Abstract
We found that insertion sequence (IS) elements are unusually abundant in the relatively recently evolved bacterial endosymbionts of maize weevils. Because multicopy elements can facilitate genomic recombination and deletion, this IS expansion may represent an early stage in the genomic reduction that is common in most ancient endosymbionts.
Mobile genetic elements that spread by transposing within and between genomes are generally considered genomic parasites (4, 17). Although insertion sequence (IS) elements are widespread in bacteria, they are typically rare within individual genomes (15). This rarity has been ascribed to the high efficiency of selection in most bacterial populations (11, 15). Consistent with this view, IS elements are most abundant in bacteria with recently evolved obligately host-restricted lifestyles (1, 15). Increased IS numbers are expected if this lifestyle shift causes many loci to be superfluous or if it decreases the effective population size, thus increasing genetic drift and in turn reducing the effectiveness of natural selection against deleterious transpositions (15). In contrast, IS elements are usually completely absent from ancient intracellular symbionts (1, 15), probably because transposition into or out of these genomes, which is crucial for the long-term spread and maintenance of mobile elements (16), is essentially impossible, and because a deletional mutation bias generally removes extraneous DNA from bacterial chromosomes (13).
The endosymbiotic bacteria living within grain weevils (Sitophilus species) represent an intermediate stage along the evolutionary pathway from free-living bacteria to ancient endosymbionts. On one hand, they are clearly obligate mutualists, provisioning nutrients to their hosts (24, 25), living exclusively within specialized host cells (bacteriocytes) and ovaries, and undergoing strict vertical transmission from mother to offspring (6). On the other hand, they lack many of the genomic hallmarks of very-long-term intimate endosymbionts (23), including a very small genome size (2), extreme A+T richness (7), and an absence of IS elements (3). Furthermore, phylogenetic analysis indicates that grain weevil symbionts colonized their hosts relatively recently, when they replaced a more-ancient symbiont still retained in other weevil groups (9). Because of the transitional status of these symbionts, we wanted to quantify the known ISs within their genomes to better understand the role that transposable elements play in the genomic evolution of long-term intimate endosymbionts. We studied symbionts of two grain weevil species, the Sitophilus oryzae primary endosymbiont, or SOPE, and the Sitophilus zeamais primary endosymbiont, or SZPE.
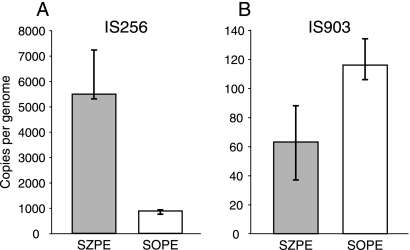
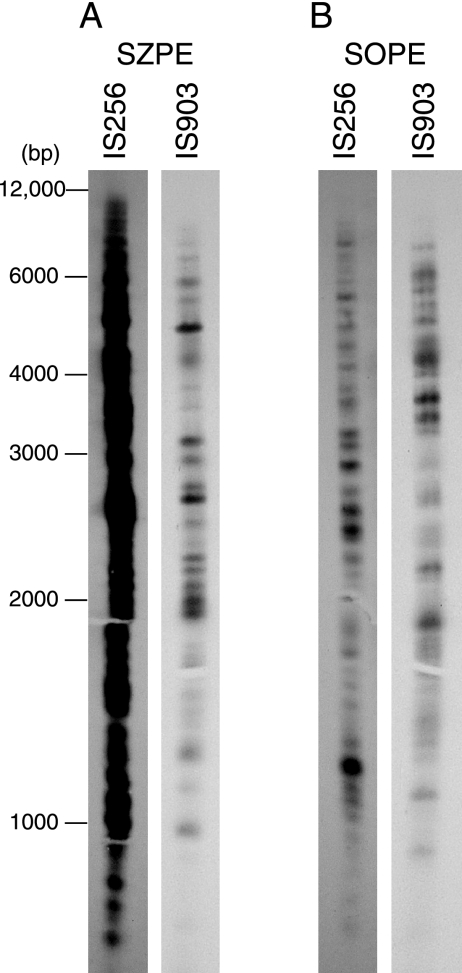
We initially encountered an IS903-like element upstream of panF in SOPE (GenBank accession no. DQ451840), and an IS256-like element was previously encountered interrupting an rRNA operon in SZPE (3). IS256 and IS903 are not present at the above loci in the alternate species, so we screened for and found these elements in each alternate species by cloning and sequencing PCR-amplified products (GenBank accession no. DQ451842 and DQ451851). We then quantified these two IS elements within the SOPE and SZPE genomes by using real-time quantitative PCR (qPCR). We estimated that SZPE has 5,489 (range, 5,300 to 7,228) and 63 (range, 37 to 88) full-length IS256 and IS903 copies, respectively, per single-copy chromosomal gene (murA) (Fig. 1), a value about 10 times greater than that in any known bacterial genome (15, 26). These approximate IS256 and IS903 quantities were confirmed for SZPE by using Southern hybridizations of digested genomic DNA (Fig. 2A). The results for SOPE indicated much lower numbers of IS256 and somewhat higher numbers of IS903 (Fig. 1 and 2B). Further details are in Methods in the supplemental material at http://www.fordham.edu/biology/plague/jbac2008/supplementary_methods, and PCR primers are listed in Table S1 at http://www.fordham.edu/biology/plague/jbac2008/table_s1.
FIG. 1.
Estimated numbers of full-length IS copies per genome for SZPE and SOPE based on qPCR. The bars represent medians, and the vertical lines indicate the ranges of estimates.
FIG. 2.
Southern hybridizations of EcoRV-digested genomic DNA (0.34 μg per lane) with IS256 and IS903 probes. Fragment sizes (in base pairs) are indicated on the left.
To check whether this unexpected result for SZPE might be attributed to a qPCR error, a second IS256 qPCR experiment using a different pair of primers (amplifying a much smaller IS256 fragment) and a different single-copy chromosomal gene (dnaN) yielded comparable IS256 estimates (5,291; range, 4,532 to 6,212). Also, none of these IS256 copies reside in (i) other bacterial inhabitants of the weevil, such as Wolbachia (8), because none were detected in exhaustive rRNA sequencing (3) or in a diagnostic Wolbachia PCR, or (ii) the S. zeamais host genome (or the S. oryzae host genome), because none were detected in PCRs with only weevil DNA. Further details and primers are in the supplemental methods (http://www.fordham.edu/biology/plague/jbac2008/supplementary_methods) and Table S1 (http://www.fordham.edu/biology/plague/jbac2008/table_s1), respectively.
These prolific ISs would produce massive genomic bloating if all were integrated into the SZPE chromosome: IS256 and IS903 together comprise ∼6.8 Mb [(5,489 IS256 copies × 1,223-bp qPCR amplicon size) + (63 IS903 copies × 1,055-bp qPCR amplicon size)]; by comparison, the largest fully sequenced gammaproteobacterial genome in GenBank is 7.2 Mb. We were unable to reliably estimate the SZPE genome size using pulsed-field gel electrophoresis but determined a minimum size of 3.5 Mb (data not shown). An alternate explanation for the large number of IS256 elements might be that many are not integrated into the chromosome but instead are present as circular extrachromosomal transposition intermediates (10, 19). To assess this, we used outward-facing primers within IS256 to amplify, clone, and sequence putative IS256 circles (see Fig. S1 in the supplemental material) (GenBank accession no. DQ451855 to DQ451869). We then used these outward-facing primers in qPCR and found only ∼14 circles per single-copy chromosomal gene (dnaN). However, IS256 circles can contain two IS256 elements if the donor elements are neighbors (19). We would not have quantified any such circles that had opposite-facing IS256 elements in the qPCRs above because the outward-facing primers would not have met, so we used a single outward-facing primer and chromosome-free DNA (to exclude the possibility of amplifying neighboring chromosomal elements) to amplify, clone, and sequence putative double IS256 circles (see Fig. S2 in the supplemental material) (GenBank accession no. EU011831). Although our data do not permit us to fully quantify the extrachromosomal IS256 copies per bacterial chromosome because we used a chromosome-free DNA template in the latter PCRs above, copies on circular transposition intermediates or on plasmids could constitute a majority of the IS256 elements within SZPE. Even if only 10% of the estimated ∼5,000 full-length IS256 elements are chromosomal, the chromosomal IS content in SZPE would be comparable to the highest yet documented for members of the domain Bacteria, that for Shigella species pathogens, which exhibit unusual genomic fluidity due to IS-mediated intragenomic recombination (26). Further details and primers are available in the supplemental methods (http://www.fordham.edu/biology/plague/jbac2008/supplementary_methods) and Table S1 (http://www.fordham.edu/biology/plague/jbac2008/table_s1), respectively.
IS element quantities are elevated in numerous bacteria that have recently evolved host-restricted pathogenic or symbiotic lifestyles and probably proliferate in these taxa for two reasons (15). First, obligate intracellular bacteria have relatively small effective population sizes (12), which decreases the effectiveness of natural selection and results in an increased likelihood of deleterious genetic changes becoming fixed (14, 18). In addition to the accumulation of slightly deleterious IS insertions, this relaxed selection could also result in the debilitation of host genes that control IS element transposition (see references 20 and 22), thus potentially facilitating IS expansion in these taxa. Second, intracellular symbionts are bathed in a generally stable, nutrient-rich environment and do not have to synthesize many of their own required metabolites (5, 21). Therefore, soon after the shift to a symbiotic lifestyle, many biosynthetic and signaling genes may be essentially selectively neutral territory for IS transposition (15).
The abundance of IS elements in SZPE is presumably not an equilibrium state. Indeed, ancient lineages of strictly clonal symbionts lack ISs entirely (1, 15), and mobile elements are typically rare in asexual taxa in general (16). Furthermore, homologous recombination between repeated sequences within a genome can lead to chromosomal inversions and deletions (see references 18 and 26). Therefore, the expansion of multicopy IS elements in recently evolved intracellular symbionts, like SZPE, may represent an initial stage in the genomic degradation and reduction that is characteristic of most ancient endosymbionts.
Nucleotide sequence accession numbers.
The sequence for the IS903-like element upstream of panF in SOPE was deposited in GenBank under accession no. DQ451840. The sequences from the IS256 and IS903 element screening for SOPE and SZPE were deposited in GenBank under accession no. DQ451851 and DQ451842. The sequences from the outward-facing primers within IS256 used to amplify, clone, and sequence putative IS256 circles were deposited in GenBank under accession no. DQ451855 to DQ451869. The sequence from the single outward-facing primer used with chromosome-free DNA to amplify, clone, and sequence putative double IS256 circles was deposited in GenBank under accession no. EU011831.
Supplementary Material
Acknowledgments
We thank B. Bishop, K. Dougherty, and B. Nankivell for technical assistance and C. Dale, K. Dougherty, and H. Ochman for insightful discussions about IS elements and comments on our results.
This work was funded by the Center for Insect Science through NIH training grant no. 1 K12 GM00708 to G.R.P. and by NSF-9978518 and NSF-0313737 to N.A.M.
Footnotes
Published ahead of print on 2 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bordenstein, S. R., and W. S. Reznikoff. 2005. Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 3688-699. [DOI] [PubMed] [Google Scholar]
- 2.Charles, H., G. Condemine, C. Nardon, and P. Nardon. 1997. Genome size characterization of the principal endocellular symbiotic bacteria of the weevil Sitophilus oryzae, using pulsed field gel electrophoresis. Insect Biochem. Mol. Biol. 27345-350. [Google Scholar]
- 3.Dale, C., B. Wang, N. Moran, and H. Ochman. 2003. Loss of DNA recombinational repair enzymes in the initial stages of genome degeneration. Mol. Biol. Evol. 201188-1194. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle, W. F., and C. Sapienza. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284601-603. [DOI] [PubMed] [Google Scholar]
- 5.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, J. L. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J.-F. Tomb, B. A. Dougherty, K. F. Bott, P.-C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270397-403. [DOI] [PubMed] [Google Scholar]
- 6.Heddi, A. 2003. Endosymbiosis in the weevil of the genus Sitophilus: genetic, physiological, and molecular interactions among associated genomes, p. 67-82. In K. Bourtzis and T. A. Miller (ed.), Insect symbiosis. CRC Press, Boca Raton, FL.
- 7.Heddi, A., H. Charles, C. Khatchadourian, G. Bonnot, and P. Nardon. 1998. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G+C content of an endocytobiotic DNA. J. Mol. Evol. 4752-61. [DOI] [PubMed] [Google Scholar]
- 8.Heddi, A., A.-M. Grenier, C. Khatchadourian, H. Charles, and P. Nardon. 1999. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl. Acad. Sci. USA 966814-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lefèvre, C., H. Charles, A. Vallier, B. Delobel, B. Farrell, and A. Heddi. 2004. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol. Biol. Evol. 21965-973. [DOI] [PubMed] [Google Scholar]
- 10.Loessner, I., K. Dietrich, D. Dittrich, J. Hacker, and W. Ziebuhr. 2002. Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J. Bacteriol. 1844709-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch, M., and J. S. Conery. 2003. The origins of genome complexity. Science 3021401-1404. [DOI] [PubMed] [Google Scholar]
- 12.Mira, A., and N. A. Moran. 2002. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb. Ecol. 44137-143. [DOI] [PubMed] [Google Scholar]
- 13.Mira, A., H. Ochman, and N. A. Moran. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17589-596. [DOI] [PubMed] [Google Scholar]
- 14.Moran, N. A. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 932873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran, N. A., and G. R. Plague. 2004. Genomic changes following host restriction in bacteria. Curr. Opin. Genet. Dev. 14627-633. [DOI] [PubMed] [Google Scholar]
- 16.Nuzhdin, S. V., and D. A. Petrov. 2003. Transposable elements in clonal lineages: lethal hangover from sex. Biol. J. Linn. Soc. 7933-41. [Google Scholar]
- 17.Orgel, L. E., and F. H. C. Crick. 1980. Selfish DNA: the ultimate parasite. Nature 284604-607. [DOI] [PubMed] [Google Scholar]
- 18.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeño-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 3532-40. [DOI] [PubMed] [Google Scholar]
- 19.Prudhomme, M., C. Turlan, J.-P. Claverys, and M. Chandler. 2002. Diversity of Tn4001 transposition products: the flanking IS256 elements can form tandem dimers and IS circles. J. Bacteriol. 184433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts, D., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1985. IS10 transposition is regulated by DNA adenine methylation. Cell 43117-130. [DOI] [PubMed] [Google Scholar]
- 21.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 40781-86. [DOI] [PubMed] [Google Scholar]
- 22.Valle, J., M. Vergara-Irigaray, N. Merino, J. R. Penadés, and I. Lasa. 2007. σB regulates IS256-mediated Staphylococcus aureus biofilm phenotypic variation. J. Bacteriol. 1892886-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wernegreen, J. J. 2005. For better or for worse: genomic consequences of intracellular mutualism and parasitism. Curr. Opin. Genet. Develop. 15572-583. [DOI] [PubMed] [Google Scholar]
- 24.Wicker, C. 1983. Differential vitamin and choline requirements of symbiotic and aposymbiotic S. oryzae (Coleoptera: Curculionidae). Comp. Biochem. Physiol. 76A177-182. [Google Scholar]
- 25.Wicker, C., and P. Nardon. 1982. Development responses of symbiotic and aposymbiotic weevils Sitophilus oryze L. (Coleoptera, Curculionidae) to a diet supplemented with aromatic amino acids. J. Insect Physiol. 281021-1024. [Google Scholar]
- 26.Yang, F., J. Yang, X. Zhang, L. Chen, Y. Jiang, Y. Yan, X. Tang, J. Wang, Z. Xiong, J. Dong, Y. Xue, Y. Zhu, X. Xu, L. Sun, S. Chen, H. Nie, J. Peng, J. Xu, Y. Wang, Z. Yuan, Y. Wen, Z. Yao, Y. Shen, B. Qiang, Y. Hou, J. Yu, and Q. Jin. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 336445-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.