Abstract
The invasion of intestinal epithelial cells by Salmonella enterica serovar Typhimurium is mediated by a type III secretion system (T3SS) encoded on Salmonella pathogenicity island 1 (SPI1). Expression of the SPI1 T3SS is tightly regulated by the combined action of HilC, HilD, and RtsA, three AraC family members that can independently activate hilA, which encodes the direct regulator of the SPI1 structural genes. Expression of hilC, hilD, and rtsA is controlled by a number of regulators that respond to a variety of environmental signals. In this work, we show that one such signal is iron mediated by Fur (ferric uptake regulator). Fur activates hilA transcription in a HilD-dependent manner. Fur regulation of HilD does not appear to be simply at the transcriptional or translational level but rather requires the presence of the HilD protein. Fur activation of SPI1 is not mediated through the Fur-regulated small RNAs RfrA and RfrB, which are the Salmonella ortholog and paralog of RyhB that control expression of sodB. Fur regulation of HilD is also not mediated through the known SPI1 repressor HilE or the CsrABC system. Although understanding the direct mechanism of Fur action on HilD requires further analysis, this work is an important step toward elucidating how various global regulatory systems control SPI1.
Salmonella serovars cause a range of diseases, ranging from self-limiting gastroenteritis to life-threatening systemic infections, in a variety of hosts, including humans. During the course of infection, Salmonella enterica serovar Typhimurium invades nonphagocytic epithelial cells in the small intestine by utilizing a type III secretion system (T3SS) encoded on Salmonella pathogenicity island 1 (SPI1). The SPI1 T3SS forms a needlelike complex that is responsible for the direct injection of bacterial effector proteins into the host cell cytosol. SPI1 effector proteins are thought to elicit a number of physiological responses leading to both the gastrointestinal symptoms of infection and engulfment of the bacterium (for a review, see reference 55).
Expression of the SPI1 T3SS is known to be tightly controlled by a large number of regulatory proteins and a specific set of environmental signals (16). It is thought that the environmental triggers tell the bacterium when it has reached the appropriate location in the host for invasion. In the laboratory, “SPI1-inducing conditions” consist of overnight static growth (low oxygen) in high-salt rich medium (5, 16, 31). HilA is the central regulator of the SPI1 T3SS. Encoded on SPI1, this protein is a transcriptional activator with a DNA binding domain belonging to the OmpR/ToxR family. HilA directly controls expression of the inv/spa and prg/org operons (1, 4, 31, 33), leading to the production of all the components necessary for a functional secretion apparatus (33). Encoded in the inv/spa operon, InvF, an AraC-like transcriptional activator, has been shown to induce expression of SPI1 effector proteins encoded on SPI1 and elsewhere in the chromosome, with the help of the chaperone SicA (12, 13).
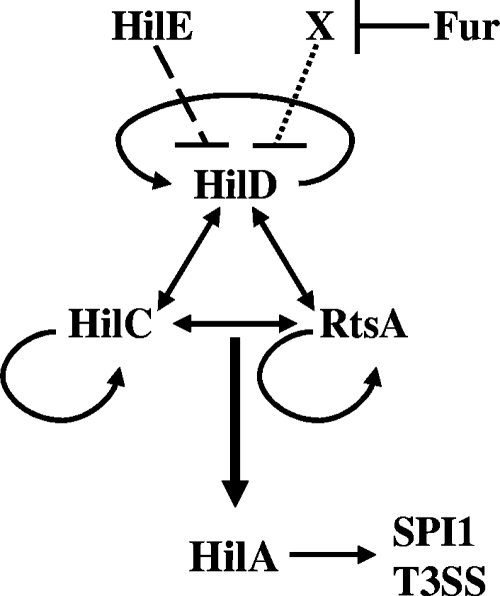
Expression of hilA is directly controlled by three AraC-like activators: HilC, HilD, and RtsA (18, 42, 47). HilC and HilD are encoded within the SPI1 locus, while RtsA is encoded in an operon with RtsB located at 93.9 centisomes. RtsB negatively regulates expression of flhDC and therefore the entire flagellar regulon (18). HilC, HilD, and RtsA have all been shown to bind directly upstream of hilA, and deletion of hilC, hilD, or rtsA leads to a lower level of hilA expression (16, 18, 42, 47, 43). HilC, HilD, and RtsA are all also capable of activating expression of hilC, hilD, and rtsA independent of each other and comprise a complex feed forward regulatory loop that controls hilA expression (Fig. 1) (16). HilD, HilC, and RtsA bind to common sites in each of the promoters and function primarily by counteracting Hns/Hha with residual activation via the alpha subunit of RNA polymerase (42, 43, 47).
FIG. 1.
Model for Fur regulation of SPI1. The arrows indicate direct activation of gene expression. HilD, HilC, and RtsA are each capable of activating hilA transcription. HilE acts by directly inhibiting HilD function. Fur presumably acts through an intermediate (indicated by X) to control HilD function. For clarity, the genes encoding HilD, HilC, RtsA, and HilA are not shown. See reference 16 for a more complete model of SPI1 regulation.
A number of additional regulators feed into the circuit to control the expression of hilA. Loss of HilE increases expression of hilA under both SPI1-inducing and SPI1-repressing conditions (7, 19). HilE has been shown via two-hybrid studies to interact with HilD, suggesting that HilE represses hilA by directly preventing HilD function (7). The PhoP/PhoQ and PhoR/PhoB two-component regulatory systems and FimZY have been shown to repress the expression of hilA by controlling expression of hilE (6, 8, 9, 44). Recently, Mlc, a global regulator of sugar utilization, has been shown to activate SPI1 by repressing hilE expression (32). Numerous other regulators have also been shown to activate expression of hilA. The EnvZ/OmpR and BarA/SirA two-component regulatory systems induce expression of hilA (34, 49), again in a manner dependent on HilD (16) but independent of HilE (unpublished data).
Fur (ferric uptake regulator) is the primary iron regulatory protein in Salmonella and Escherichia coli, and homologs of Fur have been found in other gram-negative and gram-positive organisms (27). Fur, when bound to Fe2+, binds to DNA sites in the promoters of genes which it directly represses (27). The Fur regulon includes genes involved in an array of cellular functions (27). It is thought that in the small intestine, conditions are anaerobic and therefore replete with Fe2+, leading to an active Fur protein and the repression of genes involved in uptake of iron and other metals. For example, Fur has previously been shown to repress sitABCD, encoding a manganese transport system, in Salmonella when the bacterium is in the lumen of the small intestine. The system is induced upon invasion, when iron is presumably sequestered by host iron binding proteins (25, 26).
While Fur is thought to be a simple repressor, the expression of a number of genes is decreased upon deletion of fur. One such “Fur-activated” gene is sodB (15), which encodes the superoxide dismutase with an Fe cofactor in E. coli. Under iron-limiting conditions, the small RNA RyhB is expressed. RyhB, assisted by the RNA binding protein Hfq, binds to the sodB mRNA to prevent translation (21, 38, 51). Fur activates sodB by repressing the expression of ryhB under high-iron conditions. It is thought that all “Fur-activated” genes are controlled by a similar indirect mechanism, although not necessarily via RyhB (39).
In this work, we demonstrated that Fur regulates expression of hilA via control of HilD. Fur regulation of HilD is independent of HilE and is also unaffected by RfrA and RfrB, the E. coli RyhB ortholog and paralog found in Salmonella. Both small RNAs play a role in sodB regulation in Salmonella.
MATERIALS AND METHODS
Media, reagents, and enzymatic assays.
Luria-Bertani (LB) medium was used in all experiments for growth of bacteria, and SOC was used for the recovery of transformants (36). Most bacterial strains were routinely grown at 37°C; the exceptions were strains containing temperature-sensitive plasmid pCP20 or pKD46, which were grown at 30°C. Antibiotics were used at the following concentrations: 50 μg/ml ampicillin; 20 μg/ml chloramphenicol; and 50 μg/ml kanamycin. Enzymes were purchased from Invitrogen and were used according to the manufacturer's recommendations. Primers were purchased from IDT Inc. β-Galactosidase assays were performed using a microtiter plate method described previously (48) and strains grown under the indicated conditions. β-Galactosidase activity units were determined as follows: (μmol of o-nitrophenol formed min−1) × 106/(optical density at 600 nm × ml of cell suspension). The data are expressed below as means ± standard deviations (n = 4). The cultures used in β-galactosidase assays were initially inoculated into LB medium (containing 0.5% NaCl) and grown for 8 to 12 h and then were subcultured 1/100 and grown under one of the following conditions: (i) statically overnight in 3 ml LB medium with 1% NaCl in a tube (13 by 100 mm) (referred to as SPI1-inducing conditions); (ii) SPI1-inducing conditions with ampicillin; (iii) SPI1-inducing conditions with 200 μM dipyridyl; and (iv) SPI1-inducing conditions with ampicillin and 0.2% arabinose.
Strain and plasmid construction.
Bacterial strains and plasmids are described in Table 1. All S. enterica serovar Typhimurium strains used in this study are isogenic derivatives of strain ATCC 14028 (American Type Culture Collection) and were constructed using P22 HT105/1 int-201 (P22)-mediated transduction (36). Deletion of various genes and concomitant insertion of an antibiotic resistance cassette were carried out using lambda Red-mediated recombination (14, 54) as described previously (17). The endpoints of each deletion are indicated in Table 1. In all cases, the appropriate insertion of the antibiotic resistance marker was checked by P22 linkage to known markers and/or PCR analysis. The constructs resulting from this procedure were moved into a clean wild-type background (ATCC 14028) by P22 transduction. Antibiotic resistance cassettes were removed using the temperature-sensitive plasmid pCP20 carrying the FLP recombinase (11). In some instances, the insertion mutations were converted to transcriptional or translational lac fusions using an FLP/FRT-mediated site-specific recombination method as previously described (17).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmida | Genotype or relevant characteristics | Deletion endpoints or cloned end pointsb | Source or referencec |
|---|---|---|---|
| Strains | |||
| 14028 | Wild-type serovar Typhimurium | ATCCd | |
| JS248 | ΔrtsA5 | 4561755-4560884 | 18 |
| JS251 | ΔhilA112::Cm | 3019885-3021480 | 18 |
| JS252 | ΔhilC113::Cm | 3012135-3012976 | 18 |
| JS253 | ΔhilD114::Cm | 3017865-3018730 | 18 |
| JS256 | ΔhilC-D2915::Cm | 3012135-3018730 | 18 |
| JS562 | ΔsodB111::Km | 1509924-1509449 | |
| JS564 | ΔhilD138::Km | 3017958-3018729 | |
| JS566 | Δfur-42::tetAR | 756811-756421 | |
| JS567 | ΔrfrA1::Cm | 3715394-3715551 | |
| JS568 | ΔrfrA1 | ||
| JS569 | ΔrfrB2::Cm | 1352993-1352870 | |
| JS570 | Δhfq11::Cm | 4604580-4604883 | |
| JS571 | Δhfq11::Km | 4604580-4604883 | |
| JS572 | ΔhilE115::aadA | 4763527-4764108 | |
| JS573 | ΔhilC131::Cm | 3012137-3012904 | |
| JS574 | ΔrtsA9::Cm | 4560911-4561439 | |
| JS404 | attλ::pAH125::sitA′-lac+ | 25 | |
| JS412 | Δfur-41::Cm attλ::pAH125::sitA′-lac+ | 25 | |
| JS575 | attλ::pDX1::hilA′-lac+ | ||
| JS576 | ΔhilD114::Cm attλ::pDX1::hilA′-lac+ | ||
| JS577 | ΔhilC113::Cm attλ::pDX1::hilA′-lac+ | ||
| JS578 | ΔhilC-D2915::Cm attλ::pDX1::hilA′-lac+ | ||
| JS579 | ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS580 | ΔhilD114::Cm ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS581 | ΔhilC113::Cm ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS582 | ΔhilC-D2915::Cm ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS583 | Δfur-42::tetAR attλ::pDX1::hilA′-lac+ | ||
| JS584 | Δfur-42::tetAR ΔhilD114::Cm attλ::pDX1::hilA′-lac+ | ||
| JS585 | Δfur-42::tetAR ΔhilC113::Cm attλ::pDX1::hilA′-lac+ | ||
| JS586 | Δfur-42::tetAR ΔhilC-D2915::Cm attλ::pDX1::hilA′-lac+ | ||
| JS587 | Δfur-42::tetAR ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS588 | Δfur-42::tetAR ΔhilD114::Cm ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS589 | Δfur-42::tetAR ΔhilC113::Cm ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS590 | Δfur-42::tetAR ΔhilC-D2915::Cm ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS591 | pWKS30 attλ::pDX1::hilA′ | ||
| JS592 | pFur attλ::pDX1::hilA′ | ||
| JS593 | pWKS30 ΔhilD114::Cm attλ::pDX1::hilA′ | ||
| JS594 | pFur ΔhilD114::Cm attλ::pDX1::hilA′ | ||
| JS595 | pWKS30 ΔhilC113::Cm attλ::pDX1::hilA′ | ||
| JS596 | pFur ΔhilC113::Cm attλ::pDX1::hilA′ | ||
| JS597 | pWKS30 ΔrtsA5 attλ::pDX1::hilA′ | ||
| JS598 | pFur ΔrtsA5 attλ::pDX1::hilA′ | ||
| JS613 | ΔrfrA1 attλ::pDX1::hilA′-lac+ | ||
| JS614 | Δfur-42::tetAR ΔrfrA1 attλ::pDX1::hilA′-lac+ | ||
| JS615 | ΔrfrB2::Cm ΔrfrA1 attλ::pDX1::hilA′-lac+ | ||
| JS616 | Δfur-42::tetAR ΔrfrB2::Cm ΔrfrA1 attλ::pDX1::hilA′-lac+ | ||
| JS617 | ΔrfrB2::Cm attλ::pDX1::hilA′-lac+ | ||
| JS618 | Δfur-42::tetAR ΔrfrB2::Cm attλ::pDX1::hilA′-lac+ | ||
| JS619 | Φ(sodB111′-′lac)hyb | ||
| JS620 | Δfur-42::tetAR Φ(sodB111′-′lac)hyb | ||
| JS621 | ΔrfrA1 Φ(sodB111′-′lac)hyb | ||
| JS622 | Δfur-42::tetAR ΔrfrA1 Φ(sodB111′-′lac)hyb | ||
| JS623 | ΔrfrB2::Cm ΔrfrA1 Φ(sodB111′-′lac)hyb | ||
| JS624 | Δfur-42::tetAR ΔrfrB2::Cm ΔrfrA1 Φ(sodB111′-′lac)hyb | ||
| JS625 | ΔrfrB2::Cm Φ(sodB111′-′lac)hyb | ||
| JS626 | Δfur-42::tetAR ΔrfrB2::Cm Φ(sodB111′-′lac)hyb | ||
| JS629 | Δhfq11::Cm Φ(sodB111′-′lac)hyb | ||
| JS630 | Δfur-42::tetAR Δhfq11::Cm Φ(sodB111′-′lac)hyb | ||
| JS631 | Δhfq11::Km attλ::pDX1::hilA′-lac+ | ||
| JS632 | Δhfq11::Km Δfur-42::tetAR attλ::pDX1::hilA′-lac+ | ||
| JS633 | ΔhilE115::aadA attλ::pDX1::hilA′-lac+ | ||
| JS634 | ΔhilE115::aadA ΔhilD114::Cm attλ::pDX1::hilA′-lac+ | ||
| JS635 | ΔhilE115::aadA ΔhilC113::Cm attλ::pDX1::hilA′-lac+ | ||
| JS636 | ΔhilE115::aadA ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS637 | Δfur-42::tetAR ΔhilE115::aadA attλ::pDX1::hilA′-lac+ | ||
| JS638 | Δfur-42::tetAR ΔhilE115::aadA ΔhilD114::Cm attλ::pDX1::hilA′-lac+ | ||
| JS639 | Δfur-42::tetAR ΔhilE115::aadA ΔhilC113::Cm attλ::pDX1::hilA′-lac+ | ||
| JS640 | Δfur-42::tetAR ΔhilE115::aadA ΔrtsA5 attλ::pDX1::hilA′-lac+ | ||
| JS641 | Φ(hilC′-lac+)131 | ||
| JS642 | ΔrtsA5 Φ(hilC′-lac+)131 | ||
| JS643 | ΔhilD114::Cm Φ(hilC′-lac+)131 | ||
| JS644 | ΔhilD114::Cm ΔrtsA5 Φ(hilC′-lac+)131 | ||
| JS645 | Φ(rtsA′-lac+)9 | ||
| JS646 | ΔhilD114::Cm Φ(rtsA′-lac+)9 | ||
| JS647 | ΔhilC113::Cm Φ(rtsA′-lac+)9 | ||
| JS648 | ΔhilC-D2915::Cm Φ(rtsA′-lac+)9 | ||
| JS649 | Δfur-42::tetAR Φ(hilC′-lac+)131 | ||
| JS650 | Δfur-42::tetAR ΔrtsA5 Φ(hilC′-lac+)131 | ||
| JS651 | Δfur-42::tetAR ΔhilD114::Cm Φ(hilC′-lac+)131 | ||
| JS652 | Δfur-42::tetAR ΔhilD114::Cm ΔrtsA5 Φ(hilC′-lac+)131 | ||
| JS653 | Δfur-42::tetAR Φ(rtsA′-lac+)9 | ||
| JS654 | Δfur-42::tetAR ΔhilD114::Cm Φ(rtsA′-lac+)9 | ||
| JS655 | Δfur-42::tetAR ΔhilC113::Cm Φ(rtsA′-lac+)9 | ||
| JS656 | Δfur-42::tetAR ΔhilC-D2915::Cm Φ(rtsA′-lac+)9 | ||
| JS659 | Φ(hilD′-lac+)138 | ||
| JS660 | pLS119 Φ(hilD′-lac+)138 | ||
| JS661 | pJE19 Φ(hilD′-lac+)138 | ||
| JS662 | Δfur-42::tetAR Φ(hilD′-lac+)138 | ||
| JS667 | pWKS30 Φ(sodB111′-′lac)hyb | ||
| JS668 | pFUR Φ(sodB111′-′lac)hyb | ||
| JS697 | yjeT::hilD123′::Km | ||
| JS698 | yjeT::Φ(hilD′-lac+)123 | ||
| JS699 | yjeT::PlacUV5Φ(hilD′-lac+)123 | ||
| JS700 | yjeT::Φ(hilD′-′lac)hyb123 | ||
| JS701 | yjeT::PlacUV5Φ(hilD′-′lac)hyb123 | ||
| JS702 | pWKS30 yjeT::Φ(hilD′-lac+)123 | ||
| JS703 | pFUR yjeT::Φ(hilD′-lac+)123 | ||
| JS704 | pBAD30 yjeT::Φ(hilD′-lac+)123 | ||
| JS705 | pLS119 yjeT::Φ(hilD′-lac+)123 | ||
| JS706 | pWKS30 yjeT::PlacUV5Φ(hilD′-lac+)123 | ||
| JS707 | pFUR yjeT::PlacUV5Φ(hilD′-lac+)123 | ||
| JS708 | pBAD30 yjeT::PlacUV5Φ(hilD′-lac+)123 | ||
| JS709 | pLS119 yjeT::PlacUV5Φ(hilD′-lac+)123 | ||
| JS710 | pWKS30 ΔhilD114::Cm yjeT::Φ(hilD′-lac+)123 | ||
| JS711 | pFUR ΔhilD114::Cm yjeT::Φ(hilD′-lac+)123 | ||
| JS712 | pBAD30 ΔhilD114::Cm yjeT::Φ(hilD′-lac+)123 | ||
| JS713 | pLS119 ΔhilD114::Cm yjeT::Φ(hilD′-lac+)123 | ||
| JS714 | pWKS30 ΔhilD114::Cm yjeT::PlacUV5Φ(hilD′-lac+)123 | ||
| JS715 | pFUR ΔhilD114::Cm yjeT::PlacUV5Φ(hilD′-lac+)123 | ||
| JS716 | pBAD30 ΔhilD114::Cm yjeT::PlacUV5Φ(hilD′-lac+)123 | ||
| JS717 | pLS119 ΔhilD114::Cm yjeT::PlacUV5Φ(hilD′-lac+)123 | ||
| JS718 | pWKS30 yjeT::Φ(hilD′-′lac)hyb123 | ||
| JS719 | pFUR yjeT::Φ(hilD′-′lac)hyb123 | ||
| JS720 | pBAD30 yjeT::Φ(hilD′-′lac)hyb123 | ||
| JS721 | pLS119 yjeT::Φ(hilD′-′lac)hyb123 | ||
| JS722 | pWKS30 yjeT::PlacUV5Φ(hilD′-′lac)hyb123 | ||
| JS723 | pFUR yjeT::PlacUV5Φ(hilD′-′lac)hyb123 | ||
| JS724 | pBAD30 yjeT::PlacUV5Φ(hilD′-′lac)hyb123 | ||
| JS725 | pLS119 yjeT::PlacUV5Φ(hilD′-′lac)hyb123 | ||
| JS726 | pWKS30 ΔhilD114::Cm yjeT::Φ(hilD′-′lac)hyb123 | ||
| JS727 | pFUR ΔhilD114::Cm yjeT::Φ(hilD′-′lac)hyb123 | ||
| JS728 | pBAD30 ΔhilD114::Cm yjeT::Φ(hilD′-′lac)hyb123 | ||
| JS729 | pLS119 ΔhilD114::Cm yjeT::Φ(hilD′-′lac)hyb123 | ||
| JS730 | pWKS30 ΔhilD114::Cm yjeT::PlacUV5Φ(hilD′-′lac)hyb123 | ||
| JS731 | pFUR ΔhilD114::Cm yjeT::PlacUV5Φ(hilD′-′lac)hyb123 | ||
| JS732 | pBAD30 ΔhilD114::Cm yjeT::PlacUV5Φ(hilD′-′lac)hyb123 | ||
| JS733 | pLS119 ΔhilD114::Cm yjeT::PlacUV5Φ(hilD′-′lac)hyb123 | ||
| JS734 | ΔhilD123::Km | 3018495-3018729 | |
| Plasmids | |||
| pKD46 | bla PBADgam bet exo pSC101 oriTS | 14 | |
| pCP20 | bla cat cI857 λPRflp pSC101 oriTS | 11 | |
| pKD3 | bla FRT cat FRT PS1 PS2 oriR6K | 14 | |
| pKD13 | bla FRT aph FRT PS1 PS2 oriR6K | 14 | |
| pCE40 | ahp FRT ′lacZ lacY+ thisoriR6K | 17 | |
| pCE70 | ahp FRT tnpR lacZY+ thisoriR6K | 41 | |
| pCE71 | ahp FRT tnpR lacZY+ thisoriR6K | 41 | |
| pKG137 | ahp FRT lacZY+ thisoriR6K | Y. A. Golubeva and J. M. Slauch, unpublished | |
| pWKS30 | pSC101 ori, Apr | 52 | |
| pLS119 | bla PBADhilC-myc-His pACYC184 ori | 47 | |
| pJE19 | pWKS30::fur+ | 756865-756370 | |
| pDX1 | lacZ tL3λattP oriR6K aacIV tmgB | D. Lin and J. M. Slauch, unpublished | |
| pAH125 | lacZ tL3λattP oriR6K Kan tmgB | 24 | |
| pDX4 | pDX1::hilA′ | 3019008-3019920 | |
| pBAD24 | bla araC PBADpACYC184 ori | 23 | |
| pEM1396 | pBAD24-ryhB | 37 |
All strains are isogenic derivatives of ATCC 14028.
The numbers indicate the base pairs that are deleted (strains) or cloned (plasmids) (inclusive) as defined in the S. enterica serovar Typhimurium LT2 genome sequence in the National Center for Biotechnology Information database.
The source was this study, unless otherwise noted.
ATCC, American Type Culture Collection.
Standard recombinant DNA techniques were used for construction of plasmids (46). The Salmonella fur gene was amplified using primers carrying a site for either the XhoI or XbaI restriction endonuclease (fur forward primer ACGACTCGAGGCAACAGGACAG and fur reverse primer ACGATCTAGAGTTGGCTCTTCG) and then cloned into the pWKS30 vector (52). The promoter of hilA was cloned 5′ to a promoterless lacZ gene in pDX1 (hilA promoter 5′ sequence, GTAAGGTACCGTCCAGATGACACTATC; hilA promoter 3′ sequence, TGACGAATTCTTCTGAGCGTAGCAGGG), an apramycin-resistant derivative of pAH125 (24; D. Lin and J. M. Slauch, unpublished data). The resulting fusion plasmid was integrated into the S. enterica serovar Typhimurium chromosome at the lambda attachment site using λInt produced from the CRIM helper plasmid pINT-ts (24). The integrated plasmid was tested by PCR to ensure that only a single copy was present.
Construction of hilD fusions.
Starting with a Kan insertion at position 695 in hilD, the region from position −521 through the Kan cassette was PCR amplified. The primers included 30 bp identical to nucleotides in the yjeT gene. Alternatively, hilD was PCR amplified from position 1 (42) through the Kan cassette. In this case the 5′ primer included the lacUV5 promoter, as well as a sequence identical to a yjeT sequence. The products were inserted into the yjeT locus using lambda Red-mediated recombination (14, 54). YjeT has previously been shown to have no involvement in Salmonella virulence (26), and our constructs were inserted in the orientation opposite any potential yjeT transcription. The insertions were then converted to transcriptional or translational lacZ fusions using an FLP/FRT-mediated site-specific recombination method as previously described (17).
RESULTS
Fur induces expression of hilA.
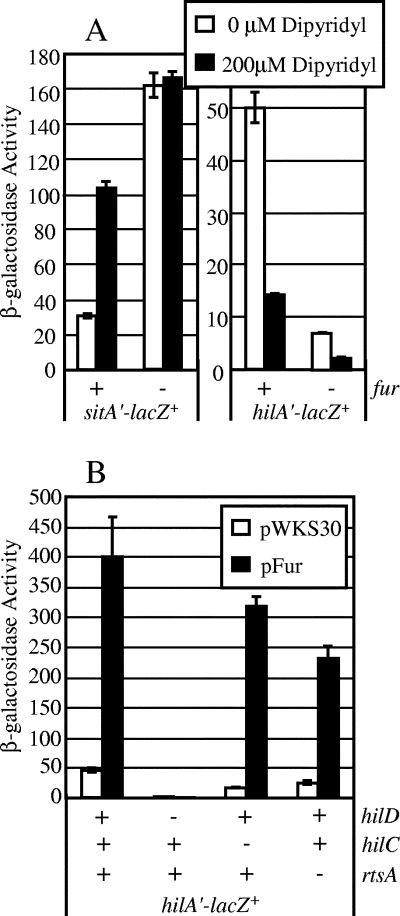
Knowing that the regulation of invasion genes in Salmonella is dependent on a specific set of environmental conditions, we wanted to observe the role, if any, of iron in the control of hilA. It has been shown that the sitABCD operon, encoding a manganese transport system repressed by Fur in the presence of high iron concentrations, is not expressed in the lumen of the small intestine of the mouse but is induced upon invasion (25, 26). Thus, iron levels are a potentially strong signal distinguishing the lumen of the small intestine from systemic sites. Using the sit system as a control, we observed the effect of the metal chelator 2,2-dipyridyl on sitA and hilA promoter-lacZ fusions in LB medium. Figure 2A shows that chelation of metals increased expression of sitA-lac, as expected. In contrast, addition of dipyridyl decreased the expression of hilA-lac about fivefold. Thus, the concentration of metals in the environment affects expression of hilA.
FIG. 2.
Fur activates expression of hilA. (A) Regulation of hilA and sitA by Fur and metal chelation. The strains contained transcriptional lacZ fusions to hilA or sitA promoters, and either the strains were fur+ or the fur gene was deleted, as indicated. The strains were grown in both SPI1-inducing conditions and SPI1-inducing conditions with 200 μM dipyridyl. The strains used were JS404, JS412, JS575, and JS583. (B) Fur overexpression induces hilA expression when HilD is present. The strains contained a transcriptional lacZ fusion to hilA, and hilC, hilD, or rtsA was deleted, as indicated. The strains also contained either pWKS30 (vector control) or pFur. All strains were grown in SPI1-inducing conditions with ampicillin. The strains used were JS591 through JS598.
To see if Fur was involved in this metal regulation, we deleted fur in the hilA-lac fusion background and monitored expression with and without dipyridyl. Deletion of fur caused a sevenfold decrease in the expression of hilA under these conditions. In the Δfur background, there was still a decrease in hilA expression upon addition of dipyridyl. As expected, sitA expression was increased in a fur background, and addition of dipyridyl did not further affect expression of sitA when fur was deleted. Taken together, these data suggest that the presence of metals in the environment induces expression of hilA and that Fur plays a major role in this metal regulation of hilA and therefore SPI1 expression. Fur is not entirely responsible for the SPI1 response to metal chelation, but given the plethora of regulatory systems that feed into SPI1, this residual effect is not surprising. For example, the PhoPQ system can be induced by low concentrations of Mg, Ca, and Mn, which in turn repress SPI1 (22).
To further examine the effects of Fur on hilA expression, we sought to overexpress Fur by cloning the gene into the pWKS30 expression vector. Figure 2B shows that overexpression of Fur caused an eightfold increase in hilA expression compared with the expression observed in the vector control strain. Taken together, these data indicate that Fur acts as a positive regulator of hilA expression. Thompson et al. (50) also noted that Fur affects SPI1 expression.
Fur regulates expression of hilA by working through HilD.
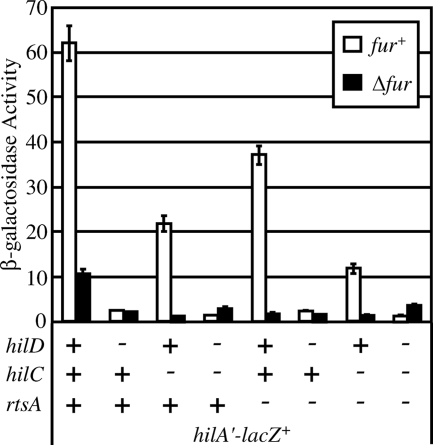
Having established that Fur controls hilA expression, we wanted to see how Fur fits into the feed forward loop model of SPI1 regulation that we previously defined (Fig. 1) (16). Therefore, we examined the effects of deleting hilC, hilD, and rtsA individually and in various combinations on expression in a hilA-lac fusion strain in fur+ and Δfur backgrounds. Figure 3 shows that Fur regulation of hilA was abrogated in any background when hilD was deleted; in all ΔhilD strains there was no significant difference in hilA expression between the fur+ and Δfur strains. As expected, based on our model (Fig. 1), deletion of hilC or rtsA affected the overall expression of hilA but did not prevent Fur regulation of hilA. This suggests that Fur regulation of SPI1 feeds into the system via HilD.
FIG. 3.
Fur regulates expression of hilA via HilD. The strains contained a transcriptional lacZ fusion to hilA, and hilC, hilD, rtsA, or fur was deleted, as indicated. The strains were grown in SPI1-inducing conditions. The strains used were JS575 through JS590.
To further test the requirement for HilD for Fur-mediated regulation of hilA, we examined the effects of Fur overexpression in strains containing the hilA-lac fusion, with and without hilC, hilD, or rtsA. Figure 2B shows that when hilD was deleted, pFur was unable to induce expression of hilA. As expected from the results described above, deletion of hilC or rtsA decreased but did not abrogate Fur-dependent induction of hilA. This provides clear genetic evidence that HilD is absolutely required for Fur regulation of hilA.
Fur control requires HilD protein.
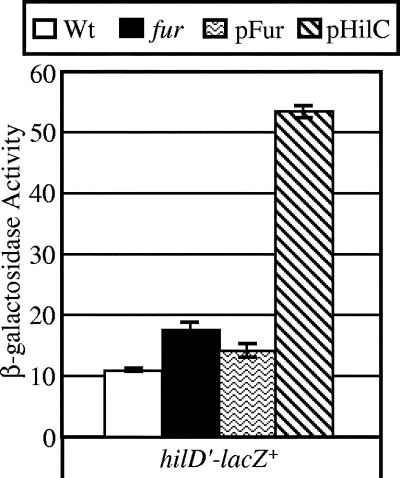
Given that HilD is required for Fur regulation of hilA, it seemed possible that Fur regulates hilD expression in some manner. We first constructed a hilD-lacZ transcriptional fusion at the hilD locus in the chromosome, making the strain hilD null. We then examined the effects of deleting fur, overproducing Fur, and overproducing HilC, which is known to independently activate hilD transcription (16). Figure 4 shows that neither deletion of fur nor overproduction of Fur had any significant effect on transcription of the hilD fusion, while overproduction of HilC clearly induced the expression of hilD, as previously reported (16).
FIG. 4.
Fur does not directly regulate hilD transcription. The strains contained a transcriptional lacZ fusion to hilD, and either fur was deleted or the strain contained pFur or pHilC, as indicated. The strains were grown in SPI1-inducing conditions. The strains used were JS659 through JS662. Wt, wild type.
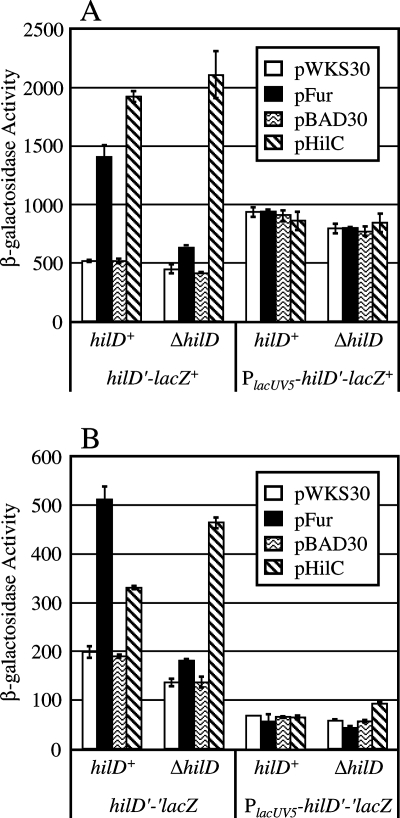
Since Fur activation of hilA is clearly dependent upon HilD, we constructed hilD-lacZ fusions in a hilD+ background. A fusion construct containing a region of the hilD gene from position −521 to position 695 located 5′ of a promoterless lacZY operon was inserted elsewhere in the chromosome. This left the wild-type copy of hilD intact and functional. We first observed the effects of Fur and HilC overproduction on the transcriptional hilD-lacZ fusion. Figure 5A shows that hilD transcription was induced by both Fur and HilC in the hilD+ background. However, when hilD was deleted in these strains, there was no longer a significant effect of Fur overproduction, while HilC overproduction still induced hilD transcription at a level that was approximately the same as the level seen in the hilD+ background. We also constructed a hilD-lacZ transcriptional fusion starting at the normal transcription start site but under control of the lacUV5 promoter. Figure 5A shows that this construct was not affected by either HilC or Fur overproduction and that the presence or absence of hilD had no effect on transcription of this fusion.
FIG. 5.
Fur regulation of hilD requires the HilD protein. The strains contained a transcriptional (A) or translational (B) lacZ fusion to hilD inserted into yjeT. The strains were either hilD+ or ΔhilD and contained pWKS30 (vector control), pFur, pBAD30 (vector control), or pHilC, as indicated. The strains were grown in SPI1-inducing conditions. The strains used were JS702 through JS731.
Known “Fur-activated” genes, like sodB, are regulated at the level of translation initiation and/or mRNA stability. To further examine potential Fur-mediated regulation of hilD, we constructed translational hilD-lacZ fusions (with the fusion joint at position 695) in which β-galactosidase production was dependent on both transcription and translation of hilD. Figure 5B shows that overproduction of HilC and Fur induced the translational hilD fusion in the hilD+ background. Again, when hilD was deleted in these strains, Fur regulation of hilD was abrogated, while overproduction of HilC still induced hilD expression. When the hilD translational fusion was placed under control of the lacUV5 promoter, overproduction of Fur or HilC had no effect.
Taken together, these data rule out the possibility that there is Fur control of SPI1 by simple activation of hilD transcription or translation. There was transcriptional induction of hilD in response to Fur, and this required the hilD promoter region. However, like hilA expression, this transcriptional induction required that HilD be present and is consistent with our model for SPI1 regulation (Fig. 1); transcription of hilD, transcription of hilC, transcription of rtsA, and transcription of hilA were all increased upon activation of the system. Thus, either Fur regulation is posttranslational, acting at the level of HilD protein, or the mechanism is more complex (see Discussion).
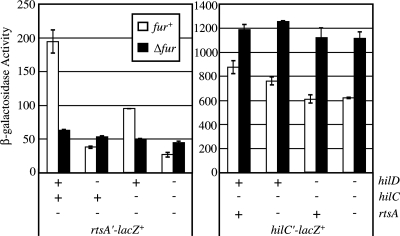
Fur regulates expression of rtsA by controlling HilD.
To show the complete impact of Fur regulation on SPI1, we observed the effect of Δfur on the expression the other two hilA regulators, HilC and RtsA. Based on the regulation of hilA-lac, we expected to see hilC and rtsA expression reduced by deletion of fur, and this regulation should have been dependent on HilD. To test this, we measured the activity of transcriptional hilC-lac and rtsA-lac fusions in fur+ and Δfur strains with hilD, hilC, and rtsA deletions in various combinations. Figure 6 shows that expression of rtsA was decreased by Δfur, but only when HilD was present. This confirmed that HilD controls expression of rtsA and that Fur works exclusively via HilD to control the expression of SPI1 genes. Transcription of hilC was actually increased by Δfur. The reason for this is not clear. However, the predominant effect of Fur is to activate SPI1, so the transcriptional effect on hilC seems to have little phenotypic effect. It is important to note that while HilD is controlled in a complex fashion, rtsA is regulated at the level of transcription via HilD, consistent with the model for SPI1 regulation (Fig. 1) (16).
FIG. 6.
Fur regulates expression of rtsA via HilD. The strains contained a transcriptional lacZ fusion to either hilC or rtsA, and hilC, hilD, or rtsA was deleted, as indicated. The strains were grown in SPI1-inducing conditions. The strains used were JS641 through JS656.
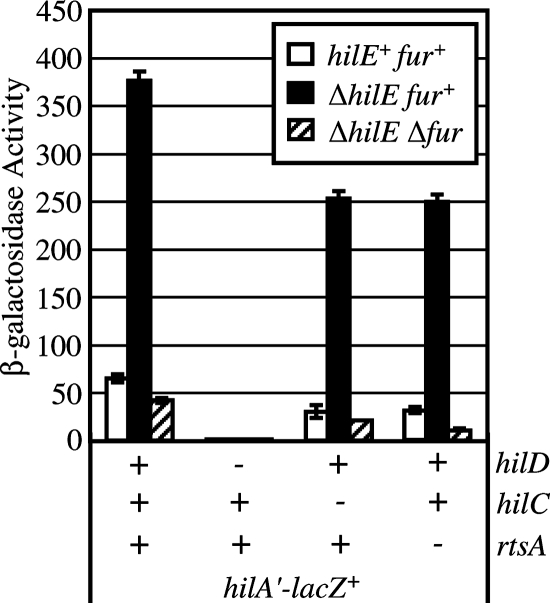
Fur regulation of HilD is not mediated through HilE.
Fur is known to be only a transcriptional repressor. Thus, one model to explain Fur regulation of SPI1 is that Fur represses a factor that negatively regulates HilD function. HilE is a known repressor of the SPI1 T3SS. Published data suggest that HilE is capable of directly binding to HilD and preventing functioning of the protein (7). Other regulatory systems that are known to repress hilA have been shown to function by inducing expression of hilE, leading to repression of hilA (6, 8). We looked at a hilA-lac transcriptional fusion in backgrounds with hilE and fur deleted. If Fur works via hilE, we would expect that deletion of fur would have no effect on the expression of hilA in the ΔhilE background. Figure 7 shows that while deletion of hilE increased expression of hilA as expected, deletion of fur in the ΔhilE background still resulted in an eightfold decrease in hilA expression. Deletion of either hilC or rtsA in this experiment decreased expression of hilA but did not block regulation by either HilE or Fur. In contrast, deletion of hilD abrogated both HilE- and Fur-mediated regulation, as expected. Based on these results, we concluded that Fur does not regulate SPI1 by repressing HilE.
FIG. 7.
Fur does not regulate hilA expression by controlling HilE. The strains contained a transcriptional lacZ fusion to hilA, and hilC, hilD, rtsA, fur, or hilE was deleted, as indicated. The strains were grown in SPI1-inducing conditions. The strains used were JS575 through JS579 and JS633 through JS640.
Fur regulates HilD in a different manner than it regulates sodB.
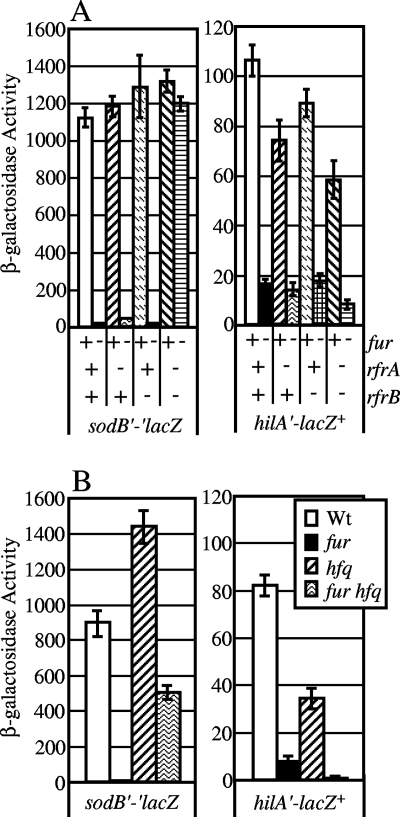
In E. coli, expression of sodB is directly controlled by a small RNA, RyhB, which binds the sodB mRNA to prevent translation (38). The expression of ryhB, in turn, is repressed by Fur under high-iron conditions (38). Thus, Fur positively controls sodB at the posttranscriptional level. Although data presented above suggest that hilD is not controlled at the translational level, we wanted to determine if RyhB was involved in Fur-mediated regulation of SPI1. The paralog of ryhB is evident in the serovar Typhimurium genome, with 75 of 95 bases identical (38). In addition, serovar Typhimurium encodes an apparent ryhB ortholog that is not found in E. coli K-12 (S. Gottesman, personal communication). The designation “ryh” indicates a small RNA with an unknown function located between centisomes 70 and 80 in the chromosome (53). Since this nomenclature is applicable to neither the ryhB paralog nor the ortholog found in serovar Typhimurium, we renamed these genes rfrA and rfrB, respectively (for “RNA for Fur response”). The gene encoding RfrB is located between STM1273 and yeaQ and exhibits 55.3% identity to the gene encoding Salmonella RfrA (RyhB). Given this apparent homology, it is possible that these RNAs have redundant functions in Salmonella. Because of this, we wanted to observe expression of a sodB′-′lacZ translational fusion as a control for Fur “activation.” If Fur represses rfrA and rfrB and allows translation of sodB, loss of rfrA and perhaps rfrB should increase expression of sodB in the Δfur background. Expression of sodB was greatly reduced in a Δfur background, as expected (Fig. 8A). Single deletions of rfrA or rfrB did not cause a significant increase in sodB expression in a Δfur background. However, when both rfrA and rfrB were deleted, expression of the sodB′-′lacZ translational fusion was restored and no longer affected by loss of Fur. Thus, as in E. coli, Fur regulation of sodB is mediated through small RNAs, but serovar Typhimurium encodes two apparently redundant small RNA molecules, RfrA and RfrB, each of which can individually repress sodB in the absence of the other and both of which contribute to Fur regulation of sodB. Note also that the sodB-lacZ translational fusion provides strong readout of translational regulation by Fur. In contrast, the hilD-lacZ translational fusions described above were clearly not regulated.
FIG. 8.
Fur regulation of sodB and Fur regulation of hilA are mechanistically different. (A) Fur controls expression of sodB, but not SPI1, via RfrA and RfrB. The strains contained a translational lacZ fusion to sodB or a transcriptional fusion to hilA, and fur, rfrA, or rfrB was deleted, as indicated. The strains were grown in SPI1-inducing conditions. The strains used were JS575, JS583, and JS613 through 626. (B) Fur regulation of SPI1 is not dependent on Hfq. The strains contained translational lacZ fusions to sodB or hilD or a transcriptional fusion to hilA, and fur or hfq was deleted, as indicated. The strains were grown in SPI1-inducing conditions. The strains used were JS575, JS583, JS619, JS620, and JS629 through JS632. Wt, wild type.
We then tested if Fur activation of SPI1 was dependent on the rfr small RNAs. We assayed expression of a hilA-lac transcriptional fusion in backgrounds with fur, rfrA, and rfrB deletions in various combinations. Figure 8A shows that hilA expression was not affected by deletion of rfrA or rfrB. Moreover, in the Δfur background, loss of both rfrA and rfrB did not increase the expression levels of hilA. Thus, Fur activation of hilA is not dependent on either of these small RNAs.
As shown above, overexpression of Fur caused an eightfold increase in hilA expression, and this effect was dependent on HilD. In contrast, overproduction of Fur in the sodB′-′lacZ strain led to a modest 1.4-fold increase in activity (sodB′-′lacZ pWKS30 activity, 763.15 ± 25.39 U; sodB′-′lacZ pFur activity, 1,044.06 ± 101.23 U). This was the predicted result. Presumably, under the iron-replete conditions of our experiment, repression of rfrA and rfrB by Fur was essentially complete and increasing the concentration of Fur had little effect. Indeed, the small increase in sodB expression is exactly what was observed upon deletion of rfrA and rfrB. Taken together, these data show that Fur activates hilA expression (HilD activity) in a manner that is mechanistically different from Fur activation of sodB.
Many small RNAs, including RyhB, are assisted by the RNA binding protein Hfq (38). We tested the effect of an hfq deletion on the expression of a sodB translational lacZ fusion and a hilA-lac transcriptional fusion. If small RNAs are involved in the Fur regulation of hilD, deletion of hfq could increase expression of hilA in the Δfur background. Figure 8B shows that sodB was indeed regulated by Hfq, as expected. There was still an effect upon deletion of fur in a Δhfq background, but a similar phenotype has been seen frequently with respect to Hfq and tests of epistasis (38, 45); RfrA and RfrB seem to be capable of acting without Hfq, just at a lower efficiency. In contrast, hfq had no effect on the Fur-mediated expression of hilA. Indeed, loss of hfq caused a slight decrease in expression, compared with the predicted increase in expression. The hfq effect was only twofold and likely can be attributed to the pleiotropic effects exhibited in a cell upon loss of Hfq (30, 35). However, the data suggest that Hfq plays little or no role in the Fur regulation of hilA, again differentiating SPI1 and sodB regulation.
The BarA/SirA two-component system activates hilA by controlling HilD expression posttranscriptionally via activation of the small RNAs CsrB and CsrC, which antagonize the CsrA RNA binding protein that presumably acts directly at the hilD mRNA (2, 3, 20, 28, 49). Although null mutations in csrA are pleiotropic and the effects on SPI1 expression are complicated (2, 28), we could clearly demonstrate that overproduction of Fur in a csrA null mutant induced a hilA-lac fusion to the same extent that was observed in a csrA+ background (data not shown). Thus, Fur does not function through the Csr system.
DISCUSSION
Multiple environmental signals impinge on the SPI1 T3SS, and much work has been done to determine the regulatory systems that mediate the environmental response (16). Expression of hilA is tightly controlled by the bacterium, apparently to designate the proper timing and location for production of the invasion system. Our study shows that Fur plays a significant role in the expression of the SPI1 T3SS by genetically activating hilA transcription. Fur regulation is mediated through HilD, but regulation does not appear to be simply at the level of hilD transcription or translation. Rather, Fur activity requires the presence of the HilD protein. Transcription of the hilD gene is activated in response to Fur, but this is also dependent on the HilD protein and is in accordance with our model for SPI1 regulation.
We propose two models to explain Fur “activation” of SPI1: (i) Fur indirectly controls HilD protein, or (ii) Fur directly or indirectly affects the threshold of HilD required to activate the hilD promoter and hence the entire SPI1 system. Because Fur is thought to be capable of acting as only a repressor (27), the first model posits that Fur represses a negative regulator to “activate” SPI1. In this model the regulator either binds to HilD and prevents its function or targets HilD for degradation. We do know that the putative negative regulator is not HilE, which functions by directly interacting with HilD to block its activity, nor is it CsrA, which presumably acts on the hilD mRNA (16, 20). We have also ruled out the known Fur-regulated small RNA RfrA (the E. coli RyhB ortholog) and its paralog RfrB. A variation of this model is that Fur indirectly regulates a factor that positively affects HilD function; e.g., Fur represses a repressor of a factor that assists HilD. Either way, Fur regulates hilA by posttranslationally affecting the HilD protein.
The second model suggests that Fur (or something regulated by Fur) acts at the level of the DNA to affect the threshold of HilD required to activate the hilD promoter. Once hilD is activated, the amplification built into the system turns all the genes on, but this requires autoinduction by the HilD protein. Fur does not directly or indirectly act as a simple activator, nor is it absolutely required for HilD to activate hilD transcription; overproduction of Fur has no effect in a hilD null background (Fig. 4), and overproduction of HilD overcomes Fur regulation (data not shown). If this model is correct, we imagine that Fur directly or indirectly affects the promoter structure (Hns?) such that HilD (and also HilC and RtsA?) works more efficiently at the hilD promoter.
Although we cannot distinguish the models described above at this time, our data do suggest certain properties of the regulatory network that need to be considered. First, overproduction of Fur significantly increases expression of hilA and hilD when HilD is present. Consequently, if Fur acts as a simple repressor of the factor that controls HilD, as it does with ryhB controlling sodB, then the Fur binding site controlling the negative regulator must be far from consensus, leading to weak binding of Fur under normal laboratory conditions; overproduction of Fur leads to further repression. This is in contrast to what has been observed for sodB regulation; overproduction of Fur had little if any effect on sodB expression, presumably because Fur repression of rfrA and rfrB is essentially complete in these conditions. Alternatively, the regulatory circuit might be more complex, with multiple components acting in succession, analogous to the Csr system (20). However, incremental action must occur even in more complex models of Fur regulation. The presumed weak Fur binding site(s) would likely be difficult to identify by bioinformatic methods.
Since one of our models proposes that Fur represses a repressor of HilD, we used Tn10dTc and performed random mutagenesis of a strain with a fur deletion and a hilA-lacZ transcriptional fusion. This strain produced white colonies on LB medium with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). If we disrupted the gene that Fur represses in order to activate hilA, we would expect two things: (i) the colonies would appear blue on LB medium with X-Gal, and (ii) expression of hilA would be unaffected by the presence or absence of Fur in the mutant background. We screened over 30,000 colonies, looking for transposon insertions which met these two criteria, and we found no such insertions. There are several possible explanations for this. First, the negative regulator could be a small RNA. Because of their small size, no small RNAs have yet been identified by insertional mutagenesis. Second, the regulator could be essential. Third, there could be redundancy, so that a single mutation would not confer a phenotype. Fourth, the model could be wrong. Further work is required to understand the mechanism by which Fur controls HilD.
Data presented here fit well with the previously described feed forward loop model of SPI1 regulation (16). For example, hilA expression, rtsA expression, and hilD expression are decreased by deletion of fur at the level of transcription via HilD, supporting the idea that HilD controls the transcription of these genes. Also, while deletion of hilC or rtsA decreased the expression of hilA and affected the fold effect of Δfur, it did not prevent Fur regulation of hilA. However, expression of hilC was actually increased by deletion of fur for reasons that are unclear; there are subtleties in the regulatory network that we do not yet understand.
The concept that Fur activates SPI1 expression in response to high iron levels is consistent with previous work that has shown that Fe2+ is abundant in the lumen of the intestine and becomes scarce following invasion. For example, we have previously shown that Fur represses the sitABCD operon in the lumen and that repression is relieved once invasion occurs (26). Thus, available iron is an ideal environmental cue for when to turn SPI1 on. Interestingly, the low iron concentrations that are found after invasion provide a mechanism to shut off the SPI1 T3SS. SPI1 is not required during acute systemic infection (16), but several studies have suggested that it has a role during long-term systemic infections (29). It remains unclear in these cases if the system is truly on in systemic sites or if the results reflect residual expression after invasion. Previous work has shown that Lon is capable of degrading SPI1 regulatory proteins after invasion (10). Lon degradation of SPI1 proteins coupled with tight posttranslational control of HilD could provide the mechanism for rapid SPI1 shutoff.
One of the strongest environmental cues for SPI1 induction in the laboratory is low oxygen (5, 16, 34). While we know that Fur does not respond to oxygen concentration directly, under high-oxygen conditions iron is oxidized to insoluble Fe3+, leading to decreased intracellular iron and a lack of Fur binding the DNA. The sit operon, for example, is dramatically regulated by oxygen, a phenomenon that is completely dependent on Fur (25). While we cannot say that Fur accounts for all of the oxygen regulation of SPI1, it certainly contributes. It is worth noting that while overproduction of Fur caused an eightfold increase in hilA expression in low-oxygen conditions, Fur overproduction induced hilA expression only threefold in high-oxygen conditions (data not shown). However, we have previously shown that the rtsA, hilC, and hilD promoters each respond to oxygen levels at the level of transcription and in the absence of all known regulators (16). Since rtsA, for example, responds to oxygen in the absence of HilD, there must be additional oxygen regulation that is independent of Fur.
In E. coli, expression of sodB is activated by Fur via repression of the small RNA RyhB. Similarly, Salmonella sodB is Fur activated. However, instead of a single regulatory RNA that controls sodB expression, Salmonella has two redundant small RNAs, which we designated RfrA and RfrB. RfrA is the RyhB ortholog, whereas RfrB is a paralog, encoded on a Salmonella-specific insert that also contains STM1273, annotated as a putative nitric oxide reductase (40). The insert is roughly 1.4 kb long and is located between yoaG and yeaQ at 27.8 centisomes on the chromosome. While both RNAs can control sodB, it is possible that they differentially control other genes, and it will be interesting to understand the complete Fur regulon in Salmonella. This regulon is potentially more complex than the Fur regulon of E. coli.
HilD is clearly the most significant contributor to the expression of hilA and therefore SPI1 in general, since many systems which control expression of hilA seem to work through HilD in some way (6, 8, 16). Here we show that Fur regulates HilD posttranslationally, leading to induction of the SPI1 T3SS in high-iron conditions. SirA and BarA work through the csr system to control HilD production at the posttranscriptional level. It will be interesting to determine how the additional factors control HilD. Is there a master regulator of HilD that controls multiple signals feeding into the system? Determining the mechanism of Fur activation of HilD is an important step in understanding the regulation of the SPI1 T3SS.
Acknowledgments
We thank Susan Gottesman for sharing unpublished data regarding rfrB and for helpful discussions. We thank Craig Altier for providing the csrA null strain. Additionally, we thank David Schlesinger and Lee Macomber for critically reading the manuscript and Craig Ellermeier and Chris Rao for many helpful discussions.
This work was supported by Public Health Service grant AI63230.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31971-982. [DOI] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 686790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35635-646. [DOI] [PubMed] [Google Scholar]
- 4.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18715-727. [DOI] [PubMed] [Google Scholar]
- 5.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22703-714. [DOI] [PubMed] [Google Scholar]
- 6.Baxter, M., and B. D. Jones. 2003. Identification of regulatory pathways that translate environmental signals into changes in expression of Salmonella motility, adherence, and invasion, abstr. D-110. Abstr. 103rd Gen. Meet. Am. Soc. Microbiol.
- 7.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 711295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baxter, M. A., and B. D. Jones. 2005. The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect. Immun. 731377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 1754475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boddicker, J. D., and B. D. Jones. 2004. Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect. Immun. 722002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1589-14. [DOI] [PubMed] [Google Scholar]
- 12.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35949-960. [DOI] [PubMed] [Google Scholar]
- 13.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 201850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 1823802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellermeier, C. D., J. R. Ellermeier, and J. M. Slauch. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57691-705. [DOI] [PubMed] [Google Scholar]
- 17.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290153-161. [DOI] [PubMed] [Google Scholar]
- 18.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1855096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fahlen, T. F., N. Mathur, and B. D. Jones. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 2825-35. [DOI] [PubMed] [Google Scholar]
- 20.Fortune, D. R., M. Suyemoto, and C. Altier. 2006. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect. Immun. 74331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 1831835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBADpromoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 1836384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda, J. S., A. Janakiraman, D. G. Kehres, M. E. Maguire, and J. M. Slauch. 2005. Transcriptional regulation of sitABCD of Salmonella enterica serovar Typhimurium by MntR and Fur. J. Bacteriol. 187912-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 351146-1155. [DOI] [PubMed] [Google Scholar]
- 27.Kadner, R. J. 2005. Regulation by iron: RNA rules the rust. J. Bacteriol. 1876870-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawhon, S. D., J. G. Frye, M. Suyemoto, S. Porwollik, M. McClelland, and C. Altier. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 481633-1645. [DOI] [PubMed] [Google Scholar]
- 29.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lease, R. A., M. E. Cusick, and M. Belfort. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. USA 9512456-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, C. A., B. D. Jones, and S. Falkow. 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. USA 891847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim, S., J. Yun, H. Yoon, C. Park, B. Kim, B. Jeon, D. Kim, and S. Ryu. 2007. Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res. 351822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 31281-1291. [DOI] [PubMed] [Google Scholar]
- 34.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1832733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 9512462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 37.Masse, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2374-2383. [DOI] [PMC free article] [PubMed]
- 38.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 994620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masse, E., C. K. Vanderpool, and S. Gottesman. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 1876962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413852-856. [DOI] [PubMed] [Google Scholar]
- 41.Merighi, M., C. D. Ellermeier, J. M. Slauch, and J. S. Gunn. 2005. Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J. Bacteriol. 1877407-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1844148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olekhnovich, I. N., and R. J. Kadner. 2007. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J. Bacteriol. 1896882-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pegues, D. A., M. J. Hantman, I. Behlau, and S. I. Miller. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17169-181. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz, N., and T. J. Silhavy. 2003. Constitutive activation of the Escherichia coli Pho regulon upregulates rpoS translation in an Hfq-dependent fashion. J. Bacteriol. 1855984-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 401289-1299. [DOI] [PubMed] [Google Scholar]
- 48.Slauch, J. M., and T. J. Silhavy. 1991. Genetic fusions as experimental tools. Methods Enzymol. 204213-248. [DOI] [PubMed] [Google Scholar]
- 49.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 1857257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, A., M. D. Rolfe, S. Lucchini, P. Schwerk, J. C. Hinton, and K. Tedin. 2006. The bacterial signal molecule, ppGpp, mediates the environmental regulation of both the invasion and intracellular virulence gene programs of Salmonella. J. Biol. Chem. 28130112-30121. [DOI] [PubMed] [Google Scholar]
- 51.Vecerek, B., I. Moll, T. Afonyushkin, V. Kaberdin, and U. Blasi. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 50897-909. [DOI] [PubMed] [Google Scholar]
- 52.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100195-199. [PubMed] [Google Scholar]
- 53.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 151637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 975978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, D., and J. Galan. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 31293-1298. [DOI] [PubMed] [Google Scholar]