Abstract
Gene amplifications have been detected as a transitory phenomenon in bacterial cultures. They are predicted to contribute to rapid adaptation by simultaneously increasing the expression of genes clustered on the chromosome. However, genome amplifications have rarely been described in natural isolates. Through DNA array analysis, we have identified two Streptococcus agalactiae strains carrying tandem genome amplifications: a fourfold amplification of 13.5 kb and a duplication of 92 kb. Both amplifications were located close to the terminus of replication and originated independently from any long repeated sequence. They probably arose in the human host and showed different stabilities, the 13.5-kb amplification being lost at a frequency of 0.003 per generation and the 92-kb tandem duplication at a frequency of 0.035 per generation. The 13.5-kb tandem amplification carried the five genes required for dihydrofolate biosynthesis and led to both trimethoprim (TMP) and sulfonamide (SU) resistance. Resistance to SU probably resulted from the increased synthesis of dihydropteroate synthase, the target of this antibiotic, whereas the amplification of the whole pathway was responsible for TMP resistance. This revealed a new mechanism of resistance to TMP involving an increased dihydrofolate biosynthesis. This is, to our knowledge, the first reported case of naturally occurring antibiotic resistance resulting from genome amplification in bacteria. The low stability of DNA segment amplifications suggests that their role in antibiotic resistance might have been underestimated.
Gene duplications in eukaryotic cells play a major role in the formation of new genes and new functions. Partial genome duplications occur frequently in yeast (17), and complete genome duplications happened in the course of the evolutionary history of the teleostean fish (16) and of the yeast Saccharomyces cerevisiae (23). Recently, a genome-scale analysis of copy number variants in the human genome emphasized their role in genetic diversity and evolution (30). In bacteria, there is no evidence for a role of large genome duplications in genome evolution. A recent analysis of 106 complete bacterial genome sequences showed that most paralogous genes probably arose from horizontal gene transfers or by small-scale duplication events (10).
Transient genome duplications allow a rapid and reversible adaptation by amplifying a set of genes, which results in a higher level of expression. This mechanism plays a major role in the resistance to drugs, by leading to the overexpression of multidrug exporter proteins, as was shown in the resistance of insects to insecticides (7), of malignant cells to anticancer drugs (13), and of Plasmodium falciparum to mefloquine (28). Gene amplification also plays a role in the abnormal differentiation and proliferation of cancer cells (21). In bacteria, duplications of chromosomal regions were shown to arise at a frequency ranging from 10−2 to 10−5 under laboratory conditions (32). It was proposed that tandem duplications contribute to the selection of functional gene clustering by allowing their simultaneous increase of expression (29). Furthermore, the increased copy number leads to increasing the number of targets for adaptive mutations (19). A broad range of experimental setups based on antibiotic or metabolic selective pressure have been devised to select for these duplications, either by increasing gene dosage (8, 14) or by placing promoterless genes under the control of an active promoter (29, 36, 42). Although bacteria encounter such adverse conditions in their natural environment, no antibiotic or antiseptic resistance resulting from chromosomal amplification has been described so far in natural isolates. Tandem duplications are unstable and, once the selective constraint is released, the amplification is rapidly lost by homologous recombination between these long direct repeated DNA regions. Only a few cases of stable gene duplications have been reported in bacteria (4, 18, 27).
Streptococcus agalactiae (group B streptococcus [GBS]) is a leading cause of infection in neonates (6). It is also a serious cause of disease and mortality in elderly and in immunocompromised adults (9). However, S. agalactiae is primarily a commensal bacterium colonizing the digestive and urogenital tracts of 20% of the human population (35). As antibiotic administration to women during labor drastically reduced the incidence of early onset disease in neonates, maternal intrapartum prophylaxis for pregnant women colonized with S. agalactiae has been recommended for several years (34). Penicillin and aminopenicillin constitute the first-line antibiotics; however, in case of allergy to these antibiotics, women would receive a macrolide or clindamycin (33). Resistances to sulfonamide (SU), to trimethoprim (TMP), or to the combination of both antibiotics (sulfamethoxazole-trimethoprim [SXT]) have been poorly characterized as these antibiotics are not recommended to treat S. agalactiae materno-fetal infections. Safe and effective alternatives are available, and adverse effects such as kernicterus in premature infants or hemolysis in neonates who have erythrocyte G6PD deficiency have been described with SU drugs.
In order to characterize the diversity of the species, isolates from various origins, including human and animal and carriage and invasive infections, have been collected. Recently, we have analyzed a collection of 75 isolates by combining multilocus sequence typing, serotyping, comparative genomics by hybridization (CGH), and sequencing of specific loci (2). In the present study, we reanalyzed the complete set of CGH array data for gene amplifications. We identified long genomic amplifications in two strains isolated from humans. These amplifications predate the isolation and probably took place in the human host. In one case, we showed that the amplification of five genes required for tetrahydrofolate biosynthesis led to both SU and TMP resistance. The present study provides new insights into naturally occurring DNA amplifications.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The 75 strains of S. agalactiae used in this work are those analyzed in a recent study (2). S. agalactiae strain CCH290 was isolated in 1998 as a vaginal carrier strain and belongs to sequence type 28 (ST28). Strain NEM969 and strain NEM318 belong to the hypervirulent ST17 and were isolated from cerebrospinal fluid of infected newborns in 1995 and in 1996, respectively. Strain NEM316 belongs to ST23 and was isolated from a fatal septicemia. Strains 61.44.98, p2779.95, and 1346.99 are ST19 strains isolated from rabbit, mare, and horse, respectively. Strains were grown overnight at 37°C in Todd Hewitt (TH) medium (Difco, Detroit, MI). Resistances to penicillin, amoxicillin, cefotaxime, kanamycin, gentamicin, tetracycline, vancomycin, erythromycin, clindamycin, rifampin, SU, TMP, and SXT were tested by the disk diffusion method on Mueller-Hinton (MH) agar according to CLSI recommendations (Clinical and Laboratory Standards Institute) (5). Etest MIC assays (AB Biodisk, Sweden) were performed on MH agar according to the manufacturer's instructions. The breakpoint values for inhibition zone diameters and MICs, respectively, were 12 to 16 mm and 4 to 8 mg/liter for TMP, 12 to 17 mm and 64 to 256 mg/liter for SU, and 15 to 19 mm and 0.5/9.5 to 4/76 mg/liter for SXT, according to the CLSI recommendations.
PCR amplification, sequence analysis, and quantitative real-time PCR.
Genomic DNA of S. agalactiae used as template for PCRs was extracted by using a DNeasy kit (Qiagen, Hilden, Germany). Sequencing reactions were performed on PCR products using a Taq BigDye terminator cycle sequencing kit and an ABI 3700 capillary DNA sequencer (Applied Biosystems, Foster City, CA). The primers used in this study are described in Table 1. The recA and dfrA genes were sequenced using primers OrecA-r, OrecA-l, OrecA-2r, OdfrA-l, and OdfrA-r. Oligonucleotides O1185-r, O1185-l, O1171-r, and O1171-l were used to analyze amplification junctions in strain CCH290. Oligonucleotides O1033-r, O1033-l, O1158-r, and O1158-l were used to analyze amplification junctions in strain NEM969. RNA concentrations were determined by real-time fluorescent-detection quantitative PCR. RNA and cDNA were prepared as previously described (20). cDNAs were amplified by using an ABI PRISM 7900 sequence detection system and SYBR green PCR kits (Applied Biosystems, Foster City, CA). All measurements were performed at least in duplicate. Only primer pairs with an efficiency of >90% were retained for further experiments. Quantitative reverse transcription-PCR was performed with primers specific for folE, folP, folK, and gyrA (Table 1). The quantity of specific cDNA was normalized to the quantity of gyrA cDNA in each sample. The relative levels of gene expression were determined by using the 2−ΔΔCT method (24).
TABLE 1.
Oligonucleotides used in this study
| Primer(s) | Sequence(s) (5′→3′) |
|---|---|
| O1185-l and O1185-r | ACTGAGGCAAGTCTTGGTT and ACAATAATTGGATGGTTGGA |
| O1171-l and O1171-r | GGTGGAGTTTTATCGTTTC and CCATTGTTTTCACAGGAGAT |
| O1033-l and O1033-r | CCTTTTTCGAAGTACAATG and AGAAAGAAGCTGGAATGACA |
| O1158-l and O1158-r | ATAGCGTTATTGGTTGTGC and GAAAATTCTGTCCCTTCACA |
| O1175-l and O1175-r | CGTTTGACTCTAATGAGGC and AGAATGTAACACCAATCG |
| OdfrA-l and OdfrA-r | TTCCAGATGGAACAGACTT and AAAGGCATGGATTGTGTTGA |
| OfolE-l and OfolE-r | TCATGGTGCTTTTTCGGAA and TCAGTTTGTGATGCTTTTGCT |
| OfolP-l and OfolP-r | CAGCAGAACGAAATGTTCC and AAAGCACGCTCTGCTCTTTC |
| OfolK-l and OfolK-r | TGGCAATAGCTGTCCAGTT and AACCGCTAAAGCCTTGAACA |
| OgyrA-l and OgyrA-r | CACAATGGTGGTCATATCG and TCCCCATCCATTGAACCAA |
| OrecA-l and OrecA-r | ACAAAAGCTTCACGGAAT and TTTAACGGGAGTTGCTGGTC |
| OrecA-2r | CACAAAAATCAAACATGGG |
Pulsed-field gel electrophoresis (PFGE), DNA transfer, labeling, and hybridization.
Agarose plugs were prepared, and BamHI restriction endonuclease digestion performed as described previously (3). From the genome sequences of the eight S. agalactiae strains, it was expected that the amplified region in strain CCH290 would not contain any recognition site for this enzyme (12, 40, 41). Agarose gels were made in 0.5× TBE buffer (45 mM Tris-borate-1 mM EDTA). An electrophoretic regime of 6 V/cm for 24 h at 16°C and a switching time from 1 to 8 s were used to resolve DNA digests. Fractionated DNA restriction fragments were transferred to a positively charged nylon membrane by the Southern method. The probe, specific to gbs1175, was synthesized using oligonucleotides O1175-l and O1175-r (Table 1). Labeling, hybridization, and detection were performed as previously described (2).
DNA array data analysis.
DNA array hybridizations carried out on an NEM316 whole-genome array carrying 1,981 probes for 75 S. agalactiae strains (2) were reanalyzed for the detection of duplicated regions. Each strain was compared to the NEM316 reference strain, and ratios higher than 1.75 were considered to correspond either to duplicated genes or to the extreme values of the Gaussian distribution of ratios centered at 1 for genes present in a single copy. After visual analysis of each strain, two were identified as having series of more than five genes with ratios higher than 1.75.
Determination of the frequencies of duplication loss.
In order to get an estimate of the frequency of recombination between duplications, strains CCH290 and NEM969 were submitted to serial passages on solid or in liquid TH medium. The presence of the tandem duplication was ascertained by PCR amplification of the junction sequence. To quantify duplication loss on solid medium, isolated colonies were streaked on TH agar. Colonies were inoculated in 2 ml of TH for DNA extraction, and the genomic organization in the founding bacterial colony and in streaked colonies was determined by PCR analysis. For strain CCH290, no recombined clone was detected among 96 colonies obtained after subcultures on TH agar. In order to enrich them in cells having lost the duplication by recombination, four CCH290 strain cultures were submitted to serial dilutions in TH broth. The culture was diluted (1:1,000, representing 10 generations) twice a day into 10 ml. Recombined clones were screened by PCR amplification of the junction after 10 dilutions, representing 100 generations.
To calculate the frequency of recombination, we estimated that the growth parameters for the recombined strain and the parental strain were not significantly different, as observed by growth in liquid culture. The distribution of the two populations follows a branching process model. At each generation, a fraction, p, of the population will recombine and will irreversibly lose the duplication, or, at each generation, the probability of conserving the duplication is 1 − p. After n generations, the proportion of cells with the duplication is expected to be (1 − p)n.
RESULTS
Identification of two long gene amplifications among S. agalactiae isolates.
By analyzing CGH array data from 75 S. agalactiae isolates, we identified two strains with several consecutive genes on the chromosome with probe ratios higher than 1.75, ST28 strain CCH290 and ST17 strain NEM969. Twenty-two hybridization ratios from strain CCH290 were higher than 1.75. Among these genes, 14 were consecutive. For these 14 probes, corresponding to genes gbs1172 to gbs1185, the hybridization ratios ranged from 2.8 to 5.2, indicating a likely multimerization of the region (Table 2).
TABLE 2.
Amplification of the gbs1171-gbs1185 region in strain CCH290
| Gene | Description of product | Hybridization ratioa for strain:
|
|||
|---|---|---|---|---|---|
| CCH290 | 61.44.98 | p2779.99 | 1346.99 | ||
| gbs1171 | Conserved hypothetical protein | 1.7 | 1.2 | 0.9 | 0.9 |
| gbs1172 | Putative hydrolase/phosphatase | 5.2 | 1.1 | 1.0 | 0.9 |
| gbs1173 | Transcriptional regulator (AraC/XylS family) | 4.5 | 0.9 | 0.7 | 0.6 |
| gbs1174 | Putative channel transporter | 3.9 | 0.9 | 0.9 | 0.9 |
| gbs1175 | Spermidine/putrescine ABC transporter (binding protein) | 4.4 | 1.1 | 1.0 | 1.1 |
| gbs1176 | Spermidine/putrescine ABC transporter (permease) | 3.2 | 1.1 | 1.2 | 1.2 |
| gbs1177 | Spermidine/putrescine ABC transporter (permease) | 2.9 | 1.1 | 1.4 | 1.5 |
| gbs1178 | Spermidine/putrescine ABC transporter (ATP-binding protein) | 2.8 | 0.9 | 1.0 | 1.1 |
| gbs1179 (murB) | UDP-N-acetylenolpyruvoylglucosamine reductase | 4.0 | 1.1 | 1.3 | 1.6 |
| gbs1180 (folC) | Hydroxymethylpterin pyrophosphokinase | 3.6 | 1.2 | 0.8 | 1.0 |
| gbs1181 (folE) | Dihydroneopterin aldolase | 3.7 | 1.0 | 1.1 | 1.4 |
| gbs1182 (folP) | Dihydropteroate synthase | 4.2 | 1.2 | 1.1 | 1.2 |
| gbs1183 (folB) | GTP cyclohydrolase | 4.3 | 1.1 | 0.8 | 0.8 |
| gbs1184 (folK) | Folyl-polyglutamate synthetase | 3.6 | 1.0 | 1.0 | 0.7 |
| gbs1185 | Conserved hypothetical protein | 3.1 | 0.6 | 0.7 | 0.6 |
Strain NEM316 was used as the reference.
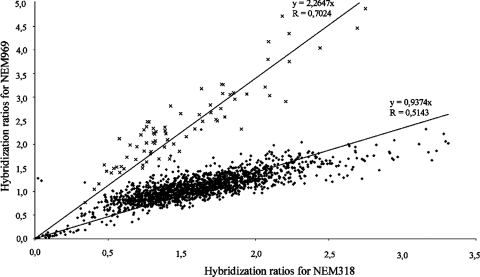
For strain NEM969, when compared to the results for the closely related ST17 strain NEM318, we observed ratios higher than 1.75 for 100 genes (Fig. 1). Of these, 78 are clustered on the NEM316 genome from gbs1033 to gbs1158. The average ratio of 2.3 for these 78 genes (Fig. 1) suggested a duplication of this chromosomal region in strain NEM969. In strain NEM316, this region encompasses two previously described genomic islands, which are different in strain NEM969, as 48 out of the 126 genes annotated in this chromosomal region in strain NEM316 were predicted as missing in strain NEM969 by CGH (2). In order to estimate the size of the duplicated region in strain NEM969, we analyzed the unfinished genome sequence of the closely related ST17 strain COH1 (40). This region in strain COH1 contains 11 genes which are missing in NEM316. These genes are likely present in NEM969, and the size of the duplicated region was therefore estimated to be 92 kb long and to carry 89 genes.
FIG. 1.
Dot matrix of normalized DNA array hybridization signals for probes corresponding to genes identified as present by CGH array in strain NEM969 and in the closely related ST17 strain NEM318. Crosses correspond to genes gbs1033-gbs1158 and dots to the remaining probes. Regression curve and correlation coefficient are indicated for both groups.
Amplifications are tandem duplications and did not involve long direct repeats.
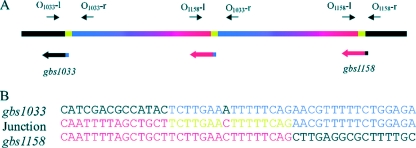
The analysis of the CGH array data provided a first estimate of the extremities of the duplicated regions: gbs1172 to gbs1185 in strain CCH290 and gbs1033 to gbs1158 in strain NEM969. DNA segment duplications in bacteria frequently correspond to tandem duplications involving direct repeats, like insertion sequences or rRNA operons (32). However, no significant long direct repeat was identified when analyzing the regions corresponding to the extremities of the amplified regions in NEM316. To test whether these regions are duplicated in tandem, we characterized the endpoints of these regions by PCR. We used divergent primers corresponding to the estimated extremities of the amplified regions: O1171-r and O1185-l for strain CCH290 and O1033-l and O1158-r for strain NEM969 (Fig. 2A and 3A). For both strains, the pairs of oligonucleotides allowed PCR amplifications of the junctions in agreement with tandem amplification of the region. The corresponding PCR products were sequenced (Fig. 2B and 3B). In strain CCH290, the amplification took place between DNA regions located within the coding sequences of gbs1171 and gbs1185. The analysis of both extremities of the amplified region did not reveal any significant sequence similarity, as only the dinucleotide AA was repeated. In strain NEM969, the duplication took place between DNA regions located in the 5′ part of the coding sequence of gbs1032 and in the 3′ part of the coding sequence of gbs1158. Alignment of the junction sequences with the sequences of both extremities of the duplicated region showed that the primary event in the duplication involved a short direct repeat of 16 bp with a single mismatch, TCTTGAA(A/C)TTTTTCAG.
FIG. 2.
(A) Schematic map of the amplified gbs1171-gbs1185 region in strain CCH290. Blue and red segments represent the fourfold-amplified DNA region. O1171-l, O1171-r, O1185-l, and O1185-r represent oligonucleotides used to amplify the extremities and the junction of the amplified DNA segment. (B) Nucleotide sequences of the junction site and of the right and left extremities (gbs1171 and gbs1185, respectively).
FIG. 3.
(A) Schematic map of the amplified gbs1033-gbs1158 region in strain NEM969. The blue and red segments represent the amplified DNA region. The green segment indicates the short repeat sequence at the junction of the duplication. O1033-l, O1033-r, O1158-l, and O1158-r represent oligonucleotides used to sequence the extremities and the junction of the amplified DNA segment. (B) Nucleotide sequences of the junction site and of the right and left extremities (gbs1033 and gbs1158, respectively).
To confirm the genetic organization of the amplification in strain CCH290, the gbs1171-gbs1185 region was analyzed by PFGE (Fig. 4A) and Southern blot hybridization (Fig. 4B). Strain CCH290 was compared to the sequenced strain NEM316 and to CCH290-derived strains having lost the amplification (see below and Fig. 4). Genomic DNAs were digested by the restriction enzyme BamHI. Hybridization with a probe specific to gbs1175 enabled the detection of a 29.5-kb band in strain NEM316, a single 68-kb band in strain CCH290, and a 28-kb band in recombined derivatives (Fig. 4B). The size of the hybridizing band in CCH290 corresponds to three additional tandem repetitions of the 13.5-kb-long region, in agreement with the DNA array hybridization ratios (Table 2). The absence of a signal corresponding to the size of the band detected in strains having lost the duplication or of bands with intermediate molecular weights indicates that tandem duplications in strain CCH290 are likely stable.
FIG. 4.
(A) PFGE analysis of BamHI-restricted genomic DNA of strains NEM316, CCH290, and CCH290-1, -2, and -3. Bacteriophage λ concatemers were used as molecular size markers and are shown in the first lane. The positions and sizes in kb are indicated on the left. (B) Southern analysis of PFGE gel showed in panel A. A 510-bp PCR product corresponding to the internal part of gbs1175 was used as a probe. Strain CCH290 harbors a fourfold DNA segment (from gbs1171 to gbs1185) leading to SU and TMP resistance. CCH290-1 and CCH290-2 correspond to CCH290 recombinant strains that have lost the DNA amplification after 10 passages. Strain CCH290-3 corresponds to a CCH290 strain isolated after 10 passages that retained the DNA amplification.
Gene amplification in strain CCH290 led to SU and TMP resistance. In strain CCH290, among the 13 genes carried by the amplified genomic region, five organized in an operon encode enzymes involved in folate biosynthesis: folCEPBK (Table 2). The amplification of the folate biosynthesis operon and especially of the folP gene was expected to lead to an increased resistance to SU (25), the dihydropteroate synthase encoded by folP being the target of this antibiotic (31). Antibiograms performed on CCH290 and NEM316 strains revealed that CCH290 was resistant to SU and, surprisingly, also to TMP in comparison to NEM316, which was susceptible to these antibiotics (Table 3).
TABLE 3.
Antibiotic sensitivity and expression level of folEPK genes for wild-type and recombined strains
| Strain | Copy no. | Druga
|
Expression level (fold)b of:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| TMP
|
Su
|
SXT ZD (mm) | |||||||
| MIC (mg/liter) | ZD (mm) | MIC (mg/liter) | ZD (mm) | ||||||
| folE | folP | folK | |||||||
| NEM316 | 1 | 8 | 20 | 0.064 | 30 | 27 | 0.2 | 0.2 | 0.2 |
| CCH290 | 4 | >32 | 6 | 0.38 | 13 | 22 | 1 | 1 | 1 |
| CCH290-1 | 1 | 5 | 20 | 0.094 | 28 | 27 | 0.2 | 0.2 | 0.2 |
| CCH290-2 | 1 | 6 | 20 | 0.094 | 27 | 27 | 0.2 | 0.2 | 0.2 |
| CCH290-3 | 4 | >32 | 6 | 0.38 | 16 | 23 | 0.9 | 1 | 1 |
| 61.44.98 | 1 | 4 | 16 | 0.125 | 24 | 22 | ND | ND | ND |
| p2779.95 | 1 | 8 | 15 | 0.25 | 12 | 22 | ND | ND | ND |
| 1346.99 | 1 | 12 | 14 | 0.25 | 17 | 18 | ND | ND | ND |
ZD, inhibition zone diameter.
Determined by quantitative PCR with strain CCH290 used as the reference. ND, not determined.
SU-insensitive dihydropteroate synthase is commonly responsible for resistance to this antibiotic in bacteria (15). As the observed resistance in CCH290 could be due to both amplification and mutation of the folP gene, we sequenced it after PCR amplification. The sequence was identical to that of folP from NEM316, and no peak heterogeneity was observed on the fluorograms. As the PCR product is a mixture of the amplification of the four folP genes, this result indicated that none of them was mutated and suggested that resistance to SU was likely due to the amplification.
Two main resistance mechanisms to TMP by chromosomal mutations have been described. Resistance results either from mutations in the promoter region leading to the overproduction of the TMP target, dihydrofolate reductase (DHFR), or from mutations in its structural gene, dfrA, making DHFR insensitive to TMP (15). In order to rule out these possibilities, we PCR amplified and sequenced dfrA and its promoter region in strain CCH290. No difference from the sequence of the dfrA gene in strain NEM316 was observed, demonstrating that the observed resistance was neither due to a TMP-resistant DHFR nor to a mutation in the dfrA promoter region in strain CCH290.
However, both SU and TMP resistance could also be due to the acquisition by horizontal gene transfer of genes encoding drug-insensitive dihydropteroate synthase and DHFR, respectively (37). In most cases, these genes are carried by plasmids and confer high-level resistance to these antibiotics. However, they have up to now never been described in the Streptococceae. To prove that the amplification was actually responsible for SU and TMP resistance, we searched for derivatives of strain CCH290 having lost the amplifications. In a first attempt to isolate bacteria that had lost the duplication, 96 isolated colonies were tested for the presence of the duplication junction by PCR. All had retained the amplification, in agreement with the stability observed by the Southern blot experiment. In order to enrich them in cells having lost the amplification by recombination, four independent clones of CCH290 were submitted to serial passages in liquid TH medium. After 10 passages, corresponding to 100 generations, cultures were plated on TH plates and 20 colonies per clone were tested by PCR for the presence of the duplication junction. Among the four strains, 4, 5, 5, and 7 colonies out of the 20 (on average, 26%) lost the duplication junction. The recombined strains were compared to the parental strain for growth in TH medium. Loss of the tandem amplification did not alter growth characteristics compared to those of the parental strain (data not shown).
The levels of TMP resistance of 10 colonies that had lost the junction sequence were compared to that of the parental strain CCH290. The derivatives that had lost the tandem amplifications became sensitive to TMP, as exemplified by strains CCH290-1 and -2 (Table 3) in comparison to the parental strain CCH290 or to strains submitted to 10 passages but retaining the amplification (CCH290-3). PFGE and Southern blot hybridization confirmed that strain CCH290-3 had retained the four copies, while strains CCH290-1 and -2 had only one copy (Fig. 4B).
The five genes involved in folate biosynthesis, folCEPBK, are the last genes of an operon of eight genes (gbs1187-gbs1180). The tandem amplifications placed the three extra copies of the folCEPBK genes under the control of the gbs1171 promoter. We compared the levels of expression of folC, folP, and folB in strain CCH290; in two recombined strains, CCH290-1 and CCH290-2; and in strain NEM316 by quantitative reverse transcription-PCR (Table 3). This analysis revealed that the amplification in CCH290 led to a fivefold-higher accumulation of the corresponding transcript and that the loss of amplification was associated with a reversion to the basal level of the transcript in the reference strain NEM316 (Table 3). The correlation between the amplification and the expression levels of the folate biosynthesis genes and the level of resistance to TMP and SU clearly demonstrated that the amplification caused the resistance to both antibiotics.
Few studies deal with resistance of S. agalactiae to SU or TMP. In order to evaluate the incidence of TMP and SU resistance among S. agalactiae strains, antibiograms to determine TMP and SU susceptibility were performed on the 75 S. agalactiae strains. We observed that only CCH290 was resistant to such levels of TMP and SU. Only three ST19 strains, of animal origin, showed a reduced susceptibility (Table 3), whereas the 71 other strains were as sensitive as NEM316 to these antibiotics (data not shown). These three ST19 strains did not show any amplification for gbs1172-gbs1185, as predicted from the analysis of the DNA array hybridizations (Table 2), and the resistance mechanisms are likely to be different than gene amplification.
Chromosomal duplication is rapidly lost in strain NEM969.
The amplified region in strain NEM969 is much longer as it carried at least 89 genes. Several loci could explain the in vivo selective advantage brought by this duplication, such as gbs1042-gbs1045, predicted to encode a ferrichrome ABC transporter, or gbs1155, predicted to encode a multidrug export protein. We also identified two gene product functions related to folate and thymidine metabolism: the thymidine kinase, encoded by gbs1110, and the formate-tetrahydrofolate ligase, encoded by gbs1089.
In order to compare the parental strain and derivatives having lost the duplication, we first tested the stability of the duplication. Analysis of isolated colonies from the original stab culture showed that only 80% of the colonies carried this duplication. In order to quantify the stability of the duplication, six colonies that had retained the duplication were streaked on TH agar. Twelve isolated colonies derived from these clones were then tested by PCR for the presence of the duplication junction. The duplication was retained, on average, in 64% of the colonies, with a broad distribution of values among the six colonies tested (from 25% to 92%). To calculate the frequency of recombination, we determined the number of generations leading to a colony. In our growth conditions, the average CFU number for five colonies was 5.107 CFU, corresponding to 25 generations. For strain NEM969, the frequency of recombination at each generation was estimated to be 3.5%. The average hybridization ratio of 2.3 for probes in the gbs1033-gbs1158 region was obtained by using a culture of strain NEM969 containing a significant proportion of recombined cells. This ratio is therefore an underestimation of the number of gene copies. We had first hypothesized that this region was duplicated in strain NEM969; however, it is likely that the number of repetitions in the original strain was higher.
To assess a possible impact of this amplification on antibiotic susceptibility, two recombined derivatives were compared to the parental strain by growing it directly from the original stock. Although this culture contained a mixture of the original strain and of derivatives having lost the duplication, it was expected that, if the duplication was to lead to antibiotic resistance, it would be detected even in a mixed culture. However, no significant difference was observed for the antibiotics tested (data not shown).
DISCUSSION
Early genetic evidence has shown that gene duplications frequently arise in bacterial populations (reviewed in reference 32). In natural environments, gene amplification is considered to be a rapid and reversible means of adaptation to adverse conditions, and it has been proposed that various stresses induce this phenomenon (38). Different metabolic- or antibiotic-based selective pressures have been used in laboratory conditions to force the maintenance of these amplifications (1, 8, 14, 26, 29). In Escherichia coli, ampicillin-resistant mutants resulting from the amplification of the ampC gene, coding for a beta-lactamase, have been isolated in laboratory conditions (8). However, ampC amplification has not been reported in natural ampicillin-resistant E. coli isolates. Bacillus Calmette-Guérin (BCG) is an example of tandem duplications arising during in vitro growth. BCG was derived from a Mycobacterium bovis strain by serial passages for 13 years on potato slices imbibed with glycerol. The BCG genome was shown to contain two duplications, of 30 and 36 kb (4). Amplification of the glycerol-3-phosphate dehydrogenase gene in one of the duplicated regions may have brought a selective advantage on the medium used for culture. DNA amplifications were also described in archival Salmonella enterica strains stored over 40 years in stabbed cultures (27). However, probably due to their instability, tandem duplications have rarely been described in natural isolates. The duplication of genes involved in capsule production in serotype b strains of Haemophilus influenzae is stabilized by a 1.2-kb deletion at one end of the duplication (18). This deletion removed most of one copy of the bexA gene which encodes the polysaccharide export protein; this made the capsulation dependent on the amplification. In this work we describe two strains of S. agalactiae with long chromosomal tandem amplifications: a fourfold amplification of 13.5 kb in strain CCH290 and a duplication of 92 kb in strain NEM969. In both cases, strains were rapidly stored frozen. It is therefore very likely that the amplifications predate the isolation and took place in the human host.
In strain CCH290, the amplified region contains the folCEPBK genes. These five genes encode proteins catalyzing the different steps of the biosynthesis of dihydrofolate from GTP, para-aminobenzoate, and glutamate. TMP and SU are two antibiotics targeting this pathway. SU, an analog of para-aminobenzoate, is a competitive inhibitor of dihydropteroate synthase, and TMP is an inhibitor of DHFR. Among the 15 antibiotics tested, strain CCH290 was resistant to SU and TMP compared to the resistance of strain NEM316. We have demonstrated that the gene amplification is responsible for the resistance to both antibiotics. Antibiotic resistance has frequently been used to select for tandem duplications under laboratory conditions (8, 26). However, this observation constitutes, to our knowledge, the first characterization of an in vivo-selected amplification leading to antibiotic resistance.
In S. agalactiae CCH290, the amplification of this genomic region led to a fivefold increase in the expression of five genes involved in dihydrofolate biosynthesis, folCEPBK. The resistance to SU could be accounted for by the increased synthesis of dihydropteroate synthase, the target of SU. However, all reported cases of resistance to this antibiotic result from mutations rendering the dihydropteroate synthase insensitive to SU or from the transfer of a gene encoding an insensitive version of this enzyme (37). To our knowledge, SU resistance due to the overexpression of dihydropteroate synthase has not been reported. We have verified that none of the four copies of the folP gene was mutated in strain CCH290. Therefore, it is possible that the increased expression of the whole pathway, and not only of the dihydropteroate synthase, is required to confer SU resistance.
Resistance to TMP has been described as resulting from mutations in the dfrA gene, from the overexpression of this gene, or from the acquisition by horizontal gene transfer of genes encoding drug-insensitive enzymes (37). Increased expression of the folCEPBK genes was not expected to lead to TMP resistance. We speculate that the amplification of the complete biosynthetic pathway leads to an increased intracellular concentration of dihydrofolate. This increased concentration could counteract the effect of TMP, a competitive inhibitor of DHFR (39). Alternatively, the intracellular concentration of dihydrofolate might reach a level at which enzymes with low DHFR reductase activity and insensitivity to TMP are sufficiently active to recycle the dihydrofolate pool and to sustain bacterial growth. Indeed, it has been shown that short-chain dehydrogenases, such as FolM in E. coli (11) or the dihydropteroate synthase from Helicobacter pylori, have TMP-independent DHFR activity (22). In agreement with this model, strain CCH290 is only weakly resistant to the combination of both antibiotics (SXT) (Table 3), SU decreasing the synthesis of dihydrofolate and rendering the strain sensitive to TMP. Therefore, this study led to the prediction of a new mechanism of TMP resistance where the concentration of the enzyme substrate is increased.
Such a mechanism of resistance to TMP and SU by gene amplification requires the clustering of the genes involved in the dihydrofolate biosynthesis pathway. To evaluate the possibility that such amplifications would take place in other clinically relevant pathogens, we analyzed complete bacterial genomes for the location of these five genes along published bacterial chromosomes. This clustering is only found in the genus Streptococcus and in the closely related firmicute genera Lactococcus and Lactobacillus. These five genes are not clustered in Staphylococcus, Listeria, Bacillus, or Enterococcus species. The rare occurrence of this gene clustering may explain why such amplification has never been reported before. Few data exist regarding the susceptibility of S. agalactiae to SU, TMP, and SXT. Because more-effective antimicrobial agents exist, these antibiotics are not considered drugs of choice for the treatment of patients with S. agalactiae infections. However, these antibiotics, and primarily SXT, are still used against other infections, like urinary tract infections, and as prophylaxis for Pneumocystis carinii infections in AIDS patients (15). We may therefore hypothesize that the amplification in strain CCH290 was selected by such a previous treatment.
Tandem duplications were first described as resulting from long direct repeats, such as copies of rrn operons or insertion sequences, and were assumed to arise by unequal RecA-dependent crossing-over between these direct repeats (32). However, a large number of studies reported the occurrence of tandem duplications independently of long direct repeats, instead involving short repeats, as short as 12 bp (8) or even 7 bp (36). Regarding both above-studied amplifications in natural isolates of S. agalactiae, one involved a 16-bp imperfect repeat and the other a 2-bp repeat. Although only two amplifications were analyzed, they are in both cases independent of long repeats, indicating that this mechanism is probably significant in natural conditions. The two duplications are nearby on the chromosome, not overlapping but separated by only 11.2 kb. Both are on the left replichore close to the terminus of replication and of the dif-like site (from 1075 kb to 1204 kb and 1215 kb and 1229 kb on the NEM316 chromosome, respectively). Although this close proximity may be accidental, it is also possible that recombination events in the course of replication termination and/or inefficient replication termination on one replichore may favor the occurrence of tandem duplication in this part of the chromosome.
Although the generation of tandem duplications has been extensively studied, the recombination events leading to the loss of the duplication have not been analyzed. This process is assumed to be RecA dependent. We observed different levels of stability between both analyzed strains. Duplication was lost at high frequency in strain NEM969: within a colony, about one-third of the bacteria lost the amplification. This high instability, estimated to be 3.5% per generation on solid media, probably led to an underestimate of the number of amplifications in the original strain. In strain CCH290, as no recombinant was identified among 96 clones, the frequency of recombination in colonies was estimated to be lower than 0.04%, i.e., 100 times less than in strain NEM969. However quantification of recombination in liquid culture led to a 10-times-higher value of 0.35%. The difference in stability between both strains is not due to a difference in the recA gene itself, since this latter is not altered in any strain (data not shown). Therefore, the difference in stability of the duplication between both strains is probably related to a difference in duplication size (13.5 kb versus 92 kb) in the number of repeats or in intrinsic recombination properties of both strains. Alternatively, the tandem duplications may bring a subtle selective advantage in the genetic background of strain CCH290.
Conclusion.
The description of tandem amplifications occurring in natural bacterial isolates is important in order to evaluate the exact role of genomic amplifications in the adaptation and evolution of bacteria in natural environments. The high throughput genome analysis of natural isolates by DNA arrays is an effective means of identifying such genomic amplifications. The rapid-frozen conservation of natural isolates after isolation is needed to ascertain that the duplication took place in the natural environment and to avoid the loss of the amplification. By analyzing CGH array data on rapidly frozen isolates, we have identified a 13.5-kb amplification leading to TMP and SU resistance. The low stability of genome amplification in vitro may explain why such amplifications might have been overlooked and their role in antibiotic resistance in the patient underestimated. Therefore, the possible role of tandem amplification in the failure of antibiotic treatment of infections deserves to be taken into account in cases where strains were identified as sensitive in vitro.
Acknowledgments
This work was supported by the Institut Pasteur: GPH9 and INSERM. M.B. is supported by a grant from the Ministère délégué à l'enseignement supérieur et à la recherche (France).
We wish to thank Carmen Buchrieser, Patrick Trieu-Cuot and Frank Kunst for their interest and critical comments.
Footnotes
Published ahead of print on 16 November 2007.
REFERENCES
- 1.Anderson, R. P., and J. R. Roth. 1978. Tandem genetic duplications in Salmonella typhimurium: amplification of the histidine operon. J. Mol. Biol. 12653-71. [DOI] [PubMed] [Google Scholar]
- 2.Brochet, M., E. Couve, M. Zouine, T. Vallaeys, C. Rusniok, M. C. Lamy, C. Buchrieser, P. Trieu-Cuot, F. Kunst, C. Poyart, and P. Glaser. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 81227-1243. [DOI] [PubMed] [Google Scholar]
- 3.Brosch, R., C. Buchrieser, and J. Rocourt. 1991. Subtyping of Listeria monocytogenes serovar 4b by use of low-frequency-cleavage restriction endonucleases and pulsed-field gel electrophoresis. Res. Microbiol. 142667-675. [DOI] [PubMed] [Google Scholar]
- 4.Brosch, R., S. V. Gordon, T. Garnier, K. Eiglmeier, W. Frigui, P. Valenti, S. Dos Santos, S. Duthoy, C. Lacroix, C. Garcia-Pelayo, J. K. Inwald, P. Golby, J. N. Garcia, R. G. Hewinson, M. A. Behr, M. A. Quail, C. Churcher, B. G. Barrell, J. Parkhill, and S. T. Cole. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. USA 1045596-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Dermer, P., C. Lee, J. Eggert, and B. Few. 2004. A history of neonatal group B streptococcus with its related morbidity and mortality rates in the United States. J. Pediatr. Nurs. 19357-363. [DOI] [PubMed] [Google Scholar]
- 7.Devonshire, A. L., and L. M. Field. 1991. Gene amplification and insecticide resistance. Annu. Rev. Entomol. 361-23. [DOI] [PubMed] [Google Scholar]
- 8.Edlund, T., and S. Normark. 1981. Recombination between short DNA homologies causes tandem duplication. Nature 292269-271. [DOI] [PubMed] [Google Scholar]
- 9.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33556-561. [DOI] [PubMed] [Google Scholar]
- 10.Gevers, D., K. Vandepoele, C. Simillon, and Y. Van de Peer. 2004. Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends Microbiol. 12148-154. [DOI] [PubMed] [Google Scholar]
- 11.Giladi, M., N. Altman-Price, I. Levin, L. Levy, and M. Mevarech. 2003. FolM, a new chromosomally encoded dihydrofolate reductase in Escherichia coli. J. Bacteriol. 1857015-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 451499-1513. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, M. M., C. A. Hrycyna, P. V. Schoenlein, U. A. Germann, and I. Pastan. 1995. Genetic analysis of the multidrug transporter. Annu. Rev. Genet. 29607-649. [DOI] [PubMed] [Google Scholar]
- 14.Hastings, P. J., H. J. Bull, J. R. Klump, and S. M. Rosenberg. 2000. Adaptive amplification: an inducible chromosomal instability mechanism. Cell 103723-731. [DOI] [PubMed] [Google Scholar]
- 15.Huovinen, P. 2001. Resistance to trimethoprim-sulfamethoxazole. Clin. Infect. Dis. 321608-1614. [DOI] [PubMed] [Google Scholar]
- 16.Jaillon, O., J. M. Aury, F. Brunet, J. L. Petit, N. Stange-Thomann, E. Mauceli, L. Bouneau, C. Fischer, C. Ozouf-Costaz, A. Bernot, S. Nicaud, D. Jaffe, S. Fisher, G. Lutfalla, C. Dossat, B. Segurens, C. Dasilva, M. Salanoubat, M. Levy, N. Boudet, S. Castellano, V. Anthouard, C. Jubin, V. Castelli, M. Katinka, B. Vacherie, C. Biemont, Z. Skalli, L. Cattolico, J. Poulain, V. De Berardinis, C. Cruaud, S. Duprat, P. Brottier, J. P. Coutanceau, J. Gouzy, G. Parra, G. Lardier, C. Chapple, K. J. McKernan, P. McEwan, S. Bosak, M. Kellis, J. N. Volff, R. Guigo, M. C. Zody, J. Mesirov, K. Lindblad-Toh, B. Birren, C. Nusbaum, D. Kahn, M. Robinson-Rechavi, V. Laudet, V. Schachter, F. Quetier, W. Saurin, C. Scarpelli, P. Wincker, E. S. Lander, J. Weissenbach, and H. Roest Crollius. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431946-957. [DOI] [PubMed] [Google Scholar]
- 17.Koszul, R., B. Dujon, and G. Fischer. 2006. Stability of large segmental duplications in the yeast genome. Genetics 1722211-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroll, J. S., B. M. Loynds, and E. R. Moxon. 1991. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol. Microbiol. 51549-1560. [DOI] [PubMed] [Google Scholar]
- 19.Kugelberg, E., E. Kofoid, A. B. Reams, D. I. Andersson, and J. R. Roth. 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc. Natl. Acad. Sci. USA 10317319-17324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamy, M. C., M. Zouine, J. Fert, M. Vergassola, E. Couve, E. Pellegrini, P. Glaser, F. Kunst, T. Msadek, P. Trieu-Cuot, and C. Poyart. 2004. CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence. Mol. Microbiol. 541250-1268. [DOI] [PubMed] [Google Scholar]
- 21.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396643-649. [DOI] [PubMed] [Google Scholar]
- 22.Levin, I., M. Giladi, N. Altman-Price, R. Ortenberg, and M. Mevarech. 2004. An alternative pathway for reduced folate biosynthesis in bacteria and halophilic archaea. Mol. Microbiol. 541307-1318. [DOI] [PubMed] [Google Scholar]
- 23.Liti, G., and E. J. Louis. 2005. Yeast evolution and comparative genomics. Annu. Rev. Microbiol. 59135-153. [DOI] [PubMed] [Google Scholar]
- 24.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 25.Nichols, B. P., and G. G. Guay. 1989. Gene amplification contributes to sulfonamide resistance in Escherichia coli. Antimicrob. Agents Chemother. 332042-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicoloff, H., V. Perreten, L. M. McMurry, and S. B. Levy. 2006. Role for tandem duplication and Lon protease in AcrAB-TolC-dependent multiple antibiotic resistance (Mar) in an Escherichia coli mutant without mutations in marRAB or acrRAB. J. Bacteriol. 1884413-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porwollik, S., R. M. Wong, R. A. Helm, K. K. Edwards, M. Calcutt, A. Eisenstark, and M. McClelland. 2004. DNA amplification and rearrangements in archival Salmonella enterica serovar Typhimurium LT2 cultures. J. Bacteriol. 1861678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price, R. N., A. C. Uhlemann, A. Brockman, R. McGready, E. Ashley, L. Phaipun, R. Patel, K. Laing, S. Looareesuwan, N. J. White, F. Nosten, and S. Krishna. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364438-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reams, A. B., and E. L. Neidle. 2004. Selection for gene clustering by tandem duplication. Annu. Rev. Microbiol. 58119-142. [DOI] [PubMed] [Google Scholar]
- 30.Redon, R., S. Ishikawa, K. R. Fitch, L. Feuk, G. H. Perry, T. D. Andrews, H. Fiegler, M. H. Shapero, A. R. Carson, W. Chen, E. K. Cho, S. Dallaire, J. L. Freeman, J. R. Gonzalez, M. Gratacos, J. Huang, D. Kalaitzopoulos, D. Komura, J. R. MacDonald, C. R. Marshall, R. Mei, L. Montgomery, K. Nishimura, K. Okamura, F. Shen, M. J. Somerville, J. Tchinda, A. Valsesia, C. Woodwark, F. Yang, J. Zhang, T. Zerjal, L. Armengol, D. F. Conrad, X. Estivill, C. Tyler-Smith, N. P. Carter, H. Aburatani, C. Lee, K. W. Jones, S. W. Scherer, and M. E. Hurles. 2006. Global variation in copy number in the human genome. Nature 444444-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roland, S., R. Ferone, R. J. Harvey, V. L. Styles, and R. W. Morrison. 1979. The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J. Biol. Chem. 25410337-10345. [PubMed] [Google Scholar]
- 32.Romero, D., and R. Palacios. 1997. Gene amplification and genomic plasticity in prokaryotes. Annu. Rev. Genet. 3191-111. [DOI] [PubMed] [Google Scholar]
- 33.Schrag, S., R. Gorwitz, K. Fultz-Butts, and A. Schuchat. 2002. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. MMWR Recomm. Rep. 51(RR-11)1-22. [PubMed] [Google Scholar]
- 34.Schrag, S. J., E. R. Zell, R. Lynfield, A. Roome, K. E. Arnold, A. S. Craig, L. H. Harrison, A. Reingold, K. Stefonek, G. Smith, M. Gamble, and A. Schuchat. 2002. A population-based comparison of strategies to prevent early-onset group B streptococcal disease in neonates. N. Engl. J. Med. 347233-239. [DOI] [PubMed] [Google Scholar]
- 35.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shyamala, V., E. Schneider, and G. F. Ames. 1990. Tandem chromosomal duplications: role of REP sequences in the recombination event at the join-point. EMBO J. 9939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skold, O. 2001. Resistance to trimethoprim and sulfonamides. Vet. Res. 32261-273. [DOI] [PubMed] [Google Scholar]
- 38.Slack, A., P. C. Thornton, D. B. Magner, S. M. Rosenberg, and P. J. Hastings. 2006. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stone, S. R., and J. F. Morrison. 1986. Mechanism of inhibition of dihydrofolate reductases from bacterial and vertebrate sources by various classes of folate analogues. Biochim. Biophys. Acta 869275-285. [DOI] [PubMed] [Google Scholar]
- 40.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 10213950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 9912391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whoriskey, S. K., V. H. Nghiem, P. M. Leong, J. M. Masson, and J. H. Miller. 1987. Genetic rearrangements and gene amplification in Escherichia coli: DNA sequences at the junctures of amplified gene fusions. Genes Dev. 1227-237. [DOI] [PubMed] [Google Scholar]