Abstract
Reducing iron (Fe) levels in a defined minimal medium reduced the growth yields of planktonic and biofilm Pseudomonas aeruginosa, though biofilm biomass was affected to the greatest extent and at FeCl3 concentrations where planktonic cell growth was not compromised. Highlighting this apparently greater need for Fe, biofilm growth yields were markedly reduced in a mutant unable to produce pyoverdine (and, so, deficient in pyoverdine-mediated Fe acquisition) at concentrations of FeCl3 that did not adversely affect biofilm yields of a pyoverdine-producing wild-type strain. Concomitant with the reduced biofilm yields at low Fe concentrations, P. aeruginosa showed enhanced twitching motility in Fe-deficient versus Fe-replete minimal media. A mutant deficient in low-Fe-stimulated twitching motility but normal as regards twitching motility on Fe-rich medium was isolated and shown to be disrupted in rhlI, whose product is responsible for synthesis of the N-butanoyl homoserine lactone (C4-HSL) quorum-sensing signal. In contrast to wild-type cells, which formed thin, flat, undeveloped biofilms in Fe-limited medium, the rhlI mutant formed substantially developed though not fully mature biofilms under Fe limitation. C4-HSL production increased markedly in Fe-limited versus Fe-rich P. aeruginosa cultures, and cell-free low-Fe culture supernatants restored the twitching motility of the rhlI mutant on Fe-limited minimal medium and stimulated the twitching motility of rhlI and wild-type P. aeruginosa on Fe-rich minimal medium. Still, addition of exogenous C4-HSL did not stimulate the twitching motility of either strain on Fe-replete medium, indicating that some Fe-regulated and RhlI/C4-HSL-dependent extracellular product(s) was responsible for the enhanced twitching motility (and reduced biofilm formation) seen in response to Fe limitation.
Pseudomonas aeruginosa is an opportunistic human pathogen that causes debilitating infections, particularly in patients with underlying diseases, such as cystic fibrosis (24, 50), where it can cause chronic infections characterized by the formation of biofilms (24, 25, 42, 50, 59, 77). These surface-associated communities of sessile bacteria embedded in a polysaccharide matrix are important features of many infectious diseases (25, 42, 45, 59), and their characteristic resistance to antimicrobials (antibiotics and biocides) and host immune responses (4, 20, 22, 32, 57, 60, 66) compromises infection control. Details of in vivo P. aeruginosa biofilm formation, structure, and properties are limited, with most of our understanding of these structures coming from the study of model biofilms formed in vitro (3, 15, 29, 30, 56, 80, 81, 84). In vitro biofilm development in P. aeruginosa is characterized by bacterial surface attachment, followed by microcolony formation by clonal expansion or motility-driven cell-to-cell aggregation and subsequent formation of a flat, uniform, confluent biofilm or heterogeneous, structured biofilms characterized by cell aggregates or “mushroom” structures separated by channels or spaces (38, 40).
A number of studies have linked quorum sensing (QS), cell density control of gene expression involving acylhomoserine lactones (acyl-HSLs) (36, 69), and biofilm formation/development in P. aeruginosa (17, 26, 38, 64), although some studies indicate that QS has little or no role, with QS mutants being proficient in biofilm formation (27, 62, 67, 68). These discrepancies are generally linked to differences in biofilm model and/or culture conditions, and indeed, a recent study confirmed that the QS dependence of biofilm formation is nutritionally conditional (i.e., QS systems are needed for biofilm formation in some growth media [e.g., succinate] but not others [e.g., glucose or glutamate]) (73). Two primary acyl-HSL systems have been reported to occur in P. aeruginosa: las, involving the 3-oxo-dodecanoyl HSL product of the LasI synthase and the 3-oxo-dodecanoyl HSL-responsive DNA-binding regulator LasR, and rhl, involving the butanoyl HSL (C4-HSL) product of the RhlI synthase and the C4-HSL-responsive DNA-binding regulator RhlR (36, 69). In some studies, lasI but not rhlI (17, 18, 64) or both lasI and rhlI (18, 84) mutants are substantially altered as regards biofilm development, while in others, las mutants form nominally wild-type biofilms (27, 62, 84).
Twitching, a form of surface motility mediated by type IV pili (46), is also implicated in biofilm development (41, 55), being involved in P. aeruginosa spreading over the surface of the substratum (in the initial stages of structured biofilm formation and in the formation of flat biofilms [41]) and in colonization of microcolony “stalks” to form the mushroom caps of structured biofilms (39). Still, stimulation of twitching motility (e.g., by Fe chelation) can also have a negative impact on structured biofilm formation, promoting the formation of thin, flat biofilms that are devoid of microcolonies (6, 75).
We examine the impacts of Fe levels on biofilm formation and twitching motility in P. aeruginosa and show that there is a switch from a structured to a flat biofilm in going from an Fe-replete to Fe-limited minimal medium that may result from enhanced twitching motility in Fe-limited medium that is dependent on RhlIR.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Routine growth of P. aeruginosa and Escherichia coli was carried out in Luria broth (LB) as before (52), with antibiotic supplementation to maintain plasmids as needed (for pUC18, ampicillin [100 μg/ml]; for mini-CTX-lacZ and its derivatives, tetracycline [10 μg/ml for E. coli and 75 to 100 μg/ml for P. aeruginosa]; for pK18mobsacB and its derivatives, kanamycin [50 μg/ml]; for pFLP2, ampicillin [100 μg/ml for E. coli] or carbenicillin [150 μg/ml for P. aeruginosa]; and for pMMB207 and its derivatives, chloramphenicol [10 μg/ml for E. coli and 75 to 100 μg/ml for P. aeruginosa]). Fe-limited BM2 succinate and BM2 glucose minimal media have been described previously (61) and were supplemented with FeCl3 as and where indicated. Plasmid pGP003, carrying the rhlI gene, was constructed by amplification of rhlI from the chromosome of P. aeruginosa K1120 by PCR using primers rhlI-F (5′-GGATCCGGATCCCTGCAATGGACCGCCGAC-3′; tandem BamHI sites underlined) and rhlI-R (5′-AAGCTTAAGCTTGCATGCGGCAGG-AGAAAGC-3′; tandem HindIII sites underlined) and cloning the gene into pMMB207. Amplification was carried out in 50-μl reaction mixtures formulated as described previously, using Vent DNA polymerase (New England Biolabs) (79), and subjected to a 5-min denaturation step at 95°C, followed by 30 cycles of 5 min at 95°C, 1 min at 61.8°C (annealing temperature), and 5 min at 95°C, before finishing with 10 min at 72°C. To introduce a deletion of the rhlR gene into P. aeruginosa K1120, a deletion construct was first prepared in plasmid pEX18Tc by cloning PCR-amplified 1-kb DNA fragments corresponding to the regions upstream and downstream of the rhlR sequences to be deleted. Sequences 5′ to the deletion were amplified using primers RhlRUp-For (5′-GATCGAATTCCAAGTTCAACGTGGCCCG-3′; EcoRI site underlined) and RhlRUp-Rev (5′-GATCGGATCCGTCATTCCTCATTGCAGTAAG-3′; BamHI site underlined). Sequences 3′ to the deletion were amplified using primers RhlRDown-For (5′-GATCGGATCCCTGGGTCTCATCTGAAGCG-3′; BamHI site underlined) and RhlRDown-Rev (5′-GATCAAGCTTGAGATGGCGGAATGACTTC-3′; HindIII site underlined). The 50-μl PCR mixtures were formulated as described previously (79) (plus 10% [vol/vol] dimethyl sulfoxide for the downstream-arm PCR) and heated for 3 min at 95°C, followed by 29 cycles of 45 s at 95°C, 45 s at 64°C (upstream fragment) or 57°C (downstream fragment), and 2 min at 72°C, before finishing with a 10-min elongation at 72°C. The upstream and downstream PCR products were sequentially cloned into pEX18Tc to yield pCG001 and sequenced to confirm the absence of mutations in the cloned DNA. Plasmid pCG001 was mobilized into P. aeruginosa K1120 from E. coli S17-1 via conjugation (78), and following selection of pCG001 transconjugants on tetracycline (50 μg/ml) and chloramphenicol (5 μg/ml; to counterselect donor E. coli), putative rhlR deletion strains were subsequently recovered on L agar supplemented with sucrose (10% [vol/vol]). Colony PCR (63) of individual sucrose-resistant colonies with primers RhlRUp-For and RhlRDown-Rev and amplification parameters for the upstream ΔrhlR arm (see above) was used to confirm the deletion.
TABLE 1.
Bacterial strains and plasmids
| Species and strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli | ||
| Sm10(λpir) | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu; Kmr λpir | 48 |
| DH5α | φ80d lacZΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 F− Δ(lacZYA-argF)U169 | 5 |
| S17-1 | thi pro hsdR recA Tra+ | 74 |
| P. aeruginosa | ||
| PAO1 | Prototroph | N. Gotoh, Kyoto Pharmaceutical University |
| K1120 | PAO1 ΔaphA; Kms | N. Gotoh, Kyoto Pharmaceutical University |
| K1203 | K1120 ΔpvdD | 72 |
| K2873 | K1120 ΔrhlR | This study |
| K2589 | K1120::PA4570-lacZ; carries a chromosomal insertion of a PA4570-lacZ transcriptional fusion | This study |
| K2590 | K2589 rhlI::mini-Tn5-tet; Tcr | This study |
| K2592 | K2589 mexI::mini-Tn5-tet; Tcr | This study |
| PAO1 | Prototroph | 44 |
| PAO1-rhlI | PAO1 rhlI::mini-Tn5-luxCDABE; Tcr | 44 |
| MPAO1 | Prototroph | 33 |
| MPAO1-rhlI | PAO1 rhlI::ISphoA/hah; Tcr | 33 |
| Plasmids | ||
| pUC18 | Cloning vector; Apr | 83 |
| Mini-CTX-lacZ | Integration vector with promoterless lacZ and oriT; Tcr | 9 |
| pGP001 | Mini-CTX::PA4570-lacZ; PA4570 promoter region cloned upstream of the promoterless lacZ gene of mini-CTX-lacZ | This study |
| pFLP2 | Source of Flp recombinase; Apr | 28 |
| pK18mobsacB | Broad-host-range gene replacement vector; sacB Kmr | 74 |
| pMMB207 | Broad-host-range low-copy-no. cloning vector; Cmr | 49 |
| pGP003 | pMMB207::rhlI | This study |
| pEX18Tc | Broad-host-range gene replacement vector; sacB Tcr | 28 |
| pCG001 | pEX18Tc::ΔrhlR | This study |
| pECP61.5 | pSW205::rhlA-lacZ; Apr | 58 |
| pMRP-1 | pUCP18::gfp; Apr Cbr | 17 |
Tcr, tetracycline resistant; Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Kms, kanamycin sensitive; Kmr, kanamycin resistant; Cbr, carbenicillin resistant.
DNA manipulations.
Standard protocols were used for restriction endonuclease digestions, ligations, transformation, plasmid isolation, preparation of electrocompetent cells, and agarose gel electrophoresis, as described by Sambrook and Russell (65). Genomic DNA of P. aeruginosa was extracted according to the protocol of Barcak et al. (7). E. coli cells were made competent using the method of Inoue et al. (31). Electroporation of P. aeruginosa with plasmid DNA was carried out as described previously (16). DNA sequencing was performed by ACGT Corporation (Toronto, Ontario, Canada) with universal and custom oligonucleotides.
Biofilm assays.
Static biofilms were grown for 12 h, based on methods described by Ceri and colleagues (15), on polyvinyl chloride pegs which protruded into the growth medium contained in the wells of a standard 96-well microtiter plate (Nunc Brand, Roskilde, Denmark). Wells were inoculated with 200 μl of P. aeruginosa (final absorbance at 600 nm [A600] of 0.05) in iron-limited BM2 succinate minimal media supplemented with the specified concentrations of FeCl3. Plates were incubated at 37°C for the specified times while shaking at 200 rpm. Biofilm mass was quantified by staining the biofilm-containing pegs on the lid with 1% (wt/vol) crystal violet solution for 20 min and then, after washing the unbound dye in water, eluting the bound dye in ethanol with gentle rocking. Both staining and eluting were performed in clean microtiter plates containing 200 μl of the respective solutions. An absorbance reading at 550 nm of the eluted dye on a plate reader spectrophotometer provided an indirect measure of the biomass (biofilm) that was attached to each peg. Alternatively, the biomasses of 12 pegs from one concentration of FeCl3 were pooled for use in β-galactosidase assays. In both cases, spectrophotometry readings at 550 nm were also performed on the planktonic fractions of cells (those remaining in culture in the wells) for comparison purposes. Flow cell biofilms were formed on a glass surface as described previously, using green fluorescence protein-expressing P. aeruginosa harboring plasmid pMRP9-1 (17). Briefly, the flow cells were inoculated with stationary-phase cells to an A600 of 0.15 in 1% (wt/vol) tryptic soy broth (TSB) with (to induce Fe limitation) and without lactoferrin (25 μg/ml), and flow was initiated after 1 h at a flow rate of 10 ml/h at room temperature. Confocal scanning laser microscopy with a MRC 1024 system (Bio-Rad) and an Axioskop microscope was used to image biofilms. For green fluorescence protein, the excitation and the emission wavelengths were 488 and 522 nm (± 35 nm), respectively. Images obtained from the confocal microscope were reconstructed to three-dimensional images by using Volocity image analysis software (Improvision).
Twitching assay.
Surface-associated twitching motility was assessed via subagar stab inoculations in various media based on a method described previously (71). Twitching media consisted of BM2 succinate, BM2 glucose, or L broth, each solidified with 1% (wt/vol) agar and, where indicated, supplemented with various concentrations of FeCl3. Colonies were transferred from solid BM2 medium and stab inoculated in the appropriate twitching medium. Twitching motility zones (occurring at the agar/petri plate interface) were allowed to develop for 40 (BM2 minimal media) or 24 (L broth) hours at 37°C, after which the agar was carefully removed, leaving the twitching zone attached to the petri plate. The twitching zones were then visualized by staining for 1 minute with 1% (wt/vol) crystal violet and their diameters measured. In some experiments, twitching plates were supplemented with C4-HSL (at 5 μM; Sigma) or cell-free culture supernatants from Fe-limited or Fe-rich cultures of wild-type and rhlI P. aeruginosa. Culture supernatants were recovered following removal of cells by centrifugation, filter sterilized (Millipore GP Express Plus membrane; 0.22-μm pore size), and used at a 1/10 dilution in the twitching medium. Where indicated, culture supernatants were heat (30 min at 95°C) or proteinase K (100 μg/ml for 1 h at 37°C) treated prior to their use in twitching plates.
Construction of a PA4570-lacZ reporter fusion.
In order to permit ready assessment of iron limitation of P. aeruginosa during planktonic and biofilm growth, reporter strain K2589, in which a known iron limitation-inducible gene (PA4570) was fused to lacZ in the chromosome, was constructed. Initially, the 5′ upstream region of PA5470 was amplified via PCR using the conditions described above for cloning of rhlI, with primers PA4570-F (5′-AAGCTTAAGCTTGACAACGAATACCGCTCC-3′; tandem HindIII sites underlined) and PA4570-R (5′-GGATCCGGATCCCGCAGGTAGGGGTGTGGC-3′; tandem BamHI sites underlined), an annealing temperature of 67°C, and 30 cycles of amplification. The PA4570 promoter-containing product was cloned upstream of the promoterless lacZ gene of plasmid mini-CTX-lacZ and the resulting vector, pGP001, mobilized into P. aeruginosa K1120 via conjugation with pGP001-carrying E. coli S17-1 (78). Transconjugants carrying pGP001 integrated into the chromosome were selected on L agar containing tetracycline (10 g/ml) and imipenem (0.5 g/ml; to counterselect donor E. coli). The mini-CTX plasmid backbone was subsequently excised from the chromosome by using the plasmid pFLP2-encoded Flp recombinase as described previously (28), leaving the PA4570-lacZ fusion behind.
β-Galactosidase assay.
β-Galactosidase assays were performed according to the method of Miller (47) on log-phase P. aeruginosa cells grown in BM2 succinate with and without FeCl3 supplementation. In the case of β-galactosidase assays of microtiter plate biofilm cells grown in the presence of various FeCl3 concentrations, pegs (12 for every different growth condition) with attached biofilms were detached from the 96-well-plate lid and vortexed with glass beads in 1 ml total of sterile phosphate-buffered saline (PBS). Following recovery of the biofilm-containing PBS and pelleting of biofilm biomass by centrifugation in a microcentrifuge, the pelleted cells were resuspended in 0.1 ml fresh PBS and assayed for β-galactosidase activity as described above with modifications. Due to limited cell recovery and, thus, minimal “signal” in this assay (which yielded substantial variation experiment to experiment), the A405 readings were normalized such that the highest A405 reading for any experiment was arbitrarily adjusted to 100 and all other A405 readings were adjusted proportionally. The corresponding planktonic cells from 12 wells were also recovered, pooled, and assayed for β-galactosidase activity by using the standard Miller assay.
C4-HSL bioassay.
Relative levels of C4-HSL in cell-free culture supernatants of P. aeruginosa were determined using a bioassay approach described previously (58) with modifications. Filter-sterilized culture supernatants (500 μl) from P. aeruginosa grown in Fe-limited or Fe-rich minimal medium were prepared as described above and mixed with equal volumes of E. coli (pECP61.5) diluted to an A600 of 0.1 in L broth supplemented with 0.1 M isopropyl-β-d-thiogalactopyranoside (IPTG). After 6 h of incubation at 30°C, β-galactosidase assays (47) were carried out, and the values obtained were normalized per ml of supernatant from a culture with an A600 of 1.0. Plasmid pECP61.5 carries the C4-HSL-responsive gene rhlA fused to lacZ, so β-galactosidase production becomes an indirect measure of supernatant C4-HSL.
Mini-Tn5-tet transposon mutagenesis.
P. aeruginosa strain K2589 was mutagenized with mini-Tn5-tet following mobilization of mini-Tn5-tet-carrying plasmid pUT from E. coli SM10 (λpir) as described previously (14). Approximately 5,000 mutants were screened for a loss of low-iron-enhanced twitching motility on iron-limited BM2 succinate twitching plates, and those showing reduced twitching motility relative to that of the K2589 parent were recovered. These were then examined on L agar and Fe-replete (100 μM FeCl3) BM2 succinate twitching plates to eliminate mutants with general defects in twitching motility (i.e., only those mutants showing reduced twitching motility on Fe-limited BM2 succinate were retained). The mini-Tn5-tet-disrupted genes from the twitching mutants were recovered following digestion of genomic DNA with PstI or NotI and shotgun cloning into pUC18. Transformants of E. coli DH5α carrying pUC18 with inserts of mini-Tn5-tet and flanking chromosomal DNA were selected on LB agar containing tetracycline (10 μg/ml), and the chromosomal DNA flanking the mini-Tn5-tet element was sequenced using primers mini-Tn5-Tc-Right and mini-Tn5-Tc-Left as before (79).
RESULTS
Impact of Fe on biofilm formation.
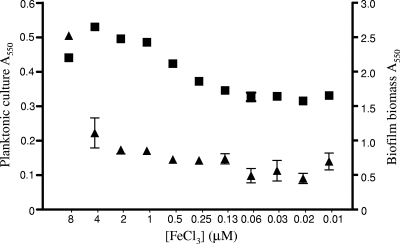
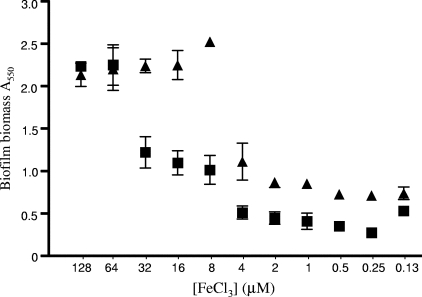
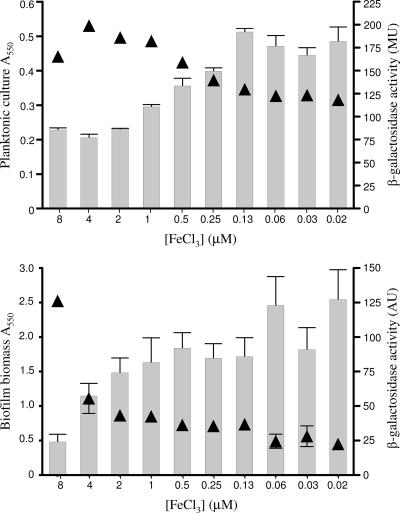
Previous studies have suggested that Fe limitation compromises biofilm formation, though these generally relied on the use of metal chelators (e.g., lactoferrin) to impose Fe restriction (75, 76). Using a defined Fe-limited minimal medium and Fe supplementation, we sought to examine the specific impacts of various medium iron levels on biofilm formation. As seen in Fig. 1, biofilm yields of P. aeruginosa K1120 declined with declining Fe supplementation, with Fe concentrations below 8 μM markedly reducing biofilm yields. Planktonic cell growth also declined with declining Fe levels, though only at concentrations below 1 μM (Fig. 1), indicating that the Fe needs of biofilm cells are greater than those of planktonic cells. A mutant of P. aeruginosa unable to synthesize pyoverdine (K1203) was even more compromised as regards the impact of Fe limitation on biofilm formation, with Fe concentrations below 64 μM markedly reducing biofilm yields (Fig. 2). This highlights both the negative impact of Fe limitation on biofilm formation by P. aeruginosa (pyoverdine-deficient mutants are less able to acquire Fe [61] and, so, more Fe limited) and the importance of pyoverdine as regards Fe acquisition in biofilm cells. To confirm that biofilm reduction was, indeed, a response to Fe limitation, a reporter P. aeruginosa strain carrying a chromosomal lacZ transcriptional fusion to a gene known to be induced in response to Fe limitation (PA4570) (54), K2589, was constructed. In planktonic cells, increasing β-galactosidase activity was clearly seen in P. aeruginosa K2589 as cell yields declined with declining Fe supplementation (Fig. 3), consistent with the known Fe limitation inducibility of PA4570. In biofilm K2589 cells, too, β-galactosidase activity increased in concert with reduced biofilm yields, confirming that biofilm cells were sensing Fe limitation and that this was likely responsible for biofilm reduction.
FIG. 1.
Impact of iron availability on biofilm formation. Results are shown for planktonic cell growth (A550; ▪) and biofilm formation (A550 of crystal violet as an indirect measure of biofilm biomass; ▴) by P. aeruginosa K1120 as a function of FeCl3 supplementation of an Fe-limited minimal medium. Cells/biofilms were grown for 12 h at 37°C. Error bars represent the standard errors of the means for a representative assay performed in triplicate.
FIG. 2.
Influence of pyoverdine on biofilm formation under iron limitation. Results are shown for biofilm formation (A550 of crystal violet as an indirect measure of biofilm biomass) by K1120 (▴) and a ΔpvdD strain, K1203 (▪), as a function of FeCl3 supplementation of an Fe-limited succinate minimal medium. Biofilms were grown for 12 h at 37°C. Error bars represent the standard errors of the means for a representative assay performed in triplicate.
FIG. 3.
Assessment of iron limitation sensed by P. aeruginosa. Results are shown for culture density (A550) (▴) and β-galactosidase activity (MU, Miller units; AU, arbitrary units) (bars) of planktonic cell culture (top) and of biofilm (bottom) P. aeruginosa K2589 (K1120::PA4570-lacZ) cells as a function of decreasing FeCl3 concentrations. Cells were grown for 12 h at 37°C. Error bars represent the standard errors of the means for a representative assay performed in at least triplicate.
Impact of Fe on twitching motility.
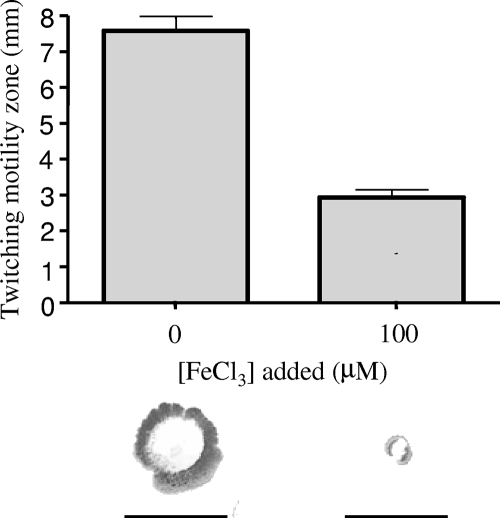
Reduced biofilm formation in response to Fe limitation has previously been attributed to enhanced twitching motility, though again, these studies were carried out using metal chelators (lactoferrin or desferrioxamine) to impose Fe restriction (75, 76), and studies of biofilm formation by other organisms (e.g., Streptococcus mutans) in response to Fe chelation have shown that lactoferrin can have effects on biofilm formation independent of Fe restriction (10). To directly examine the impact of Fe on twitching motility, we again used an Fe-limited minimal medium with or without Fe supplementation. While limited twitching motility of P. aeruginosa K1120 was observed in Fe-replete (100 μM FeCl3) medium, this increased markedly in Fe-limited (i.e., non-Fe-supplemented) medium (Fig. 4). Moreover, twitching motility remained limited at all levels of Fe supplementation above 4 μM, with concentrations at or below 4 μM only promoting enhanced twitching motility (data not shown). The pyoverdine-deficient mutant K1203 showed increased (relative to that of K1120) twitching motility on Fe-limited medium (the twitching zone increased from 7.6 to 14 mm), presumably because it is less able to transport Fe and, so, is more Fe limited than its parent strain. The effect of Fe on twitching motility was not explainable by a negative impact of Fe on twitching and/or growth on twitching plates, inasmuch as addition of 100 μM FeCl3 to L agar twitching plates did not adversely affect twitching motility on this medium (data not shown), where P. aeruginosa displays substantial twitching motility (Fig. 5c). The enhanced twitching motility with decreasing Fe thus parallels the reduction in biofilm formation, consistent with low-Fe-stimulated twitching motility contributing to loss of biofilm formation.
FIG. 4.
Influence of iron availability on twitching motility. P. aeruginosa K1120 was inoculated onto succinate minimal agar with or without Fe supplementation, and following incubation at 37°C for 40 h, the twitching zones were stained (bottom) and their diameters measured (top). Error bars represent the standard errors of the means for a representative assay performed in triplicate. Bars under twitching zones (bottom) represent 10 mm. Similar results were observed using glucose minimal medium.
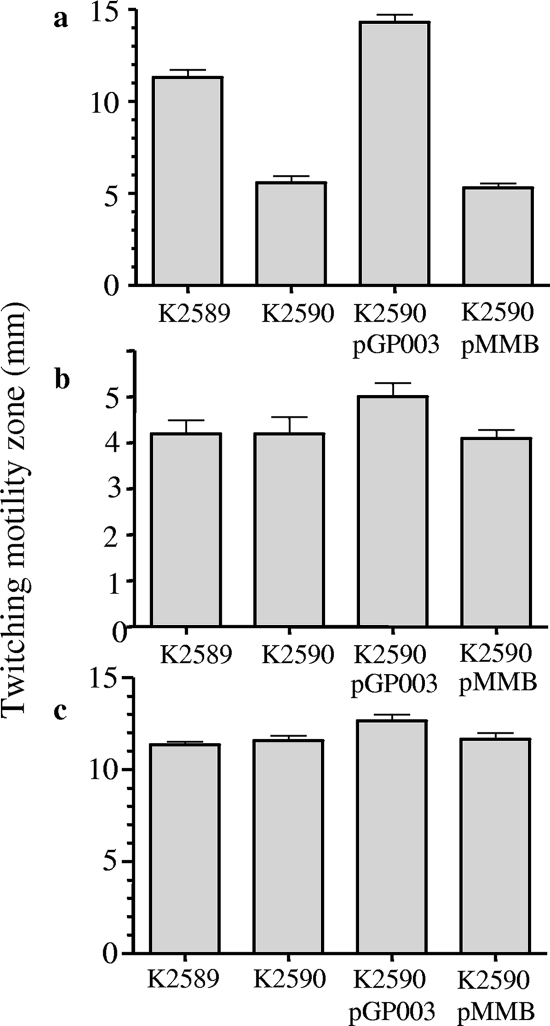
FIG. 5.
Involvement of rhlI in low-iron-stimulated twitching motility. P. aeruginosa strains K2589 (RhlI+), K2590 (RhlI−), K2590 harboring rhlI on low-copy-number plasmid pGP003, and K2590 with pMMB207 alone were inoculated onto iron-limited BM2 glucose (a), iron-replete (100 μM FeCl3) BM2 glucose (b), and L agar (c) twitching plates and twitching zones (diameter) measured after 24 (L agar) or 40 (minimal media) h of incubation at 37°C. Error bars represent the standard errors of the means for a representative assay performed at least in triplicate.
Low-Fe-stimulated twitching motility requires RhlI.
To gain insight into the mechanism of low-Fe-stimulated twitching motility and its apparently negative impact on biofilm formation, we sought to isolate mutants of P. aeruginosa specifically defective in the enhanced twitching motility seen in response to Fe limitation. Thus, P. aeruginosa K2589 was mutagenized using the mini-Tn5-tet transposon and mutants showing normal twitching motility on L agar and Fe-replete (100 μM FeCl3) minimal medium (i.e., same as K2589) but lacking the enhanced twitching motility on Fe-limited minimal medium were selected. Of the 5,000 mutants screened, there was one, K2590, that consistently showed a loss of enhanced twitching motility on Fe-limited medium (i.e., reduced relative to that of K2589) (Fig. 5a) but showed the same twitching motility as the K2589 parent on Fe-replete minimal medium (Fig. 5b) and L agar (Fig. 5c), indicating that it did not have a general defect in twitching motility. The disrupted gene in K2590 was subsequently identified as rhlI, encoding the C4-HSL synthase (43), which affects a number of genes in P. aeruginosa whose expression is cell density dependent (i.e., regulated by QS) (69). Introduction of the cloned rhlI gene into K2590 restored enhanced twitching motility on Fe-limited minimal medium (Fig. 5a), consistent with the rhlI mutation being responsible for the low-Fe-stimulated twitching motility defect of this mutant. In agreement with this, rhlI mutants obtained from two different transposon mutant libraries (33, 44) (Table 1) also showed the same twitching motility defect on Fe-limited medium (data not shown).
Impact of RhlI on low-Fe-abrogated biofilm formation.
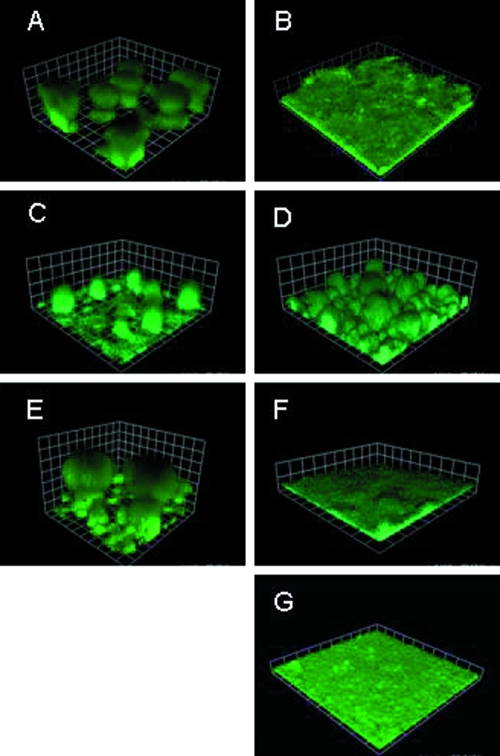
If the enhanced twitching motility seen in P. aeruginosa in response to growth in Fe-limed medium is responsible for reduced biofilm formation, loss of this low-Fe-stimulated twitching motility in the rhlI mutant should largely restore biofilm formation. With a flow cell biofilm reactor, RhlI+ strain K2589 was shown to form well-developed three-dimensional mushroom-like biofilms during growth under Fe-replete conditions (Fig. 6A), with Fe limitation yielding flat, undeveloped biofilms (Fig. 6B), as reported previously (6). In the rhlI mutant strain K2590, structured biofilm formation was largely restored during growth in Fe-limited medium (Fig. 6D), though mushroom “caps” were generally lacking, and this was also true of K2590 grown in Fe-rich medium (Fig. 6C). Introduction of the cloned rhlI gene on plasmid pGP003 into K2590 yielded wild-type biofilms in both media (Fig. 6E and F). Addition of exogenous C4-HSL to K2590 also restored the wild-type biofilm phenotype in low-Fe medium (Fig. 6G). Thus, while an rhlI mutant is somewhat defective as regards mature biofilm formation under either growth condition, biofilm formation by the mutant is not negatively affected by Fe limitation as it is in the RhlI+ parent strain.
FIG. 6.
Impact of rhlI mutation on biofilm formation in Fe-rich and Fe-limited media. The P. aeruginosa RhlI+ parent strain K2589 in iron-rich (1% TSB) (A) and iron-limited (1% TSB plus 25 μg/ml lactoferrin) (B) media, the rhlI mutant K2590 in iron-rich (C) and iron-limited (D) media, K2590 carrying rhlI plasmid pGP003 in iron-rich (E) and iron-limited (F) media, and K2590 in Fe-limited medium supplemented with 2 μM C4-HSL (G) were cultured in flow cells for 6 days. P. aeruginosa cells carried the gfp plasmid pMRP9-1. Squares are 30 μm on a side.
Impact of C4-HSL on twitching motility.
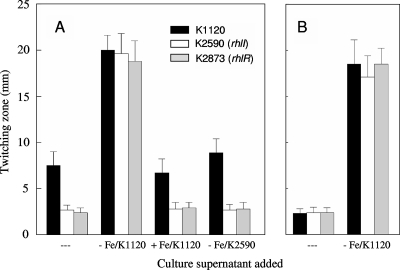
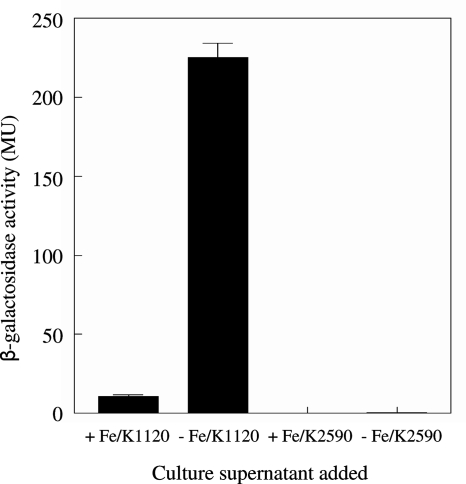
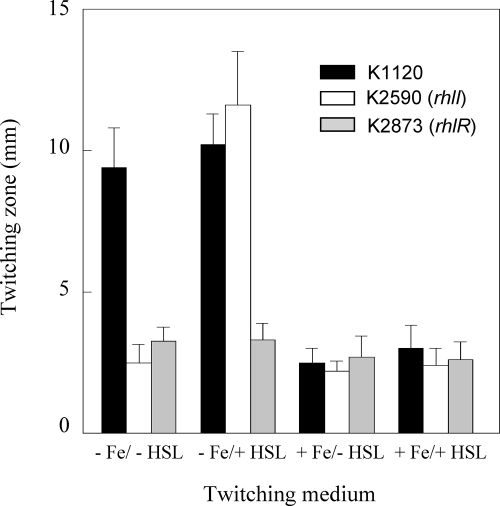
The impact of an rhlI mutation on twitching motility and biofilm formation in Fe-limited medium might simply be explained by loss of C4-HSL production, which may be needed for expression of one or more genes involved in twitching motility. Initially, cell-free spent culture supernatants from RhlI+ P. aeruginosa K1120 grown in Fe-limited minimal medium (as a crude source of C4-HSL) were tested for their abilities to restore the twitching motility of the rhlI mutant K2590 on Fe-limited medium. As seen in Fig. 7A, the Fe-limited culture supernatant of P. aeruginosa K1120 promoted substantial twitching motility of K2590 on Fe-limited medium and, indeed, enhanced the twitching motility of RhlI+ K1120 on this medium as well (Fig. 7A). In contrast, cell-free supernatants from Fe-replete (10 μM FeCl3) cultures of P. aeruginosa K1120 did not promote the twitching motility of K2590 on Fe-limited medium (Fig. 7A). This concentration of Fe was chosen to represent Fe-replete conditions in this study, because unlike supernatants from 100 μM FeCl3 cultures, which adversely affected the twitching motility of K1120 on Fe-limited plates (likely owing to Fe contamination of the twitching plates), supernatants from 10 μM FeCl3 cultures did not (Fig. 7A). Thus, the absence of an effect of the Fe-replete K1120 supernatant on K2590 twitching on Fe-limited plates could not attributed to excess Fe (from the culture supernatant) introduced into the Fe-limited twitching medium interfering with any twitching motility-promoting component that might have been present in the supernatant. As expected, supernatants prepared from Fe-limited K2590 did not promote the twitching motility of K2590 (or any strain tested) on Fe-limited medium (Fig. 7A). These data were consistent with C4-HSL being produced under Fe-limited but not Fe-replete conditions and, so, specifically stimulating twitching motility in the former instance. Using a bioassay involving E. coli carrying a C4-HSL-responsive lacZ reporter fusion to measure C4-HSL in spent culture supernatants of P. aeruginosa K1120, we confirmed that β-galactosidase activity (and, so, C4-HSL levels) increased markedly in Fe-limited versus Fe-replete minimal media (Fig. 8). Moreover, C4-HSL supplementation (5 μM) of Fe-limited twitching medium stimulated the twitching motility of K2590 (Fig. 9). Enhanced C4-HSL production by P. aeruginosa under Fe-limiting conditions is consistent with previous reports of low-Fe stimulation of rhlIR expression (21, 34) and suggests that the observed low-Fe stimulation of twitching motility results from the C4-HSL/RhlR-promoted expression of a gene(s) required for twitching. Consistent with this, a ΔrhlR mutant (K2873) was also defective in low-Fe-stimulated twitching motility (Fig. 7A) and this was not reversed upon C4-HSL supplementation (Fig. 9).
FIG. 7.
Influence of Fe-rich and Fe-limited spent culture supernatants on twitching motility under Fe-limiting (A) and Fe-replete (B) conditions. P. aeruginosa strains K1120 (RhIR+), K2590 (RhlI−), and K2873 (RhlR−) were inoculated onto Fe-limited (A) or Fe-replete (100 μM FeCl3) (B) glucose minimal agar medium without (---) or with added (10% final concentration) spent culture supernatants from P. aeruginosa strains K1120 grown in Fe-rich (10 μM FeCl3) glucose minimal medium (+ Fe/K1120), K1120 grown in Fe-limited glucose minimal medium (− Fe/K1120), and K2590 grown in Fe-limited glucose minimal medium (− Fe/K2590) as indicated. Following incubation at 37°C for 40 h, the twitching zones were stained and their diameters measured. Results shown represent the means ± standard deviations for a representative assay (16 individual twitching zones measured per strain and/or growth conditions on two separate plates) performed in duplicate. Results for RhlIR+ K2589 were indistinguishable from those for K1120 and are not shown.
FIG. 8.
Bioassay of C4-HSL production by P. aeruginosa during growth under Fe-limited and Fe-replete conditions. Aliquots (500 μl) of cell-free spent culture supernatants of P. aeruginosa K1120 (RhlI+) and K2590 (RhlI−) grown overnight in Fe-limited (− Fe/K1120 and − Fe/K2590) or Fe-replete (100 μM FeCl3) (+ Fe/K1120 and + Fe/K2590) glucose minimal medium were incubated with E. coli harboring the C4-HSL-responsive rhlA-lacZ fusion plasmid. After 6 h of incubation at room temperature, E. coli was assayed for β-galactosidase activity (reporter in Miller units [MU]) as an indirect measure of C4-HSL. The values shown represent the means ± standard deviations for two independent experiments performed in duplicate and are normalized to 1 ml of supernatant from a culture with an A600 of 1.
FIG. 9.
Influence of exogenous C4-HSL on twitching motility. P. aeruginosa K1120, H2590 (RhlI−), and K2784 (RhlR−) were inoculated onto Fe-limited (− Fe) or Fe-replete (100 μM FeCl3) (+ Fe) glucose minimal agar without (− HSL) or supplemented with (+ HSL) 5 μM C4-HSL. Following incubation at 37°C for 40 h, the twitching zones were stained and their diameters measured. Results shown represent the means ± standard deviations for a representative assay (16 individual twitching zones measured per strain and/or growth condition on two separate plates) performed in duplicate.
Low-Fe-promoted C4-HSL production does not explain low-Fe-stimulated twitching motility.
If the Fe-regulated production of C4-HSL could explain the enhanced twitching motility of Fe-limited P. aeruginosa K1120 and the general lack of twitching motility for Fe-replete K1120, addition of exogenous C4-HSL would promote twitching of this strain on Fe-replete medium. Exogenous C4-HSL failed, however, to promote the twitching motility of K1120 and, indeed, the rhlI and rhlR mutant strains on Fe-replete medium (Fig. 9). Interestingly, however, spent culture supernatants from Fe-limited (but not Fe-replete) K1120 strongly stimulated the twitching motilities of all three strains on Fe-replete medium (Fig. 7B) and, unlike C4-HSL, also stimulated twitching motility by the ΔrhlR strain K2873 on Fe-limited medium (Fig. 7A). Heat and proteinase K treatment of the K1120 Fe-limited culture supernatant did not abrogate its twitching motility-stimulating activity (data not shown). These data suggest that some soluble, nonproteinaceous secreted factor whose production is both Fe regulated and C4-HSL/RhlIR dependent is required for twitching motility and either promotes the process directly or positively influences a gene(s) whose product is necessary for twitching motility.
DISCUSSION
Apparent Fe influences on P. aeruginosa biofilm formation (6) and twitching motility (75, 76) have been reported previously, although given the general use of rich media in these studies (supplemented with metal chelators to effect Fe limitation) and the known medium/environmental dependence of these processes (73), we sought to assess the impact of this metal on biofilm formation and twitching motility directly by using a minimal medium with/without Fe supplementation. Our observation that Fe limitation compromises biofilm formation is in agreement with previous studies of P. aeruginosa (6) and, interestingly, contrasts with studies of biofilms in another organism, Staphylococcus aureus, where Fe limitation appears to stimulate biofilm formation (35). The negative impact of Fe limitation on biofilm formation coincides with its stimulation of twitching motility, again in agreement with previous studies showing that Fe-binding lactoferrin also promotes the twitching motility of P. aeruginosa (75, 76). The observation here and elsewhere (6) that a defect in siderophore (pyoverdine)-mediated Fe acquisition compromises biofilm formation seems also to be linked to enhanced twitching, presumably because such mutants are more Fe limited than their wild-type counterparts.
While these results indicate that limited Fe negatively affects biofilm formation by P. aeruginosa, other studies clearly show that elevated levels of Fe also compromise biofilm formation (51, 82) and, indeed, promote biofilm dispersal (51). The suggestion in one instance that low Fe thus favors P. aeruginosa biofilm formation (82), however, is incorrect and misleading, inasmuch as the Fe supplementation that negatively affected biofilm formation in these studies was typically >50 μM (51, 82) (a modest effect was seen at 10 μM, and concentrations below this were not tested [82]). In our studies, Fe levels below 8 μM were needed to effect Fe limitation and the attendant negative impact on biofilm formation, so clearly, Fe limitation and Fe excess both adversely affect biofilm formation in P. aeruginosa. Indeed, we also noted a reduction in biofilm formation, usually at levels of FeCl3 supplementation above 32 to 64 μM (G. Patriquin, unpublished results).
A significant finding of these studies is an RhlIR/C4-HSL requirement for the twitching motility seen in Fe-limited minimal medium. While an earlier report suggested that rhlI mutants were compromised for twitching on a rich medium (23), the authors of this study could not complement this defect with the cloned rhlI gene. A subsequent study revealed that rhlI mutants were not, in fact, altered with respect to their abilities to migrate via twitching motility on rich medium (a finding also confirmed here), although twitching motility-defective variants of this mutant did accumulate during culturing, as a consequence of secondary mutations in algR (8). This suggested that secondary algR mutations may well explain the earlier report of a twitching defect in the rhlI mutant. The observation that the RhlIR requirement for twitching is thus nutritionally conditional is in line with earlier observations that the QS dependence of other processes is also dependent on the growth medium/environmental conditions. In one report, for example, a negative impact of rhl and las mutations on biofilm formation and swarming was seen during growth on succinate but not on glucose or glutamate (73).
A link between Fe and QS is now well established, with lasIR (11, 21, 37) and rhlIR (12, 21, 34) expression both reportedly enhanced by Fe limitation and/or repressed by Fe supplementation. One explanation, then, for the general lack of twitching on Fe-replete minimal medium and the enhanced twitching on Fe-limited medium is the increased rhlIR expression and concomitant C4-HSL production observed in Fe-limited medium, with the latter upregulating a gene(s) required for twitching. Still, the failure of exogenous C4-HSL to stimulate twitching of Rhl+ or Rhl− P. aeruginosa under Fe-replete conditions argues that, while RhlIR/C4-HSL are clearly required for low-Fe-stimulated twitching (rhlIR mutants are twitching defective on Fe-limited minimal medium, and C4-HSL restores twitching of an rhlI mutant), other Fe-regulated factors are also involved. Additional screening of our mini-Tn5-tet mutant library for mutants defective in low-Fe-promoted twitching (to identify possible Fe- and RhlI-regulated genes that might be responsible for this twitching phenomenon) yielded a mutant, K2592, showing a more modest reduction (from ca. 8 mm to 5 mm) in twitching motility on Fe-limited medium than its K2589 parent. The twitching motility of K2592 was unaltered relative to that of K2589 on Fe-replete medium or L agar (data not shown). This mutant was disrupted in the mexI gene of the mexHI-opmD multidrug efflux operon (2, 70). Still, given previous observations that mutants defective in this efflux system show markedly reduced synthesis of C4-HSL (1, 2), it is likely that the twitching defect seen in K2592 on Fe-limited medium again relates to defects in C4-HSL production.
The observation that spent culture supernatants from Fe-limited but not Fe-replete RhlIR+ P. aeruginosa are able to promote twitching of RhlIR+ and RhlIR− P. aeruginosa under Fe-replete conditions is consistent with some soluble, extracellular factor(s) which is low Fe inducible (and RhlIR dependent; supernatants from an rhlI mutant do not promote twitching) positively influencing twitching. Rhamnolipids are extracellular glycolipids implicated in another form of surface motility, swarming (19), and interestingly, expression of the rhlA rhamnolipid biosynthetic gene is both RhlIR dependent (13, 53) and Fe regulated (Fe limitation yielded an 80-fold increase in rhlAB expression in M9 medium but not TSB [54], while Fe supplementation of a glucose-nutrient broth strongly reduced rhlA expression [19]). Moreover, rhamnolipid production by P. aeruginosa is typically observed in Fe-limited media (19), making rhamnolipids a possible candidate for the twitching motility-promoting component of Fe-limited P. aeruginosa culture supernatants. The observation that heat and protease treatment did not adversely affect the twitching motility-promoting activities of these supernatants is consistent with this, though the role, if any, for rhamnolipids in twitching motility remains to be seen.
Acknowledgments
This work was supported by operating grants from the Canadian Cystic Fibrosis Foundation (to K.P.), the United States Cystic Fibrosis Foundation (to E.P.G.), and the National Institute of Allergy and Infectious Disease (grant number A130040 to E.P.G.). E.B. was supported by operating and equipment grants from the Israel Science Foundation.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Aendekerk, S., S. P. Diggle, Z. Song, N. Hoiby, P. Cornelis, P. Williams, and M. Camara. 2005. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 1511113-1125. [DOI] [PubMed] [Google Scholar]
- 2.Aendekerk, S., B. Ghysels, P. Cornelis, and C. Baysse. 2002. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 1482371-2381. [DOI] [PubMed] [Google Scholar]
- 3.Anwar, H., T. van Biesen, M. Dasgupta, K. Lam, and J. W. Costerton. 1989. Interaction of biofilm bacteria with antibiotics in a novel in vitro chemostat system. Antimicrob. Agents Chemother. 331824-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armitage, G. C. 2004. Basic features of biofilms—why are they difficult therapeutic targets? Ann. R. Australas. Coll. Dent. Surg. 1730-34. [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, NY.
- 6.Banin, E., M. L. Vasil, and E. P. Greenberg. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 10211076-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204321-342. [DOI] [PubMed] [Google Scholar]
- 8.Beatson, S. A., C. B. Whitchurch, A. B. Semmler, and J. S. Mattick. 2002. Quorum sensing is not required for twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 1843598-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. BioTechniques 29948-950, 952. [DOI] [PubMed] [Google Scholar]
- 10.Berlutti, F., M. Ajello, P. Bosso, C. Morea, A. Petrucca, G. Antonini, and P. Valenti. 2004. Both lactoferrin and iron influence aggregation and biofilm formation in Streptococcus mutans. Biometals 17271-278. [DOI] [PubMed] [Google Scholar]
- 11.Bollinger, N., D. J. Hassett, B. H. Iglewski, J. W. Costerton, and T. R. McDermott. 2001. Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 1831990-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bredenbruch, F., R. Geffers, M. Nimtz, J. Buer, and S. Haussler. 2006. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol. 81318-1329. [DOI] [PubMed] [Google Scholar]
- 13.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 1777155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC type multidrug resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 531423-1436. [DOI] [PubMed] [Google Scholar]
- 15.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 371771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64391-397. [DOI] [PubMed] [Google Scholar]
- 17.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280295-298. [DOI] [PubMed] [Google Scholar]
- 18.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 671865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deziel, E., F. Lepine, S. Milot, and R. Villemur. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 1492005-2013. [DOI] [PubMed] [Google Scholar]
- 20.Drenkard, E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 51213-1219. [DOI] [PubMed] [Google Scholar]
- 21.Duan, K., and M. G. Surette. 2007. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J. Bacteriol. 1894827-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert, P., T. Maira-Litran, A. J. McBain, A. H. Rickard, and F. W. Whyte. 2002. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 46202-256. [PubMed] [Google Scholar]
- 23.Glessner, A., R. S. Smith, B. H. Iglewski, and J. B. Robinson. 1999. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J. Bacteriol. 1811623-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez, M. I., and A. Prince. 2007. Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr. Opin. Pharmacol. 7244-251. [DOI] [PubMed] [Google Scholar]
- 25.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 295-108. [DOI] [PubMed] [Google Scholar]
- 26.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 14887-102. [DOI] [PubMed] [Google Scholar]
- 27.Heydorn, A., B. Ersboll, J. Kato, M. Hentzer, M. R. Parsek, T. Tolker-Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 682008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 4359-72. [DOI] [PubMed] [Google Scholar]
- 29.Hodgson, A. E., S. M. Nelson, M. R. Brown, and P. Gilbert. 1995. A simple in vitro model for growth control of bacterial biofilms. J. Appl. Bacteriol. 7987-93. [DOI] [PubMed] [Google Scholar]
- 30.Huang, C. T., K. D. Xu, G. A. McFeters, and P. S. Stewart. 1998. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ. Microbiol. 641526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 9623-28. [DOI] [PubMed] [Google Scholar]
- 32.Jackson, K., R. Keyser, and D. J. Wozniak. 2003. The role of biofilms in airway disease. Semin. Respir. Crit. Care Med. 24663-670. [DOI] [PubMed] [Google Scholar]
- 33.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 10014339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen, V., D. Lons, C. Zaoui, F. Bredenbruch, A. Meissner, G. Dieterich, R. Munch, and S. Haussler. 2006. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J. Bacteriol. 1888601-8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, M., A. Cockayne, P. H. Williams, and J. A. Morrissey. 2005. Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves Fur-dependent and Fur-independent mechanisms. J. Bacteriol. 1878211-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juhas, M., L. Eberl, and B. Tummler. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7459-471. [DOI] [PubMed] [Google Scholar]
- 37.Kim, E. J., W. Wang, W. D. Deckwer, and A. P. Zeng. 2005. Expression of the quorum-sensing regulatory protein LasR is strongly affected by iron and oxygen concentrations in cultures of Pseudomonas aeruginosa irrespective of cell density. Microbiology 1511127-1138. [DOI] [PubMed] [Google Scholar]
- 38.Kirisits, M. J., and M. R. Parsek. 2006. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell. Microbiol. 81841-1849. [DOI] [PubMed] [Google Scholar]
- 39.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 5061-68. [DOI] [PubMed] [Google Scholar]
- 40.Klausen, M., M. Gjermansen, J. U. Kreft, and T. Tolker-Nielsen. 2006. Dynamics of development and dispersal in sessile microbial communities: examples from Pseudomonas aeruginosa and Pseudomonas putida model biofilms. FEMS Microbiol. Lett. 2611-11. [DOI] [PubMed] [Google Scholar]
- 41.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 481511-1524. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi, H. 2005. Airway biofilms: implications for pathogenesis and therapy of respiratory tract infections. Treat. Respir. Med. 4241-253. [DOI] [PubMed] [Google Scholar]
- 43.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17333-343. [DOI] [PubMed] [Google Scholar]
- 44.Lewenza, S., R. K. Falsafi, G. Winsor, W. J. Gooderham, J. B. McPhee, F. S. Brinkman, and R. E. Hancock. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindsay, D., and A. von Holy. 2006. Bacterial biofilms within the clinical setting: what healthcare professionals should know. J. Hosp. Infect. 64313-325. [DOI] [PubMed] [Google Scholar]
- 46.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56289-314. [DOI] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria, p. 72-74. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 48.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 9739-47. [DOI] [PubMed] [Google Scholar]
- 50.Murray, T. S., M. Egan, and B. I. Kazmierczak. 2007. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr. Opin. Pediatr. 1983-88. [DOI] [PubMed] [Google Scholar]
- 51.Musk, D. J., D. A. Banko, and P. J. Hergenrother. 2005. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem. Biol. 12789-796. [DOI] [PubMed] [Google Scholar]
- 52.Nehme, D., X. Z. Li, R. Elliot, and K. Poole. 2004. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 1862973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 926424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip(R) expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 451277-1287. [DOI] [PubMed] [Google Scholar]
- 55.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30295-304. [DOI] [PubMed] [Google Scholar]
- 56.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 57.Otto, M. 2006. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr. Top. Microbiol. Immunol. 306251-258. [DOI] [PubMed] [Google Scholar]
- 58.Parsek, M. R., A. L. Schaefer, and E. P. Greenberg. 1997. Analysis of random and site-directed mutations in rhII, a Pseudomonas aeruginosa gene encoding an acylhomoserine lactone synthase. Mol. Microbiol. 26301-310. [DOI] [PubMed] [Google Scholar]
- 59.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57677-701. [DOI] [PubMed] [Google Scholar]
- 60.Patel, R. 2005. Biofilms and antimicrobial resistance. Clin. Orthop. Relat. Res. 43741-47. [DOI] [PubMed] [Google Scholar]
- 61.Poole, K., S. Neshat, and D. Heinrichs. 1991. Pyoverdine-mediated iron transport in Pseudomonas aeruginosa: involvement of a high-molecular-mass outer membrane protein. FEMS Microbiol. Lett. 621-5. [PubMed] [Google Scholar]
- 62.Purevdorj, B., J. W. Costerton, and P. Stoodley. 2002. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 684457-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rédly, G. A., and K. Poole. 2003. Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: involvement of a probable extracytoplasmic-function sigma factor, FpvI. J. Bacteriol. 1851261-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakuragi, Y., and R. Kolter. 2007. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 1895383-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 66.Saye, D. E. 2007. Recurring and antimicrobial-resistant infections: considering the potential role of biofilms in clinical practice. Ostomy Wound Manage. 5346-48, 50, 52. [PubMed] [Google Scholar]
- 67.Schaber, J. A., A. Hammond, N. L. Carty, S. C. Williams, J. A. Colmer-Hamood, B. H. Burrowes, V. Dhevan, J. A. Griswold, and A. N. Hamood. 2007. Diversity of biofilms produced by quorum-sensing-deficient clinical isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 56738-748. [DOI] [PubMed] [Google Scholar]
- 68.Schaber, J. A., W. J. Triffo, S. J. Suh, J. W. Oliver, M. C. Hastert, J. A. Griswold, M. Auer, A. N. Hamood, and K. P. Rumbaugh. 2007. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect. Immun. 753715-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schuster, M., and E. P. Greenberg. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 29673-81. [DOI] [PubMed] [Google Scholar]
- 70.Sekiya, H., T. Mima, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Functional cloning and characterization of a multidrug efflux pump, MexHI-OpmD, from a Pseudomonas aeruginosa mutant. Antimicrob Agents Chemother. 472990-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Semmler, A. B., C. B. Whitchurch, and J. S. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 1452863-2873. [DOI] [PubMed] [Google Scholar]
- 72.Shen, J. S., V. Geoffroy, S. Neshat, Z. Jia, A. Meldrum, J. M. Meyer, and K. Poole. 2005. FpvA-mediated ferric pyoverdine uptake in Pseudomonas aeruginosa: identification of aromatic residues in FpvA implicated in ferric pyoverdine binding and transport. J. Bacteriol. 1878511-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 621264-1277. [DOI] [PubMed] [Google Scholar]
- 74.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1784-791. [Google Scholar]
- 75.Singh, P. K. 2004. Iron sequestration by human lactoferrin stimulates Pseudomonas aeruginosa surface motility and blocks biofilm formation. Biometals 17267-270. [DOI] [PubMed] [Google Scholar]
- 76.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417552-555. [DOI] [PubMed] [Google Scholar]
- 77.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407762-764. [DOI] [PubMed] [Google Scholar]
- 78.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 473202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sobel, M. L., K. Poole, and S. Neshat. 2005. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 1871246-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413860-864. [DOI] [PubMed] [Google Scholar]
- 81.Wolfaardt, G. M., J. R. Lawrence, R. D. Robarts, S. J. Caldwell, and D. E. Caldwell. 1994. Multicellular organization in a degradative biofilm community. Appl. Environ. Microbiol. 60434-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang, L., K. B. Barken, M. E. Skindersoe, A. B. Christensen, M. Givskov, and T. Tolker-Nielsen. 2007. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 1531318-1328. [DOI] [PubMed] [Google Scholar]
- 83.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 84.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3593-603. [DOI] [PubMed] [Google Scholar]