Abstract
AcrAB-TolC from Escherichia coli is a multidrug efflux complex capable of transenvelope transport. In this complex, AcrA is a periplasmic membrane fusion protein that establishes a functional connection between the inner membrane transporter AcrB of the RND superfamily and the outer membrane channel TolC. To gain insight into the mechanism of the functional association between components of this complex, we replaced AcrB with its close homolog MexB from Pseudomonas aeruginosa. Surprisingly, we found that AcrA is promiscuous and can form a partially functional complex with MexB and TolC. The chimeric AcrA-MexB-TolC complex protected cells from sodium dodecyl sulfate, novobiocin, and ethidium bromide but failed with other known substrates of MexB. We next identified single and double mutations in AcrA and MexB that enabled the complete functional fit between AcrA, MexB, and TolC. Mutations in either the α-helical hairpin of AcrA making contact with TolC or the β-barrel domain lying on MexB improved the functional alignment between components of the complex. Our results suggest that three components of multidrug efflux pumps do not associate in an “all-or-nothing” fashion but accommodate a certain degree of flexibility. This flexibility in the association between components affects the transport efficiency of RND pumps.
Proteins belonging to the membrane fusion protein (MFP) family are broadly represented in both gram-negative and gram-positive bacteria (6, 13). In gram-negative bacteria, MFPs are located in the periplasm and act on both the inner membranes (IMs) and the outer membranes (OMs) to enable the transport of diverse substrates across the whole-cell envelope directly into the medium (42, 4). On the IM, MFPs associate specifically with transporters and stimulate their activities. On the OM, they recruit OM channels and possibly enable the expulsion of substrates into the medium. The three components form large multiprotein assemblies that traverse both the IMs and the OMs of gram-negative bacteria. Working together as a well-coordinated team, they achieve the direct extrusion of substrates across two membranes and into the medium.
The multidrug efflux transporters AcrAB-TolC from Escherichia coli and MexAB-OprM from Pseudomonas aeruginosa are the best-characterized examples of three-component pumps capable of transenvelope transport (28, 29). In these complexes, the periplasmic proteins AcrA and MexA belong to the MFP family and share 57.7% amino acid sequence identity. Structural analyses showed that these proteins have modular structures. The central domains of AcrA/MexA share symmetrical coiled-coil and lipoyl-binding motifs. These domains fold back on themselves at the gap between the helical regions, forming an α-helical hairpin (3, 14, 24). The lipoyl domain connects the α-helical hairpin with the six-stranded and the short α-helix β-barrel domains (the α-plus-β-domain of MexA, as described in reference 3). This modular organization befits the function of AcrA/MexA to establish a functional link between the IM transporter and the OM channel.
AcrB and MexB (with 70% identity), which belong to the resistance-nodulation cell division (RND) superfamily of proteins, are integral IM proteins with 12 transmembrane segments (TMs) and large periplasmic domains (26). The periplasmic domains are the sites of substrate recognition and binding, as well as the sites of stable associations with MFPs (40, 39, 25). RNDs are drug/proton antiporters and function as trimers. Structural and genetic studies suggested that AcrB and possibly other RND proteins transport their substrates across the OM by the “alternate occupancy” mechanism (25, 31). In this mechanism, each protomer of AcrB cycles through three consecutive conformations in a sequential manner such that at any given time, each one of them exists in a different phase. The result of this cycling is the alternate opening of the drug-binding pocket, either into the periplasm or into the funnel connected to TolC. These alternations are possibly coupled to proton transport and the motion of MFPs.
The OM channel proteins TolC and OprM do not share a high degree of sequence similarity, but their three-dimensional structures are strikingly similar: both proteins are inserted into the OM by their β-barrel domains and extend ∼70 Å into the periplasm to bind the respective IM complex (17, 2). It is believed they associate with the IM complexes through binding of the periplasmic extensions of the OM channels to the α-helical hairpins of the respective MFPs (33, 19). TolC and OprM are not interchangeable between AcrAB and MexAB (36). However, both proteins are reported to function with multiple IM complexes in E. coli and in P. aeruginosa. TolC demonstrated extreme versatility by supporting the functions of several multidrug efflux pumps, e.g., AcrAB, AcrEF, and EmrAB (34), as well as the type I protein secretion transporters HlyBD and CvaAB. Furthermore, recombinant multidrug transporters from various gram-negative bacteria can also recruit TolC for their activities (38, 5).
Several models of the tripartite association between MFP-RND complexes and OM channels have been proposed based on both structural and mutagenesis studies (10, 14). In all models, MFPs provide a physical link between RNDs and OM channels that barely touch each other. The major uncertainties in all these models are the conformation, the oligomerization state, and the molecular contacts of MFPs with IM and OM proteins. Mutagenesis studies of AcrA and MexA placed the amino acid residues that were important for the interaction with the respective TolC and OprM channels into the α-helical hairpins (7, 33, 19). The loss-of-function mutations in MexA that also affect the interaction with MexB were mapped predominantly to the C-terminal residues of the β-barrel domain of MexA (27). However, the overlapping set of residues was also found to be important for functional interactions between AcrA and TolC (11). In addition, chimeric analysis of AcrA narrowed the choice of the protein domain responsible for the specificity of association with AcrB to the unstructured, distal C-terminal region of the protein (8).
In this study, we found that AcrA interacts with the noncognate MexB transporter and that the AcrA-MexB IM complex can recruit TolC. This chimeric complex confers partial resistance to the detergent sodium dodecyl sulfate (SDS), the antibiotic novobiocin (NOV), and the dye ethidium bromide (EtBr) but fails to pump out other substrates of MexB. We then used chemical mutagenesis of acrA and mexB to identify amino acid substitutions that could enable the complete functional fit between AcrA, MexB, and TolC and extend the spectrum of antibiotic resistance. We found that single-amino-acid substitutions in AcrA are sufficient to improve the multidrug activity of MexB. We conclude that the reduced activity of MexB is due to the partial misfit between components of the complex.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli strains and plasmids used in this study are described in Table 1. Strain W4680AD is a derivative of W4680A, obtained by the P1vir-mediated transduction of acrD::tet from the JZM320 strain (30). The pAMexB plasmid was constructed by replacing the acrB gene in the pAHisB plasmid with the PCR-amplified mexB gene. For this purpose, mexB was PCR amplified using P. aeruginosa (PAO1) chromosomal DNA as the template and primers 5′-CGGCGCTCTAGACAGGAGCCGTTAAGACATGTCGAAGTTTTTCATTG-3′ (the XbaI site is underlined) and 5′-CCGGCAAGCTTTCATTGCCCCTTTTCGACGGAC-3′ (the Hind III site is underlined). To construct pAMexB, the pAHisB plasmid was treated with XbaI and HindIII. The pAcrAHis fragment was purified and ligated with the mexB gene PCR product pretreated with XbaI and HindIII. To construct the pAMexBO plasmid, the MluI-HindIII DNA fragment from pMABO, which contains 1,003 bp of mexB and the entire oprM gene, was recloned into the same sites of pAMexB. All constructs and point mutations were verified by DNA sequencing.
TABLE 1.
Bacterial strains and plasmids
| Strain | Description | Source or reference |
|---|---|---|
| W4680A | K-12 ΔacrAB::kan | 21 |
| W4680AE | K-12 ΔacrAB::kan ΔacrEF::spe | Gift from H. Nikaido |
| W4680AD | K-12 ΔacrAB::kan ΔacrD::Tet | This study |
| ECM 2112 | MC4100 ΔacrAB::kan ΔtolC::Tn10 | Gift from O. Lomovskaya |
| pAhisB | pUC19 carrying His6-tagged acrA and acrB; Ampr | 37 |
| pMABO | pUC18 carrying mexA, mexB, and oprM; Ampr | 36 |
| pAMexB | pAhisB derivative carrying mexB | This study |
| pAMexBO | pAMexB derivative carrying oprM | This study |
| pAMexB D408A | pAMexB derivative carrying D408A mutation in mexB | This study |
| pMABO D408A | pMABO derivative carrying D408A mutation in mexB | This study |
| pAD111N-MexB | pAMexB carrying D111N mutation in acrA | This study |
| pAG240S-MexB | pAMexB carrying G240S mutation in acrA | This study |
| pAV244M-MexB | pAMexB carrying V244M mutation in acrA | This study |
| pAS249N-MexB | pAMexB carrying S249N mutation in acrA | This study |
| pA-MexBT329I | pAMexB carrying T329I mutation in mexB | This study |
| pA-MexBT489I | pAMexB carrying T489I mutation in mexB | This study |
| pA-MexBT557I | pAMexB carrying T557I mutation in mexB | This study |
| pA-MexBA802V | pAMexB carrying A802V mutation in mexB | This study |
| pAV244M-MexBT557I | pAMexB carrying mutations V244M in acrA and T557I in mexB | This study |
| pAV244M-MexBT489I | pAMexB carrying mutations V244M in acrA and T489I in mexB | This study |
| pA-MexBT329I/A802V | pAMexB carrying mutations T329I and A802V in mexB | This study |
| pAD111N-AcrB | pAD111N-MexB derivative carrying acrB | This study |
| pAG240S-AcrB | pAG240S-MexB derivative carrying acrB | This study |
| pAV244M-AcrB | pAV244M-MexB derivative carrying acrB | This study |
| pAS249N-AcrB | pAS249M-MexB derivative carrying acrB | This study |
Bacterial cultures were grown in Luria-Bertani (LB) medium (20 g/liter Bacto tryptone, 10 g/liter Bacto yeast extract, and 10 g/liter NaCl, and pH was adjusted to 7.0 with 1 ml of 1 N NaOH) or LB agar (LB medium containing 15 g/liter agar) at 37°C. Antibiotics ampicillin (100 μg/ml), kanamycin (34 μg/ml), spectinomycin (50 μg/ml), and tetracycline (10 μg/ml) were used for the selection of respective plasmids and strains when indicated.
MIC.
Exponentially growing cell cultures (A600 of 0.6) were inoculated at a density of 105 cells per ml into 96-well microtiter plates containing LB broth with the twofold serially diluted antimicrobials under investigation. Cell growth was determined visually after incubation at 37°C for at least 16 h. The MIC was determined as the concentration of the drug that completely inhibited bacterial growth. Each determination of MIC was repeated at least three times to ensure reproducibility of results.
Chemical mutagenesis and selection of mutants.
Chemical mutagenesis of pAMexB was carried out as described previously (23). Briefly, 15 μg of the pAMexB plasmid was incubated at 70°C for 3 h in 44 mM potassium phosphate buffer (pH 6.0) containing 460 mM hydroxylamine hydrochloride and 5 mM EDTA. Equal aliquots of DNA (5 μg) were removed every hour, and the reaction was stopped by the addition of Tris-HCl (pH 8.0) and EDTA, to final concentrations of 90 mM and 9 mM, respectively. The DNA was precipitated by the addition of sodium acetate (0.3 mM final concentration, pH 4.8) and ethanol (70% final concentration) at −20°C overnight. The precipitated DNA was washed with 70% (vol/vol) ethanol, dried under laminar airflow, and resuspended in 30 μl of sterile water. To select for multidrug-resistant mutants, the hydroxylamine-treated pAMexB plasmids (2 μl) were electroporated into DH5α cells and plated onto LB agar plates containing 100 μg/ml of ampicillin. After overnight incubation, colonies were washed from plates into 5 ml of LB-ampicillin broth and incubated for 5 h at 37°C. Subsequently, plasmids were purified and electroporated into E. coli W4680AD cells. Drug-resistant colonies were selected on LB agar plates containing one of the antibiotics erythromycin (ERY; 8 and 16 μg/ml), lincomycin (LIN; 156 and 312 μg/ml), or chloramphenicol (CF) (1.6 and 3.2 μg/ml). All plasmids isolated from drug-resistant colonies were retransformed into W4680AD cells, and the multidrug resistance phenotype was verified by determining the MIC for the selected antimicrobials. Determination of the DNA sequence of the promoter region and the coding sequences of acrA and mexB was carried out at the Oklahoma Medical Research Foundation DNA Sequencing Facility. In addition to amino acid substitutions, some plasmids also contained the following silent mutations: plasmid E1 contained the base substitution C1137T in the coding sequence of acrA, plasmid E23 contained the substitution C2668T in the coding sequence of mexB, plasmid E70 contained substitutions C981T, C984T, and G3105A in mexB, plasmid E82 contained substitution G1815A in mexB, and plasmid E88 contained the substitution G714A in acrA.
Site-directed mutagenesis.
All point mutations were introduced using a QuikChange-XL site-directed mutagenesis kit (Stratagene) as recommended by the manufacturer. For each mutant, at least two plasmids were purified and sequenced to ascertain the presence of the desired mutation. To construct plasmids carrying a single substitution in AcrA and AcrB, the acrA gene from the pAD111N-MexB, pAG240S-MexB, pAV244M-MexB, and pAS249N-MexB plasmids was restricted with SacI and XbaI and subcloned into the corresponding sites of pUC19. Subsequently, the resulting plasmids were treated with XbaI and SphI and ligated with the PCR-amplified acrB gene pretreated with the same enzymes to yield pAD111N-AcrB, pAG240S-AcrB, pAV244M-AcrB, and pAS249N-AcrB. Primers 5′-CAACTCTAGACAGGAGCCGTTAAGA-3′ (the XbaI site is underlined) and 5′-CCGCCGCATGCTCGTAGGTTATGCATA-3′ (the SphI site is underlined) were used to amplify acrB by PCR.
Protein assays, affinity purification, and immunoblotting.
To assess protein-protein interactions in chimeric and native complexes, E. coli cells carrying various constructs were grown until A600 reached ∼0.6 in 0.2 liters of LB broth containing ampicillin. Cells were harvested by centrifugation at 20°C and washed twice in phosphate-buffered saline. Cross-linking with DSP (dithio-bis-succinimidyl-propionate) and AcrAHis (AcrA containing a C-terminal six-histidine tag) purification were carried out as described previously (37).
Samples for denaturing SDS-polyacrylamide gel electrophoresis (PAGE) were prepared by boiling the samples in 1× SDS-PAGE sample buffer for 5 min. For analysis of MexB, boiling of samples prior to SDS-PAGE was omitted.
Immunoblotting analyses were carried out using standard procedures. Proteins were separated by SDS-PAGE and then transferred onto polyvinylidene difluoride membranes in transfer buffer (25 mM Tris base, 192 mM glycine, 10% methanol). Specific proteins were visualized after incubation with primary polyclonal rabbit antibodies, followed by incubation with secondary alkaline phosphatase-conjugated anti-rabbit antibody. The 5-bromo-4-chloro-3-indoyl phosphate/nitro blue tetrazolium substrates were used to visualize protein bands.
RESULTS
AcrA, MexB, and TolC form a partially functional drug efflux pump.
To gain insight into the mechanism of functional association between periplasmic MFPs and RND transporters, we cloned MexB into a single operon with AcrAHis and expressed the AcrA-MexB hybrid pump in the multidrug-susceptible strain W4680AD (ΔacrAB ΔacrD) of E. coli. Unexpectedly, cells producing AcrA-MexB demonstrated an elevated resistance to SDS, NOV, and EtBr but remained susceptible to other tested antimicrobial agents (Table 2). This result suggested that AcrA is able to interact with MexB and supports efflux of a restricted range of compounds. No resistance was detected in the TolC deficient strain, suggesting that the third component of the complex is TolC (data not shown). Previous studies showed that the MexAB-TolC complex is nonfunctional in E. coli (32, 36). We also tested the functionality of the MexA-AcrB-TolC and AcrA-MexB-OprM combinations. However, these hybrid efflux pumps also did not provide elevated drug resistance in E. coli (data not shown and Table 2).
TABLE 2.
Antibiotic susceptibility of E. coli W4680AD expressing multidrug efflux pumps AcrAB and MexAB-OprM and their derivatives
| MDR pumpa | Plasmid | MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|---|
| SDS | ERY | NOV | CF | NAL | LIN | EtBr | ||
| AcrA-AcrB | pAhisB | >10,240 | 32 | 256 | 0.8 | 3.125 | 312.5 | 200 |
| MexAB-OprM | pMABO | >10,240 | 8 | 128 | 6.4 | 6.25 | 312.5 | 50 |
| AcrA-MexB | pAMexB | 5,120 | 0.5 | 32 | 0.2 | 0.78 | 19.5 | 12.5 |
| AcrA-MexBD408A | pAMexB D408A | 20 | 0.5 | 0.5 | 0.1 | 0.39 | 19.5 | 1.56 |
| MexABD408A-OprM | pMABO D408A | 20 | 0.5 | 0.5 | 0.1 | 0.39 | 19.5 | 1.56 |
| AcrA-MexB-OprMb | pAMexBO | 20 | 0.5 | 1 | 0.2 | 0.39 | 19.5 | 1.56 |
| None | 20 | 0.5 | 1 | 0.2 | 0.39 | 19.5 | 1.56 | |
MDR, multidrug resistance.
Susceptibility was determined with E. coli ECM2112 (ΔacrAB ΔtolC).
Immunoblotting analysis showed that the amounts of AcrA and MexB produced from pAMexB were similar to those of native AcrAB and MexAB-OprM transporters produced from the plasmids pAhisB and pMABO, respectively (Fig. 1A). Therefore, differences in protein amounts cannot account for differences in MICs. We next used the chemical cross-linking and affinity copurification approaches to confirm interactions between AcrAHis and MexB. As shown in Fig. 1B, MexB is copurified with AcrAHis on a metal-affinity column. In addition, immunoblotting with anti-TolC antibodies showed that the chromosomally encoded TolC also copurifies with the AcrAHis-MexB complex. Consistent with the previous results, TolC also copurifies with AcrAHis alone (Fig. 1B, lane 4) (15, 37). Thus, AcrA and MexB associate with each other and TolC in a drug transporter that efficiently extrudes some but not other substrates of MexB.
FIG. 1.
AcrA, MexB, and TolC form a tripartite multidrug efflux complex. (A) Expression of AcrA and MexB in E. coli W4680AD cells. Membrane fractions were isolated from W4680AD cells (lane 1), W4680AD cells producing chimeric AcrA-MexB (lane 2), wild-type AcrA-AcrB cells (lane 3), and wild-type MexAB-OprM cells (lane 4). Equal amounts of the total membrane protein were separated on 10% SDS-PAGE and analyzed by immunoblotting with anti-AcrA (top panel) and anti-MexB (bottom panel) antibodies. Arrows indicate AcrA and MexB proteins. (B) MexB and TolC were copurified with AcrA. AcrA was purified using metal-affinity chromatography from membrane fractions of cells producing AcrA-AcrB (lane 1), MexAB-OprM (lane 2), AcrA-MexB (lane 3), or AcrA only (lane 4). Samples were analyzed by SDS-PAGE, followed by immunoblotting with anti-AcrA (top panel), anti-MexB (middle panel), and anti-TolC antibodies (bottom panel).
Previously, AcrA was reported to form functional complexes with various RND-type transporters including AcrB, AcrD, and AcrF (9, 18). Among these transporters, only AcrB is expressed at levels that are sufficiently high enough to contribute to the intrinsic resistance of E. coli (34). In the absence of AcrB, the overproduction of the AcrD or AcrEF transporter leads to high levels of resistance (9, 18).
Two control experiments were carried out to confirm that the increased resistance to NOV, SDS, and EtBr originates from the activity of the AcrA-MexB hybrid pump and not from the association of AcrA with the chromosomally encoded RND transporters. First, MexB was inactivated by the D408A substitution in the putative proton translocation channel of TMIV (12). In agreement with previous reports, the D408A substitution led to a total loss of drug resistance by cells carrying the mexB D408A mutation in combination with acrA (pAMexB) as well as in the native mexAB-oprM gene arrangement (pMABOD408A) (Table 2). Thus, MexB is required for the increased levels of resistance to SDS, NOV, and EtBr.
In the second control, the AcrA-MexB chimera was expressed with E. coli lacking AcrAB alone or lacking combinations of either AcrEF or AcrD. We also constructed a W4680BDF mutant lacking all three Acr transporters. Although W4680BDF was viable in LB medium, this strain could not be transformed with plasmids (data not shown). MIC measurements showed that the resistance to SDS and NOV provided by AcrA-MexB was not affected by the lack of AcrF or AcrD (Table 3). Thus, these transporters do not contribute to the increased resistance of cells producing the AcrA-MexB transporter.
TABLE 3.
Activity of AcrA-MexB chimera does not depend on genetic background of E. coli
| E. coli strain | MIC (μg/ml) provided by:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AcrAB
|
AcrA-MexB
|
MexAB-OprM
|
|||||||
| SDS | ERY | NOV | SDS | ERY | NOV | SDS | ERY | NOV | |
| W4680AD | >10,240 | 32 | 64 | 5,120 | 0.5 | 32 | >10,240 | 4 | 128 |
| W4680AE | >10,240 | 64 | 64 | 5,120 | 1 | 32 | >10,240 | 8 | 64 |
| W4680A | >10,240 | 64 | 128 | 2,560 | 0.5 | 32 | >10,240 | 8 | 64 |
We conclude that the chimeric AcrA-MexB-TolC transporter is responsible for the elevated resistance to SDS, NOV, and EtBr in cells carrying the pAMexB plasmid. Furthermore, neither the AcrA-MexB-OprM nor the MexAB-TolC chimeric complex is capable of multidrug efflux. This result suggests that interactions between AcrA and TolC enable the partial activity of AcrA-MexB, whereas the lack of full multidrug efflux activity is due to the misfit between AcrA and MexB.
Functional alignment of AcrA and MexB does not require large structural changes in proteins.
We next carried out chemical mutagenesis of pAMexB to identify the gain-of-function mutations that can expand the substrate spectrum of the AcrA-MexB pump. A total of 90 plasmids were isolated from colonies on LB plates containing four- and eightfold increases in MICs of LIN, CF, or ERY. Immunoblotting with anti-AcrA and anti-MexB antibodies showed that nine plasmids produced AcrA with aberrant electrophoretic mobilities, and these plasmids were excluded from further analysis (data not shown). DNA sequences of acrA and mexB were determined for 19 plasmids. Among the sequenced plasmids, seven plasmids did not contain any substitutions in the coding sequences of acrA and mexB, and two plasmids were present in multiple copies.
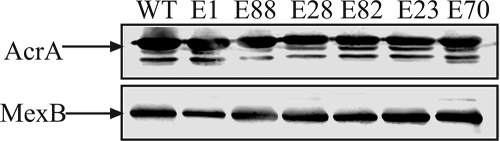
Six plasmids were selected for further studies (Table 4). Figure 2 shows the immunoblotting analyses of AcrA and MexB produced from these plasmids. In all cases, the amounts of AcrA and MexB produced from mutated plasmids were comparable to those of the parent plasmids, suggesting that the mutations did not lead to altered expression levels of proteins. DNA sequencing of the complete acrA and mexB genes showed that three plasmids, E1, E28 and E88, contained single-amino-acid substitutions in AcrA, D111N, S249N, and G240S, respectively (Table 4). E70 contained two mutations, T329I and A802V, both in mexB. Two plasmids, E23 and E82, contained the same V244M mutation in acrA but different mutations, T557I and T489I, in the mexB gene. The finding that single- or double-amino-acid substitutions are sufficient to restore the multidrug efflux activity suggested that fitting AcrA to MexB does not require major changes in protein structures.
TABLE 4.
MIC ratios of E. coli W4680AD containing the plasmid-encoded chimeric AcrA-MexB and its derivatives
| Plasmida | Mutation(s)
|
MDR pumpc | Fold change in MICsb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AcrA | MexB | SDS | ERY | NOV | CF | NAL | LIN | EtBr | ||
| E1 | D111N | AcrA-MexB | 2 | 4 | 8 | 4 | 8 | 4 | 4 | |
| AcrAB | 1 | 0.06 | 1 | 1 | 2 | 0.5 | 0.25 | |||
| MexAB | 1 | 0.25 | 2 | 0.25 | 1 | 0.5 | 1 | |||
| E23 | V244M | T557I | AcrA-MexB | 2 | 1 | 2 | 8 | 8 | 4 | 2 |
| AcrAB | 1 | 0.02 | 0.25 | 1 | 2 | 0.5 | 0.125 | |||
| MexAB | 1 | 0.06 | 0.5 | 0.25 | 1 | 0.5 | 0.5 | |||
| E28 | S249N | AcrA-MexB | 2 | 2 | 2 | 4 | 8 | 4 | 4 | |
| AcrAB | 1 | 0.03 | 0.5 | 0.5 | 2 | 0.5 | 0.25 | |||
| MexAB | 1 | 0.125 | 0.5 | 0.125 | 1 | 0.5 | 1 | |||
| E70 | T329I, A802V | AcrA-MexB | 2 | 4 | 4 | 8 | 8 | 8 | 4 | |
| AcrAB | 1 | 0.06 | 0.5 | 1 | 2 | 1 | 0.25 | |||
| MexAB | 1 | 0.25 | 1 | 0.25 | 1 | 1 | 1 | |||
| E82 | V244M | T489I | AcrA-MexB | 2 | 2 | 4 | 16 | 8 | 8 | 8 |
| AcrAB | 1 | 0.03 | 0.5 | 2 | 1 | 1 | 0.5 | |||
| MexAB | 1 | 0.25 | 1 | 0.5 | 1 | 1 | 2 | |||
| E88 | G240S | AcrA-MexB | 2 | 2 | 4 | 8 | 8 | 8 | 4 | |
| AcrAB | 1 | 0.03 | 0.5 | 1 | 2 | 1 | 0.25 | |||
| MexAB | 1 | 0.125 | 1 | 0.25 | 1 | 1 | 1 | |||
Plasmids are pAMexB derivatives isolated after chemical mutagenesis with hydroxylamide.
Relative MICs were calculated as ratios between the MICs for W4680AD containing the plasmid-encoded mutants and those for W4680AD containing the wild-type chimeric AcrA-MexB, AcrAB, or MexAB-OprM.
The changes in MICs were calculated relative to these MDR pumps.
FIG. 2.
Isolated AcrA-MexB mutants are expressed at the same level in E. coli W4680AD cells. Equal amounts of total membrane protein isolated from cells carrying mutant AcrA-MexB and wild-type AcrA-MexB variants were separated on 10% SDS-PAGE and analyzed by immunoblotting with anti-AcrA (top panel) and anti-MexB (bottom panel) antibodies. Arrows indicate AcrA and MexB proteins.
All plasmids isolated from antibiotic-containing plates, upon transformation into drug-susceptible strains, conferred high levels of resistance not only to selection markers but also to other unrelated drugs. In comparison to the MICs of E. coli cells carrying pAMexB, the mutated plasmids enabled up to a 16-fold increase in the MICs of not only NOV and SDS but also of CF, LIN, nalidixic acid (NAL), EtBr, and ERY (Table 4). However, levels of resistance differed between the plasmids and were at or below those of the native assemblies (Table 4). Thus, mutations enabled a better fit between AcrA and MexB, but the chimeric transporters still were less efficient than the native assemblies in multidrug efflux.
Single-amino-acid substitutions in AcrA and MexB are sufficient to reconstruct the fully functional multidrug efflux pump.
We next used site-directed mutagenesis to investigate the contribution of each substitution to the functionality of the AcrA-MexB pump. The four point mutations D111N, G240S, V244M, and S249N were introduced into AcrA. The D111N mutation is located in the α-helical hairpin of AcrA and was previously reported to directly contact TolC (19). The other three mutations are all located in the β-barrel domain of AcrA, the site of possible interactions with MexB (Fig. 3A).
FIG. 3.
Positions of amino acid substitutions in AcrA and MexB that enhance the activity of chimeric AcrA-MexB. (A) Three-dimensional structure of monomeric AcrA (24), with mutated residues shown as red spheres. (B) The homology model of MexB based on the AcrB crystal structure (26) was generated using Swiss-Model software (http://swissmodel.expasy.org//SWISS-MODEL.html), with the mutated residues shown as spheres.
The four point mutations T329I, T489I, T557I, and A802V were introduced into MexB. In the crystal structure of the homologous AcrB (26), the T329I, T557I, and A802V mutations are accessible from the periplasm (Fig. 3B). The A802V mutation is located in the TolC docking domain, whereas T329I and T557I are located on the periplasmic end of TM II and TM VII, respectively. The three mutated residues T329I, T557I, and A802V could all potentially be in a direct contact with AcrA. The last substitution, T489I, is located in the TM III of MexB and is unlikely to contact AcrA directly.
Site-directed mutagenesis confirmed that single mutations in AcrA and MexB are sufficient to significantly improve multidrug function of the chimeric AcrA-MexB pump (Table 5). The susceptibility profiles of cells carrying the AcrA mutations D111N, G240S, and S249N showed the four- to eightfold increase in resistance to multiple drugs. Interestingly, only the D111N mutation was advantageous with ERY resistance, whereas the S249N and G240S mutations had no effect. Single substitutions in MexB also improved the activity of the AcrA-MexB chimera, albeit to a lesser extent than the mutations in AcrA. The T557I and A802V mutations increased resistance by only two- to fourfold, whereas the T329I and T489I mutations had very little if any effect on AcrA-MexB activity.
TABLE 5.
Antibiotic susceptibility of E. coli W4680AD containing plasmid-encoded AcrA-MexB and its derivatives
| Plasmid | MIC value (μg/ml) or range
|
||||||
|---|---|---|---|---|---|---|---|
| SDS | ERY | NOV | CF | NAL | LIN | EtBr | |
| Control | 20 | 0.5 | 1 | 0.2 | 0.39 | 19.5 | 1.56 |
| pAMexB | 5,120 | 0.5 | 32 | 0.2-0.4 | 0.78 | 19.5 | 12.5 |
| pAD111N-MexB | >10,240 | 2 | 128 | 0.8 | 6.25 | 78 | 50 |
| pAG240S-MexB | 5,120 | 0.5 | 128 | 1.6 | 12.5 | 156 | 50 |
| pAV244M-MexB | 320 | 0.5 | 64 | 0.4 | 3.125 | 39 | 25 |
| pAS249N-MexB | >10,240 | 1 | 128 | 0.8 | 3.125 | 78 | 50 |
| pA-MexBT557I | >10,240 | 1 | 128 | 0.4 | 3.125 | 78 | 25 |
| pA-MexBT489I | >10,240 | 0.5 | 64 | 0.4 | 3.125 | 19.5-39 | 12.5 |
| pA-MexBT329I | 10,240 | 0.5 | 64 | 0.4 | 3.125 | 39 | 25 |
| pA-MexBA802V | >10,240 | 0.5 | 64-128 | 0.4 | 3.125 | 39 | 25 |
| pAV244M-MexBT489I | >10,240 | 0.5 | 64 | 0.8 | 3.125-6.25 | 156 | 25 |
| pAV244M-MexBT557I | >10,240 | 1 | 128 | 0.8 | 6.25 | 156 | 50 |
| pA-MexB T329I/A802V | >10,240 | 0.5 | 64 | 0.8 | 3.125-6.25 | 78 | 25-50 |
| pAD111N-AcrB | >10,240 | 32 | 64 | 1.6 | 3.125 | 312.5 | 200 |
| pAG240S-AcrB | >10,240 | 32 | 128 | 0.8 | 3.125 | 312.5 | 200 |
| pAV244M-AcrB | >10,240 | 16 | 64-128 | 0.8 | 3.125 | 312.5 | 200 |
| pAS249N-AcrB | >10,240 | 16 | 64 | 1.6 | 3.125 | 312.5 | 200 |
Only the AcrA D111N and S249N mutations matched the susceptibility profiles of the respective E1 and E28 mutants (Table 4). The correlation between phenotypes of other mutants isolated by site-directed and chemical mutagenesis was only partial. In the case of MexB substitutions, this result is expected since, in the original mutants, these substitutions are paired either with each other (e.g., T329I and A802A in E70) or with mutations in AcrA (e.g., E23 and E82). Indeed, when mutations were combined by site-directed mutagenesis, the MICs increased (see below and Table 5). In contrast, the G240S mutation was the only amino acid substitution identified in E88. However, cells carrying the AcrA G240S mutation generated by site-directed mutagenesis did not have an elevated resistance to SDS and ERY. DNA sequencing of the coding region revealed that the E88 mutant contained, in addition to the G240S substitution in AcrA, one silent base substitution, G714A, leading to the change from the GAG codon for GLU to that of GAA. Recent studies suggested that the silent mutations could affect the rate of protein synthesis, leading to changes in folding and activity of the protein (16). Alternatively, mutations in the noncoding region of the plasmid could contribute to the E88 phenotype.
The V244M substitution was identified in the AcrA protein of the E23 and E82 mutants (Table 4), where it was paired with the T557I and T489I mutations in MexB, respectively. Cells producing AcrA(V244M)-MexB showed a unique drug susceptibility profile. The V244M mutation alone improved the efflux of several drugs by two- to fourfold (i.e., NOV, CF, NAL, LIN, and EtBr). However, this mutation impaired SDS efflux by 16-fold. The dependent role of this mutation in the functional alignment of AcrA and MexB was confirmed by the finding that the T557I substitution alone notably increased the MIC of SDS, with a small effect (twofold increase) on other tested drugs. Similarly, the beneficial effect of the T489I substitution was notable with SDS, whereas the MICs of other tested drugs increased only by twofold. However, when either the T557I or the T489I substitution was combined with the AcrA V244M mutation by site-directed mutagenesis, the MICs of multiple drugs increased by 4- to 16-fold. Thus, the V244M mutation in AcrA and the T557I or the T489I mutation in MexB have an additive effect on drug susceptibility of cells carrying these mutations.
We also tested how the substitutions identified in AcrA affected the activity of the native AcrAB-TolC complex. For this purpose, AcrA containing the single substitutions D111N, G240S, V244M, and S249N was subcloned and expressed with the native AcrB pump. MIC measurements showed that these substitutions in AcrA did not change the activity of the native complex (Table 5). Thus, the effect of the AcrA substitutions is specific to MexB.
Taken together, site-directed mutagenesis studies confirmed that single-amino-acid substitutions in either AcrA or MexB can notably improve the functional alignment of these two noncognate subunits. Judging by the MICs, paired mutations play an additive rather than a synergistic role in drug efflux by the AcrA-MexB transporter, suggesting that these mutations act on the same function.
Protein-protein interactions in the chimeric AcrA-MexB-TolC multidrug efflux pump.
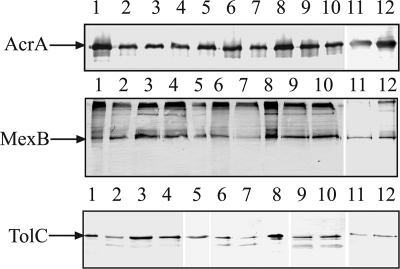
The ability to confer a multidrug resistance phenotype varies broadly between chimeric AcrA-MexB and its mutant derivatives. To investigate whether these differences in MICs could be correlated with the ability to form complexes, we investigated the effects of mutations on interactions between AcrA and MexB and on the association of the IM complex with the OM channel TolC. Immunoblotting analyses of AcrAHis-containing complexes purified from cells carrying single and double mutations showed that tripartite complexes are assembled in all cases (Fig. 4). The amounts of copurified complexes were similar for all constructs tested and did not correlate with MIC values. We conclude that the gain-of-function mutations do not improve interactions between components of complexes but contribute to functionally fitting three components into a complex.
FIG. 4.
MICs do not correlate with the amounts of MexB and TolC copurified with AcrAHis. AcrAHis was purified from cells producing the wild-type and the AcrA-MexB-TolC mutant complexes. The amounts of copurified proteins were analyzed by immunoblotting with anti-AcrA (top panel), anti-MexB (middle panel), and anti-TolC (bottom panel) antibodies. Lanes 1, AcrA-MexB; 2, AcrA(D111N)-MexB; 3, AcrA(G240S)-MexB; 4, AcrA(V244M)-MexB; 5, AcrA(S249N)-MexB; 6, AcrA-MexB(T489I); 7, AcrA-MexB(T557I); 8, AcrA-MexB(A802V); 9, AcrA(V244M)-MexB(T489I); 10, AcrA(V244M)-MexB(T557I); 11, AcrA-MexB(T329I); 12, AcrA-MexB(T329I/A802V).
DISCUSSION
The mechanism of the three-component multidrug efflux pumps such as AcrAB-TolC is highlighted by two signature features: (i) the binding of substrates in the periplasm and (ii) the concerted action of proteins located in two different membranes. Here we show that the interactions between three components are not rigid and can accommodate a certain degree of flexibility. AcrA interacts with the noncognate MexB and forms a tripartite complex, which also includes TolC. This interaction is sufficient to confer resistance to SDS, NOV, and EtBr. However, the chimeric AcrA-MexB-TolC pump fails to extrude other known substrates of MexB such as NAL or CF. Since each of the proteins is functionally competent, the changes in the transport activity are likely to be caused by the misalignment between MexB and AcrA and/or TolC, which in turn, could affect the overall efficiency of the complex. Indeed, drugs transported by the chimeric AcrA-MexB-TolC, SDS, NOV, and EtBr, are also the best substrates of the native AcrAB-TolC and MexAB-OprM assemblies. Thus, the defective AcrA-MexB-TolC pump could provide some protection against these compounds but fail with other “poor” substrates. On the other hand, analysis of the single-substitution mutants of AcrA suggests that accessory proteins could also affect the substrate specificity of MexB. The AcrA G240S substitution enables increased resistance to NOV and LIN and other drugs but fails with SDS and ERY. Similarly, the V244M mutation decreases resistance to SDS with the slight improvement in resistance to other drugs. Further biochemical analyses are required to establish the possible contribution of accessory proteins to substrate specificities of inner membrane transporters.
Amino acid substitutions that enable functional interactions in AcrA-MexB-TolC map to the α-helical hairpin (D111N) and β-barrel (G240S, V244M, and S249N) domains of AcrA (Fig. 3A). Previous domain-swapping studies suggested that the α-helical hairpins of AcrA and its homolog MexA are responsible for the association with TolC and OprM, respectively (7, 33). In fact, the cysteine cross-linking studies showed that D111 of AcrA interfaces directly with TolC (the D87C substitution described in reference 19). Mutagenesis studies showed that the A108T substitution in MexA (the G110 of AcrA) disrupts interactions with OprM (27). Thus, the D111N substitution affects AcrA-TolC interactions. However, AcrA and TolC are the two natural partners and are suppose to fit perfectly with each other. It is possible, therefore, that the skewed position of AcrA on MexB causes a partial misfit of the recruited TolC, either with AcrA or with the funnel of MexB. In this case, the D111N substitution adjusts the alignment of TolC in the complex.
The other three substitutions, G240S, V244M, and S249N, located in the β-barrel domain of AcrA, presumably mend the placement of AcrA on MexB. In models of the tripartite complex, this domain of AcrA is placed on the “Porter” domain of AcrB (10, 14, 20). Additional support for the involvement of this domain in interactions with MexB comes from a previous finding that the R221H substitution in MexA, which corresponds to the Q237 position in AcrA, disrupts interactions between MexA and MexB (27). Thus, the G240S, V244M, and S249N substitutions possibly adjust the functional interface between AcrA and MexB. Indeed, the AcrA V244M substitution alone provides only partial activity, and increased resistance is achieved when the V244M mutation is paired with the T489I and T557I mutations in MexB (Table 5). Given their location in the transmembrane domain, the mutations T489I and T557I probably have a long-range effect on the overall conformation of MexB, making it more compatible with the AcrA V244M mutation. Interestingly, the G240S substitution (G216S, as described in reference 11) and other mutations in the β-barrel domain of AcrA were also identified as suppressors of the instability of the TolC mutant variant (11). Since this domain of AcrA is unlikely to directly contact TolC, these mutations probably change the positioning of AcrA on the transporter and, as a result, the interface with TolC. The α-helical hairpin and the β-barrel domain of AcrA are connected by the lipoyl or hinge domain, which allows conformational flexibility in the protein (Fig. 3A) (24). The hinge domain could act as a conformational switch and adjust the respective orientations of the α-helical hairpin and the β-barrel domains of AcrA during the assembly or function of the complex.
The finding that mutations in MexB itself are sufficient to improve multidrug efflux is consistent with the notion that the mutations identified adjust the functional fit between AcrA and MexB. The A802V substitution located on the tip of the “TolC-docking” domain of MexB significantly improves the functional fit between components of the complex. The A802V substitution goes together with the T329I substitution on the periplasmic end of the longest TM II of MexB. Both mutations are located away from the drug binding sites identified in structural and genetic studies (22, 40, 25) and are unlikely to affect MICs by changing interactions between MexB and substrates. Thus, the effect of these mutations is likely to be structural and to affect the interactions with accessory proteins. Previous studies suggested that the interactions between RND transporters and OM channels are very limited (35) and that reconstituted transporters are functional in the absence of OM channels (1, 41). It seems more likely that mutations in MexB affect interactions with AcrA rather than with TolC.
Taken together, these results suggest that in the chimeric AcrA-MexB-TolC complex, the interacting subunits are misaligned and that the identified mutations mend interactions between proteins. The functional misalignment between subunits strongly affects the efficiency of multidrug efflux and possibly the substrate specificity of the complex.
Acknowledgments
This study was supported by National Institutes of Health grant AI052293.
Footnotes
Published ahead of print on 16 November 2007.
REFERENCES
- 1.Aires, J. R., and H. Nikaido. 2005. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 1871923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akama, H., M. Kanemaki, M. Yoshimura, T. Tsukihara, T. Kashiwagi, H. Yoneyama, S. Narita, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 27952816-52819. [DOI] [PubMed] [Google Scholar]
- 3.Akama, H., T. Matsuura, S. Kashiwagi, H. Yoneyama, S. Narita, T. Tsukihara, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 27925939-25942. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, C., C. Hughes, and V. Koronakis. 2001. Protein export and drug efflux through bacterial channel-tunnels. Curr. Opin. Cell Biol. 13412-416. [DOI] [PubMed] [Google Scholar]
- 5.Dastidar, V., W. Mao, O. Lomovskaya, and H. I. Zgurskaya. 2007. Drug-induced conformational changes in multidrug efflux transporter AcrB from Haemophilus influenzae. J. Bacteriol. 1895550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinh, T., I. T. Paulsen, and M. H. Saier, Jr. 1994. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 1763825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eda, S., H. Maseda, E. Yoshihara, and T. Nakae. 2006. Assignment of the outer-membrane-subunit-selective domain of the membrane fusion protein in the tripartite xenobiotic efflux pump of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 254101-107. [DOI] [PubMed] [Google Scholar]
- 8.Elkins, C. A., and H. Nikaido. 2003. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J. Bacteriol. 1855349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 1846490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Recio, J., F. Walas, L. Federici, J. Venkatesh Pratap, V. N. Bavro, R. N. Miguel, K. Mizuguchi, and B. Luisi. 2004. A model of a transmembrane drug-efflux pump from gram-negative bacteria. FEBS Lett. 5785-9. [DOI] [PubMed] [Google Scholar]
- 11.Gerken, H., and R. Misra. 2004. Genetic evidence for functional interactions between TolC and AcrA proteins of a major antibiotic efflux pump of Escherichia coli. Mol. Microbiol. 54620-631. [DOI] [PubMed] [Google Scholar]
- 12.Guan, L., and T. Nakae. 2001. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 1831734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harley, K. T., G. M. Djordjevic, T. Tseng, and M. H. Saier, Jr. 2000. Membrane-fusion protein homologues in Gram-positive bacteria. Mol. Microbiol. 36516-517. [DOI] [PubMed] [Google Scholar]
- 14.Higgins, M. K., E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA 1019994-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain, F., M. Humbard, and R. Misra. 2004. Interaction between the TolC and AcrA proteins of a multidrug efflux system of Escherichia coli. J. Bacteriol. 1868533-8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimchi-Sarfaty, C., J. M. Oh, I. W. Kim, Z. E. Sauna, A. M. Calcagno, S. V. Ambudkar, and M. M. Gottesman. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315525-528. [DOI] [PubMed] [Google Scholar]
- 17.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405914-919. [DOI] [PubMed] [Google Scholar]
- 18.Lau, S. Y., and H. I. Zgurskaya. 2005. Cell division defects in Escherichia coli deficient in the multidrug efflux transporter AcrEF-TolC. J. Bacteriol. 1877815-7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobedanz, S., E. Bokma, M. F. Symmons, E. Koronakis, C. Hughes, and V. Koronakis. 2007. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl. Acad. Sci. USA 1044612-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomovskaya, O., H. I. Zgurskaya, M. Totrov, and W. J. Watkins. 2007. Waltzing transporters and “the dance macabre” between humans and bacteria. Nat. Rev. Drug Discov. 656-65. [DOI] [PubMed] [Google Scholar]
- 21.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 1645-55. [DOI] [PubMed] [Google Scholar]
- 22.Mao, W., M. S. Warren, D. S. Black, T. Satou, T. Murata, T. Nishino, N. Gotoh, and O. Lomovskaya. 2002. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 46889-901. [DOI] [PubMed] [Google Scholar]
- 23.Middlemiss, J. K., and K. Poole. 2004. Differential impact of MexB mutations on substrate selectivity of the MexAB-OprM multidrug efflux pump of Pseudomonas aeruginosa. J. Bacteriol. 1861258-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikolosko, J., K. Bobyk, H. I. Zgurskaya, and P. Ghosh. 2006. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami, S., R. Nakashima, E. Yamashita, T. Matsumoto, and A. Yamaguchi. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443173-179. [DOI] [PubMed] [Google Scholar]
- 26.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419587-593. [DOI] [PubMed] [Google Scholar]
- 27.Nehme, D., X. Z. Li, R. Elliot, and K. Poole. 2004. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 1862973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido, H., and H. I. Zgurskaya. 2001. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3215-218. [PubMed] [Google Scholar]
- 29.Poole, K. 2007. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39162-176. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg, E. Y., D. Ma, and H. Nikaido. 2000. AcrD of Escherichia coli Is an aminoglycoside efflux pump. J. Bacteriol. 1821754-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seeger, M. A., A. Schiefner, T. Eicher, F. Verrey, K. Diederichs, and K. M. Pos. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 3131295-1298. [DOI] [PubMed] [Google Scholar]
- 32.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB- OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 4265-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stegmeier, J. F., G. Polleichtner, N. Brandes, C. Hotz, and C. Andersen. 2006. Importance of the adaptor (membrane fusion) protein hairpin domain for the functionality of multidrug efflux pumps. Biochemistry 4510303-10312. [DOI] [PubMed] [Google Scholar]
- 34.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 451126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura, N., S. Murakami, Y. Oyama, M. Ishiguro, and A. Yamaguchi. 2005. Direct interaction of multidrug efflux transporter AcrB and outer membrane channel TolC detected via site-directed disulfide cross-linking. Biochemistry 4411115-11121. [DOI] [PubMed] [Google Scholar]
- 36.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 1846499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tikhonova, E. B., and H. I. Zgurskaya. 2004. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 27932116-32124. [DOI] [PubMed] [Google Scholar]
- 38.Vediyappan, G., T. Borisova, and J. A. Fralick. 2006. Isolation and characterization of VceC gain-of-function mutants that can function with the AcrAB multiple-drug-resistant efflux pump of Escherichia coli. J. Bacteriol. 1883757-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, E. W., J. R. Aires, G. McDermott, and H. Nikaido. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 1876804-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300976-980. [DOI] [PubMed] [Google Scholar]
- 41.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 967190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37219-225. [DOI] [PubMed] [Google Scholar]