Abstract
The peptidoglycan structure of Mycobacterium spp. has been investigated primarily with the readily cultivable Mycobacterium smegmatis and Mycobacterium tuberculosis and has been shown to contain unusual features, including the occurrence of N-glycolylated, in addition to N-acetylated, muramic acid residues and direct cross-linkage between meso-diaminopimelic acid residues. Based on results from earlier studies, peptidoglycan from in vivo-derived noncultivable Mycobacterium leprae was assumed to possess the basic structural features of peptidoglycans from other mycobacteria, other than the reported replacement of l-alanine by glycine in the peptide side chains. In the present study, we have analyzed the structure of M. leprae peptidoglycan in detail by combined liquid chromatography and mass spectrometry. In contrast to earlier reports, and to the peptidoglycans in M. tuberculosis and M. smegmatis, the muramic acid residues of M. leprae peptidoglycan are exclusively N acetylated. The un-cross-linked peptide side chains of M. leprae consist of tetra- and tripeptides, some of which contain additional glycine residues. Based on these findings and genome comparisons, it can be concluded that the massive genome decay in M. leprae does not markedly affect the peptidoglycan biosynthesis pathway, with the exception of the nonfunctional namH gene responsible for N-glycolylmuramic acid biosynthesis.
The success of Mycobacterium tuberculosis and Mycobacterium leprae as pathogens has been linked to their ability to survive in the host, particularly in the macrophage (6, 30). An important element in intracellular survival and consequent pathogenesis is the unique composition of the bacterial cell envelope of pathogenic mycobacteria, consisting of a highly complex array of distinctive lipids, glycolipids, proteins, and polymers (3, 4, 7), of which the mycolyl-arabinogalactan-peptidoglycan complex (MAPc) is the major structural component (7). The peptidoglycan (PG) layer of MAPc forms the backbone of the cell envelope, maintaining cell shape and size. In addition, the C6 of some of the muramic acid residues of PG serves as the linkage site for arabinogalactan (23), which in turn provides the site for the attachment of mycolic acids through the esterification of terminal arabinose residues (24).
The individual components of the MAPc have been subjected to structural and biosynthetic studies in pursuit of the discovery of novel enzymatic activities that may be exploited as drug targets (2). Previous structural analyses of the PGs of, mainly, Mycobacterium smegmatis and M. tuberculosis have identified several unusual features (18, 25, 26), including the occurrence of N-glycolylmuramic acid (MurNGlyc), direct diaminopimelic acid (DAP)-DAP cross-links, and modifications at the free carboxylic acid functions of DAP and d-Glu. The data on these diverse structural features of a few members of the Mycobacterium genus have been partially extended to other mycobacteria, including M. leprae (9, 10). There are direct and a priori reasons to expect a distinctive PG structure in M. leprae. The genome, in comparison to those of M. tuberculosis and other mycobacteria, has undergone an exceptional degree of self-deletion and rearrangement such that only about 50% coding capacity remains, with only about 1,600 complete open reading frames (ORFs), compared to about 4,000 for M. tuberculosis (5) and about 7,000 for M. smegmatis (Comprehensive Microbial Resource database [http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi]). Although the three species have similar clusters of mur genes, organized as that in Escherichia coli, the M. tuberculosis cluster contains four additional ORFs encoding hypothetical proteins between pbpB and murE that are missing from the M. leprae or M. smegmatis cluster (20).
Draper et al. (10) demonstrated that the amino acid in the first position of the tetrapeptide side chain of the PG of M. leprae is Gly rather than the d-Ala of all other mycobacteria. Thus, in light of a dramatically rearranged and reduced genome marked by few complete ORFs, evidence for chemical changes, the obligate intracellular nature and origins of M. leprae, and interest in the molecular basis of leprosy pathogenesis, we analyzed the PG of M. leprae, applying sensitive analytical means in accord with the paucity of in vivo-derived material, and drew comparisons with the more thoroughly studied products from M. tuberculosis and M. smegmatis.
MATERIALS AND METHODS
Preparation of PGs from M. leprae, M. tuberculosis, and M. smegmatis.
M. leprae cells were purified from armadillo spleens and livers as described previously (16). M. tuberculosis H37Rv (ATCC 25618; American Type Culture Collection, Manassas, VA) was grown in a glycerol-alanine-salt medium. M. smegmatis MC2155 (ATCC 700084; American Type Culture Collection, Manassas, VA) was grown in nutrient broth (EM Science, Gibbstown, NJ). In all cases, cells at mid-log phase were harvested by centrifugation and washed with phosphate-buffered saline (PBS) to remove growth medium before the isolation of PG.
The bacilli were resuspended in 10 mM NH4HCO3 containing 1 mM phenylmethylsulfonyl fluoride and disrupted by intermittent probe sonication with an MSE Soniprep 150 (MSE-Sanyo; Integrated Services, Palisades Park, NJ) for 30 cycles (60-s bursts separated by 60 s of cooling). The sonicate was digested with 10 μg each of DNase and RNase/ml for 1 h at 4°C. A cell wall-enriched fraction was obtained by centrifugation at 27,000 × g for 30 min. PG was prepared from the cell wall fraction as reported previously, with some modifications (10, 18, 21). The pellet containing cell walls was resuspended in PBS containing 2% sodium dodecyl sulfate (SDS), the suspension was incubated for 1 h at 50°C with constant stirring and recentrifuged at 27,000 × g for 30 min, and the supernatant was discarded. This process was repeated twice. The resulting pellet was resuspended in PBS containing 1% SDS and 0.1 mg of self-digested proteinase K/ml, and the suspension was incubated at 45°C for 1 h with constant stirring. The mixture was then heated at 90°C for 1 h before centrifugation at 27,000 × g for 30 min. The supernatant was discarded, and the 1% SDS extraction procedure was repeated twice to remove proteinase K. The pelleted material was washed twice with PBS and four times with deionized water to remove SDS. The resulting MAPc was extracted with ethanol-diethyl ether (1:1) and dried under a vacuum. In order to hydrolyze the mycolic acids, the MAPc was resuspended in 0.5% KOH in methanol and stirred at 37°C for 4 days. The mixture was centrifuged, and the pellet was washed twice with methanol and twice with diethyl ether and dried under a vacuum. The resulting arabinogalactan-PG was digested with 0.05 N H2SO4 at 37°C for 5 days to remove the arabinogalactan. The resulting insoluble PG was washed four times by centrifugation in deionized water and dried under a vacuum.
Solubilization of PG and purification and analysis of muropeptides.
The purified PG (2 mg) was suspended in 0.5 ml of 10 mM sodium acetate (pH 5.0) containing 25 μg of purified muramidase from a Chalaropsis sp., prepared as described previously (14), and the suspension was incubated at 37°C for 16 h with stirring (32). Digests were centrifuged at 27,000 × g for 30 min, and the supernatant was filtered through a 10-kDa-cutoff ultrafiltration membrane (Millipore) to remove muramidase and dried under a vacuum. The muropeptides were resuspended in 0.5 M sodium-borate buffer (pH 9.0), and sodium borohydride was added to achieve a final concentration of 8 mg/ml. The mixture was incubated for 30 min at room temperature to reduce the sugar moieties. The reaction was stopped by the addition of orthophosphoric acid, and the pH was adjusted to 4.0 prior to fractionation by size exclusion chromatography on a Superdex peptide 10/300 GL column (Amersham Biosciences, Piscataway, NJ) with a model 600 controller connected to a model 600 pump and a model 2487 UV detector (all from Waters, Milford, MA). The column was equilibrated and eluted with 30% acetonitrile (Burdick and Jackson, Muskegon, MI) containing 0.1% trifluoroacetic acid (Supelco, Bellefonte, PA) with a flow rate of 0.5 ml/min. The absorbance of the effluent at 214 nm was monitored. The fractions containing muropeptides were dried under a vacuum and resuspended in high-performance liquid chromatography (HPLC)-grade water at an approximate concentration of 10 μM. An aliquot (20 μl) was applied to a 2-by-150-mm Hypersil octyldecyl silane (C18) column (Phenomenex, Torrance, CA) connected to an Agilent 1100 HPLC system (Agilent Technologies, Palo Alto, CA). The muropeptides were eluted with a 2 to 30% linear gradient of acetonitrile containing 0.5% formic acid at 320 μl/min. The eluate was monitored at 214 nm and introduced directly into an LCQ Duo electrospray mass spectrometer (Finnigan-Thermoquest, San Jose, CA), and the muropeptides were analyzed by mass spectrometry (MS) and tandem mass spectrometry (MS-MS). MS-MS was performed on the fly of the most dominant ion from the previous MS scan. A standard preparation of N-acetylmuramyl-l-alanyl-d-glutamyl-meso-diaminopimelyl-d-alanyl-d-alanine (MurNAc pentapeptide) was prepared from enzymatically synthesized UDP-MurNAc pentapeptide and was analyzed by liquid chromatography (LC)-MS to establish the pattern of MS fragmentation, as described previously (21, 22).
Analysis of amino sugars and amino acids.
Aliquots of the Chalaropsis sp. muramidase-solubilized muropeptides obtained after fractionation by size exclusion chromatography were resuspended in 200 μl of 50 mM 2-(N-morpholino)ethanesulfonate buffer (pH 6.0; Sigma, St. Louis, MO) containing 1 mM MgCl2 and 10 U of mutanolysin (Sigma, St. Louis, MO), and the suspensions were incubated for 16 h at 37°C. The resulting hydrolysate was further digested with β-N-acetylhexosaminidase (Sigma, St. Louis, MO) to obtain the amino sugars. The reaction mixture was deproteinated by ethanol precipitation, and the supernatant was transferred into a 13-by-100-mm glass tube and dried under a vacuum. Scyllo-inositol was added as an internal standard. To prepare trimethylsilane derivatives (21), samples were resuspended in 3 N methanolic HCl (Supelco, Bellefonte, PA) in a tightly capped tube, heated at 80°C for 1 h, cooled to room temperature, and dried under a stream of N2. Tri-Sil reagent (Pierce, Rockford, IL) was added, the tightly capped tube was heated at 70°C for 20 min and cooled to room temperature, and excess reagent was evaporated under a stream of N2. The derivatized products were dissolved in a small volume of hexane and analyzed in a Trace 2000 gas chromatograph (Finnigan-Thermoquest, San Jose, CA) fitted with a DB-5 column (10 m by 0.18 mm; Agilent Technologies, Palo Alto, CA) linked to a Polaris mass detector (Finnigan-Thermoquest). Analytical runs were programmed at an initial temperature of 80°C, held for 1 min, and the initial temperature was first raised at a rate of 30°C min−1 to 130°C and then at a rate of 10°C min−1 to a final temperature of 280°C, which was held for 10 min. Muramic acid and glucosamine standards were prepared in the same way.
For amino acid analysis, an aliquot (0.2 mg) of purified PG was transferred into a glass tube, 0.1 ml of 6 N HCl (Pierce, Rockford, IL) was added, and the tube was flushed with N2. The tightly capped tube was heated on a heat block at 110°C for 18 h. The amino acid compositions of the PGs obtained from M. leprae, M. tuberculosis, and M. smegmatis were determined using the EZ:faast gas chromatography (GC)-MS kit according to the instructions supplied by the manufacturer (Phenomenex, Torrance, CA). The amino acid EZ:faast derivatives were analyzed on a Trace 2000 gas chromatograph fitted with a DB-5 column (10 m by 0.18 mm) connected to a Polaris mass detector. Analytical runs were programmed at an initial temperature of 110°C, which was held for 2 min and then raised to 285°C at 15°C min−1. This highly sensitive method allowed us to analyze samples smaller than those that can be analyzed by the conventional methods, which facilitated the analysis of M. leprae PG.
RESULTS
Analysis of amino acids and amino sugars of PG.
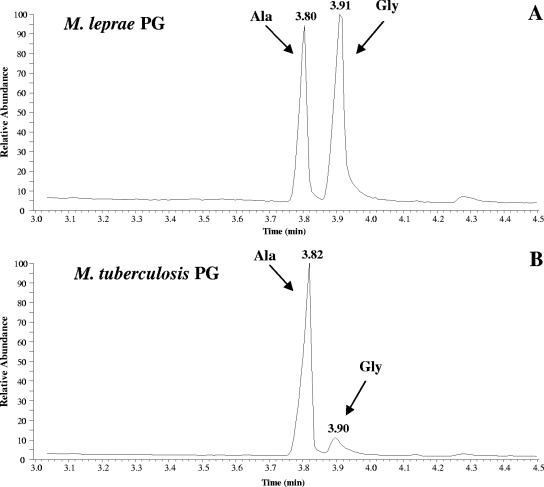
The amino acid compositions of M. tuberculosis and M. smegmatis PGs were similar in including Ala, Glu, and DAP (data not shown). However, as previously reported (10), M. leprae PG also contained Gly (Fig. 1).
FIG. 1.
Analysis of the amino acid contents of the PGs from M. leprae and M. tuberculosis. Amino acids from PGs were released by acid hydrolysis and subsequently analyzed by GC-MS following derivatization, as described in Materials and Methods.
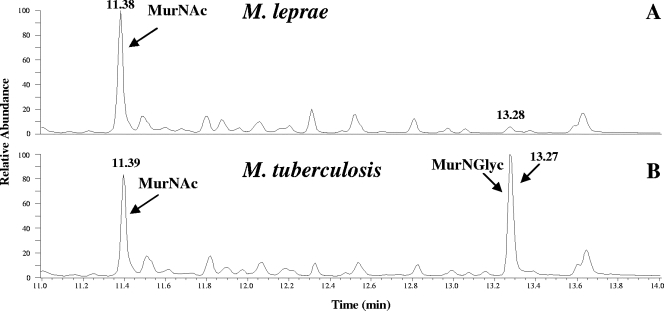
Previously, we established that the muramic acid residues of M. tuberculosis and M. smegmatis PGs comprised of a mixture of MurNGlyc and MurNAc (21). However, the M. leprae PG was different in that there was no detectable MurNGlyc; MurNAc was the only form of muramic acid present in M. leprae (Fig. 2). All of the Glc-NH2 residues from the PGs of these three organisms were N acetylated (data not shown).
FIG. 2.
Analysis of the muramic acid residues of PGs from M. tuberculosis and M. leprae. Shown are the total-ion chromatograms for M. leprae and M. tuberculosis. The trimethylsilane derivatives of the muramic acids were analyzed by GC-MS as described in Materials and Methods. The peak at 13.28 min in panel A does not correspond to MurNGlyc, as determined by MS.
Solubilization of PG and purification and analysis of muropeptides.
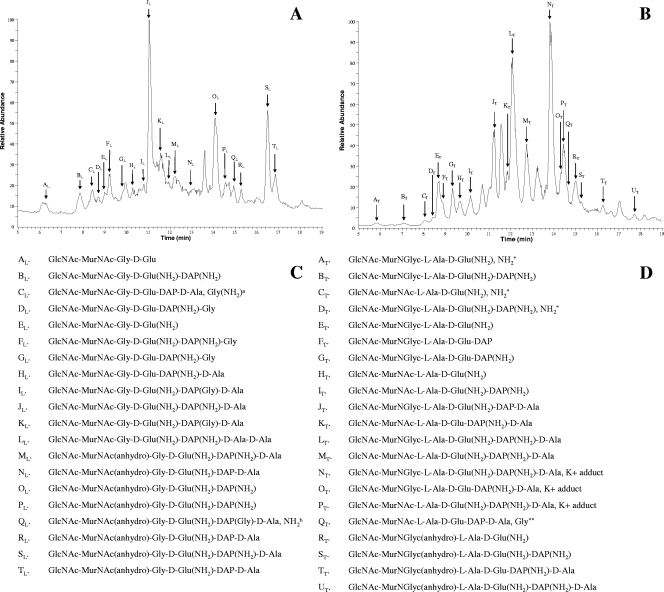
PGs obtained from M. leprae, M. tuberculosis, and M. smegmatis were digested with Chalaropsis sp. muramidase, resulting in the solubilization of about 80% of the starting material. Further digestion of the insoluble material yielded only a marginal increase in soluble products. The soluble material was deproteinated by ultrafiltration and subjected to size exclusion chromatography on a Superdex peptide 10/300 GL column. The size-fractionated muropeptides obtained from the PGs from the three species were analyzed by LC-MS. The initial MS analysis of the samples was used to identify the dominating molecular ions of the muropeptides. The structures of these dominating ions were subsequently analyzed by MS-MS. The secondary ions generated by MS-MS were compared with those from the MS-MS analysis of the standard MurNAc pentapeptide prepared as described previously (21), enabling the identification of the major monomers and the nature and locations of the modifications present (Fig. 3). The results indicate that all of the muropeptides from the M. leprae PG contained MurNAc residues, in agreement with the results of amino sugar analysis. Almost all of the DAP residues and the majority of the d-Glu residues were amidated. Muropeptides with different lengths of peptide side chains ranging from GlcNAc-MurNAc-Gly-d-Glu(NH2)-DAP(NH2)-d-Ala (tetrapeptide) to GlcNAc-MurNAc-Gly-d-Glu (dipeptide) were observed. However, the relative abundances of these molecules differed, with GlcNAc-MurNAc-Gly-d-Glu(NH2)-DAP(NH2)-d-Ala being dominant (Fig. 3). Significant amounts of muropeptide with anhydomuramic acid residues were also identified, indicative of the terminal unit of the PG chain. A large number of M. leprae muropeptides containing additional Gly residues, predominantly attached to the DAP residues, were observed in peaks CL, DL, FL, GL, IL, KL, and QL (peaks were designated in alphabetical order with a subscript indicating M. leprae [L]or M. tuberculosis [T]) (Fig. 3). When a Gly residue was attached to a DAP residue in the presence of amidation, it was not possible to determine which of the residues was actually amidated. However, considering the extent of DAP amidation in mycobacterial PG, it was most likely the DAP residue that was amidated. Significantly higher amounts of anhydromuramic acid-containing muropeptides in the M. leprae sample than in the M. tuberculosis and M. smegmatis samples were observed.
FIG. 3.
LC-MS analysis of the muropeptides obtained from M. leprae and M. tuberculosis PGs. (A) Total-ion chromatogram of the positive-ion mass spectrum of monomeric muropeptides from M. leprae PG and the dominating molecular ions (peaks AL to TL) whose structure has been determined with subsequent MS-MS analysis. The arrows indicate approximate retention times for the molecular ions. Some of the peaks corresponded to more than one molecular species, and in such cases, the structure of the dominant ion was determined and presented. (B) Total-ion chromatogram of the positive-ion mass spectrum of monomeric muropeptides and the dominating molecular ions (peaks AT to UT). (C) Inferred structures of the dominant molecular ions of peaks AL to TL. a, the position of this Gly(NH2) was not determined; b, the position of this NH2 was not determined. (D) Inferred structures of the ions of peaks AT to UT based on MS-MS analysis. As in the case of M. leprae, only the structures of the dominant molecular ions were determined when more than one molecular ion corresponded to a peak.*, the position of this NH2 was not determined; **, the position of this Gly was not determined.
The monomeric muropeptides of M. tuberculosis were found to contain both MurNAc and MurNGlyc, again in agreement with the results of amino sugar analysis. The monomeric muropeptide profile from M. tuberculosis was dominated by tetrapeptides, with some tripeptides and traces of dipeptides (Fig. 3). The DAP residues of the peptide sides chains were often amidated (peaks GT, KT, OT, and TT) and, in some cases, were amidated in combination with the carboxylic acid group of the d-Glu (peaks BT, DT, IT, LT, MT, NT, PT, ST, and UT). The amidation of peptide side chains and the occurrence of the two forms of muramic acid occurred in all combinations. Muropeptides with additional Gly residues were virtually absent from the M. tuberculosis PG, with the exception of peak QT (Fig. 3); the exact location of the Gly was not determined due to the extremely low abundance of the molecular ion. The monomeric muropeptides obtained from the M. smegmatis PG were similar to those of M. tuberculosis (data not shown).
DISCUSSION
In the present work, we have compared the detailed structural features of the PGs of M. tuberculosis, M. leprae, and M. smegmatis as representatives of pathogenic and nonpathogenic laboratory strains of the genus Mycobacterium. M. leprae is noncultivable in vitro, and the difficulty in obtaining sufficient PG was a major obstacle in the analysis of the structural details. We have partially resolved this problem by the isolation of PG by means of a previously described protocol (10) developed specifically for M. leprae, followed by solubilization, purification, and analysis of the muropeptides and amino sugars by sensitive GC-MS- and LC-MS-based analytical methods. For ease of analysis, we have separated the monomeric muropeptides from the cross-linked muropeptides and used the former for subsequent MS analysis, as indicated in Fig. 3. This strategy simplified otherwise very complex mass spectra of unfractionated muropeptides while providing details of the PG building blocks.
Recently, analyses of the amino sugar compositions of the PGs from M. tuberculosis and M. smegmatis showed the presence of two forms of muramic acid, MurNAc and MurNGlyc (21). M. leprae PG had previously been reported to contain exclusively MurNGlyc (9, 10) based on the estimation of the glycolic acid content. However, according to our present analysis, the only form of muramic acid present in M. leprae PG is MurNAc (Fig. 2). The gene responsible for the synthesis of MurNGlyc (namH) has been identified previously (28) in M. smegmatis, and the mutant with the knockout of this gene harbors exclusively MurNAc in PG (28). The orthologue of namH in M. tuberculosis (Rv3818) has been identified. However, the M. leprae orthologue of namH (ML0085c) was reported to be a possible pseudogene (5), and thus, this is the most likely explanation for the absence of MurNGlyc in M. leprae PG. Although the presence of MurNGlyc in PG has been attributed to the increased lysozyme resistance of mycobacterial PG (28), the absence of MurNGlyc in M. leprae demonstrates that the N glycolylation of muramic acid is not essential for survival in the host.
An amino acid analysis of the purified PG was performed to check on purity. PG purified using proteinase K treatment showed the presence of other amino acids (Fig. 1), most of which have been reported previously (18). However, the subsequent MS analysis indicated that these amino acids were not constituents of PG, as previously reported (18), but were most probably derived from contaminating PG-associated proteins (15), possibly remnants of partially digested covalently attached proteins. The almost equal abundances of Gly and Ala from the M. leprae sample were in agreement with the previous reports (9, 10), in which it was assumed that the l-Ala had been replaced by Gly. This assumption was subsequently confirmed by MS analysis of the muropeptides. The reason for the occurrence of Gly in M. leprae is unknown, and previous attempts to answer this question through the analysis of the properties of the enzyme most likely to be involved in the addition of this residue provided ambiguous results (20). Whether this change benefits M. leprae in its obligate intracellular presence and its particular pathogenesis is unknown; however, it may potentially increase resistance to host lytic enzymes that cleave the bond between the lactoyl group of MurNAc and the l-Ala residue.
The Chalaropsis sp. muramidase has previously been used for the solubilization of mycobacterial PG (25). The conventional approach to the analysis of the structure of PG involves solubilization, reduction, and reversed-phase HPLC separation of the numerous populations of the muropeptides (12), an approach highly successful in the analysis of PGs from a range of organisms (1, 8, 13, 27). However, the application of this method was not an option for the analysis of the PG of M. leprae due to the paucity of armadillo-derived M. leprae samples. This problem was addressed by the introduction of size exclusion chromatography, which allowed the effective separation of monomers and cross-linked muropeptides prior to reversed-phase HPLC and direct analysis by MS or MS-MS. The separation of the monomers from cross-linked muropeptides by size exclusion chromatography allowed improved separation of the muropeptides by reversed-phase HPLC, in addition to generating a relatively simple mass spectrum that eased subsequent data analysis. The method was initially used for the structural analysis of M. tuberculosis and M. smegmatis PGs, and the results were compared and validated with the published structures (18, 19, 33). Using this methodology, we were able to analyze the structure of M. leprae PG from a sample of about 2 mg.
The monomeric muropeptides from M. leprae PG were a complex mixture of disaccharide peptides with various peptide lengths and compositions (Fig. 3). Unlike M. tuberculosis, M. leprae did not have the added complexity generated by two forms of muramic acid in the muropeptides (Fig. 3). However, complexity was caused by the presence of additional Gly residues in a large number of M. leprae muropeptides (Fig. 3). These additional Gly residues, in most instances, were associated with DAP; however, in some cases they were also linked to d-Glu, indicating the presence of additional enzymes in the M. leprae PG biosynthesis pathway. The M. leprae PG also had higher amounts of tri- and dipeptides than the PGs of the other Mycobacterium species, possibly due to higher-level dd-carboxypeptidase activity in M. leprae. The amidation of the carboxylic acid functions of d-Glu and DAP was extensive in all three species. Almost all the DAP residues found in the PGs from all three species were amidated, along with the majority of d-Glu residues. The extent of amidation observed in the PG was comparable to that observed in the lipid II intermediates of M. smegmatis (22). In this analysis, we have not seen any evidence of the presence of methylated DAP or d-Glu residues or muramic acid residues with a free amino group, which was observed previously in M. smegmatis lipid II (22). We hypothesize that these modifications may be part of a regulatory mechanism which maintains the overall degree of cross-linking in mycobacterial PG without directly covering the sites.
Although estimates of glycan chain lengths of the PGs from any of these organisms based on the data were not possible, the presence of significantly higher amounts of anhydromuramic acid-containing muropeptide in the M. leprae samples than in the M. tuberculosis and M. smegmatis samples suggests the existence of shorter glycan chains. The physiological significance of this feature is a matter of speculation.
Therefore, although the PG biosynthetic pathways of M. tuberculosis and M. leprae are very similar based on the analysis of the genomes (5, 20, 31), the actual fine structures differ. Whether these differences are due to the lack of some key enzymes, as in the case of the NamH hydroxylase, or to the in vivo growth conditions, as has been assumed for the replacement of l-Ala with Gly, is subject to further investigation.
Although the physiological implications of amidated residues in PG are unknown, involvement in pathogenesis can be hypothesized. Epithelial cells and antigen-presenting cells play an important role in the innate immune response, which is considered to be the first line of defense against pathogens. The nucleotide binding oligomerization domain (NOD) receptors present in these cells, NOD1 and NOD2, recognize moieties of bacterial PG (19) and initiate the immune response. The minimum PG derivative structure recognized by NOD1 is a muramyl tripeptide containing meso-DAP (11). In addition, it has been demonstrated previously that NOD1 has a reduced capacity to recognize muramyl peptides with amidated meso-DAP compared to nonamidated peptides (11, 29). Therefore, the amidation of the meso-DAP of PG may play an important role in the pathogenesis of M. leprae and M. tuberculosis. Furthermore, the muramyl dipeptide MurNAc-l-Ala-d-iso-Glu(NH2) is recognized by NOD2 and the replacement of l-Ala with d-Ala eliminates the ability of muramyl dipeptide to stimulate NOD2, indicating stereoselectivity (17). Accordingly, M. leprae, with its PG containing amidated DAP and Gly residues, may escape the NOD1- and NOD2-mediated innate immune response of the host. The additional Gly in the peptide side chains and the higher level of anhydromuramic acid may also contribute to lytic enzyme resistance and host-pathogen interaction.
Acknowledgments
This work was supported by grants (AI18357, AI033706, and AI49151) contract NO1-AI025469 from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health.
Footnotes
Published ahead of print on 16 November 2007.
REFERENCES
- 1.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 1813956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besra, G. S., and P. J. Brennan. 1997. The mycobacterial cell envelope: a target for novel drugs against tuberculosis. J. Pharm. Pharmacol. 4925-30.9178204 [Google Scholar]
- 3.Brennan, P. J., and G. S. Besra. 1997. Structure, function and biogenesis of the mycobacterial cell wall. Biochem. Soc. Trans. 25188-194. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 6429-63. [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 4091007-1011. [DOI] [PubMed] [Google Scholar]
- 6.Collins, H. L., and S. H. Kaufmann. 2001. The many faces of host responses to tuberculosis. Immunology 1031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crick, D. C., S. Mahapatra, and P. J. Brennan. 2001. Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11107R-118R. [DOI] [PubMed] [Google Scholar]
- 8.de Jonge, B. L., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 26711248-11254. [PubMed] [Google Scholar]
- 9.Draper, P. 1976. Cell walls of Mycobacterium leprae. Int. J. Lepr. Other Mycobact. Dis. 4495-98. [PubMed] [Google Scholar]
- 10.Draper, P., O. Kandler, and A. Darbre. 1987. Peptidoglycan and arabinogalactan of Mycobacterium leprae. J. Gen. Microbiol. 1331187-1194. [DOI] [PubMed] [Google Scholar]
- 11.Girardin, S. E., L. H. Travassos, M. Herve, D. Blanot, I. G. Boneca, D. J. Philpott, P. J. Sansonetti, and D. Mengin-Lecreulx. 2003. Peptidoglycan molecular requirements allowing detection by NOD1 and NOD2. J. Biol. Chem. 27841702-41708. [DOI] [PubMed] [Google Scholar]
- 12.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172451-464. [DOI] [PubMed] [Google Scholar]
- 13.Glauner, B., J. V. Holtje, and U. Schwarz. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 26310088-10095. [PubMed] [Google Scholar]
- 14.Hash, J. H., and M. V. Rothlauf. 1967. The N,O-diacetylmuramidase of Chalaropsis species. I. Purification and crystallization. J. Biol. Chem. 2425586-5590. [PubMed] [Google Scholar]
- 15.Hirschfield, G. R., M. McNeil, and P. J. Brennan. 1990. Peptidoglycan-associated polypeptides of Mycobacterium tuberculosis. J. Bacteriol. 1721005-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter, S. W., B. Rivoire, V. Mehra, B. R. Bloom, and P. J. Brennan. 1990. The major native proteins of the leprosy bacillus. J. Biol. Chem. 26514065-14068. [PubMed] [Google Scholar]
- 17.Inohara, N., Y. Ogura, A. Fontalba, O. Gutierrez, F. Pons, J. Crespo, K. Fukase, S. Inamura, S. Kusumoto, M. Hashimoto, S. J. Foster, A. P. Moran, J. L. Fernandez-Luna, and G. Nunez. 2003. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 2785509-5512. [DOI] [PubMed] [Google Scholar]
- 18.Kotani, S., I. Yanagida, K. Kato, and T. Matsuda. 1970. Studies on peptides, glycopetides and antigenic polysaccharide-glycopeptide complexes isolated from an L-11 enzyme lysate of cell walls of Mycobacterium tuberculosis strain H37Rv. Biken J. 13249-275. [PubMed] [Google Scholar]
- 19.Lederer, E., A. Adam, R. Ciorbaru, J. F. Petit, and J. Wietzerbin. 1975. Cell walls of Mycobacteria and related organisms; chemistry and immunostimulant properties. Mol. Cell. Biochem. 787-104. [DOI] [PubMed] [Google Scholar]
- 20.Mahapatra, S., D. C. Crick, and P. J. Brennan. 2000. Comparison of the UDP-N-acetylmuramate:l-alanine ligase enzymes from Mycobacterium tuberculosis and Mycobacterium leprae. J. Bacteriol. 1826827-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahapatra, S., H. Scherman, P. J. Brennan, and D. C. Crick. 2005. N-glycolylation of the nucleotide precursors of peptidoglycan biosynthesis of Mycobacterium spp. is altered by drug treatment. J. Bacteriol. 1872341-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahapatra, S., T. Yagi, J. T. Belisle, B. J. Espinosa, P. J. Hill, M. R. McNeil, P. J. Brennan, and D. C. Crick. 2005. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J. Bacteriol. 1872747-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNeil, M., M. Daffe, and P. J. Brennan. 1990. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J. Biol. Chem. 26518200-18206. [PubMed] [Google Scholar]
- 24.McNeil, M., M. Daffe, and P. J. Brennan. 1991. Location of the mycolyl ester substituents in the cell walls of mycobacteria. J. Biol. Chem. 26613217-13223. [PubMed] [Google Scholar]
- 25.Petit, J. F., A. Adam, J. Wietzerbin-Falszpan, E. Lederer, and J. M. Ghuysen. 1969. Chemical structure of the cell wall of Mycobacterium smegmatis. I. Isolation and partial characterization of the peptidoglycan. Biochem. Biophys. Res. Commun. 35478-485. [DOI] [PubMed] [Google Scholar]
- 26.Petit, J. F., J. Wietzerbin, B. C. Das, and E. Lederer. 1975. Chemical structure of the cell wall of Mycobacterium tuberculosis var. bovis, strain BCG. Z. Immunitatsforsch. Exp. Klin. Immunol. 149118-125. [PubMed] [Google Scholar]
- 27.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J. Bacteriol. 1786451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raymond, J. B., S. Mahapatra, D. C. Crick, and M. S. Pavelka, Jr. 2005. Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J. Biol. Chem. 280326-333. [DOI] [PubMed] [Google Scholar]
- 29.Roychowdhury, A., M. A. Wolfert, and G. J. Boons. 2005. Synthesis and proinflammatory properties of muramyl tripeptides containing lysine and diaminopimelic acid moieties. Chembiochem 62088-2097. [DOI] [PubMed] [Google Scholar]
- 30.Vergne, I., J. Chua, S. B. Singh, and V. Deretic. 2004. Cell biology of Mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 20367-394. [DOI] [PubMed] [Google Scholar]
- 31.Vissa, V. D., and P. J. Brennan. 2001. The genome of Mycobacterium leprae: a minimal mycobacterial gene set. Genome Biol. 2REVIEWS1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wietzerbin, J., B. C. Das, J. F. Petit, E. Lederer, M. Leyh-Bouille, and J. M. Ghuysen. 1974. Occurrence of D-alanyl-(D)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria. Biochemistry 133471-3476. [DOI] [PubMed] [Google Scholar]
- 33.Wietzerbin-Falszpan, J., B. C. Das, I. Azuma, A. Adam, J. F. Petit, and E. Lederer. 1970. Isolation and mass spectrometric identification of the peptide subunits of mycobacterial cell walls. Biochem. Biophys. Res. Commun. 4057-63. [DOI] [PubMed] [Google Scholar]