- 63.Sprang, S. R. 1997. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 66639-678. [DOI] [PubMed] [Google Scholar]
- 64.te Riele, H., B. Michel, and S. D. Ehrlich. 1986. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 832541-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Traut, R. R., D. Dey, D. E. Bochkariov, A. V. Oleinikov, G. G. Jokhadze, B. Hamman, and D. Jameson. 1995. Location and domain structure of Escherichia coli ribosomal protein L7/L12: site specific cysteine crosslinking and attachment of fluorescent probes. Biochem. Cell Biol. 73949-958. [DOI] [PubMed] [Google Scholar]
- 66.Uicker, W. C., L. Schaefer, and R. A. Britton. 2006. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 59528-540. [DOI] [PubMed] [Google Scholar]
- 67.Vetter, I. R., and A. Wittinghofer. 2001. The guanine nucleotide-binding switch in three dimensions. Science 2941299-1304. [DOI] [PubMed] [Google Scholar]
- 68.Walleczek, J., D. Schuler, M. Stoffler-Meilicke, R. Brimacombe, and G. Stoffler. 1988. A model for the spatial arrangement of the proteins in the large subunit of the Escherichia coli ribosome. EMBO J. 73571-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, B., and H. K. Kuramitsu. 2003. Assessment of the utilization of the antisense RNA strategy to identify essential genes in heterologous bacteria. FEMS Microbiol. Lett. 220171-176. [DOI] [PubMed] [Google Scholar]
- 70.Wittinghofer, A., and E. F. Pai. 1991. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem. Sci. 16382-387. [DOI] [PubMed] [Google Scholar]
- 71.Wittinghofer, A., K. Scheffzek, and M. R. Ahmadian. 1997. The interaction of Ras with GTPase-activating proteins. FEBS Lett. 41063-67. [DOI] [PubMed] [Google Scholar]
- 72.Wout, P., K. Pu, S. M. Sullivan, V. Reese, S. Zhou, B. Lin, and J. R. Maddock. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 1865249-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wower, I. K., J. Wower, and R. A. Zimmermann. 1998. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J. Biol. Chem. 27319847-19852. [DOI] [PubMed] [Google Scholar]
- 74.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292883-896. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, S., and W. G. Haldenwang. 2004. Guanine nucleotides stabilize the binding of B. subtilis Obg to ribosomes. Biochem. Biophys. Res. Commun. 322565-569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.