Abstract
Enterocin AS-48 production and immunity characters are encoded by 10 genes (as-48ABCC1DD1EFGH) of the pMB2 plasmid from the Enterococcus faecalis S-48 strain. Among these, as-48A, encoding the AS-48 peptide, and the as-48BC genes constitute a cluster required for AS-48 biogenesis and full immunity. In this study, the levels of expression of this cluster have been altered by insertion and site-directed mutagenesis as well as by expression coupled to trans complementation. Phenotypic studies of the mutants have indicated cotranscription of the three genes and revealed that the inactivation of as-48B prevents the production of AS-48, thus confirming its essentiality in AS-48 biogenesis. These studies have also supported the involvement of as-48C in enterocin immunity. In addition, they established that the intergenic region between the as-48A and as-48B genes is decisive for AS-48 expression, since a 3-bp substitution, which should disrupt a potential 47-nucleotide complex secondary structure, resulted in a hypoproducing phenotype. Transcriptional analyses of the E. faecalis wild-type and mutant strains supports the possibility that the as-48ABC genes are transcribed from the PA promoter located upstream of as-48A. Moreover, analysis and bioinformatic predictions of RNA folding indicate that as-48ABC mRNA is processed at the secondary structure located between as-48A and as-48B. Thus, synthesis of the AS-48 peptide appears to be controlled at the posttranscriptional level and is uncoupled from as-48BC translation. This mechanism of genetic regulation has not been previously described for the regulation of bacteriocin expression in enterococci.
The production of antimicrobial peptides (bacteriocins) directed against competitive microorganisms is widespread among gram-positive bacteria (21). A large variety of these are secreted by lactic acid bacteria, and the peptides have been extensively studied due to their potential use as natural preservatives in food applications (21). Enterocin AS-48 is a broad-spectrum, circular peptide of hydrophobic and cationic nature, which is ribosomally synthesized and posttranslationally circularized by a head-to-tail peptide bond (19). The well-defined, three-dimensional structure of AS-48, recently solved by nuclear magnetic resonance and X-ray diffraction techniques (reviewed in reference 30), is a reference for numerous studies of molecules structured in α helices (39, 42). As is usual for bacteriocin determinants, the genes encoding processing, secretion, and immunity functions are clustered. In fact, the as-48 gene cluster encoding the AS-48 trait was located on the 68-kb pMB2 conjugative pheromone response plasmid identified in Enterococcus faecalis S-48 (31) and functionally transferred to E. faecalis JH2-2 (34). The full expression of AS-48 and immunity depends on the coordinated expression of the as-48ABCC1DD1EFGH genes (12, 34). Among these, as-48A is the structural gene and as-48D1, which encodes a small cationic hydrophobic peptide, provides some degree of bacteriocin resistance by itself and has thus been identified as the immunity determinant (34). Interestingly, two overlapping genes (as-48BC) are located 73 nucleotides (nt) downstream of the TAA termination codon of as-48A and the intergenic region includes a long inverted repeat (IR). This repeat was previously proposed to be a transcriptional terminator or attenuator, since Northern hybridization revealed an abundant RNA species containing the structural gene (TA) and a minor species designated T1, which could include either as-48ABC or as-48BC (34). The as-48 cluster also contains two ABC transporters encoded by the as-48C1D and as-48EFGH genes, which are cotranscribed in a polycistronic mRNA, together with as-48D1 (12). As-48C1D seems to be devoted to exporting the newly synthesized bacteriocin and provides low levels of immunity. The function of this transporter cannot be replaced by the second multicomponent ABC transporter, As-48EFGH, which is involved in self-protection against exogenously administered AS-48 and works as a complementary immunity mechanism (12). We have postulated that As-48B, alone or in association with the ABC transporter As-48C1D, may form a pore that is specific for the secretion of AS-48 molecules. The ultimate activity of the multicomponent proteins, As-48BC1D, would be the removal of the leader peptide promoting head-to-tail circularization concomitantly with the export of the molecule (34). However, the mechanisms underlying AS-48 processing and its transport to the cell surface remain unknown.
The expression of most of the bacteriocin systems so far characterized (lantibiotics [reviewed in reference 28] or nonlantibiotics [reviewed in reference 15]) is controlled by a so-called, three-component regulatory system, which is an autoregulatory model that includes the secreted bacteriocin as peptide pheromone and the signal transduction by the corresponding two-component regulatory signaling system. We have previously shown that genes encoding transduction signals do not exist in the as-48 cluster and that the cloning of this DNA region into the pAM401 plasmid results in full expression of the AS-48 character (production and immunity) (12). Here we present a detailed transcriptional analysis of as-48ABC genes reporting the posttranscriptional mechanism that regulates their expression and the essential role that as-48B plays in the biogenesis of AS-48.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study, together with the Tn5 insertion mutants of the pAM401-81 enterocin determinant, are listed in Table 1. Enterococcal cells were grown without aeration at 37°C in brain heart infusion broth (BHI-B; Oxoid). Escherichia coli strains were grown with shaking in Luria broth at 37°C. The plasmid-free strain E. faecalis JH2-2 (49) was used in cloning experiments of the as-48ABC cluster and as an indicator strain to detect enterocin activity. When necessary, the indicated antibiotics (Sigma-Aldrich, Madrid, Spain) were added as follows: ampicillin (Ap), 50 μg ml−1; chloramphenicol (Cm), 20 μg ml−1; erythromycin (Ery), 200 μg ml−1 for E. coli and 6 μg ml−1 for E. faecalis; and kanamycin (Km), 30 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| E. faecalis JH2-2 | Plasmid-free derivative of E. faecalis JH-2 | 49 |
| E. coli DH5α | supE44 ΔlacU169(φ80lacZΔM15)hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| E. coli RYC1000 | araD139 ΔlacU169 rpsL Δrib7 thiA gyrA recA56 | 7 |
| pAM401 | Bifunctional E. coli-E. faecalis cloning vector; Cmr Tcr | 17 |
| pMB2 | AS-48+ Imm+; 68-kb as-48 cluster (as-48ABCC1DD1EFGH) | 32 |
| pMG36e | Lactoccocal cloning and expression vector, replicative in E. coli; Eryr | 48 |
| pGEM-3Zf(+) | Cloning vector; Ampr | Promega |
| pSL1180 | Cloning vector; Ampr | Pharmacia |
| pAM401-76 | AS-48−, partially Imm+; 1.9-kb SphI-BglII (as-48A promoter (PA) and as-48A structural gene) and 6.6-kb BglII (as-48H) DNA fragments from pMB2 cloned into pAM401; Cmr | 12 |
| pAM401-81 | AS-48+ Imm+; complete 25-kb as-48 region (PA and as-48ABCC1DD1EFGH) from pMB2 cloned into pAM401; Cmr | 12 |
| pAM401-52 | AS-48+ partially Imm+; 7.8-kb SphI-PstI (PA and as-48ABCC1DD1EF) DNA fragment from pMB2 cloned into pAM401; Cmr | 34 |
| pAM401-58 | AS-48− and Imm+ 0.16.7 kb of the as-48 region (carries the 3′ region of as-48A, the intergenic region between as-48A and as-48B, and as-48BCC1DD1EF); derivative of pAM401-52 by deletion of the 1.7-kb SphI-PstI DNA fragment; Cmr Tcr | 34 |
| pAM401-81:: Tn5PA | Tn5 insert in PA (as-48A promoter); Cmr Tcr | This work |
| pAM401-81::Tn5B | Tn5 insert in as-48B gene; Cmr Tcr | This work |
| pAM401-81::Tn5C | Tn5 in as-48C gene; Cmr Tcr | This work |
| pMG36-A | PA-as-48A cloned into pMG36e; Eryr | This work |
| pMG36-BC | as-48BC cloned into pMG36e under control of P32 promoter; Eryr | This work |
| pAM401-76X | pAM401-76 containing XhoI restriction site 22 nt downstream of as-48A gene; Cmr | This work |
| pBgD12S | 1.9-kb SphI-BglII D fragment from pMB2 cloned into pSL1180; Ampr | This work |
| pAM401-81X | pAM401-81 containing XhoI restriction site 22 nt downstream of the as-48A gene; Cmr | This work |
Cmr, chloramphenicol resistant; Tcr, tetracycline resistant; Ampr, ampicillin resistant; Eryr, erythromycin resistant; AS-48+, AS-48 production; AS-48−, no AS-48 activity detected; AS-48r, AS-8 resistant; AS-48s, AS-48 sensitive.
Antimicrobial activity assays.
AS-48 production by different E. faecalis transformants was assayed by spotting 2 μl from a liquid overnight culture onto BHI agar (1.5%), followed by incubation at 37°C for 16 h. We then overlaid the plate with 6 ml of BHI soft agar (0.75%, with or without antibiotics) containing a 2% inoculum of indicator strain and incubated at 37°C for 12 to 18 h before reading the results. To test AS-48 sensitivity, 5-μl spots containing different quantities of purified AS-48 were applied to lawns seeded with E. faecalis JH2-2 (harboring different plasmids) and the plates were examined for inhibition halos after 18 h of incubation at 37°C.
Prediction genetic analysis.
Mfold (50) and Fold programs (Genetics Computer Group package; Wisconsin University), based on energy minimization, were used for predicting RNA secondary structures and folding, respectively. Calculation of the predicted termination efficiency of the secondary structure located between as-48A and as-48B was performed by using the algorithm developed by d'Aubenton Carafa et al. for E. coli (10) and optimized by De Hoon et al. (11) for low G+C content bacteria, in which the d value for a given terminator is calculated from the following equation: d = 7.9(ΔG/nSL) + 2.67T − 14.61, where ΔG is the Gibbs free energy of stem-loop formation in kilocalories per mole (calculated with the Mfold program), nSL is the number of nucleotides of the entire stem-loop structure, and T is a parameter that depends on the number of consecutive U residues in the 14-nt stretch adjacent to the 3′ end of the stem-loop and for which an exponential decay weight from the 5′ to 3′ direction is applied. The condition d > 0 establishes a threshold that separates transcriptional terminators from intracistronic structures and shows that a positive value for d can be taken as strong indication of a ρ-independent terminators. This d score has a sensitivity of 96.93% for the detection of the predicted terminators in the E. faecalis V583 entire genome, and these terminators have average values for ΔG, stem length, and uracil stretch of −18.2, 11.9, and 9.9, respectively (11), which yield an average d value of 3.07. An analysis of different regions of structure I (see Fig. 5A) revealed that the entire structure (47 nt) has a d value of −11.85, and the best score corresponds to its central region (tA), which has a negative value of −1.2. tA and includes a stem of 11 nt, followed by a UUUUUUAAUU stretch close to the average for E. faecalis terminators, but has a ΔG of −7.5, which would minimize its formation efficiency and, consequently, its attenuation effect on transcription by the RNA polymerase in this host.
FIG. 5.
Prediction of the secondary structures present in the processing region of as-48ABC mRNAs and of their 5′ end processed species folding. Secondary structures present between the TAA translation termination codon of as-48A and the fifth codon of as-48B of the as-48ABC mRNAs encoded by pAM401-81 (A) and pAM401-81X (B). Prediction was made with the Mfold program. Folding of the 5′ RNA species generated from pAM401-81 (C) and pAM401-81X (D) transcripts by mRNA processing at the major cleavage sites detected in Fig. 4C and indicated here in panels A and B by arrowheads. The folding was performed with the Fold program.
Plasmids and DNA manipulations.
DNA cloning and E. coli transformations were performed according to standard protocols (44). E. faecalis plasmid DNA was extracted according to the method of Anderson and McKay (4). E. faecalis cells were transformed by electroporation according to the method of Fiedler and Wirth (16). Antibiotic-resistant transformants were screened for AS-48 production by replica plating and overlaid with a sensitive strain. DNA was sequenced with the ABI Prism Dye Terminator cycle sequencing ready reaction kit (Perkin-Elmer, Applied Biosystems). Synthetic oligonucleotides were from Amersham Biosciences Europe GmBH (Cerdanyola, Barcelona, Spain).
PCR and DNA sequencing.
All primers used in PCRs were synthesized by Amersham Pharmacia Biotech (Uppsala, Sweden) and were based on the published DNA sequence of the as-48 locus of E. faecalis (EMBL X79542). Total DNA was prepared as described elsewhere (38) and used as a DNA template for PCR amplifications carried out with Biotools DNA polymerase (B&M Labs, Madrid, Spain) in an iCycler 170-8720 thermal cycler (Bio-Rad Laboratories, Madrid, Spain). The presence and molecular size of the PCR products were determined by electrophoresis on 1% (wt/vol) agarose gels using λ DNA digested with HindIII and/or the 100-bp ladder (Gibco-BRL, Invitrogen, United Kingdom) as the molecular weight standard. All PCR products were purified with the Perfectprep Gel Cleanup kit (Eppendorf AG, Hamburg, Germany) and sequenced.
Generation of transposon insertional mutants.
The transposition of Tn5 into pAM401-81 was performed by introducing the plasmid into E. coli RYC1000 according to the method of Martínez-Bueno et al. (34). Selected pAM401-81::Tn5 mutagenized DNAs were transferred to E. faecalis JH2-2 by electroporation, and the new recombinant strains were screened for AS-48 production and immunity.
Site-directed DNA mutagenesis.
An XhoI restriction site in the intergenic region between the as-48A and as-48B genes was generated by overlap extension PCR mutagenesis (24). The external oligonucleotides used were the M13 universal (5′-GTAAAACGACGGCCAGT-3′) and reverse (5′-AACAGCTATGACCAT-3′) primers. The internal primers, mutXh3 (5′-TTTAACAATATGATAAAAAAACTCGAGATTTTCTAGAGATATTCTGTTTT-3′) and mutXh4 (5′-AAAACAGAATATCTCTAGAAAATCTCGAGTTTTTTTATCATATTGTTAAA-3′) (underlined sequences show the added restriction sites, and bold nucleotides are the point mutations), exhibited three mismatches relative to the wild-type sequence, generating the new XhoI site. The template for mutagenesis was the pBgD12S construction, a pSL1180 derivative bearing the D (SphI-BglII) fragment from pMB2 (Table 1). Amplification conditions were as described by Higuchi (23), and the resulting 2-kb amplified fragment containing the XhoI restriction site was SphI-BglII digested and cloned into pAM401-76 (12) by replacing the equivalent SphI-BglII fragment [pAM401-76X]. Finally, ligation in the frame of the C fragment (BglII) of 6.4 kb from pMB2 into pAM401-76X, previously digested with the same enzyme, yielded the plasmid pAM401-81X (25 kb). The proper orientation of the D, C, and B fragments was established by restriction analysis, and the presence of the XhoI restriction site was confirmed by DNA sequencing.
Complementation plasmid constructions and compatibility test.
The lactococcal expression vector pMG36e (Eryr) was chosen to construct complementation plasmids. This broad host range plasmid is a derivative of pWV01, which replicates via rolling circle (48), and consequently, in E. faecalis it should be compatible with pAM401 (Cmr), a very stable theta-replicating plasmid able to accommodate large inserts (such as the 14.8 kb of the as-48 cluster) while maintaining high segregational stability. To test this compatibility, the JH2-2[pAM401-81] and JH2-2[pAM401] strains were electroporated with pMG36e plasmid and transformants were selected on plates containing chloramphenicol and/or erythromycin. Several transformants of each transformation were grown in either the presence or the absence of selective antibiotics over several generations, and the stability of both plasmids was confirmed by electrophoretic DNA plasmid analysis (data not shown). On the basis of these experiments, pMG36e was used to clone the as-48A and as-48BC genes independently. To construct pMG36-A, a 644-bp EcoRV/EcoRI fragment containing the structural gene and its PA promoter was obtained from pAM401-81. This fragment was cloned in pMG36e, previously digested with SmaI/EcoRI enzymes, which delete the P32 promoter. Plasmid pMG36-BC was constructed by PCR amplification of the coding regions of the as-48BC overlapping genes without a promoter by using appropriate primers as-48BXma (5′-CGCCCGGGGATGAATCTCTTTGGAATTCTA-3′) and as-48AC2 (5′-CTGCAGCATGCGGATCCGATATCGTAATACCGATGCACTTTTTCAA-3′). The PCR product (2,620 bp) was digested with SmaI/SphI and cloned into the pMG36e vector, previously digested with the same restriction enzymes, downstream of the P32 promoter (18). Therefore, pMG36-BC carries a P32-as-48BC transcriptional fusion. These constructions were made in E. coli DH5α and then transferred to E. faecalis JH2-2, harboring different plasmids.
RNA isolation.
Cultures of E. faecalis JH2-2, harboring the indicated plasmids (see Fig. 2, 3, and 4), were grown to an optical density at 600 nm (OD600) of 0.4 for semiquantitative reverse transcription-PCR (RT-PCR) and primer extension analysis or to OD600 values of 0.4 and 0.9 for Northern hybridization analysis. Cells were harvested by centrifugation, and their total RNAs were extracted by use of the Fast RNA Pro Blue kit (Q-Biogen, MP Biomedicals, Illkirch, France), following the manufacturer's instructions. For RT, total RNAs were treated with 5 U of RNase-free DNase I (Roche Diagnostics, San Cugat del Vallés, Spain) for 1 h at 37°C. The total RNA preparations were checked for the integrity and yield of the rRNAs by analysis in agarose gels stained with ethidium bromide. The total RNA concentrations were determined by UV spectrophotometry.
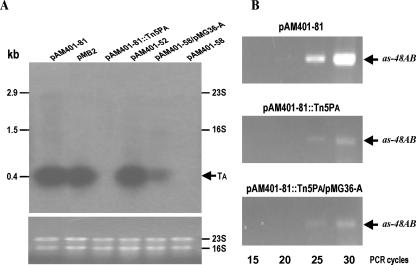
FIG. 2.
Analysis of the as-48AB cotranscription. (A) Northern blot analysis. RNA was isolated from E. faecalis S-48[pMB2] and JH2-2 carrying the indicated pAM401-derived plasmid at the end of the exponential phase. The upper panel shows an autoradiogram of the blotting. The lines labeled 16S rRNA and 23S rRNA indicate the locations of these rRNA species, which are known to trap smaller RNA species (25). The lower panel shows the 23S rRNA and 16S rRNA loaded in the gel and stained with ethidium bromide. TA, RNA species containing the as-48A gene (see the legend of Fig. 5 for details). (B) Semiquantitative RT-PCR detection. RNA was isolated from E. faecalis JH2-2 carrying pM401-81 wild-type and mutant plasmids. The specific primers used and reaction conditions are described in Materials and Methods. The plasmid carried by the JH2-2 strain and the number of cycles of each sample are indicated.
FIG. 3.
Transcriptional analysis of the as-48ABC genes in JH2-2[pAM401-81] and JH2-2[pAM401-81X] strains. (A) Northern blotting detection using specific probes derived from the as-48A gene. RNA was extracted at the indicated OD600. O.D., optical density. (B) Semiquantitative RT-PCR analysis of as-48A (upper panel), as-48AB (middle panel), and as-48BC (lower panel) mRNAs in the indicated strains. The plasmid carried by the JH2-2 strain and the number of cycles of each sample are indicated.
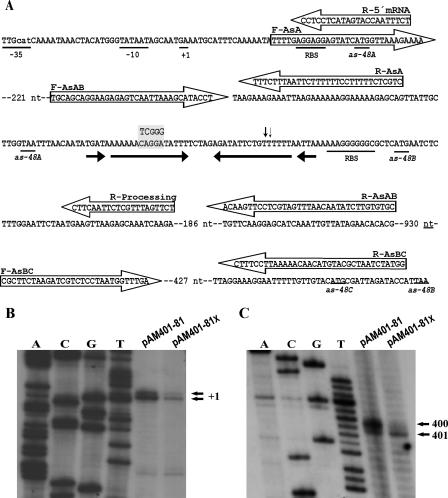
FIG. 4.
Relevant DNA sequences for transcription and translation of the as-48ABC operon and determination of the initiation of transcription and mRNA processing. (A) The PA promoter of the as-48ABC operon and RBSs, as well as the initiation and termination translational codons, are underlined. The long IR is marked by arrows. The three bases changed to generate the XhoI site are in the gray box. Transcription initiation sites of as-48A (inferred from results presented in panel B) are underlined and labeled as +1. The processing sites in the mRNA (inferred from results presented in panel C and corresponding to positions 401 and 402 of the mRNA) are indicated by arrowheads. Also indicated are the location, sequence, and polarity of all primers used for RT-PCR experiments depicted in Fig. 2 (F-AsAB and R-AsAB) and 3 (F-AsA, F-AsAB, F-AsBC, R-AsA, R-AsAB, and R-AsBC) and for primer extensions depicted in panel B (R-5′ mRNA) or in panel C (R-processing). F, forward; R, reverse. Determination of the initiation of transcription from PA (B) and mRNA processing (C) by primer extension in the wild-type and pAM401-81X mutant strains. Autoradiograms of the gels are depicted, and the detected extended products are labeled with the corresponding coordinates in the as-48ABC mRNA. The coordinates were inferred from the length of the products (95 nt for +1 in panel B and 70 and 69 nt for 400 and 401 in panel C) that were estimated by the use of unrelated DNA sequence (A, C, T, and G) ladders.
Semiquantitative RT-PCR.
Semiquantitative analysis of RNA levels containing regions of as-48A, as-48AB, as-48BC, and 16S genes was performed by two-step RT-PCR assays. For RT assays, total RNAs (500 ng) were added to 20-μl RT reaction mixtures containing 4 μl of cDNA synthesis buffer, 5 mM dithiothreitol, 40 U of RNaseOUT (Invitrogen), 1 mM deoxynucleoside triphosphate mix, a 10 μM concentration of the appropriate gene-specific primer, and 15 U of ThermoScript RT (Invitrogen) and then incubated for 30 min at 56°C (as-48A), 60°C (as-48BC), or 61°C (as-48AB). Reaction mixtures containing RNA instead of cDNA were included in the analysis to confirm the absence of DNA contamination in RNA preparations. The primers used were R-16S (5′-CTGCTGCCTCCCGTA-3′), R-AsA (5′-CTGCTCTTTTTCCTTTTTTCTTAATTTCTTT-3′), R-AsAB (5′-CGTGTGTTCTATAACAATTTGATGCTCCTTGAACA-3′), and R-AsBC (5′-GGTATCTAATCGCATGTACAACAAAAATTCCTTTC-3′) for synthesis of 16S, as-48A, as-48AB, and as-48BC cDNAs, respectively. RT was terminated by incubation at 37°C for 20 min in the presence of 2 U of RNase H (Invitrogen).
For the amplification of either cDNA, 10% of the cDNA synthesis reaction mixture, 50 pmol of each primer, 500 μM of each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), 3.5 mM MgCl2, 20 mM Tris-HCl (pH 8.4), and 50 mM KCl were used for each 50-μl PCR, which was performed with 2 U of Taq DNA polymerase (Invitrogen). The cycling conditions were 1 cycle (96°C for 2 min) and 15, 20, 25, and 30 cycles (94°C for 1 min, 56°C, 60°C, or 61°C for 1 min for as-48A, as-48BC, or as-48AB, respectively, and 72°C for 1 min). Six microliters of the RT-PCR product was analyzed by 1% agarose gel electrophoresis and visualized with the Quantity One Gel Doc 2000 software (Bio-Rad Laboratories). The primers used for PCR amplification were F-AsA (5′-TTTTGAGGAGGAGTATCATGGTTAAAGAAAA-3′) and R-AsA for synthesis of a 321-nt amplicon containing the 100 first codons of as-48A for the detection of as-48A mRNA (TA), F-AsAB (5′-TGCAGCAGGAAGAGAGTCAATTAAAGCATACCT-3′) and R-AsAB for synthesis of a 420-nt amplicon containing the last 26 codons of as-48A and the first 88 codons of as-48B for the detection of as-48AB mRNA, F-AsBC (5′-CGCTTCTAAGATCGTCTCCTAATGGTTTGA-3′) and R-AsBC for synthesis of a 492-nt amplicon containing the 164 last codons of B and the 5 first codons of C for the detection of as-48BC mRNA, and F-16S (5′-AGAGTTTGATCCTGGCTCAG-3′) and R-16S for synthesis of a 368-nt amplicon containing the 5′ region of the gene encoding the 16S rRNA.
Northern blot hybridization.
For the Northern blot analysis, 5 μg of total RNAs were denatured in 30 μl of MOPS (morpholinepropanesulfonic acid) (40 mM MOPS, 10 mM sodium acetate, 2 mM EDTA) buffer containing 50% deionized formamide, 17% formaldehyde, and 0.4% bromophenol. Then they were fractionated on a 1% agarose-formaldehyde gel run at 23 V at room temperature for 14 h. Staining of the gel with ethidium bromide allowed us to check the integrity of rRNAs after fractionation and their proper transfer to the membrane after blotting. Nucleic acids were transferred to a nylon membrane (HybondN+; Amersham Pharmacia) by capillary blotting for 16 h using 20× SSC (150 mM NaCl plus 15 mM sodium citrate) buffer. The RNAs were fixed to the membrane by exposing the filters to UV light for 4 min. The DNA probes used were those described above: the 321-nt amplicon (containing the first 100 codons of as-48A) and the 492-nt amplicon (containing the last 164 codons of B and the first 5 codons of C). The amplicons were PCR synthesized as described above and purified by use of the FavorPrep kit (Favorgen Biotech Corp., Kaohsiung, Taiwan). Then they were used as substrates for radioactive synthesis and labeling of the probe in a thermocycling reaction mixture using 20 ng of template DNA, 25 pmol of each primer, 1.5 mM MgCl2, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 150 μM of each deoxynucleoside triphosphate (dATP, dGTP, and dTTP), 50 μM dCTP, and 150 nM of [α-32P]dCTP (3,000 Ci/mmol; Amersham). Free [α-32P]dCTP was removed by filtration on a MicroSpin G25 column (Amersham Pharmacia). Hybridization was performed overnight at 50°C in ULTRAhyb buffer (Ambion, Applera Hispania S.A., Madrid, Spain) that contains 50% formamide. For autoradiography, the blotted filter was exposed to Agfa CURIX RP2 Plus films. The radioactivity present in the filter bands was quantified directly with a PhosphorImager (Fujifilm, Barcelona, Spain).
Primer extension analysis and DNA sequencing.
Primer extension analysis was performed as previously described (20). The primer used to detect the start site of as-48ABC mRNA was F-5′ mRNA (5′-TCTTTAACCATGATACTCCTCC-3′), which is complementary to the transcript from nucleotides 46 to 25. One picomole of primer was annealed to 3.8 μg of total RNA from the JH2-2[pAM401-81] or JH2-2[pAM401-81X] strain. The primer used to determine the processing site of as-48ABC mRNA was R-Processing (5′-TCTTGATTTGCTCTTAACTTC-3′), which is complementary to the mRNA from 470 to 450. One picomole of primer was annealed to 7.5 μg or 15 μg of total RNA from the JH2-2[pAM401-81] or JH2-2[pAM401-81X] strain. The primers were previously labeled at their 5′ ends by using [γ-32P]ATP and T4 polynucleotide kinase. Primer extension reactions were performed by incubating the annealing mixture with 15 U of ThermoScript RT (Invitrogen) at 50°C for 30 min. The sizes of the reaction products were determined by using an 8% polyacrylamide gel containing 7 M urea. Bands labeled with 32P were detected by autoradiography on Kodak X-Omat S film. The length of the extended products was inferred by the use of an unrelated DNA sequence ladders. The double-stranded plasmid pFS21 (47), which harbors the lactococcal citQRP operon heterologous sequence, and unlabeled primer R-Pcit (5′-CGGGTATCAAGTCATGG-3′) were used for sequencing reactions. The nucleotide sequence of the lactococcal citQRP operon has the accession number S77101 in the EMBL data bank. DNA sequencing was determined by the dideoxy chain terminator method (45) by using the T7 polymerase sequencing kit (Amershan-Pharmacia) and labeled [α-32P]dCTP.
Accession numbers for nucleotide sequences are as follows: as-48A, X79542 (33); as-48ABCC1DD1, Y12234 (34); and as-48EFGH from fragment B BglII cloned in plasmid pAM401-81, AJ438950 (12).
RESULTS
Mutational insertion analysis of the as-48ABC cluster.
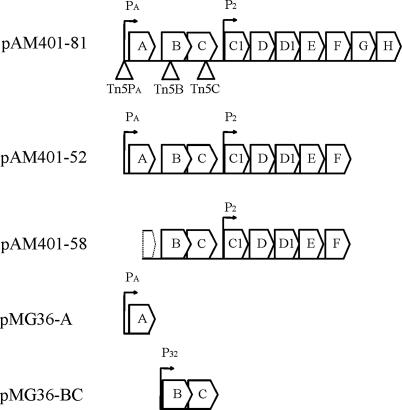
E. faecalis JH2-2 is a plasmid-free strain sensitive to AS-48 enterocin, and we have previously constructed, and transferred to it, the pAM401-81 recombinant plasmid (12) carrying the as-48ABCC1DD1EFGH wild-type cluster from pMB2 and its derivatives pAM401-52 and pAM401-58 (Fig. 1 and Table 1). Both of these deletion plasmids lack the last two coding genes of the ABC transporter As-48EFGH (34). In addition, pAM401-58 lacks the 5′ region of the as-48A structural gene and its PA promoter (Fig. 1) (34). Consequently, this plasmid cannot express the structural as-48A gene, but still carries the 3′ region of as-48A, the intergenic region between the as-48A and as-48B and as-48BCC1DD1EF genes. Here, to characterize expression of the as-48ABC genes, different Tn5 insertions were randomly introduced into pAM401-81 and, subsequently, insertions at three different locations were selected, namely, three nucleotides upstream of the −10 region of the putative PA promoter of as-48A (Tn5PA) or within either the as-48B (Tn5B) or the as-48C (Tn5C) gene (Fig. 1). The new insertion plasmids were transferred independently to the JH2-2 strain, and subsequently, the bacteriocin production and immunity of the transposition mutants were analyzed in comparison with those of JH2-2[pAM401-81], JH2-2[pAM401-52], and JH2-2[pAM401-58] (Table 2).
FIG. 1.
Physical map of plasmids carrying as-48 genes. A schematic representation of the inserts present in the plasmids is depicted. The locations of the Tn5 insertions in the mutants constructed in this work are indicated in pAM401-81. Putative promoters (PA, P2, and P32) are shown for each plasmid.
TABLE 2.
AS-48 production and resistance from E. faecalis JH2-2 harboring the plasmids used in this work
| Plasmid | AS-48 production zone of inhibition (mm)a | Sensitivity to indicated amt (μg 5 μl−1) of AS-48b
|
|||||
|---|---|---|---|---|---|---|---|
| 0.5 | 1 | 3 | 6 | 9 | 18 | ||
| pMB2 (positive control) | 16 | R | R | R | R | R | R |
| pAM401 (negative control) | ND | S | S | S | S | S | S |
| pAM401-81 | 16 | R | R | R | R | R | R |
| pAM401-81::Tn5PA | ND | R | R | R | R | S | S |
| pAM401-81::Tn5B | ND | R | R | R | R | S | S |
| pAM401-81::Tn5C | 8 | R | R | R | R | S | S |
| pAM401-81X | 11 | R | R | R | R | R | R |
| pAM401-52 | 11 | R | R | R | S | S | S |
| pAM401-58 | ND | R | R | S | S | S | S |
| pAM401-81::Tn5PA/pMG36-Ac | 7 | R | R | R | R | S | S |
| pAM401-58/pMG36-Ac | ND | R | R | S | S | S | S |
| pAM401-81::Tn5B/pMG36-BCc | 11 | R | R | R | R | R | R |
AS-48 production was assayed in plates against the indicator strain E. faecalis JH2-2[pAM401] by using AS-48[pMB2] as a control (wild-type phenotype). ND, no detectable production.
To test AS-48 sensitivity, 5-μl spots containing different amounts of purified AS-48 were applied on lawns seeded with a standard inoculum of E. faecalis JH2-2 harboring different plasmids. The plates were examined for inhibition halos after 18 h of incubation at 37°C. R, resistant; S, sensitive.
Erythromycin was present during growth of the strains in liquid and solid media, and JH2-2[pAM401/pMG36e] was used as an indicator for AS-48 production.
The JH2-2[pAM401-81] transformant produces AS-48 and has resistance levels to this enterocin similar to those of the original isolate S-48[pMB2] (12) and the recombinant bacteria JH2-2[pMB2] (32, 34) (Table 2). Nonproducer phenotypes were detected in JH2-2[pAM401-81::Tn5PA], JH2-2[pAM401-81::Tn5B], and JH2-2[pAM401-58]. By contrast, the interruption of as-48C in pAM401-81::Tn5C only decreased the levels of AS-48 production (hypoproducing phenotype) in the JH2-2 background. These results supported an essential function for not only as-48A but also as-48B in the expression of the AS-48 character (production and immunity). Moreover, the results suggested that PA is a transcriptional promoter and, indeed, primer extension experiments (see below) showed this to be the case.
Unexpectedly, the three insertion mutants displayed the same reduced levels of resistance to AS-48 that JH2-2[pAM401-81] did. In addition, their levels of immunity were higher than that of JH2-2[pAM401-58] (resistance to 6 μg 5 μl−1 versus 1 μg 5 μl−1). Interestingly, JH2-2[pAM401-58] showed higher sensitivity to AS-48 than JH2-2[pAM401-52] (Table 2) did, despite the fact that the only difference between them was the absence of a functional as-48A gene and of its putative PA promoter (Fig. 1; Table 1). Therefore, these results suggested that as-48C could be involved in immunity and that this gene is cotranscribed with as-48AB from PA.
Complementation analysis between genes from the as-48ABC cluster.
To test the above hypothesis, plasmid trans complementation experiments were performed after independently cloning the PA-as-48A transcriptional unit and the as-48BC cluster, which lacks a known promoter. Independent cloning of as-48B and as48C was not attempted, since the overlapping of the genes (see Fig. 4) and the lack of an apparent RBS for as-48C indicated a coupled translation of both genes. For cloning, the pMG36e (Eryr) plasmid was used (48). This expression vector carries the strong constitutive lactococcal P32 promoter (18), which could support the expression of as-48BC, and it was shown to be compatible in E. faecalis with pAM401-81 (Cmr), which harbors the as-48 cluster (see Materials and Methods for details). Two recombinant plasmids, pMG36-BC and pMG36-A, were generated (Fig. 1); the first carries the as-48BC genes under the control of P32, whereas the second lacks this promoter and harbors the as-48A structural gene coupled to its own natural PA promoter. Subsequently, the JH2-2[pAM401-58/pMG36-A], JH2-2[pAM401-81::Tn5PA/pMG36-A], and JH2-2[pAM401-81::Tn5B/pMG36-BC] strains were constructed by plasmid transfer and their production of AS-48 as well as immunity to the enterocin were investigated (Table 2).
Remarkably, the expression of as-48A in trans from pMG36-A in the deletion and insertion mutants had different effects. The introduction of pMG36-A in JH2-2[pAM401-58] did not result in an AS-48-proficient producer phenotype, whereas the presence of pMG36-A in JH2-2[pAM401-81::Tn5PA] partially restored the AS-48-producing capacity (7 versus 16 mm of halo in the isogenic wild-type strain) of the enterococcal cells that had been lost due to the Tn5 insertion into the PA. This differential complementation in the two mutants did not appear to be due to gene dosage effects, as the transcription of as-48A (the expected TA RNA of approximately 400 nt) from pMG36-A was detected by Northern blot hybridization in both, although at lower levels than in JH2-2[pAM401-81] (Fig. 2A and data not shown). These results suggested that the nonproducer AS-48 phenotype of JH2-2[pAM401-58/pMG36-A] was due to a lack of as-48BC expression, and indeed, it was undetectable in the pAM401-58 background by RT-PCR (data not shown). They results also indicated that the transcriptional read-through from the Tn5 carried in pAM401-81::Tn5PA allowed low levels of synthesis of As-48B, which was detectable by RT-PCR analysis of the intergenic region between as-48A and as-48B (Fig. 2B). Consequently, it seems that in the wild-type situation, as-48B cotranscribed with as-48A from PA was involved in AS-48 biogenesis. Supporting this hypothesis, in JH2-2[pAM401-81::Tn5B/pMG36-BC], expression from pMG36-BC under the P32 promoter partially restored the AS-48-producing capacity (Table 2).
In addition, the presence of pMG36-A did not increase the immunity levels of either JH2-2[pAM401-81::Tn5PA] or JH2-2[pAM401-58], suggesting the absence of the necessary expression of the as-48BC accompanying genes. However, expression from pMG36-BC in JH2-2[pAM401-81::Tn5B/pMG36-BC] conferred wild-type levels of resistance to the strain. This total complementation of the immunity phenotype, coupled with the overall results demonstrated that indeed, the low resistance against AS-48 of Tn5B and Tn5C insertion mutants is not due to any polar effects in expression of the as-48D1EFGH downstream genes involved in the double-immunity mechanisms previously described (12, 34). Consequently, as-48C must constitute a third determinant involved in that process.
Transcriptional analysis of the as-48ABC region.
The expression of the as-48ABC gene cluster in JH2-2, carrying the natural pMB2 plasmid or its pAM401-based derivatives, was investigated by Northern blot analysis, and the results obtained were corroborated by RT-PCR analysis. Total RNA was extracted at the end of the exponential growth phase and hybridized with specific probes derived from the as-48A and as-48B genes (see Materials and Methods for details). The detection of RNA hybridization with the as-48A probe by autoradiogram and of rRNAs by ethidium bromide stain is depicted in Fig. 2A. A single and prominent band corresponding to the as-48A transcript (TA) was detected in RNA preparations from S-48[pMB2] and JH2-2 carrying either pAM401-81 or pAM401-52 (Fig. 2A), correlating with their AS-48 producer phenotype. As expected, in JH2-2[pAM401-58] or JH2-2[pAM401-81::Tn5PA] lacking a functional PA promoter, TA was not detected in the blotting. In addition, TA was revealed (albeit at low levels) in RNA samples from JH2-2[pAM401-58/pMG36-A], in which expression of the structural gene is driven from the PA-as-48A transcriptional unit carried by pMG36-A (Fig. 2A). Furthermore, two faint and smeared bands larger than TA were also detected only in strains carrying a functional PA promoter (Fig. 2A and 3A and data not shown). However, when an as-48B probe was used, no clear hybridization bands could be detected in any of the blottings obtained; only weak marks could be detected just below the 23S (2,905 nt) and 16S (1,522 nt) rRNAs (data not shown). The overall results suggest that transcripts carrying as-48B had a very short life span, which impaired its detection by Northern hybridization.
Therefore, to confirm the cotranscription of as-48A and as-48B genes, their 73-nt intergenic region was analyzed by RT-PCR using specific primers (F-AsAB and R-AsAB) (Fig. 4) deduced from the coding regions of these genes. The expected 420-nt amplicon was detected in the three strains analyzed (Fig. 2B), JH2-2[pAM401-81], JH2-2[pAM401-81::Tn5PA], and JH2-2[pAM401-81::Tn5PA/pMG36-A]. The existence of RNA substrate for the RT-PCR amplification in the Tn5PA mutant at low level but not affected by the presence of pMG36-A, demonstrated transcriptional read-through from pAM401-81::Tn5PA, presumably driven from the aminoglycoside phosphotransferase promoter of Tn5. The detection of this amplicon in JH2-2[pAM401-81] and the fact that as-48B and as-48C overlap (Fig. 4A) suggested that the as-48ABC genes constitute a transcriptional unit driven from the PA promoter. Moreover, the results indicated that TA could be a specific and stable degradation product of the unstable T1 transcript (TABC) previously detected in JH2-2[pAM401-52] as a minor band, migrating at the position of the 23S RNA and encompassing the as-48ABC genes (34).
Analysis of the intergenic region between as-48A and as-48BC genes.
No obvious promoter sequences were observed in the 73-nt intergenic region between as-48A and as-48B. Moreover, the AS-48 nonproducer phenotype of JH2-2[pAM401-58], not complemented by pMG36-A and its reduced immunity (Table 2), rules out the existence of a promoter for the as-48BC genes in this region. However, analysis of this region with the Mfold program revealed the existence of a long IR with potential to form a complex secondary structure having an estimated ΔG free energy of −22.4 (Fig. 4A and 5A). This kind of structure is characteristic of substrates for endoribonucleases, suggesting that it could be implicated in mRNA processing and be decisive in the expression of the as-48BC genes (1, 13, 26, 31). To test this hypothesis, we analyzed the effect of replacing three bases within the IR in a site-directed mutagenesis experiment, creating an XhoI restriction site 21 nt downstream of the as-48A gene (Fig. 4A). For this purpose, the pAM401-81X plasmid, exhibiting three mismatches relative to the wild-type sequence, was constructed by overlap extension PCR mutagenesis. This plasmid should encode an mRNA with an altered secondary structure (Fig. 5B). When this plasmid was transferred to the JH2-2 strain, the transformants produced smaller inhibition zones than those of the wild-type strains (11 versus 16 mm), but they were fully resistant to 18 μg per 5 μl of pure AS-48 added exogenously (Table 2).
A comparative analysis by Northern blotting of as-48A expression in JH2-2[pAM401-81X] and its isogenic JH2-2[pAM401-81] wild-type control during the exponential phase of growth revealed that the mutation introduced in the intergenic region between as-48A and as-48B had important consequences at the mRNA level. The most significant was the almost undetectable amounts of TA present in RNA preparations of the mutant (Fig. 3A), which is most likely related to the conformational changes provoked by the mutation at the mRNA level (see below). In fact, a degraded mRNA, visualized as faint and smeared bands above the TA position, was observed for the mutant and not for the wild-type strain (Fig. 3A), in spite of the fact that rRNAs of both strains were intact (Fig. 3A, bottom panel). Furthermore, when levels of RNA species carrying as-48A were analyzed by RT-PCR (Fig. 3B) using the F-AsA and R-AsA primers (Fig. 4A), higher levels of as-48A mRNA were detected in the wild type than in the mutant, correlating with the results obtained by Northern blotting and in accordance with the hypoproducer phenotype of the mutant strain (Table 2). By contrast, the opposite situation was observed for as-48AB and as-48BC mRNAs, since their levels were higher in the mutant, in accordance with its full resistance against AS-48 and correlating with the detection of hybridization bands longer than as-48A by Northern blot analysis that could be a substrate for RT-PCR amplification. These results strongly indicated that the intergenic region between as-48A and as-48B plays an important role in the fate of the unstable as-48ABC mRNA transcribed from the PA promoter.
Therefore, the initiation of transcription from PA (Fig. 4B), as well as the processing of mRNA (Fig. 4C), was investigated in the wild-type and pAM401-81X mutant strains by primer extension. The detection of the 5′ end of the mRNA revealed in both strains, two extended products, corresponding to transcript starts with an adenosine and a guanidine residue located 38 and 39 nt upstream of the start codon of as-48A and at the correct position if transcription is driven from the PA promoter (Fig. 4A). This promoter (Fig. 4A) has a canonical TATAAT −10 region and a TTGctA (lowercase letters indicate bases that do not conform to the consensus sequence) −35 region that are separated from each other by 17 nt, which is the correct spacing for binding of the vegetative sigma factor of the bacterial RNA polymerase without the need of an activator. Moreover, inverted or direct repeats characteristic of transcriptionally regulated operators are present neither within nor surrounding the PA promoter. Consequently, under physiological conditions, transcription from PA in the wild type occurs during exponential growth (Fig. 2 to 4). Unexpectedly, a lower amount of the extended DNA fragments was detected in the pAM401-81X mutant (Fig. 4B). This result could be due to a shorter half-life of the TA in the mutant than in the wild-type strain, since lower levels of as-48A mRNA were also detected in the mutant by Northern blot and RT-PCR analyses (Fig. 3) and DNA sequencing of this region revealed no rearrangements in the mutant plasmid.
In addition, the use of a primer located proximal to the 3′ end of the secondary structure revealed that in the wild-type strain, there exist two RNA species (one major and one minor) (Fig. 4C) having in their 5′ ends two adjacent uridine residues located within the right arm of the secondary structure (Fig. 5A). Interestingly, only the minor RNA species was detected in the mutant strain, which is predicted to encode an mRNA with an altered secondary structure (Fig. 5B). Moreover, folding of the processed RNA species carrying as-48A in the wild-type strain (Fig. 5C) revealed protective secondary structures at the 5′ and 3′ ends that were not present in the mutant RNA species (Fig. 5D). These predictions, together with the results of the transcriptional analysis (Fig. 3), indicated that alteration of the 3′ end of the processed TA species in the mutant should result in its being unstable and being a good substrate for fast degradation by 3′ exonucleases without accumulating specific degradation products.
Finally, the overall results from RT-PCR and primer extension experiments (Fig. 3B and 4) indicate that the mutation at the secondary structure in pAM401-81X causes a decrease of mRNA processing in the JH2-2 background. However, this decrease is accompanied by an increase of species longer than TA but not TABC (Fig. 3A), indicating that degradation from the 3′ end of TABC is still very efficient in the mutant and that the detected degradation products could be the substrate for the increased RT-PCR amplification of as-48BC observed in JH2-2[pAM401-81X] (Fig. 3B).
DISCUSSION
In this work, we present evidence that a constitutive PA promoter drives transcription of the as-48ABC operon that includes the as-48A bacteriocin structural gene, the as-48B gene with an essential function for AS-48 biogenesis, and a putative cognate immunity gene (as-48C). Moreover, we have shown that the expression of as-48ABC is controlled at the posttranscriptional level.
RT-PCR analysis showed that as-48A and as-48B, as well as as-48B and as-48C, could be linked (Fig. 3B). Primer extension analysis revealed that an RNA species exists, carrying as-48B and having the 5′ end located in a putative secondary structure (Fig. 4 and 5A). Northern blotting analysis suggests that the as-48ABC operon mRNA transcript, TABC, is rapidly degraded by endonucleases, leading to a stable mRNA product, TA, and a TBC with a short life span, which makes its detection almost impossible (Fig. 2 and 3). The detection of RNA species containing the overlapping 3′ end of as-48B and 5′ end of as-48C by the more sensitive RT-PCR assay (Fig. 2 and 3) supports this hypothesis.
The computation of the secondary structure of the as-48AB intergenic region and the first five codons of as-48B with the Mfold program predicted two stem-loop secondary structures (I and II) (Fig. 5A). Structure I includes the two detected processing sites. The putative RBS of as-48B is located in the stem of structure II, which could modulate the translation of As-48B. The processing sites are two adjacent uracils located in a double-stranded region of structure I. The ubiquitous endoribonuclease RNase III identified in lactic acid bacteria (3) cleaves in double-stranded regions located in stem-loop structures (14, 43), and it is therefore possible that it is responsible for the processing of the as-48ABC transcript. Interestingly, the specificity of RNase III of E. faecalis may also depend on the mRNA sequence, since the TABC transcript encoded by the JH2-2[pAM401-81X] mutant is processed at the second uracil residue (Fig. 4 and 5B). The computation predicted that this U is also located in a double-stranded region of the mutant mRNA, but in structure III rather than structure I (Fig. 5B). The ΔGs predicted for the formation of the secondary structure of the intergenic regions of the wild type (−22.4) and the mutant (−16.6) indicate that processing should occur in the latter with less frequency, as was indeed detected (Fig. 4).
Here, we have reported that mRNA processing yields an abundant TA in the wild-type strain. The prediction of the folding of this TA (Fig. 5C) revealed that its 3′ end is located in a small secondary structure similar to the ρ-independent transcriptional terminators, which can act as a barrier to 3′-5′ exoribonucleases. Consequently, the prominent 0.4-kb TA RNA species could result from a combination of endoribonucleolytic cleavage of the primary TABC transcript in the region between as-48A and as-48B and the subsequent resistance of the resulting product to 3′-5′ exoribonucleolytic degradation (6, 22). Supporting this proposed role of the small secondary structure, the TA encoded by the pAM401-81X mutant is much less abundant than the wild type (Fig. 3) and does not seem to possess this structure at its 3′ end (Fig. 5D). The high level of TA in the wild type may be due to the synthesis of an independent as-48A transcript from PA, because the upper part of structure I (tA) includes a 3-nt loop and an 11-nt stem, followed by the sequence UUUUUUAAUU, as occurs in the weak transcriptional terminators that act as attenuators (36). This resembles the model described for the ltnA1A2 transcript in the production of lacticin 3147 and its immunity (37). However, the predicted ρ-independent terminators of E. faecalis have an average d value of 3.07 (see Materials and Methods for details) (11), whereas tA has a negative d score of −1.2, which makes it very unlikely that tA acts as an attenuator. Nevertheless, even if the termination of transcription occurs at low efficiency at this location, it is predicted that as in the case of processing, it will yield a stable as-48A transcript with a protective secondary structure at its 3′ end (data not shown).
Thus, it appears that RNA processing is the main mechanism that uncouples the transcription and translation of as-48A and as-48BC (8). The presence of a stem-loop structure between the structural gene and the downstream biosynthetic genes is a feature of a number of regulated bacteriocinogenic systems, such as mersacidin (2), sakacin A (5), sakacin P (35), nisin (29), or lacticin 3147 (9), where notable differences in concentrations between transcripts of the structural gene and of those genes involved in biosynthetic machinery have also been reported. It has been established that in addition to allowing partial read-through to downstream synthetic genes, the stem-loop structure in Pep5 is also required for mRNA stability (40). Our results demonstrate that the mutagenesis of such structures is not a reliable way to improve read-through without adversely impacting mRNA stability and thus reducing bacteriocin production.
With regard to as-48ABC operon translation, as-48A and as-48B are preceded by putative RBSs (5′-tgAGGAGGag-3′, where lowercase letters indicate bases that do not conform to the consensus sequence, and 5′-AAAGGGGGGC-3′) complementary to the 3′ end of the E. faecalis 16S rRNA (3′-UUUCCUCCCGAGUG-5′) (41) and at the correct spacing from their AUG translation initiation codons. These RBSs have computed ΔG free-energy values of −12.1 and −14.2, respectively, for a strong binding to the 3′ end of the enterococcal 16S rRNA according with the rules of Schur et al. (46). By contrast, as-48C does not possess sequences that correspond to a competent RBS and the overlapping of its 5′ end with the 3′ end of as-48B suggests that coupled translation of both genes occurs, constituting a second mechanism of posttranscriptional control. This is also the case for sakacin Q, in which the translation of the structural and the immunity genes are coupled (35).
It is frequently postulated that the bacteriocin-encoding and the immunity protein-encoding genes must be cotranscribed and expressed to ensure that the producer strain is not killed by its own bacteriocin. The results presented here support the fact that transcription of as-48BC takes place only from the PA linked to the as-48A structural gene. The precise function of As-48B and As-48C is still unknown, although both have a significant homology with CirB (58%) and CirC (65%), which are required for the production of circularin A (27). The reason why the genes of as-48ABC are cotranscribed must be investigated through the functional analysis of their gene products in the cell, and according to our results, As-48B is indispensable for AS-48 biogenesis, while the integral membrane protein As-48C, with four postulated transmembrane domains (34), could be involved in protection functions, together with the other immunity determinants as-48D1 and as-48EFGH, which are located in an independent operon (12, 34).
Acknowledgments
This study has been supported by the Spanish Dirección General de Investigación Científica y Técnica (projects BIO98-0908-CO2-01 and BIO2001-3237). M. Fernández and M. Sánchez-Hidalgo were recipients of fellowships from the Spanish Ministry of Education, Culture, and Sports.
We thank Stephen Elson for critical reading of the manuscript.
Footnotes
Published ahead of print on 2 November 2007.
REFERENCES
- 1.Allenby, N. E. E., N. O'Connor, Z. Prágai, N. M. Carter, M. Miethke, S. Engelmann, M. Hecker, A. Wipat, A. C. Ward, and C. R. Harwood. 2004. Post-transcriptional regulation of the Bacillus subtilis pst operon encoding a phosphate-specific ABC transporter. Microbiology 1502619-2628. [DOI] [PubMed] [Google Scholar]
- 2.Altena, K., A. Guder, C. Cramer, and G. Bierbaum. 2000. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol. 662565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amblar, M., S. C. Viegas, P. López, and C. M. Arraiano. 2004. Homologous and heterologous expression of RNase III from Lactococcus lactis. Biochem. Biophys. Res. Commun. 323884-890. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, D. G., and L. L. McKay. 1983. A simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axelsson, L., and A. Holck. 1995. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Bacteriol. 1772125-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belasco, J. C., and C. F. Higgins. 1988. Mechanisms of mRNA decay in bacteria: a perspective. Gene 7215-23. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and mu. J. Mol. Biol. 104541-555. [DOI] [PubMed] [Google Scholar]
- 8.Condon, C. 2003. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67157-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotter, P. D., L. H. Deegan, E. M. Lawton, L. A. Draper, P. M. O'Connor, C. Hill, and R. P. Ross. 2006. Complete alanine scanning of the two-component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol. Microbiol. 62735-747. [DOI] [PubMed] [Google Scholar]
- 10.d'Aubenton Carafa, Y., E. Brody, and C. Thermes. 1990. Prediction of rho-independent Escherichia coli transcription terminators. J. Mol. Biol. 216835-858. [DOI] [PubMed] [Google Scholar]
- 11.De Hoon, M. J., Y. Makita, K. Nakai, and S. Miyano. 2005. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 1212-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Díaz, M., E. Valdivia, M. Martínez-Bueno, M. Fernández, A. S. Soler-González, H. Ramírez-Rodrigo, and M. Maqueda. 2003. Characterization of a new operon, as-48EFGH, from the as-48 gene cluster involved in immunity to enterocin AS-48. Appl. Environ. Microbiol. 691229-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drider, D., J. M. Santos, C. M. Arraiano, and P. López. 1998. RNA processing is involved in the post-transcriptional control of the citQRP operon from Lactococcus lactis biovar diacetylactis. Mol. Gen. Genet. 2589-15. [DOI] [PubMed] [Google Scholar]
- 14.Drider, D., and C. Condon. 2004. The continuing story of endoribonuclease III. J. Mol. Microbiol. Biotechnol. 8195-200. [DOI] [PubMed] [Google Scholar]
- 15.Eijsink, V. H., L. Axelsson, D. B. Diep, L. S. Håvarstein, H. Holo, and I. F. Nes. 2002. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie van Leeuwenhoek 81639-654. [DOI] [PubMed] [Google Scholar]
- 16.Fiedler, S., and Wirth R. 1991. Transformation of Enterococcus faecalis and Enterococcus faecium by electroporation, p. 301. In G. M. Dunny, P. P. Clearly, and L. L. McKay (ed.), Genetics and molecular biology of streptococci, lactococci, and enterococci. American Society for Microbiology, Washington, DC.
- 17.Fujimoto, S., and Ike Y. 2001. pAM401-based shuttle vectors that enable overexpression of promoterless genes and one-step purification of tag fusion proteins directly from Enterococcus faecalis. Appl. Environ. Microbiol. 671262-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaeng, S., S. Scherer, H. Neve, and M. J. Loessner. 2000. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl. Environ. Microbiol. 662951-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gálvez, A., E. Valdivia, M. Martínez-Bueno, and M. Maqueda. 1989. Purification and amino acid composition of peptide antibiotic AS-48 produced by Streptococcus (Enterococcus) faecalis subsp. liquefaciens S-48. Antimicrob. Agents Chemother. 33437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Quintáns, N. C. Magni, D. de Mendoza, and P. López. 1998. The citrate transport system of Lactococcus lactis subsp. lactis biovar diacetylactis is induced by acid stress. Appl. Environ. Microbiol. 64850-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heng, N. C. K., P. A. Wescombe, J. P. Burton, R. W. Jack, and J. R. Tagg. 2007. The diversity of bacteriocins produced by gram-positive bacteria, p. 45-92. In M. A. Riley and M. A. Chavan (ed.), Bacteriocins—ecology and evolution. Springer, Heidelberg, Germany.
- 22.Higgins, C. F., S. W. Peltz, and A. Jacobson. 1992. Turnover of mRNA in prokaryotes and lower eukaryotes. Curr. Opin. Genet. Dev. 2739-747. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.
- 24.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap-extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 25.Homuth, G., S., Masuda, A. Mogk, Y. Kobayashi, and W. Schumann. 1997. The dnaK operon of Bacillus subtilis is heptacistronic. J. Bacteriol. 1791153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homuth, G., A. Mogk, and W. Schumann. 1999. Post-transcriptional regulation of the Bacillus subtilis dnaK operon. Mol. Microbiol. 321183-1197. [DOI] [PubMed] [Google Scholar]
- 27.Kemperman, R., M. Jonker, A. Nauta, O. P. Kuipers, and J. Kok. 2003. Functional analysis of the gene cluster involved in production of the bacteriocin circularin A by Clostridium beijerinckii ATCC 25752. Appl. Environ. Microbiol. 695839-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleerebezem, M. 2004. Quorum sensing controls of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 251405-1414. [DOI] [PubMed] [Google Scholar]
- 29.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. De Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216281-291. [DOI] [PubMed] [Google Scholar]
- 30.Maqueda, M., A. Gálvez, M. Martínez-Bueno, M. J. Sánchez-Barrena, C. González, A. Albert, M. Rico, and E. Valdivia. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr. Protein Pept. Sci. 5399-416. [DOI] [PubMed] [Google Scholar]
- 31.Marqués, S., J. L. Ramos, and K. N. Timmis. 1993. Analysis of the mRNA structure of the Pseudomonas putida TOL meta fission pathway operon around the transcription initiation point, the xylTE and the xylFJ regions. Biochim. Biophys. Acta 1215227-236. [DOI] [PubMed] [Google Scholar]
- 32.Martínez-Bueno, M., A. Gálvez, E. Valdivia, and M. Maqueda. 1990. A transferable plasmid associated with AS-48 production in Enterococcus faecalis. J. Bacteriol. 1722817-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martínez-Bueno, M., M. Maqueda, A. Gálvez, B. Samyn, J. van Beeumen, J. Coyette, and E. Valdivia. 1994. Determination of the gene sequence and molecular structure of the enterococcal peptide antibiotic AS-48. J. Bacteriol. 1766334-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martínez-Bueno, M., E. Valdivia, A. Gálvez, J. Coyette, and M. Maqueda. 1998. Analysis of the gene cluster involved in production and immunity of the peptide antibiotic AS-48 in Enterococcus faecalis. Mol. Microbiol. 27347-358. [DOI] [PubMed] [Google Scholar]
- 35.Mathiesen, G., K. Huehne, L. Kroeckel, L. Axelsson, and V. G. H. Eijsink. 2005. Characterization of a new bacteriocin operon in sakacin P-producing Lactobacillus sakei, showing strong translational coupling between the bacteriocin and immunity genes. Appl. Environ. Microbiol. 713565-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merino, E., and C. Yanofsky. 2005. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 21260-264. [DOI] [PubMed] [Google Scholar]
- 37.McAuliffe, O., T. O'Keeffe, C. Hill, and R. P. Ross. 2001. Regulation of immunity to the two-component lantibiotic, lacticin 3147, by the transcriptional repressor LtnR. Mol. Microbiol. 39982-993. [DOI] [PubMed] [Google Scholar]
- 38.Mora, D., M. G. Fortina, C. Parini, D. Daffonchio, and P. L. Manachini. 2000. Genomic subpopulations within the species Pediococcus acidilactici detected by multilocus typing analysis: relationships between pediocin AcH/PA-1 producing and non-producing strains. Microbiology 1462027-2038. [DOI] [PubMed] [Google Scholar]
- 39.Murzin, A. G., and A. Bateman. 2001. CASP2 knowledge-based approach to distant homology recognition and fold prediction in CASP4. Proteins 45(Suppl. 5)76-85. [DOI] [PubMed] [Google Scholar]
- 40.Pag, U., C. Heidrich, G. Bierbaum, and H.-G. Sahl. 1999. Molecular analysis of expression of the lantibiotic Pep5 immunity phenotype. Appl. Environ. Microbiol. 65591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulsen, I., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. D. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2992071-2074. [DOI] [PubMed] [Google Scholar]
- 42.Pillardy, J., C. Czaplewski, A. Liwo, J. Lee, D. R. Ripoll, R. Kazmierkiewicz, S. Oldziej, W. J. Wedemeyer, K. D. Gibson, Y. A. Arnautova, J. Saunders, Y. J. Ye, and H. A. Scheraga. 2001. Recent improvements in prediction of protein structure by global optimization of a potential energy function. Proc. Natl. Acad. Sci. USA 982329-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regnier, P., and M. C. Arraiano. 2000. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays 22235-244. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Nat. Acad. Sci. USA 745463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schur, T., E. Nadir, and H. Margalit. 1993. Identification and characterization of the Escherichia coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 214019-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sesma, F., D. Gardiol, A. P. de Ruiz Holgado, and D. de Mendoza. 1990. Cloning and expression of the citrate permease gene of Lactococcus lactis subsp. lactis biovar diacetylactis in Escherichia coli. Appl. Environ. Microbiol. 562099-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Guchte, M., J. M. B. M. Van der Vossen, J. Kok, and G. Venema. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis IL1403. Appl. Environ. Microbiol. 621689-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]