Abstract
A unique heterotrimeric caffeine dehydrogenase was purified from Pseudomonas sp. strain CBB1. This enzyme oxidized caffeine to trimethyluric acid stoichiometrically and hydrolytically, without producing hydrogen peroxide. The enzyme was not NAD(P)+ dependent; coenzyme Q0 was the preferred electron acceptor. The enzyme was specific for caffeine and theobromine and showed no activity with xanthine.
Caffeine (1,3,7-trimethylxanthine) is readily found in many plant species. It is a major human dietary ingredient that can be found in common beverages and food products, such as coffee, tea, and chocolates. In humans, dietary caffeine is metabolized by the well-characterized hepatic cytochrome P450 1A2 system (2). Although high concentrations of caffeine are regarded as toxic for bacteria, bacteria capable of degrading caffeine have been isolated from soil with and without enrichment procedures (4, 12). Bacteria utilize diverse caffeine catabolic pathways, which can be classified into two groups: demethylating pathways and oxidative pathways. Several Pseudomonas putida strains use a demethylating pathway to metabolize caffeine, producing theobromine (3,7-dimethylxanthine) or paraxanthine (1,7-dimethylxanthine). Subsequent demethylation of these dimethylxanthines is via 7-methyxanthine, which is further demethylated to xanthine. Finally, xanthine is oxidized to urea by xanthine dehydrogenase/oxidase (4, 12). Attempts to purify caffeine N-demethylases from these bacteria were unsuccessful; the nature of these N-demethylases is currently unclear.
Oxidative degradation of caffeine by a Rhodococcus sp.-Klebsiella sp. mixed-culture consortium has been reported (10). Caffeine is oxidized at the C-8 position to form 1,3,7-trimethyluric acid (TMU). An 85-kDa, flavin-containing caffeine oxidase has been purified from this consortium (11). An in vitro coupling study showed that cytochrome c is the best electron acceptor for this enzyme, while oxygen is a poor electron acceptor, resulting in a 10-fold decrease in specific activity. However, concomitant production of hydrogen peroxide and the stoichiometry of TMU formation by the purified caffeine oxidase were not reported. Hence, it is not clear whether this enzyme really metabolizes caffeine by an oxidase mechanism. A 65-kDa caffeine oxidase has been purified from Alcaligenes sp. strain CF8 (13). Although production of hydrogen peroxide was reported, oxygen is a poor electron acceptor for this enzyme because the specific enzyme activity was eight times higher when dichlorophenol indophenol was used as an electron acceptor than when oxygen was used as an electron acceptor. In addition, TMU production by this enzyme was not confirmed. Finally, a caffeine-degrading P. putida strain (ATCC 700097) has been isolated from domestic wastewater (14). When this bacterium was grown on caffeine, a fourfold increase in a cytochrome P450 absorption spectrum signal was observed, compared to the signal for cells that were grown on glucose. However, there is no direct evidence to support the hypothesis that there is oxidative caffeine catabolism in this bacterium by cytochrome P450, since a caffeine- degrading enzyme has not been purified.
These issues prompted us to examine the caffeine degradation pathway in detail. We previously reported isolation of a new caffeine-degrading bacterium, Pseudomonas sp. strain CBB1, by a soil enrichment procedure with caffeine as the sole carbon and nitrogen source (21). TMU was detected as the initial metabolite of caffeine degradation in CBB1 cultures and resting cell suspensions. Here, we describe the purification, preliminary biochemical characterization, and complete stoichiometry of a caffeine-specific, non-NAD(P)+-dependent caffeine dehydrogenase that hydrolytically oxidizes caffeine to TMU.
Detection of caffeine dehydrogenase activities in CBB1 cell extracts.
CBB1 grown in M9 medium (15) with 0.25% (wt/vol) caffeine and 0.4% (wt/vol) yeast nitrogen base (ForMedium, Norfolk, United Kingdom) was harvested at late log phase by centrifugation (13,800 × g for 10 min at 4°C). Yeast nitrogen base did not suppress caffeine utilization but did enhance biomass yield. About 8 g (wet weight) of cells was suspended in 30 ml KD buffer (50 mM potassium phosphate buffer [pH 7.5] with 1.0 mM dithiothreitol) with 10 μg/ml DNase I. The cells were broken by passing them through a chilled French press cell twice at 138 MPa. Unbroken cells and cell debris were removed from the lysate by centrifugation (20,400 × g for 20 min at 4°C). The clear supernatant was saved as the cell extract. Caffeine dehydrogenase in cell extracts could be detected by an activity staining procedure after proteins in cell extracts were resolved by native polyacrylamide gel electrophoresis (PAGE) on 4 to 15% Tris-HCl gels (Bio-Rad, Hercules, CA). The staining solution contained 50 mM potassium phosphate buffer (pH 7.5), 0.5 mM caffeine, and 0.25 mM nitroblue tetrazolium (NBT). Caffeine-dependent reduction of NBT by caffeine dehydrogenase on the gel resulted in a stained band (Fig. 1A). If the caffeine in the staining solution was replaced with xanthine, there was no stained band, suggesting that the caffeine dehydrogenase of CBB1 is caffeine specific and is not a fortuitous activity of xanthine dehydrogenase/oxidase systems. When CBB1 was grown in M9 medium with 0.25% (wt/vol) caffeine plus 0.4% (wt/vol) soytone, caffeine was utilized very slowly compared to the utilization by cells that were grown in caffeine plus yeast nitrogen base. Caffeine dehydrogenase activity was barely detectable in the cell extracts prepared from caffeine-soytone-grown cells, suggesting that alternative carbon and/or nitrogen sources in soytone can repress caffeine dehydrogenase expression.
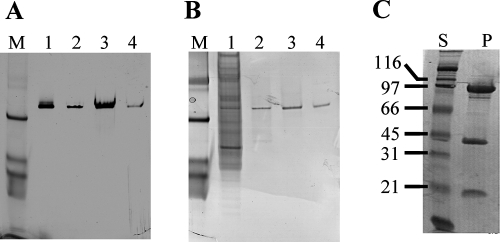
FIG. 1.
Native PAGE of preparations obtained during purification of Pseudomonas sp. strain CBB1 caffeine dehydrogenase. The PAGE gel was stained either (A) for caffeine-dependent NBT reduction activity or (B) for total proteins using Invitrogen SimplyBlue Safe stain. Lane M, prestained broad-range sodium dodecyl sulfate-PAGE standards (Bio-Rad) for monitoring electrophoresis; lane 1, cell extract; lane 2, phenyl Sepharose eluant; lane 3, gel filtration eluant; lane 4, hydroxyapatite eluant. Replacement of the caffeine in the activity staining solution with xanthine did not result in any stained band on PAGE gels. (C) Purified caffeine dehydrogenase resolved on a sodium dodecyl sulfate-PAGE gel (lane P). Lane S, broad-range molecular mass standards (Bio-Rad) (the molecular masses [in kDa] are indicated on the left).
A spectrophotometric activity assay was developed based on caffeine-specific reduction of NBT. A typical 1-ml reaction mixture contained an appropriate amount of enzyme, 0.5 mM caffeine, and 0.5 mM NBT in 50 mM potassium phosphate buffer (pH 7.5). Caffeine dehydrogenase activity was determined by monitoring the increase in absorbance at 566 nm (Δɛ566 = 15,500 M−1·cm−1) due to formazan production with a UV/visible spectrophotometer (Shimadzu UV-2450). One unit of enzyme activity was defined as the reduction of 1 nmol NBT per min under the defined conditions. The activity staining procedure and the spectrophotometric assay facilitated monitoring of the purification of the caffeine dehydrogenase.
Purification of CBB1 caffeine dehydrogenase.
All purification procedures were performed at 4°C using an automated fast protein liquid chromatography system (AKTA Design, Amersham Pharmacia Biotech, Piscataway, NJ). A 4.0 M ammonium sulfate solution was added to cell extracts to obtain a final ammonium sulfate concentration of 0.8 M with constant stirring. After 1 h, the mixture was centrifuged at 16,000 × g for 15 min. The supernatant was loaded onto a 40-ml (bed volume) phenyl Sepharose high-performance column (Amersham) preequilibrated with KD buffer containing 0.8 M ammonium sulfate. Bound proteins were eluted with a 200-ml reverse gradient of ammonium sulfate (0.8 to 0 M in KD buffer) at a rate of 1 ml/min. The column was then washed with 40 ml KD buffer, followed by 40 ml water at the same flow rate. The activity staining procedure and the spectrophotometric assay showed that caffeine dehydrogenase was eluted from the column during the final wash step with water, indicating that this enzyme is hydrophobic. Fractions containing activity were combined, concentrated, and loaded onto an 80-ml (bed volume) Sephacryl S-300 HR gel filtration column (Amersham). Proteins were eluted from the column with KD buffer containing 0.1 M NaCl at an isocratic flow rate of 1 ml/min. Fractions containing activity were combined, concentrated, and exchanged into 5 mM potassium phosphate buffer (pH 7.5) using a YM50 ultrafiltration membrane (Millipore, Bedford, MA). The protein was then loaded onto a 40-ml (bed volume) hydroxyapatite column (Bio-Rad) that had been preequilibrated with 5 mM potassium phosphate buffer (pH 7.5). Caffeine dehydrogenase did not bind to the column. This purification scheme is summarized in Table 1. Purified caffeine dehydrogenase precipitated out of solution and lost activity after a few days of storage at −80°C. Therefore, freshly purified enzyme was immediately used in subsequent biochemical characterization experiments.
TABLE 1.
Purification of caffeine dehydrogenase from Pseudomonas sp. strain CBB1
| Purification step | Total protein (mg)a | Total activity (U) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude cell extract | 468.0 | 8,200 | 18 | 1 | 100 |
| Phenyl Sepharose high-performance liquid chromatography | 18.3 | 2,630 | 144 | 8 | 32 |
| Sephacryl S-300 HR | 3.1 | 1,820 | 586 | 33 | 22 |
| Hydroxyapatite | 0.6 | 875 | 1,530 | 87 | 11 |
Determined by using the Bradford assay (Bio-Rad) with bovine serum albumin as the standard.
Biochemical characterization of caffeine dehydrogenase.
A native PAGE gel showed that purified caffeine dehydrogenase migrated as a single band (Fig. 1B, lane 4). The molecular weight of caffeine dehydrogenase was estimated to be 158,000 by gel filtration chromatography using an 80-ml (bed volume) Sephacryl S-300 HR column equilibrated with KD buffer containing 0.1 M NaCl. Ferritin (440 kDa), catalase (232 kDa), amylase (200 kDa), adolase (158 kDa), alcohol dehydrogenase (150 kDa), and bovine serum albumin (67 kDa) were used to calibrate the column. Sodium dodecyl sulfate-PAGE of the purified enzyme resulted in three major bands, with apparent molecular masses of 90, 40, and 20 kDa (Fig. 1C), suggesting that CBB1 caffeine dehydrogenase is a heterotrimer with an αβγ structure. This αβγ subunit structure differs from the monomeric structure of bacterial caffeine oxidases from Alcaligenes sp. strain CF8 (65 kDa) and a Rhodococcus sp.-Klebsiella sp. mixed-culture consortium (85 kDa) (11, 13). However, the CBB1 caffeine dehydrogenase subunit molecular masses and αβγ subunit structure are similar to those of Veillonella atypica xanthine dehydrogenase (6). Eubacterium barkeri xanthine dehydrogenase is also composed of three subunits with similar molecular masses, but it has an α4β4γ4 structure (16). An annotated xanthine dehydrogenase of Bacillus subtilis also consisted of three catalytic subunits (PucCDE) with theoretical molecular masses of 30, 80, and 19 kDa, respectively (17). A heterotrimeric xanthine oxidase (71, 65.6, and 61.8 kDa) has been purified from the caffeine-degrading strain P. putida L (20). This xanthine oxidase metabolizes xanthine, hypoxanthine, 3-methylxanthine, and theophylline (1,3-dimethylxanthine), but caffeine is not a substrate. Xanthine dehydrogenases/xanthine oxidases belong to the family of molybdenum hydroxylases, and none of them has been reported to use caffeine as a substrate. Molybdopterin, flavin adenine dinucleotide (FAD), and [2Fe-2S] clusters are cofactors that are usually associated with these enzymes (5, 7). The gene encoding the caffeine dehydrogenase 90-kDa subunit (designated cdhA) was amplified from CBB1 genomic DNA using degenerate PCR primers designed from N-terminal protein sequences of the 90- and 40-kDa subunits (data not shown). A BLASTP (1) search revealed that the deduced protein sequence of cdhA has significant homology to molybdopterin-binding subunits of xanthine dehydrogenases, aldehyde dehydrogenases, and carbon monoxide dehydrogenases, suggesting that a molybdopterin cofactor is probably associated with the 90-kDa subunit. The UV/visible absorbance spectrum of purified caffeine dehydrogenase showed an absorption maximum at 275 nm and broad double peaks at around 360 and 450 nm (data not shown), which are characteristic of enzymes containing FAD and iron-sulfur clusters and are similar to the peaks of xanthine dehydrogenases (9, 16). We are in the process of obtaining the gene sequences for the 40- and 20-kDa subunits of caffeine dehydrogenases, which would be helpful in predicting which of these two subunits contains the binding sites for the iron-sulfur clusters and FAD cofactor.
Despite these similarities to xanthine dehydrogenases, a substrate preference assay showed that caffeine is the preferred substrate for CBB1 caffeine dehydrogenase and that the enzyme is not a xanthine dehydrogenase. The substrate preference assay was monitored spectrophotometrically at room temperature using 54 μg of partially purified caffeine dehydrogenase (eluted from the phenyl Sepharose column) in 50 mM phosphate buffer (pH 7.5) containing 0.5 mM test substrates and 0.5 mM NBT as the electron acceptor. The concomitant formation of product in each reaction was also determined by comparison to the retention times and absorption spectra of authentic standards, using a Shimadzu LC-10AD high-performance liquid chromatography system equipped with a BDS Hypersil C18 column (4.6 by 50 mm) and a photodiode array detector. TMU was stoichiometrically produced from caffeine at a 1:1 ratio (data not shown). In the absence of NBT, no enzyme activity, caffeine consumption, or TMU formation was detected. Hydrogen peroxide was not detected by either a QuantiChrome peroxide assay kit (BioAssay Systems, Hayward, CA) or a peroxidase/2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)-based assay kit (Sigma, St. Louis, MO) coupled to TMU formation. With theobromine and theophylline as the substrates, the enzyme activities as determined by the NBT reduction assay were 46 and 0.6%, respectively, of the activity with caffeine as the substrate. No NBT reduction or corresponding product formation occurred when xanthine, 3-methylxanthine, 7-methylxanthine, TMU, and 3,7-dimethyl uric acid were used as substrates.
The maximal caffeine dehydrogenase activity was observed at pH 7.0 in 50 mM potassium phosphate buffer. In the same buffer at pH 6.0, 6.5, 7.5, and 8.0, the activities were 52, 62, 61, and 47% of the maximum activity, respectively, in 50 mM potassium phosphate buffer. When enzyme activity was assayed at various temperatures in 50 mM potassium phosphate buffer (pH 7.0) with 0.5 mM caffeine and 0.5 mM NBT, the caffeine dehydrogenase activity increased linearly from 25 to 66°C, which was the highest temperature allowed by our thermostat-equipped spectrophotometer. This was unexpected, but not unusual. Trimethylamine N-oxide reductase of Escherichia coli (3) and chlorate reductase of Pseudomonas chloritidismutans (18), both of which are molybdoenzymes in mesophilic bacteria, have optimal temperatures of 80 and 75°C, respectively. To assess caffeine dehydrogenase thermostability, the enzyme was incubated at 42 and 70°C for various lengths of time before residual enzyme activity was quantified at 25°C. While a 30-min incubation at 70°C drastically reduced the enzyme activity to 11% of the initial value, 77% of the activity remained after 30 min of incubation at 42°C. Because the optimal temperature for caffeine dehydrogenase has not been established due to instrument limitations, kinetic parameters were determined in 50 mM potassium phosphate buffer (pH 7.0) at 35°C using purified caffeine dehydrogenase. The apparent Km and kcat values for caffeine were 3.7 ± 0.9 μM and 10 ± 0.3 min−1, respectively.
Coenzyme Q0 was the preferred electron acceptor for caffeine dehydrogenase.
The lack of enzyme activity based on caffeine transformation and TMU production in the absence of NBT suggested that CBB1 caffeine dehydrogenase could not utilize oxygen as an electron acceptor. This property is unique. Oxygen is a poor electron acceptor for the two known bacterial caffeine oxidases (11, 13), although they are defined as “oxidases.” Oxygen is also the preferred electron acceptor for mammalian xanthine oxidases, while mammalian and bacterial xanthine dehydrogenases prefer NAD+ as the oxidant (8, 9, 19). This prompted us to test various electron acceptors with CBB1 caffeine dehydrogenase, and the results are summarized in Table 2. Unlike xanthine dehydrogenases, NAD(P)+ did not serve as an electron acceptor for CBB1 caffeine dehydrogenase. Coenzyme Q0 was the preferred electron acceptor, with concomitant formation of TMU. The specific activity decreased about 14-fold when coenzyme Q2 was used. Coenzymes Q4 and Q10 were not active, which could have been due to limited solubility of these hydrophobic molecules in phosphate buffer, steric hindrance by the side chain with repeating isoprene units, or the cofactors being in free form. Cytochrome c did not serve as a direct electron acceptor for caffeine dehydrogenase, although it was the best electron acceptor for caffeine oxidase isolated from a mixed culture of Klebsiella sp. and Rhodococcus sp. (11). However, cytochrome c was reduced in the presence of coenzymes Q0 and Q2, with concomitant formation of TMU. Cytochrome c and coenzyme Q with a side chain containing multiple isoprene units are common electron carriers in a bacterial respiratory electron transport chain. Although CBB1 caffeine dehydrogenase is hydrophobic in nature, no detergent was used during enzyme purification, suggesting that it is unlikely to be an integral membrane protein. However, we cannot completely exclude the possibility that caffeine dehydrogenase may be a peripheral membrane protein. How electron transfer occurs from caffeine dehydrogenase to coenzyme Q and cytochrome c in vivo cannot be elaborated satisfactorily at this time.
TABLE 2.
Caffeine dehydrogenase activity with various electron acceptorsa
| Electron acceptor(s)b | Detection wavelength (nm) | Δɛ (M−1·cm−1) | Sp act (nmol electron acceptor reduced/min/mg)c |
|---|---|---|---|
| NBT | 566 | 15,500 | 747 ± 38 |
| DCPIP | 600 | 21,000 | 1,313 ± 12 |
| DCPIP + phenazine methosulfate | 600 | 21,000 | 1,751 ± 37 |
| Coenzyme Q0 | 410 | 700 | 2,882 ± 75 |
| Coenzyme Q2 | 410 | 700 | 206 ± 26 |
| Coenzyme Q0 + cytochrome c | 550 | 21,000 | 1,205 ± 45 |
| Coenzyme Q2 + cytochrome c | 550 | 21,000 | 37 ± 17 |
Partially purified caffeine dehydrogenase with no TMU degradation activity (eluted from the phenyl Sepharose column) was used in this study. The enzyme activity was assayed at 25°C in 50 mM potassium phosphate buffer (pH 7.0).
Each of the electron acceptors was used at a concentration of 0.5 mM. When 0.5 mM NAD(P)+, 0.5 mM cytochrome c, 0.1 mM coenzyme Q4, and 0.1 mM coenzyme Q10 were used as electron acceptors, enzyme activity could not be detected spectrophotometrically. TMU production was also not detected by high-performance liquid chromatography. DCPIP, dichlorophenol indophenol.
Stoichiometric formation of TMU from caffeine was detected by high-performance liquid chromatography when caffeine dehydrogenase was coupled with the electron acceptors. The values are averages ± standard deviations for triplicate determinations.
Water is the source of the oxygen atom in TMU.
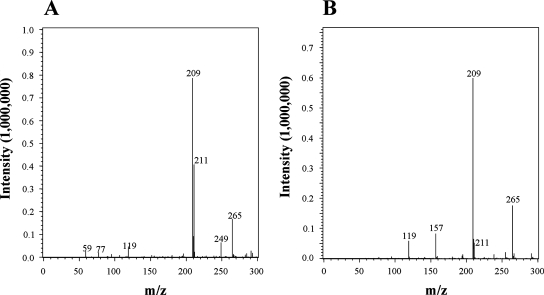
The source of the oxygen atom incorporated at position C-8 of caffeine during the formation of TMU was examined in an H218O (Cambridge Isotope Laboratories, Inc., Andover, MA) incorporation experiment. Partially purified caffeine dehydrogenase with no TMU degradation activity (0.1 mg; eluted from the phenyl Sepharose column), 0.25 mM NBT, and 0.5 mM caffeine were mixed together in 300 μl of 70 mM potassium phosphate buffer (pH 7.4) prepared with 30% (vol/vol) H218O. After incubation at 30°C for 30 min, an aliquot of the reaction mixture was mixed with an equal volume of acetonitrile to precipitate proteins. The supernatant was then analyzed by a high-performance liquid chromatography system equipped with a Shimadzu LCMS-2010EV single-stage quadrupole mass analyzer. In a control reaction with a mixture prepared with regular water, the M-1 parent ion for TMU (m/z 209.0) was detected (Fig. 2B). This parent ion peak was also detected in the reaction mixture prepared with 30% (vol/vol) H218O (Fig. 2A). An m/z 211.0 peak, which corresponded to the parent ion for [18O]TMU, was detected in the reaction mixture prepared with H218O, while this peak in the control reaction was at the background level. Furthermore, the ratio of the m/z 209.0 peak to the m/z 211.0 peak was about 2:1 (Fig. 2A) and corresponded well with the ratio of H216O to H218O (70:30, vol/vol) present in the reaction mixture. Incubation of 0.5 mM TMU in 70 mM potassium phosphate (pH 7.4) buffer prepared with 30% (vol/vol) H218O for 30 min yielded only a parent ion with an m/z 209.0 peak, indicating that there was no exchange of oxygen between TMU and water (data not shown). These results strongly suggest that the oxygen atom in TMU is derived exclusively from water. NAD+-dependent xanthine dehydrogenases also utilize water rather than molecular oxygen (7).
FIG. 2.
Liquid chromatography-mass spectrometry analyses of TMU produced from caffeine by caffeine dehydrogenase in reaction mixtures containing either (A) 30% (vol/vol) H218O or (B) H216O.
Summary.
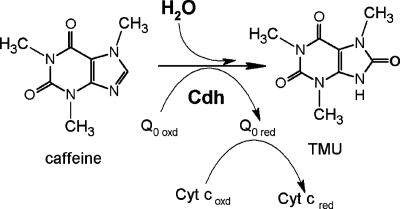
Pseudomonas sp. strain CBB1 uses a unique caffeine dehydrogenase to oxidize caffeine to TMU (Fig. 3). Oxygen is not an electron acceptor, and hydrogen peroxide is not formed during oxidation. This caffeine dehydrogenase differs from previously reported bacterial caffeine oxidases in subunit molecular weights, subunit structure, the inability to use oxygen as an electron acceptor, and the preference for reducing coenzyme Q0 in vitro. Additionally, this enzyme is caffeine specific and could not transform xanthine. The inability to use NAD(P)+ as an electron acceptor also distinguishes it from xanthine dehydrogenases. However, the protein sequence deduced from cdhA revealed significant homology between the caffeine dehydrogenase 90-kDa subunit and the molybdopterin-binding subunit of xanthine dehydrogenases. Furthermore, the αβγ subunit structure of caffeine dehydrogenase, the subunit molecular weights, and the ability to utilize water as the source of oxygen atoms incorporated into the product are properties similar to those of xanthine dehydrogenases. This suggests that caffeine dehydrogenase may be a new member of the xanthine dehydrogenase/oxidase family. Sequencing of the genes encoding the 40- and 20-kDa subunits plus the crystallographic structure of caffeine dehydrogenase could provide insight into the evolutionary and mechanistic relationship between caffeine dehydrogenase and xanthine dehydrogenases.
FIG. 3.
Caffeine oxidation to TMU by Pseudomonas sp. strain CBB1 caffeine dehydrogenase (Cdh). Q0 oxd, oxidized coenzyme Q0; Q0 red, reduced coenzyme Q0; Cyt coxd, oxidized cytochrome c; Cyt cred, reduced cytochrome c.
Acknowledgments
This research was supported by University of Iowa research funds.
We thank William Liechty for his assistance during the course of this study and Shuvendu Das for his help with the liquid chromatography-mass spectrometry experiment.
Footnotes
Published ahead of print on 2 November 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthou, F., B. Guillois, C. Riche, Y. Dreano, E. Jacqz-Aigrain, and P. Beaune. 1992. Interspecies variations in caffeine metabolism related to cytochrome P4501A enzymes. Xenobiotica 22671-680. [DOI] [PubMed] [Google Scholar]
- 3.Buc, J., C. L. Santini, R. Giordani, M. Czjzek, L. F. Wu, and G. Giordano. 1999. Enzymatic and physiological properties of the tungsten-substituted molybdenum TMAO reductase from Escherichia coli. Mol. Microbiol. 32159-168. [DOI] [PubMed] [Google Scholar]
- 4.Dash, S. S., and S. N. Gummadi. 2006. Catabolic pathways and biotechnological applications of microbial caffeine degradation. Biotechnol. Lett. 281993-2002. [DOI] [PubMed] [Google Scholar]
- 5.Fetzner, S. 2000. Enzymes involved in the aerobic bacterial degradation of N-heteroaromatic compounds: molybdenum hydroxylases and ring-opening 2,4-dioxygenases. Naturwissenschaften 8759-69. [DOI] [PubMed] [Google Scholar]
- 6.Gremer, L., and O. Meyer. 1996. Characterization of xanthine dehydrogenase from the anaerobic bacterium Veillonella atypica and identification of a molybdopterin-cytosine-dinucleotide-containing molybdenum cofactor. Eur. J. Biochem. 238862-866. [DOI] [PubMed] [Google Scholar]
- 7.Hille, R. 2005. Molybdenum-containing hydroxylases. Arch. Biochem. Biophys. 433107-116. [DOI] [PubMed] [Google Scholar]
- 8.Hille, R., and T. Nishino. 1995. Flavoprotein structure and mechanism. 4. Xanthine oxidase and xanthine dehydrogenase. FASEB J. 9995-1003. [PubMed] [Google Scholar]
- 9.Leimkuhler, S., R. Hodson, G. N. George, and K. V. Rajagopalan. 2003. Recombinant Rhodobacter capsulatus xanthine dehydrogenase, a useful model system for the characterization of protein variants leading to xanthinuria I in humans. J. Biol. Chem. 27820802-20811. [DOI] [PubMed] [Google Scholar]
- 10.Madyastha, K. M., and G. R. Sridhar. 1998. A novel pathway for the metabolism of caffeine by a mixed culture consortium. Biochem. Biophys. Res. Commun. 249178-181. [DOI] [PubMed] [Google Scholar]
- 11.Madyastha, K. M., G. R. Sridhar, B. B. Vadiraja, and Y. S. Madhavi. 1999. Purification and partial characterization of caffeine oxidase—a novel enzyme from a mixed culture consortium. Biochem. Biophys. Res. Commun. 263460-464. [DOI] [PubMed] [Google Scholar]
- 12.Mazzafera, P. 2004. Catabolism of caffeine in plants and microorganisms. Front. Biosci. 91348-1359. [DOI] [PubMed] [Google Scholar]
- 13.Mohapatra, B. R., N. Harris, R. Nordin, and A. Mazumder. 2006. Purification and characterization of a novel caffeine oxidase from Alcaligenes species. J. Biotechnol. 125319-327. [DOI] [PubMed] [Google Scholar]
- 14.Ogunseitan, O. A. 2002. Caffeine-inducible enzyme activity in Pseudomonas putida ATCC 700097. World J. Microbiol. Biotechnol. 18423-428. [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor laboratory Press, Cold Spring Harbor, NY.
- 16.Schräder, T., A. Rienhöfer, and J. R. Andreesen. 1999. Selenium-containing xanthine dehydrogenase from Eubacteria barkeri. Eur. J. Biochem. 264862-871. [DOI] [PubMed] [Google Scholar]
- 17.Schultz, A. C., P. Nygaard, and H. H. Saxild. 2001. Functional analysis of 14 genes that constitute the purine catabolic pathway in Bacillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator. J. Bacteriol. 1833293-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolterink, A. F. W. M., E. Schiltz, P. L. Hagedoorn, W. R. Hagen, S. W. M. Kengen, and A. J. M. Stams. 2003. Characterization of the chlorate reductase from Pseudomonas chloritidismutans. J. Bacteriol. 1853210-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang, Q., and D. E. Edmondson. 1996. Purification and characterization of a prokaryotic xanthine dehydrogenase from Comamonas acidovorans. Biochemistry 355441-5450. [DOI] [PubMed] [Google Scholar]
- 20.Yamaoka-Yano, D. M., and P. Mazzafera. 1999. Catabolism of caffeine and purification of a xanthine oxidase responsible for methyluric acids production in Pseudomonas putida L. Rev. Microbiol. 3062-70. [Google Scholar]
- 21.Yu, C., K. Yogesh, S. Gopishetty, and M. Subramanian. 2007. Bacterial degradation of caffeine via demethylation and oxidation, abstr. Q-023. Abstr. 107th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.