Abstract
CodY is a nutritional regulator mainly involved in amino acid metabolism. It has been extensively studied in Bacillus subtilis and Lactococcus lactis. We investigated the role of CodY in gene regulation and virulence of the human pathogen Streptococcus pneumoniae. We constructed a codY mutant and examined the effect on gene and protein expression by microarray and two-dimensional differential gel electrophoresis analysis. The pneumococcal CodY regulon was found to consist predominantly of genes involved in amino acid metabolism but also several other cellular processes, such as carbon metabolism and iron uptake. By means of electrophoretic mobility shift assays and DNA footprinting, we showed that most of the targets identified are under the direct control of CodY. By mutating DNA predicted to represent the CodY box based on the L. lactis consensus, we demonstrated that this sequence is indeed required for in vitro DNA binding to target promoters. Similar to L. lactis, DNA binding of CodY was enhanced in the presence of branched-chain amino acids, but not by GTP. We observed in experimental mouse models that codY is transcribed in the murine nasopharynx and lungs and is specifically required for colonization. This finding was underscored by the diminished ability of the codY mutant to adhere to nasopharyngeal cells in vitro. Furthermore, we found that pcpA, activated by CodY, is required for adherence to nasopharyngeal cells, suggesting a direct link between nutritional regulation and adherence. In conclusion, pneumococcal CodY predominantly regulates genes involved in amino acid metabolism and contributes to the early stages of infection, i.e., colonization of the nasopharynx.
Bacteria encounter various environmental cues during their life cycle to which they need to respond appropriately in order to survive. Different niches within a host are likely to provide different nutritional challenges to the microorganism. Such alterations in bacterial gene expression in response to environmental changes are under the strict control of transcriptional regulators.
CodY is a global nutritional repressor that is highly conserved in low-G+C bacteria and has been investigated extensively in Bacillus subtilis, Lactococcus lactis, Streptococcus pyogenes, and very recently in Listeria monocytogenes (3, 16, 33, 35, 39, 42). B. subtilis CodY represses the transcription of more than 100 genes during exponential growth which are involved in different metabolic pathways and cellular processes, such as peptide uptake, development of genetic competence, branched-chain amino acid (BCAA) biosynthesis, motility, and sugar uptake (35, 38). There is one exception, where CodY functions as an activator in B. subtilis; the gene that encodes acetate kinase is positively regulated by CodY and the carbon regulator CcpA, possibly ensuring that an overflow pathway for carbon metabolism is active (40).
Functional studies have shown that the DNA-binding activity of B. subtilis CodY is enhanced by both GTP and the BCAAs isoleucine, leucine, and valine (23, 41). The crystal structure of two fragments of B. subtilis CodY, containing its cofactor and DNA-binding domains, revealed that the regulatory protein interacts with DNA as a dimer (31). L. lactis CodY-DNA binding to its recently identified binding consensus is enhanced by BCAAs but not by GTP (13, 14, 17, 36). Differences between these needs for cofactors of B. subtilis and L. lactis CodY might reflect the physiology of these bacteria. For instance, GTP plays a major role in the development of sporulation in B. subtilis, a process absent from the life cycle of L. lactis. Moreover, in B. subtilis, low levels of GTP induce the development of competence by relieving CodY repression of comK, a critical competence regulator (20, 38). In contrast, natural transformation has never been observed in L. lactis. Interestingly, the lactococcal genome sequence revealed the presence of orthologues of several genes involved in natural transformation in other bacteria (6, 29), but none of these putative competence genes belong to the lactococcal CodY regulon (14).
Another process in which GTP plays a major part is the stringent response. This is a response of a bacterium to amino acid starvation, during which the signal molecule ppGpp is accumulated, resulting in a shutdown of the synthesis of many rRNAs and tRNAs (21, 42). An essential factor in the accumulation of ppGpp is the ribosome-bound protein RelA, which converts GTP to ppGpp. In S. pyogenes, a RelA-independent response to amino acid starvation is found that is, at least in part, regulated by CodY (44, 45). Among the genes repressed during this response are virulence factors such as those encoded by graB, speB, and speH (33). Interestingly, CodY induced the expression of pel/sagA and mga, genes that encode regulatory proteins that themselves positively affect the expression of numerous other virulence factors. This observation suggests a link between nutritional regulation and virulence in S. pyogenes (33).
Streptococcus pneumoniae (the pneumococcus) is a human pathogen that causes diseases such as meningitis, pneumonia, and otitis media in the young, elderly, and immunocompromised (5). Pneumococcal disease is preceded by colonization of the nasopharynx, which is asymptomatic. From there, it can develop into disease under the appropriate conditions. Analysis of the genomes of S. pneumoniae R6 (19), D39 (30), and TIGR4 (46) revealed that CodY orthologs are present on the chromosomes of these strains (spr1439 for R6, spd1412 for D39, and sp1584 for TIGR4). Here, we report on the physiological role of CodY in S. pneumoniae D39 in global transcription, translation, and DNA binding. We show that the pneumococcal CodY regulon consists mainly of genes that are involved in amino acid metabolism, biosynthesis, and uptake. Binding of CodY to its target promoters requires a 15-bp recognition site and is enhanced by BCAAs but not by GTP. Furthermore, we demonstrate that CodY is required for optimal levels of in vitro adherence and colonization of the murine nasopharynx.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are listed in Table 1. All of the pneumococcal strains used in this study were grown in Todd-Hewitt yeast broth at 37°C or on Columbia base agar supplemented with 5% sheep blood (Biotrading). Pneumococcal strains were maintained in 10% glycerol-10% skim milk at −80°C. Escherichia coli DH5α (Table 1) was grown in Luria broth at 37°C while shaking or on Luria broth agar supplemented with appropriate antibiotics (50 mg ampicillin and/or 20 mg trimethoprim).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Antibiotic resistance | Reference |
|---|---|---|
| E. coli | ||
| BL21(DE3) | Stratagene | |
| DH5α | Stratagene | |
| S. pneumoniae | ||
| D39 wild type | NCTC 7466 | |
| D39ΔcodY | Trimethoprim | This study |
| D39Δcps | Kanamycin | 7 |
| D39ΔcpsΔcodY | Kanamycin, trimethoprim | This study |
| D39ΔcpsΔpcpA | Erythromycin, kanamycin | This study |
| D39ΔcpsΔcodYΔpcpA | Kanamycin, trimethoprim, erythromycin | This study |
| Plasmids | ||
| pBlueScript KS+ | Stratagene | |
| pCR2.1 | Invitrogen | |
| pET11C | New England BioLabs | |
| pR412T7 | 4 | |
| pKOT | This study | |
| pKOCOD | This study |
Construction of mutant strains.
The gene that encodes CodY (spd1412) was deleted from strain D39 by allelic replacement with the dfr13 cassette, which confers trimethoprim resistance (2). To this end, codY with 1,000 bp of upstream and downstream flanking sequences was amplified from chromosomal D39 DNA with primers CodSacFwd and CodKpnRv (Table 2). This amplicon was cloned into pBlueScript KS+. Coding DNA of codY was deleted from the plasmid by performing an inverse PCR with primers CodHindFwdinv and CodPstRvinv, amplifying the codY-flanking sequences and pBlueScript KS+ and introducing HindIII and PstI restriction sites for further cloning. This amplicon was ligated with the dfr13 cassette excised from pKOT with HindIII and PstI to create the knockout construct pKOCOD and transformed into E. coli DH5α. A 2,660-bp linear DNA fragment containing codY-flanking DNA and dfr13 was amplified from pKOCOD with primers CodSacFwd and CodKpnRv. This PCR product was used to delete codY from the genome of S. pneumoniae D39 by CSP-1-induced (100 ng/ml) transformation. Transformants were selected on the basis of trimethoprim resistance and were checked by PCR for recombination at the desired location on the chromosome. Wild-type D39 was subsequently transformed with chromosomal DNA isolated from these transformants to rule out the possibility of any additional mutations on the chromosome.
TABLE 2.
Oligonucleotide primers used in this study
| Primer application and name | Nucleotide sequence 5′-3′e | Restriction site |
|---|---|---|
| Cloning | ||
| CodSacFwd | GCGCGCCCGCGGAGGTCGTGCTGGTAAGTCAG | SacII |
| CodKpnRv | GCGCGCGGTACCGATGATTTTCAGGCCAGATG | KpnI |
| CodHindFwdinv | TACGAAGCTTTGAGTCTGCGGGGATTATTG | HindIII |
| CodPstRvinv | ATGCCTGCAGGTGCCATTTTTCACCTCGAA | PstI |
| CodKOrv | CATATTAGCCCCTTGAACGTAGTC | |
| Cod 5′ chrom | CATGTCAAGATTGCGGCTAA | |
| pcpA_L1 | GTTTCCCAGAAAGACTCTGG | |
| pcpA_L2a | CCACTAGTTCTAGAGCGGCGGAGTATCTGCTAGGATTGG | |
| pcpA_R1 | ATAAGAATACGGATTCGGACG | |
| pcpA_R2a | GCGTCAATTCGAGGGGTATCGATGGCTATCGAGTCAATGC | |
| pR412_L | GCCGCTCTAGAACTAGTGG | |
| pR412_R | GATACCCCTCGAATTGACGC | |
| Overexpression | ||
| CodY-NheI-H6-Fwb | CGGCTAGCCATCACCATCACCATCACGCACATTTATTAGAAAAAACT | NheI |
| CodYBamRv | CGCGCGGGATCCTTAGTAATCTCTTTTCTT | BamHI |
| EMSA | ||
| codY EMSA fw | TGCAAGTCAATAAGGAATTTTCA | |
| codY EMSA rv | GCCATTTTTCACCTCGAATT | |
| gdhA EMSA fw | CCAAAAACTGAATTGAAAGAATTT | |
| gdhA EMSA rv | CTTTAGCAGATGTCATATCGTTCTCC | |
| amiA EMSA fw | GACACTTCGAACGACAATTTG | |
| amiA EMSA rv | TGACAACCATTATATCACATTATCCA | |
| ilvB EMSA fw | CATAAATAAACGTTAAAATAGAAAATTCAG | |
| ilvB EMSA rv | CCCTTTCTTTCCTCTTAAAAATAAC | |
| ilvE EMSA fw | GAAATGAAGAATCAGTTCTAAGATGG | |
| ilvE EMSA rv | TCCCAATCAATCGTTACTGTCA | |
| livJ EMSA fw | CCCTTTGTGGGCAATCTTTA | |
| livJ EMSA rv | CAAGCGCCACAAACGATA | |
| psaR EMSA fw | TGAAAGAAGAGCTATTTTCGTCAT | |
| psaR EMSA rv | CTTTGTTTGGGGTCATTCGT | |
| livH EMSA fw | TCAACGTCGCCTTGGATTAT | |
| livH EMSA rv | CGAGGGTTTTCCCTCACTTT | |
| acuB EMSA fw | TTCAGAGCTCTTTTTGCTAGCTT | |
| acuB EMSA rv | CCTTGCGGGTCATAAAATCT | |
| asd EMSA fw | CCCTAGTCTAGCGACTGGGATT | |
| asd EMSA rv | GCGCCGACTACAGCAACT | |
| gapN EMSA fw | CGCCTTGACGTAGTGGATTT | |
| gapN EMSA rv | TCGGATGATTTCCATTTTCC | |
| aliB EMSA fw | TTGAACAATCTTTTAGGAGAACTTGA | |
| aliB EMSA rv | CATTTCCAGAACCTCCTGCT | |
| fatD EMSA fw | TCCCTCGAAGATATTATTTTATCAGA | |
| fatD EMSA rv | TTTCATACCCCGTCCTTTCA | |
| spr0140 EMSA fw | GACATTCTATTTAGAACGAGGATTGA | |
| spr0140 EMSA rv | ATTCCCCCAGTTCCATTTTT | |
| spr0141 EMSA fw | TCTATCAAAATCGCAAATAAGAAA | |
| spr0141 EMSA rv | TTCCATTGTTTCTGCAAATTGT | |
| spr0157 EMSA fw | AACAATGTTTTAGAAGCAAAGGTG | |
| spr0157 EMSA rv | GCTTGCGACATGATAAATACTCC | |
| spr0788 EMSA fw | GAGGAAGGCCTTGTCCAGTT | |
| spr0788 EMSA rv | CCCATAGAGCAACCTGTCGT | |
| spr1436 EMSA fw | TGCGTAAACTACGTGAGCAA | |
| spr1436 EMSA rv | TGACCTGCTTCTGACATTTGA | |
| spr1934 EMSA fw | CTCCGGTTGCTTGTCTCAGT | |
| spr1934 EMSA rv | AGCAGTCCCTCCACGTGATA | |
| spr1945 EMSA fw | TGTGTTTATGGAGAGATGACAATTT | |
| spr1945 EMSA rv | AACCGCAGCTGTAGTTAATGA | |
| spr1982 EMSA fw | CTTGGTCAGGGTCAAGGAAG | |
| spr1982 EMSA rv | TCGCCATAAGTGTGTTCCTG | |
| fake gdhA EMSA fw2c | GAATTGAAAGGGTCTCGAGCTGCTATCTGTTTTTTC | |
| psaR EMSA codY fw2d | GCTATTTTCAATTTTTAGAAAATTTCGTTTTTTC | |
| Footprinting | ||
| codY FP fw | GCAACTTGTCAATAGAAAAGGAA | |
| fake gdhA FP fw | GATATTTCCAAGAAAAACGTTCG | |
| RT-PCR | ||
| codYF | GATTGCCAGTACCGTTGT | |
| codYR | CACGGAGTTCGGAGTAAG | |
| gyrAF | TCTTGATTGCGCTAGACC | |
| gyrAR | ACGACGAAGACGCATATC |
The overlap with primers pR412_L and pR412_R is in bold.
The six-His tag is in bold.
The underlined sequence indicates random nucleotides used to replace the putative CodY box of the gdhA promoter.
The underlined sequence indicates the CodY box introduced into the psaR promoter.
Restriction sites are underlined.
The pcpA (spd1965) deletion mutants were constructed by allelic replacement with the spectinomycin resistance cassette of plasmid pR412T7 as follows. Primers pcpA_L1/pcpA_L2 and pcpA_R1/pcpA_R2 were used to generate PCR products of the left and right flanking regions of pcpA (approximately 500 bp each) (Table 2). These PCR products were fused to the spectinomycin resistance gene amplified with primers pR412_L and pR412_R by means of overlap extension PCR. The resulting PCR product was transformed into S. pneumoniae D39Δcps, and transformants were checked for the presence of the mutation by PCR.
Transcriptional profiling of D39ΔcodY.
Microarray analysis was performed essentially as previously described (18). In short, 500 ml of Todd-Hewitt yeast broth was inoculated with 10 to 20 colonies from agar plates and these cultures were statically grown at 37°C. In all experiments, the D39 wild-type and ΔcodY strains displayed comparable growth characteristics. Samples for RNA isolation were taken when the cultures reached an optical density at 600 nm (OD600) of either 0.1 or 0.2 (early and mid-log growth, respectively). RNA was isolated and purified with the High Pure RNA isolation kit (Roche diagnostics) as previously described (18). Contaminating genomic DNA was removed by treatment with RNase-free DNase I (Roche Diagnostics). RNA was isolated from three replicate cultures. Synthesis, subsequent labeling of cDNA, and microarray hybridization were performed as previously described (18, 48). In all cases, dye swapping was performed with one of the three biological replicates. The microarrays used in this study were constructed as previously described (18, 28) and contain PCR amplicons representing 2,087 open reading frames of S. pneumoniae TIGR4 and 184 open reading frames unique for S. pneumoniae R6, all spotted in duplicate.
DNA microarray data analysis.
Dual-channel array images were acquired with a GeneTac LS IV confocal laser scanner (Genomics Solutions) and analyzed with ArrayPro 4.5 software (Media Cybernetics Inc.). Spots were screened visually to identify those of low quality. Slide data were processed with MicroPreP as previously described (15, 18, 49). Prior to analysis, automatically and manually flagged spots and spots with very low background subtracted signal intensity (5% of the weakest spots [sum of Cy3 and Cy5 net signals]) were filtered out of all data sets. Net signal intensities were calculated by grid-based background subtraction. A grid-based Lowess transformation was performed for slide normalization, negative and empty values were removed, and outliers were removed by the deviation test. Further analysis was performed with a Cyber-T Student t test for paired data (32). For identification of differentially expressed genes, only genes with a minimum of six reliable measurements, a Bayesian P value of <0.001, a false discovery rate of <0.05, and a standard deviation less than the ratio were included. Since these criteria are purely a statistical measurement of differential gene expression and reproducibility across replicates, an additional fold change cutoff of 2 was applied. Initial analysis indicated that the sets of genes regulated by CodY at OD600 values of 0.1 and 0.2 are similar; i.e., 33 genes were upregulated at both optical densities, four genes upregulated at 0.1 and not at 0.2 and three genes upregulated at 0.2 and not at 0.1. Because growth at these phases is also more or less the same (exponential growth), data sets for both ODs were combined. Sequences of several differentially expressed genes were analyzed by TMHMM on the CBS Prediction Server for transmembrane domains (www.cbs.dtu.dk/services/TMHMM/).
Two-dimensional (2D) differential gel electrophoresis (DIGE).
The three independent pneumococcal cultures used for transcriptional profiling (described above) were also used for proteome analysis. Pneumococcal cells were harvested by centrifugation at 4°C and washed four times with cold phosphate-buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride. The pellet was resuspended in 500 μl of milliQ and stored at −20°C until further use. Sample preparation and Cy labeling of proteins were performed according to the manufacturer's protocol (www.amershambiosciences.com). In short, 50 μg of protein of both the wild-type and mutant strains was labeled with Cy3 and Cy5, respectively. After labeling, an additional 200 μg of protein of the corresponding strain was added to have sufficient material for spot identification by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. Fluorescently labeled protein samples were combined, and the total of 500 μg of protein was isoelectrically focused on 18-cm immobilized pH 4 to 7 gradient strips (Amersham). For separation in the second dimension, 12 to 20% gradient polyacrylamide gels were used. Gels were scanned on a Typhoon 9410 imager (Amersham Biosciences) and analyzed with Z3 software (Compugen). Spots that showed at least a twofold change in protein abundance were selected and cut out of the gel after visualization by Coomassie staining. Tryptic digests of proteins were analyzed by MALDI-TOF analysis with the UltraFlex mass spectrometer (Bruker). Mascot Search software (Matrix Science) was used for identification of the proteins. Ratios were calculated from duplicate gels of the three biological replicates. Average ratios were only calculated from spots showing at least a twofold change in abundance in at least four out of six gels.
Overexpression of pneumococcal CodY in E. coli and purification of CodY.
The gene codY was PCR amplified with primers CodY-NheI-H6-Fw and CodYBamRv (Table 2) and cloned into pCR2.1. With the restriction sites NheI and BamHI introduced into the PCR product, N-terminally His6-tagged CodY (H6-CodY) was then cloned in the NheI/BamHI site of pET11C and transformed into E. coli BL21(DE3) for overexpression. The codY coding sequence was confirmed by sequencing. Purification of H6-CodY was performed as previously described, with the HisTrap Kit (Amersham Biosciences) by means of Ni affinity chromatography (1). The purified protein was dialyzed against 60 mM NH4HCO3, freeze-dried, and stored at −20°C until further use. The identity of the purified protein was confirmed by MALDI-TOF analysis, and the concentration was determined by the bicinchoninic acid assay (Bio-Rad).
Electrophoretic mobility shift assay (EMSA).
Gel mobility shift assays were performed essentially as previously described (13). Briefly, DNA fragments of several upstream regions were PCR amplified in the presence of [α-32P]dATP (10 μCi, 3,000 Ci/mmol/50-μl reaction volume; MP Biomedicals) with primers shown in Table 2. Subsequently, 0.4 ng of radioactive amplicon was added to a 50-μl reaction mixture containing binding buffer (20 mM Tris-HCl [pH 8], 8.7% [vol/vol] glycerol, 1 mM EDTA, 5 mM MgCl2, 250 mM KCl, 0.5 mM dithiothreitol, 2 μg bovine serum albumin) and purified CodY at a concentration of 0, 100, 250, 500, 1,000, or 2,000 nM. BCAAs (leucine, isoleucine, and valine) were added up to a concentration of 10 mM each. GTP was added to a concentration of 5 mM. To reduce nonspecific binding, poly(dI-dC) (Amersham) was added to a final concentration of 40 μg/ml. Immediately after incubation for 30 min at 37°C, samples were loaded onto an 8 to 10% (depending on the size of the PCR product) nondenaturing polyacrylamide gel. Gel electrophoresis was performed initially at 100 V for 60 min, after which the voltage was lowered to 50V. Gels were air dried, and X-ray films were developed and scanned after autoradiography. Intensities of free probe (amplicon) were quantified with Imagequant software (Molecular Dynamics). The Kd was calculated by interpolation. Kd is defined as the concentration of CodY at which 50% of the probe has shifted. The psaR promoter region was used as a negative control, since this gene is not regulated by CodY.
DNase I footprinting.
DNA fragments were end labeled with the fmol DNA Cycle Sequencing System kit (Promega). Ten nanograms of end-labeled DNA was incubated for 30 min with 0, 2, 5, 10, 20, or 40 μM purified CodY in 50 μl of binding buffer (EMSA), after which 2 μl of DNase reaction buffer (31.3 U/ml of DNase [Roche], 52 mM CaCl2, 1 mM Tris [pH 7.6]) was added. DNase treatment was stopped after 105 s by the addition of 100 μl of stop buffer (2.5 M ammonium acetate, 20 mM EDTA, 10 μg/μl herring sperm DNA), and DNA was precipitated by ethanol precipitation (in the presence of 20 μg of glycogen). Pellets were washed with 70% ethanol, air dried, and resuspended in 10 μl of formamide loading buffer (containing bromophenol blue and xylene blue). These samples were heated for 1 min at 99°C and applied to a preheated 8% polyacrylamide denaturing sequencing gel. Gels were air dried, and autoradiography was performed by exposing the gel to an X-ray film.
In vitro pneumococcal adherence assay.
Adherence of pneumococci to epithelial cells was studied essentially as described previously (7, 27, 28). In short, monolayers of the human pharyngeal cell line Detroit 562 (ATCC CCL-138) were washed twice with 1 ml of PBS. Aliquots of bacteria (grown to mid-log phase in Todd-Hewitt yeast broth) stored at −80°C were thawed rapidly, harvested by centrifugation, and resuspended in RPMI 1640 medium without phenol red (Invitrogen) supplemented with 1% fetal calf serum to 1 × 107 CFU/ml. One milliliter of bacterial suspension was allowed to adhere for 2 h, after which nonadherent bacteria were removed by three washes with 1 ml of PBS. For quantification of adherence, epithelial cells were subsequently detached by treatment with 25% trypsin-1 mM EDTA in PBS and lysed by the addition of ice-cold 0.025% Triton X-100 in PBS. Serial 10-fold dilutions were plated on blood agar plates to count the adherent bacteria, and the counts were corrected mathematically to account for small differences in the counts of the initial inocula. The wild-type and mutant strains grew comparably in RMPI medium without Detroit 562 cells.
Experimental mouse models.
Nine-week-old female outbred CD-1 mice (Harlan, Horst, The Netherlands) were used for all infection models. Prior to infection, the D39 wild-type and ΔcodY strains were passaged in mice to maintain virulence as described previously (25). Cultures of S. pneumoniae D39 or ΔcodY were grown to an OD600 of 0.3 and stored in aliquots at −80°C in 10% glycerol. Prior to infection, these aliquots were spun down and bacteria were resuspended in sterile PBS. Mice were lightly anesthetized with 2.5% (vol/vol) isoflurane-O2, and infected intranasally with 106 CFU of bacteria as described previously (24). At predetermined time points after infection, groups of mice were sacrificed by cervical dislocation and samples of various sites were taken to determine the bacterial loads. In the colonization model, five mice per group were infected with 10 μl of PBS containing 106 CFU of either the D39 wild-type or ΔcodY strain, a volume small enough to only infect the noses (nasopharynxes) of the mice. Bacteria were recovered from the nasopharynx by flushing the nostrils with 2 ml of sterile PBS (26), and lungs were removed from the body and homogenized in 2 ml of sterile PBS with a hand-held homogenizer (polytron PT 1200; Kinematica AG). Viable bacteria in nasopharyngeal lavage fluid (NPLF) and homogenized lungs were counted by plating serial 10-fold dilutions on Columbia blood agar (Oxoid) supplemented with 5% (vol/vol) defibrinated sheep blood (Biotrading). Time points for sampling were 0, 24, 48, 96, and 192 h postinfection. For the pneumonia model, five mice per group were infected with 50 μl of PBS containing 106 CFU of the D39 wild-type or ΔcodY strain. Viable bacteria were recovered from the different sites and quantified as described above. In addition, a blood sample was removed by cardiac puncture with a 1-ml syringe. Time points for sampling were 0, 12, 24, and 36 h postinfection. In the sepsis model, six mice per group were infected by the tail vein with 106 CFU resuspended in 100 μl of sterile PBS. Bacteria were recovered from the blood by a lateral tail vein puncture from the same mouse at 0, 12, 24, and 36 h postinfection and quantified as described above. Bacteriology results are expressed as the geometric mean ± the standard errors of the mean. Comparison of bacterial loads was performed with Student's t test. In all analyses, P < 0.05 was considered statistically significant. All experiments were performed with approval of the Animal Experimentation Committee (DEC) of the Erasmus Medical Centre.
In vivo expression of pneumococcal codY.
Female outbred CD-1 mice were infected with 107 CFU of wild-type D39 according to the pneumonia model described above. Control mice were inoculated with sterile PBS only. At 40 h postinfection, mice were sacrificed by cervical dislocation and NPLF and bronchoalveolar lavage fluid (BALF) were collected. Upon collection of 2 ml of NPLF and BALF, 20 μl was used for determination of the bacterial load and the remaining fluid was mixed with 4 ml of RNAprotect (Qiagen) and incubated for 5 min at room temperature. Bacteria were collected by centrifugation (5 min at 16,000 × g and 4°C) and pellets were snap-frozen in liquid nitrogen. RNA from NPLF and BALF was isolated with the RNeasy kit (Qiagen) by on-column DNase treatment (Qiagen). Subsequently, 200 to 250 ng of total RNA was amplified with the SenseAmp kit (Genisphere). The product of this amplification was reverse transcribed by Superscript III reverse transcriptase (Invitrogen). To confirm the absence of genomic DNA, reactions without reverse transcriptase were performed. Of the obtained cDNA, 1 μl of a 1:2 dilution served as a template for a PCR with codY-specific primers (Table 2). The gyrA (sp1219) amplicon was used as an internal control. Gene expression was assessed in samples obtained from three individual mice.
Nucleotide sequence accession number.
The microarray data obtained in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) under GEO Series accession number GSE7350.
RESULTS
The CodY regulon.
To identify genes regulated by CodY, we compared the transcriptional profiles of wild-type D39 and its codY mutant by DNA microarray analysis. Western blotting of wild-type and codY mutant cell lysates with anti-H6-CodY antibodies showed that CodY is present in exponentially growing (OD600 values of 0.1 and 0.2) wild-type D39 cells and not in D39ΔcodY (data not shown). The microarray analysis showed that pneumococcal CodY functions mainly as a transcriptional repressor, as 43 of the 47 differentially expressed genes were found to be upregulated in the codY mutant at both of the ODs measured (Table 3 ). These included mainly genes predicted to encode proteins involved in amino acid metabolism, such as the oligopeptide permease AliA/B-Ami (aliA, sp0366; amiACDEF, sp1887 to sp1891), a putative BCAA transporter (liv operon, sp0749 to sp0753), acuB (sp0754), the ilv operon (sp0445 to sp0450), and a putative operon that includes ilvE, a BCAA aminotransferase gene, genes for two hypothetical proteins, and pcp (sp0856 to sp0860). Additional CodY-regulated genes involved in amino acid metabolism were gdhA, asd, and dapA (sp1306, sp1013, and sp1014, respectively). The repressed gene set also contained genes predicted to be involved in other cellular processes, such as the fat locus that encodes an iron transport system (sp1869 to sp1872) that has been shown to contribute to pneumococcal virulence (8, 9) and gapN (sp1119), which encodes NADP-dependent glyceraldehyde-3-phosphate dehydrogenase. Interestingly, a cluster of five genes that encode a putative transcriptional regulator, a putative bacteriocin (12), and three putative membrane proteins (sp0141 to sp0145) was also strongly derepressed.
TABLE 3.
Genes differentially expressed in D39ΔcodY
| Gene name | ID
|
Annotationa | MA ratiob | 2D ratioc | EMSAd |
Kd (nM)e
|
||
|---|---|---|---|---|---|---|---|---|
| TIGR4 | D39 | CodY | CodY (10 nM ILV) | |||||
| mutR | sp0141 | spd0144 | Positive transcriptional regulator of MutA | 2.51 | + | 1,115 | 439 | |
| sp0142 | spd0145 | Hypothetical protein (bacteriocin) | 2.24 | + | >2,000 | 1,938 | ||
| sp0143 | spd0146 | Hypothetical protein | 2.48 | |||||
| sp0144 | spd0147 | Hypothetical protein | 2.84 | |||||
| sp0145 | spd0148 | Hypothetical protein | 2.43 | |||||
| sp0159 | spd0161 | Hypothetical protein | 2.46 | + | 834 | 73 | ||
| cps2K | spd0326 | UDP-glucose 6-dehydrogenase | 1.47 | |||||
| aliA | sp0366 | spd0334 | Oligopeptide-binding protein | 2.18 | 2.98 | |||
| pbpA | sp0369 | spd0336 | Penicillin-binding protein 1A | −0.09 | 1.92 | |||
| ilvB | sp0445 | spd0404 | Acetolactate synthase large subunit | 3.02 | + | >2,000 | 284 | |
| ilvN | sp0446 | spd0405 | Acetolactate synthase small subunit | 3.24 | ||||
| ilvC | sp0447 | spd0406 | Ketol-acid reductoisomerase | 3.03 | 2.28 | |||
| sp0448 | spd0407 | Hypothetical protein | 2.98 | |||||
| sp0449 | spd0408 | Hypothetical protein | 3.70 | |||||
| ilvA | sp0450 | spd0409 | Threonine dehydratase | 2.95 | 1.70 | |||
| grpE | sp0516 | spd0459 | Heat shock protein GrpE | 0.22 | 1.26 | |||
| livJ | sp0749 | spd0652 | ABC transporter substrate-binding protein, BCAA transport | 1.88 | + | 597 | 52 | |
| livH | sp0750 | spd0653 | ABC transporter membrane-spanning permease, BCAA transport | 1.58 | ||||
| livM | sp0751 | spd0654 | ABC transporter membrane-spanning permease, BCAA transport | 1.90 | ||||
| livG | sp0752 | spd0655 | ABC transporter ATP-binding protein, BCAA transport | 2.25 | ||||
| livF | sp0753 | spd0656 | ABC transporter ATP-binding protein, BCAA transport | 2.30 | ||||
| acuB | sp0754 | spd0657 | Acetoin utilization protein | 1.08 | + | >2,000 | 773 | |
| pnp | sp0831 | spd0726 | Purine-nucleoside phosphorylase | 0.35 | 1.44 | |||
| ilvE | sp0856 | spd0749 | BCAA aminotransferase | 2.08 | —f | + | 482 | 68 |
| sp0857 | spd0750 | ABC-SBP-internal deletion, AliB like (blastN) | 2.09 | |||||
| sp0858 | spd0751 | Hypothetical protein | 2.15 | |||||
| sp0859 | spd0752 | Membrane protein | 2.07 | |||||
| pcp | sp0860 | spd0753 | Pyrrolidone-carboxylate peptidase | 1.39 | 1.75 | |||
| sp0882 | spr0778 | Putative esterase, S. suis (blastN) | 1.64 | |||||
| sp0884 | spd0780 | Putative esterase S. suis (blastN) | 1.78 | − | ||||
| sp0885 | spd0781 | Putative carbamoylphosphate synthase large subunit (blastN) | 1.94 | |||||
| asd | sp1013 | spd0900 | Aspartate-semialdehyde dehydrogenase | 2.06 | 1.40 | + | >2,000 | 120 |
| dapA | sp1014 | spd0901 | Dihydrodipicolinate synthase | 1.69 | ||||
| gapN | sp1119 | spd1004 | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | 1.05 | + | 752 | 211 | |
| glxK | sp1126 | spd1011 | Glycerate kinase | 1.33 | ||||
| gdhA | sp1306 | spd1158 | Glutamate dehydrogenase (NADP+) | 1.71 | 2.80 | + | 726 | 156 |
| rplL | sp1354 | spd1187 | 50S ribosomal protein L7/L12 | −0.17 | 2.20 | |||
| rplJ | sp1355 | spd1188 | 50S ribosomal protein L10 | −0.10 | 1.43 | |||
| nagB | sp1415 | spd1246 | N-Acetylglucosamine-6-phosphate isomerase | −0.19 | 1.37 | |||
| sp1578 | spd1408 | Methyltransferase, putative | 1.16 | + | >2,000 | 368 | ||
| dctA | sp1753 | spd1563 | Dicarboxylate/amino acid:cation (Na+ or H+) symporter | 2.81 | ||||
| sp1754 | spd1564 | Hypothetical protein (integral membrane protein) | 2.84 | |||||
| fatD | sp1869 | spd1649 | Iron compound ABC transporter, permease protein | 2.80 | + | >2,000 | >2,000 | |
| fatC | sp1870 | spd1650 | Iron compound ABC transporter, permease protein | 2.44 | ||||
| fecE | sp1871 | spd1651 | Iron compound ABC transporter, ATP-binding protein | 1.37 | ||||
| fatB | sp1872 | spd1652 | Iron compound ABC transporter, iron-compound-binding protein | 1.07 | ||||
| amiD | sp1889 | spd1669 | Oligopeptide ABC transporter, permease protein | 1.20 | ||||
| amiC | sp1890 | spd1670 | Oligopeptide ABC transporter, permease protein | 1.72 | ||||
| amiA | sp1891 | spd1671 | Oligopeptide ABC transporter, oligopeptide-binding protein | 1.21 | 1.41 | + | 677 | 433 |
| sp2125 | spd1954 | Hypothetical protein | 2.18 | + | 754 | 630 | ||
| rpoA | sp0236 | spd0218 | DNA-directed RNA polymerase subunit α | −0.12 | −1.86 | |||
| ABC-NBD | sp0483 | spd0434 | ABC transporter ATP-binding protein, cobalt transport | −0.34 | −1.96 | |||
| fba | sp0605 | spd0526 | Fructose-bisphosphate aldolase | −0.20 | −1.64 | |||
| pepV | sp0623 | spd0542 | Dipeptidase | −0.21 | −2.42 | |||
| sp1429 | spd1258 | Putative peptidase, U32 family | −1.29 | −1.81 | ||||
| pgm | sp1498 | spd1326 | Phosphoglucomutase | 0.11 | −1.57 | |||
| atpB | sp1513 | spd1340 | Proton-translocating ATPase, F0 sector, subunit a | −0.15 | −1.58 | |||
| dpr | sp1572 | spd1402 | DNA binding protein starved-cell-like peroxide resistance protein | −2.69 | −1.55 | |||
| codY | sp1584 | spd1412 | Transcriptional pleiotropic repressor CodY | −1.68 | + | >2,000 | 382 | |
| gapA | sp2012 | spd1823 | Glyceraldehyde-3-phosphate dehydrogenase | −0.20 | −1.93 | |||
| pcpA | sp2136 | spd1965 | Choline-binding protein PcpA | −1.36 | + | >2,000 | 903 | |
| pgsA | sp2222 | spd2049 | CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase | 0.14 | −1.67 | |||
Annotation is according to the Kyoto Encyclopedia of Genes and Genomes database (www.kegg.com).
Microarray (MA) ratios are given as follows: expression of ΔcodY/expression of wild type (log2 transformed).
2D DIGE (2D) ratios are given as follows: expression of ΔcodY/expression of wild type (log2 transformed).
In the EMSA column, a plus sign indicates a shift, while a minus sign indicates no shift.
The Kds of the promoter region (concentration of CodY at which 50% of the probe is shifted) without addition of BCAAs and in the presence 10 mM BCAAs are shown.
—, only expressed in the codY mutant.
In addition to codY itself, three genes were found to be downregulated in the codY mutant, i.e., sp1429, which is predicted to encode a peptidase; sp2136, which encodes the choline-binding protein PcpA; and dpr (sp1572), which codes for a putative starvation-induced protein.
To examine if the transcriptional differences corresponded with changes in protein expression, 2D DIGE was performed with protein isolated from the D39 wild-type and ΔcodY strains. Fifteen proteins were identified as being significantly more abundant in the codY mutant, and of these, eight of the corresponding genes had also been identified as differentially expressed by microarray analysis (Table 3). Upregulated proteins identified solely by 2D DIGE included penicillin-binding protein PbpA (sp0369), heat shock protein GrpE (sp0516), and glucosamine-6-phosphate isomerase (sp1415). Ten proteins were found to be significantly more abundant in the wild type than in the mutant, two of which were identified by transcriptional analysis as well (Table 3). Proteins only detected by 2D-DIGE included fructose bisphosphate aldolase (Fba, sp0605), glyceraldehyde-3-phophate dehydrogenase (GapA, sp2012), F-type H+-transporting ATPase α chain (AtpB, sp1513), and the dipeptidase PepV (sp0623). Interestingly, L. lactis PepV belongs to the proteolytic system regulated by CodY, although direct regulation of pepV by CodY has never been shown (16).
Binding of CodY to target promoters.
To discriminate between direct and indirect regulation by CodY, EMSAs were performed. Upstream regions containing putative promoter regions of genes identified by microarray analysis, 2D DIGE analysis, or in silico screening with the L. lactis binding consensus sequence (AATTTTCWGAAAATT) (14) were amplified and incubated with purified H6-CodY. Also, the effect of addition of BCAAs and GTP on DNA binding to these promoters was investigated. The upstream region of codY was bound by purified H6-CodY, especially in the presence of BCAAs, suggesting that CodY regulates its own expression (Table 3). Five promoter regions of genes or operons predicted to be involved in BCAA biosynthesis or transport showed a shift, namely, PilvB, PilvE, PlivJ, PamiA (Table 3), and PaliB (in microarray analysis, it was 1.8-fold upregulated in ΔcodY). This binding of CodY was enhanced by addition of BCAAs with a factor 2 to 20. For instance, the Kds (the CodY concentration at which 50% of the DNA probe is shifted) of PilvE and PlivJ ranged from 480 to 600 nM CodY without the addition of BCAAs and from 50 to 70 nM CodY in the presence of BCAAs (Table 3). No effect of GTP on DNA binding was observed (data not shown). Binding of CodY to the other three promoters was also enhanced by BCAAs, although to a lower extent. Other promoter regions to which CodY was able to bind were PgdhA, PfatD, PacuB, Pasd, PgapN, Psp0141, Psp0142, Psp0159, Psp1578, and Psp2125 (Fig. 1A and Table 3). Several of these promoters had Kds (without BCAAs) higher than the highest CodY concentration tested (Table 3), indicating that their Kds were greater than 2,000 nM. Again, the affinity of CodY for the promoters was enhanced by the presence of BCAAs (Kds ranging from 150 to 1,950 nM, depending on the promoter region). All but two of these promoter regions (Psp0141 and Psp0142) contained a sequence resembling the CodY-binding box. Finally, CodY also bound to the upstream region of pcpA, one of the genes downregulated in the codY mutant, and this binding was enhanced by the addition of BCAAs (Table 3). In addition, a sequence resembling the CodY box is present 105 bp upstream of the pcpA start codon (5′-AATTTATAAAATGTA-3′). This suggests that CodY might positively regulate the expression of PcpA, a choline-binding protein suggested to be involved in adherence (37).
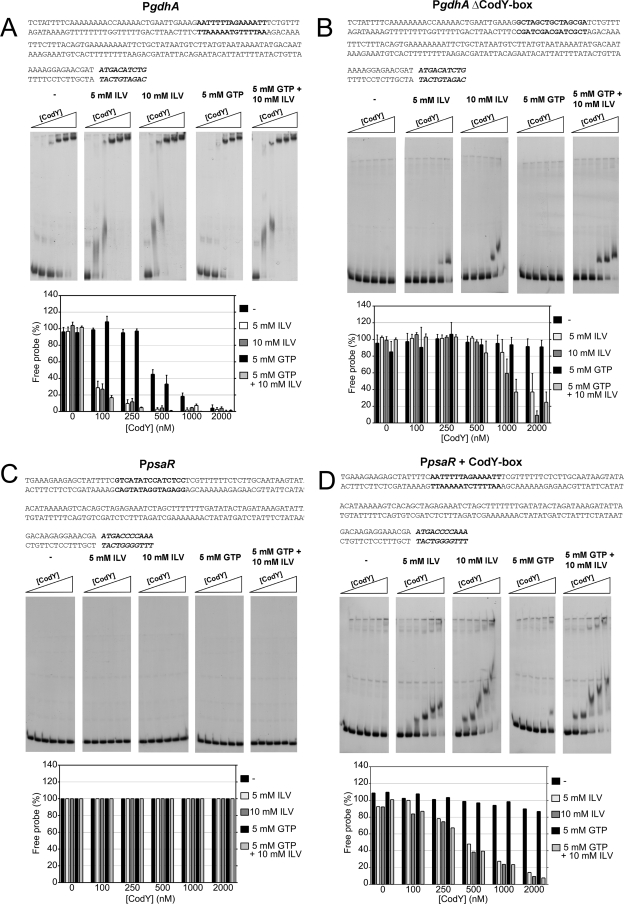
FIG. 1.
EMSA with H6-CodY. DNA binding of CodY to the gdhA promoter region (140 bp) and the psaR promoter region (147 bp) in the absence or presence of BCAA (5 or 10 mM ILV) and/or GTP (5 mM). In each series, successive lanes contained increasing amounts of CodY (0, 100, 250, 500, 1,000, and 2,000 nM). In the sequences (top), bold letters indicate the CodY recognition site (or its position in the mutated sequence) and the start of the coding sequence is in bold and italic letters. Histograms (bottom) show the percentages of free probe (DNA fragment) left. The wild-type gdhA promoter region (A); the mutated gdhA promoter with the CodY box replaced with random nucleotides (B); the wild-type psaR promoter, not regulated by CodY (C); and the mutated psaR promoter with a CodY box introduced 100 bp upstream of the coding sequence (D) are shown.
EMSA of the gdhA promoter revealed the presence of two retarded protein-DNA complexes, a major one with the lowest mobility seen at intermediate to high concentrations of CodY and one with intermediate mobility seen with lower concentrations of CodY only in the presence of BCAAs (Fig. 1A). Similar patterns were observed for the other promoters tested (not shown). To gain further insight into the binding of CodY to the gdhA promoter region, the sequence matching the L. lactis consensus sequence was replaced with random nucleotides and tested by EMSA (Fig. 1B). The distance of this consensus sequence to the start codon is 79 bp. Without its putative binding domain, CodY did bind the DNA, but only at high concentrations, i.e., 1,000 and 2,000 nM. Furthermore, only the intermediately shifted band was seen. The major complex might consist of the promoter region to which two (or more) CodY dimers have bound, one to the CodY box, and one to a secondary binding site (Fig. 1A). This putative secondary binding site would still be present in the mutated gdhA promoter fragment, resulting in an intermediate shift. Next, the upstream sequence of the gene psaR (sp1638), to which CodY is not able to bind under any conditions (Fig. 1C), was mutated in such a way that the CodY box consensus sequence was introduced. Although no CodY binding to this DNA fragment was observed without BCAAs, a clear shift was observed upon the addition of BCAAs (Kd of 400 nM), indicating that this sequence is indeed involved in protein-DNA interaction (Fig. 1D). Only a complex with intermediate mobility was seen, indicative of binding of one CodY dimer to the introduced CodY box. In comparison to the PgdhA with a CodY box, this intermediate shift occurs at lower CodY concentrations, suggesting that CodY has a higher affinity for the CodY box than for the putative secondary binding site.
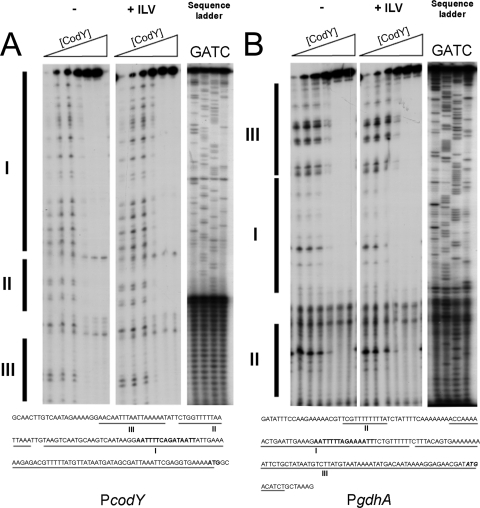
DNase I footprinting of the gdhA and codY promoters.
To determine the CodY binding sites in more detail, we performed DNase I footprint assays. First, H6-CodY was incubated with a 160-bp DNA fragment containing the promoter region of codY, both without and with the addition of BCAAs (10 mM isoleucine, leucine, and valine V [ILV]) or BCAAs (10 mM ILV) and GTP (5 mM). Increased protection from DNase I was observed with increasing concentrations of H6-CodY (Fig. 2A). This protection was not enhanced by addition of GTP (data not shown) or, surprisingly, BCAAs (Fig. 2A). Three protected regions were identified, one of which contained the putative CodY box (region I, Fig. 2A). Additional protected sequences were found to be AT-rich DNA sequences (regions II and III, Fig. 2A). Next, the 178-bp region upstream of gdhA was incubated with H6-CodY. Again, we did not observe any effect of addition of BCAA (Fig. 2B) or GTP (data not shown). Three regions were protected from DNase I degradation. One large region, similar to that observed in the codY promoter region, contained the putative CodY box (region I, Fig. 2B). The second region was a T-rich domain (region II, Fig. 2B). The third region was an additional region downstream of region I. At higher CodY concentrations regions I and III appeared to form a large protected DNA sequence of approximately 120 bp (Fig. 2B). Apart from the CodY box, the codY and gdhA promoter regions bear no clear similarities.
FIG. 2.
DNase I footprinting with H6-CodY. DNase I footprint of the codY (A) and gdhA (B) promoter regions in the absence or presence of BCAA (10 mM ILV). In each series, successive lanes contained increasing amounts of CodY (0, 2, 5, 10, 20, and 40 μM). Protected regions are marked by vertical bars and underlined in the DNA sequence (bottom). The CodY boxes are in bold letters, and the start codons are in italic and bold letters.
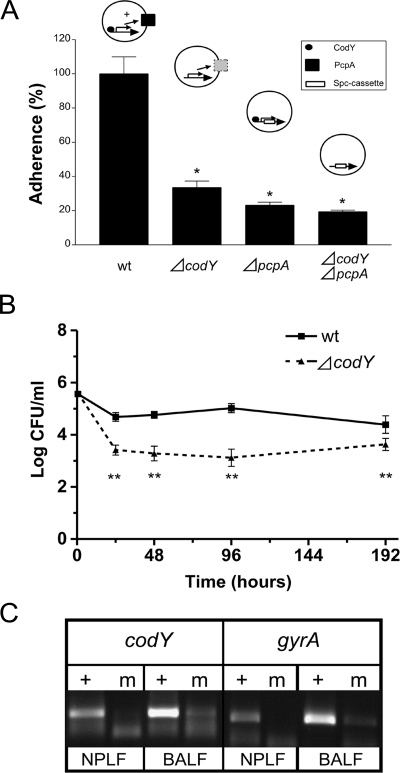
Adherence of the codY mutant to pharyngeal cells.
Colonization of the nasopharynx is mediated by adherence of the bacterium to respiratory epithelial cells, a process that likely needs to be tightly regulated. To examine the role of CodY in this process, we tested the ability of D39ΔcodY to adhere to the human pharyngeal epithelial cell line Detroit 562 in vitro. Since encapsulated strains tend to adhere to a lower extent compared to unencapsulated strains, a codY mutant in a capsule-negative genetic background (D39Δcps) was constructed (7). Clearly, D39ΔcpsΔcodY was less capable of adhering to human pharyngeal cells than was D39Δcps (P < 0.0001, Fig. 3A). As previously mentioned, pcpA, which encodes a putative adhesin (37), was found to be downregulated in the codY mutant, meaning that this gene is activated by CodY. To investigate the role of CodY activation in adherence through PcpA, we constructed a D39ΔcpsΔpcpA strain and a D39ΔcpsΔcodYΔpcpA strain and tested them for the ability to adhere to human epithelial cells. D39ΔcpsΔpcpA showed significantly lower levels of adherence than D39Δcps (P < 0.0001, Fig. 3A). This difference was even greater than that between the D39Δcps and D39ΔcpsΔcodY strains (Fig. 3A).
FIG. 3.
Involvement of CodY in adherence and colonization. (A) In vitro adhesion of D39Δcps, D39ΔcpsΔcodY, D39ΔcpsΔpcpA, and D39ΔcpsΔcodYΔpcpA to Detroit 562 nasopharyngeal cells. The adherence of the codY mutant is given as a percentage of that of the wild type (wt). A possible model of the regulation of pcpA by CodY is shown above the histogram. For an explanation, see text. (B) Intranasal challenge with 106 CFU of the D39 wild-type or ΔcodY strain. (C) Expression of codY during experimental virulence. Transcripts of codY and gyrA were identified in bacterial RNA isolated from NPFL and BALF at 40 h postinfection. A plus sign indicates a reaction with reverse transcriptase, and m indicates the negative control (without reverse transcriptase). *, P < 0.0001 (Mann-Whitney U test); **, P < 0.05 (Student's t test).
Contribution of CodY to experimental virulence.
To assess the contribution of codY to pneumococcal virulence, the D39 wild-type and ΔcodY strains were tested in three mouse models of pneumococcal infection. No significant differences in bacterial loads were observed with the pneumonia and sepsis model of infection (data not shown). In the colonization model of infection, however, a clear phenotype for D39ΔcodY was seen, i.e., a consistently and significantly lower bacterial load compared to the wild-type strain for the duration of the infection (P < 0.0015, Fig. 3B).
To assess whether codY is actually expressed at the different sites during infection, we isolated bacterial RNA from the NPLF and BALF obtained from mice infected with wild-type D39. Indeed, a clear codY transcript could be detected in the nasopharynx and lungs at 40 h postinfection (Fig. 3C).
DISCUSSION
CodY has been described as a nutritional repressor in various bacteria, in which it represses genes that are involved in the biosynthesis and uptake of amino acids, as well as genes that are typically expressed during late exponential or stationary phase (14, 35). The aim of the present study was to elucidate the role of CodY in the physiology of S. pneumoniae. Transcriptome and proteome analyses identified several genes previously shown to be part of the CodY regulon in other bacteria (Table 3). Ten of the genes and proteins were identified as CodY targets by both techniques. Discrepancies observed between transcriptional and proteome analyses could be due to several factors, such as low levels of gene expression, instability of proteins, or specific regulation at the translational level.
The pneumococcal CodY regulon predominantly consists of genes and operons involved in BCAA metabolism and general amino acid metabolism, such as the ilv operon (ilvBNC) and the genes ilvA and ilvE, which were found to be strongly upregulated in the codY mutant. Interestingly, ilvA, ilvD, and ilvE are strongly upregulated in a ciprofloxacin-resistant S. pneumoniae strain compared to its ciprofloxacin-sensitive parental strain after induction with this antibiotic (34). However, both the D39 wild-type and codY mutant strains were found to be sensitive for ciprofloxacin, suggesting that these enzymes are not directly involved in ciprofloxacin resistance (data not shown). In B. subtilis, the ilvBNC operon is tightly regulated by three regulators, CcpA, TnrA, and CodY. These regulators can activate or repress the transcription of the ilv operon, depending on the nutritional conditions (47). No TnrA homologue is found in the genome of the pneumococcus, and regulation of ilvBNC by CcpA has not been investigated yet in S. pneumoniae. Another example of a gene whose expression is controlled by multiple regulators is gdhA (glutamate dehydrogenase). Apart from its repression by CodY, the expression of this gene is also regulated by the nitrogen regulatory protein GlnR (28), suggesting that GdhA plays a central role in pneumococcal nitrogen metabolism.
Preliminary results indicate that the intracellular amino acid pool is indeed affected in the codY mutant; lower intracellular glutamate and higher NH3 concentrations were measured (unpublished results). This is probably due to the higher abundance of GdhA, which catalyzes the deamination of glutamate into α-ketoglutarate and NH3. The higher NH3 concentration may also be the result of the higher abundance of threonine dehydratase (IlvA), as this enzyme catalyzes the conversion of threonine to 2-oxobutanoate and NH3. Accordingly, the butanoate concentration was also higher in the codY mutant (unpublished results).
By DNase I footprinting, we identified the pneumococcal CodY-binding box, similar to the consensus sequence described for L. lactis. Furthermore, EMSAs showed that most of the genes upregulated in the codY mutant are under the direct control of CodY, although CodY does not have the same affinity for all promoters. As in L. lactis, BCAAs but not GTP, enhanced the binding of H6-CodY to its target promoters. For instance, the Kd without BCAAs for PcodY was >2,000 nM whereas with BCAAs it was 382 nM. This suggests that when high concentrations of BCAAs are present, CodY might repress itself stringently, whereas the affinity of CodY for its own promoter is relatively low in the presence of low levels of BCAAs. On the other hand, the overall affinity of CodY for the gdhA promoter region was much higher, indicating that even at very low BCAA concentrations, CodY might still able to repress gdhA expression.
CodY targets whose expression was affected most in the codY mutant (6- to 10-fold upregulation) were genes that encode products predicted to be involved in BCAA metabolism. Their promoter regions also displayed the highest affinity for CodY, especially in the presence of BCAAs. Among these was the previously mentioned ilvBNC operon, which encodes enzymes that condense threonine and pyruvate or two pyruvates into branched-chain keto acids, precursors of the BCAAs. Derepression of this operon might therefore result in an alteration of the pyruvate pool (40). Previous studies have shown that a mutant for pyruvate oxidase, encoded by spxB, is affected in the ability to adhere to type II lung cells and epithelial cells (43). It was found that upon addition of acetate, adherence of the spxB mutant was restored to wild-type levels, indicating that SpxB is not an adhesin and that acetyl-CoA influences the adhesive properties of pneumococci. Metabolites, which pneumococcus produces during colonization, could potentially play a role in creating a favorable environment for adhesion. Normally, bacteria use the transport of acids like acetate and lactate out of the cell to build up an electrochemical gradient (proton motive force). As a result of the codY mutation, S. pneumoniae might no longer be able to adequately maintain this proton motive force. From a nutritional point of view, without cellular active CodY, the cell is considered to be in a “hunger state,” which is in agreement with the hypothesis of Spellerberg and coworkers, who proposed that pneumococci adhere in a nutrient-rich, but not in a nutrient-poor environment (43). This suggests that adhesins are preferentially expressed under nutrient-rich conditions. In line with this, expression of the choline-binding protein PcpA, a putative adhesin (37), was downregulated in the codY mutant, and CodY bound to the pcpA upstream region, suggesting a possible link between nutritional regulation and adhesion. Our in vitro adhesion assays showed that PcpA is indeed required for wild-type levels of adherence in D39. With a cps-codY-pcpA triple mutant, we showed that there is an additional effect when pcpA is deleted in a codY mutant. This could indicate that in the case of a codY mutation, no induction of pcpA transcription occurs, but there is still some background transcription, allowing suboptimal adherence compared to that of the isogenic wild type. The cps-codY-pcpA triple mutant adhered at levels comparable to those of the cps-pcpA mutant, suggesting that induction of pcpA expression by CodY might be required for efficient adherence to Detroit 562 cells. In this way, expression of adhesins might be controlled by nutritional regulation (Fig. 3A). However, PcpA was shown not to be required for colonization in another strain (22), so the observed effect of the codY mutation on adherence and colonization could be indirect. In contrast, the oligopeptide permease AliA/B-Ami complex, which is strongly upregulated in the codY mutant, has been shown to be either directly or indirectly involved in interaction with type II lung cells and epithelial cells (11). Furthermore, an aliA/B-ami mutant has been shown to be attenuated for colonization in a murine model of infection (25). Because the codY mutant was also attenuated for colonization, while overexpressing AliA/B-Ami, it is likely that this oligopeptide permease is indirectly involved in adherence and colonization (i.e., by modulating adhesins), although further experiments are needed to verify this.
By identifying the pneumococcal CodY regulon, we were able to confirm its role as a nutritional regulator described for other gram positives and at the same time show species-specific targets, such as the putative bacteriocin system. In contrast to B. subtilis, no genes directly involved in competence appeared to be under the control of CodY. The only link between CodY and competence is the regulation of the oligopeptide permease AliA/B-Ami, which has been indirectly implicated with the development of competence (10). Furthermore, we have shown that CodY is required for colonization of the nasopharynx, in particular, through adherence to epithelial cells, as demonstrated in vitro. However, it remains unclear which factor(s) (adhesins, proton motive force, intracellular or extracellular metabolites) is the main player during colonization and adhesion.
In conclusion, the CodY regulon of S. pneumoniae is of profound importance for the adaptation of this bacterium to nutrients. As such, this regulon is considered to contribute to the early stage of infection, i.e., colonization of the nasopharynx.
Acknowledgments
We thank Peter Burghout and Gulistan Akin for work on the overexpression and purification of H6-CodY; Christa de Jongh for the construction of the D39ΔcpsΔpcpA and D39ΔcpsΔcodYΔpcpA mutants; Ron Wevers, Udo F. H. Engelke, and Angela van Diepen for the measurements of intracellular amino acid concentrations of the S. pneumoniae wild-type and codY mutant strains; and Donald A. Morrison for the generous gift of competence-stimulating peptide.
This work was supported by the Sophia Foundation for Medical Research, Rotterdam, The Netherlands (SSWO 356).
Footnotes
Published ahead of print on 16 November 2007.
REFERENCES
- 1.Adrian, P. V., D. Bogaert, M. Oprins, S. Rapola, M. Lahdenkari, T. Kilpi, R. de Groot, H. Käyhty, and P. W. Hermans. 2004. Development of antibodies against pneumococcal proteins α-enolase, immunoglobulin A1 protease, streptococcal lipoprotein rotamase A, and putative proteinase maturation protein A in relation to pneumococcal carriage and Otitis Media. Vaccine 222737-2742. [DOI] [PubMed] [Google Scholar]
- 2.Adrian, P. V., C. J. Thomson, K. P. Klugman, and S. G. Amyes. 2000. New gene cassettes for trimethoprim resistance, dfr13, and streptomycin-spectinomycin resistance, aadA4, inserted on a class 1 integron. Antimicrob. Agents Chemother. 44355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, H. J., D. M. Pearce, S. Glenn, C. M. Taylor, M. Kuhn, A. L. Sonenshein, P. W. Andrew, and I. S. Roberts. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 631453-1467. [DOI] [PubMed] [Google Scholar]
- 4.Bijlsma, J. J., P. Burghout, T. G. Kloosterman, H. J. Bootsma, A. de Jong, P. W. Hermans, and O. P. Kuipers. 2007. Development of genomic array footprinting for identification of conditionally essential genes in Streptococcus pneumoniae. Appl. Environ. Microbiol. 731514-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogaert, D., R. De Groot, and P. W. Hermans. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4144-154. [DOI] [PubMed] [Google Scholar]
- 6.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bootsma, H. J., M. Egmont-Petersen, and P. W. Hermans. 2007. Analysis of the in vitro transcriptional response of human pharyngeal epithelial cells to adherent Streptococcus pneumoniae: evidence for a distinct response to encapsulated strains. Infect. Immun. 755489-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, J. S., S. M. Gilliland, and D. W. Holden. 2001. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40572-585. [DOI] [PubMed] [Google Scholar]
- 9.Brown, J. S., A. D. Ogunniyi, M. C. Woodrow, D. W. Holden, and J. C. Paton. 2001. Immunization with components of two iron uptake ABC transporters protects mice against systemic Streptococcus pneumoniae infection. Infect. Immun. 696702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claverys, J. P., B. Grossiord, and G. Alloing. 2000. Is the Ami-AliA/B oligopeptide permease of Streptococcus pneumoniae involved in sensing environmental conditions? Res. Microbiol. 151457-463. [DOI] [PubMed] [Google Scholar]
- 11.Cundell, D. R., B. J. Pearce, J. Sandros, A. M. Naughton, and H. R. Masure. 1995. Peptide permeases from Streptococcus pneumoniae affect adherence to eucaryotic cells. Infect. Immun. 632493-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jong, A., S. A. van Hijum, J. J. Bijlsma, J. Kok, and O. P. Kuipers. 2006. BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res. 34W273-W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.den Hengst, C. D., P. Curley, R. Larsen, G. Buist, A. Nauta, D. van Sinderen, O. P. Kuipers, and J. Kok. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.den Hengst, C. D., S. A. van Hijum, J. M. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 28034332-34342. [DOI] [PubMed] [Google Scholar]
- 15.García de la Nava, J., S. van Hijum, and O. Trelles. 2003. PreP: gene expression data pre-processing. Bioinformatics 192328-2329. [DOI] [PubMed] [Google Scholar]
- 16.Guédon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 401227-1239. [DOI] [PubMed] [Google Scholar]
- 17.Guédon, E., B. Sperandio, N. Pons, S. D. Ehrlich, and P. Renault. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 1513895-3909. [DOI] [PubMed] [Google Scholar]
- 18.Hendriksen, W. T., N. Silva, H. J. Bootsma, C. E. Blue, G. K. Paterson, A. R. Kerr, A. de Jong, O. P. Kuipers, P. W. Hermans, and T. J. Mitchell. 2007. Regulation of gene expression in Streptococcus pneumoniae by response regulator 09 is strain dependent. J. Bacteriol. 1891382-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inaoka, T., and K. Ochi. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 1843923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain, V., M. Kumar, and D. Chatterji. 2006. ppGpp: stringent response and survival. J. Microbiol. 441-10. [PubMed] [Google Scholar]
- 22.Johnston, J. W., D. E. Briles, L. E. Myers, and S. K. Hollingshead. 2006. Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect. Immun. 741171-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joseph, P., M. Ratnayake-Lecamwasam, and A. L. Sonenshein. 2005. A region of Bacillus subtilis CodY protein required for interaction with DNA. J. Bacteriol. 1874127-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadioglu, A., N. A. Gingles, K. Grattan, A. Kerr, T. J. Mitchell, and P. W. Andrew. 2000. Host cellular immune response to pneumococcal lung infection in mice. Infect. Immun. 68492-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerr, A. R., P. V. Adrian, S. Estevao, R. de Groot, G. Alloing, J. P. Claverys, T. J. Mitchell, and P. W. Hermans. 2004. The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect. Immun. 723902-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr, A. R., J. J. Irvine, J. J. Search, N. A. Gingles, A. Kadioglu, P. W. Andrew, W. L. McPheat, C. G. Booth, and T. J. Mitchell. 2002. Role of inflammatory mediators in resistance and susceptibility to pneumococcal infection. Infect. Immun. 701547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kharat, A. S., and A. Tomasz. 2003. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 712758-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloosterman, T. G., W. T. Hendriksen, J. J. Bijlsma, H. J. Bootsma, S. A. van Hijum, J. Kok, P. W. Hermans, and O. P. Kuipers. 2006. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 28125097-25109. [DOI] [PubMed] [Google Scholar]
- 29.Kuipers, O. P. 2001. Complete DNA sequence of Lactococcus lactis adds flavor to genomics. Genome Res. 11673-674. [DOI] [PubMed] [Google Scholar]
- 30.Lanie, J. A., W. L. Ng, K. M. Kazmierczak, T. M. Andrzejewski, T. M. Davidsen, K. J. Wayne, H. Tettelin, J. I. Glass, and M. E. Winkler. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 18938-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levdikov, V. M., E. Blagova, P. Joseph, A. L. Sonenshein, and A. J. Wilkinson. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J. Biol. Chem. 28111366-11373. [DOI] [PubMed] [Google Scholar]
- 32.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 27619937-19944. [DOI] [PubMed] [Google Scholar]
- 33.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296259-275. [DOI] [PubMed] [Google Scholar]
- 34.Marrer, E., A. T. Satoh, M. M. Johnson, L. J. Piddock, and M. G. Page. 2006. Global transcriptome analysis of the responses of a fluoroquinolone-resistant Streptococcus pneumoniae mutant and its parent to ciprofloxacin. Antimicrob. Agents Chemother. 50269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 1851911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petranovic, D., E. Guedon, B. Sperandio, C. Delorme, D. Ehrlich, and P. Renault. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol. 53613-621. [DOI] [PubMed] [Google Scholar]
- 37.Sánchez-Beato, A. R., R. Lopez, and J. L. Garcia. 1998. Molecular characterization of PcpA: a novel choline-binding protein of Streptococcus pneumoniae. FEMS Microbiol. Lett. 164207-214. [DOI] [PubMed] [Google Scholar]
- 38.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 1785910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serror, P., and A. L. Sonenshein. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20843-852. [DOI] [PubMed] [Google Scholar]
- 40.Shivers, R. P., S. S. Dineen, and A. L. Sonenshein. 2006. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol. Microbiol. 62811-822. [DOI] [PubMed] [Google Scholar]
- 41.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53599-611. [DOI] [PubMed] [Google Scholar]
- 42.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in gram-positive bacteria. Curr. Opin. Microbiol. 8203-207. [DOI] [PubMed] [Google Scholar]
- 43.Spellerberg, B., D. R. Cundell, J. Sandros, B. J. Pearce, I. Idanpaan-Heikkila, C. Rosenow, and H. R. Masure. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19803-813. [DOI] [PubMed] [Google Scholar]
- 44.Steiner, K., and H. Malke. 2000. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol. Microbiol. 381004-1016. [DOI] [PubMed] [Google Scholar]
- 45.Steiner, K., and H. Malke. 2001. relA-independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 1837354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293498-506. [DOI] [PubMed] [Google Scholar]
- 47.Tojo, S., T. Satomura, K. Morisaki, J. Deutscher, K. Hirooka, and Y. Fujita. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 561560-1573. [DOI] [PubMed] [Google Scholar]
- 48.van Hijum, S. A. F. T., A. de Jong, R. J. S. Baerends, H. A. Karsens, N. E. Kramer, R. Larsen, C. D. den Hengst, C. J. Albers, J. Kok, and O. P. Kuipers. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Hijum, S. A., J. Garcia de la Nava, O. Trelles, J. Kok, and O. P. Kuipers. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2241-244. [PubMed] [Google Scholar]