Abstract
This work describes a regulatory network of Pseudomonas putida controlled in response to nitrogen availability. We define NtrC as the master nitrogen regulator and suggest that it not only activates pathways for the assimilation of alternative nitrogen sources but also represses carbon catabolism under nitrogen-limited conditions, possibly to prevent excessive carbon and energy flow in the cell.
Nitrogen is one of the most limiting elements in the environment. In bacteria, a number of systems have evolved which are induced to scavenge nitrogen from alternative sources when the preferential sources are scarce. In enterobacteria, NtrC is the global activator of alternative nitrogen assimilation pathways under nitrogen-limited growth conditions (for reviews, see references 17, 19, and 23). A uridylyl transferase/uridylyl-removing enzyme senses the nitrogen status and modulates the function of protein PII accordingly, which in turn transduces the signal to NtrB, which finally modulates NtrC function via phosphorylation (1, 21).
Pseudomonas is a model free-living organism and the paradigmatic organism used in a diverse range of biotechnological applications, such as bioremediation or biotransformation processes (8, 14, 24, 27-29). However, very little is known about its ability to adapt to recurrent changes in nitrogen availability in the environment. NtrC has recently been identified as a protein involved in nitrogen regulation of atrazine biodegradation in Pseudomonas putida (9, 10), although its possible effects on other operons have not been described. Also, analysis of the P. putida genome in silico revealed interesting differences compared to nitrogen control genes in Escherichia coli, which belongs to the same phylum but a different order. P. putida has only one gene coding for a PII homologue, in contrast to enterobacteria, which have two paralogs coding for GlnK and GlnB. Also, in enterobacteria, the nitrogen assimilation control (Nac) protein activates transcription of σ70-dependent genes, whereas NtrC activates transcription of σ54-dependent genes (16, 33). Thus, Nac serves as a link between NtrC and σ70-dependent operons (33). There is no Nac homologue in P. putida; therefore, activation of σ70-dependent operons by nitrogen in Pseudomonas must operate differently.
The transcription profiles of P. putida strains KT2440 (20) and KT2442, a rifampin-resistant derivative, were compared by using DNA microarrays (32) in a minimal medium containing 25 mM sodium succinate as a carbon source, ammonium chloride (1 g liter−1) and l-serine (1 g liter−1) for nitrogen-excess conditions, and l-serine alone (1 g liter−1) for nitrogen-limited conditions. The ΔntrC deletion mutant MPO201 (9) was also used in order to compare its expression pattern to that of the isogenic wild-type strain in nitrogen-limited conditions. For each experiment, a minimum of three independent RNA extracts were analyzed at least twice as described previously (32). After background subtraction, the signal intensities for each replica were normalized and statistically analyzed using the Lowess intensity-dependent normalization method (30) included in the Almazen System software (Alma Bioinformatics S.L.). P values were calculated with Student's test algorithm based on the differences between log2 ratio values for each replicate. Genes were considered differentially expressed when they fulfilled the following filter parameters: expression ratio of ≥2 or ≤−2 and an adjusted P value of ≤0.05. The P values were adjusted for multiple tests as described previously (2). Confirmation by quantitative reverse transcription (RT)-PCR of the expression of selected open reading frames was performed as described previously (32). Primer sequences are shown in Table S1 in the supplemental material. Strains KT2440 and KT2442 had almost identical expression profiles, suggesting that the altered RNA polymerase was not highly pleiotropic (see Table S2 in the supplemental material).
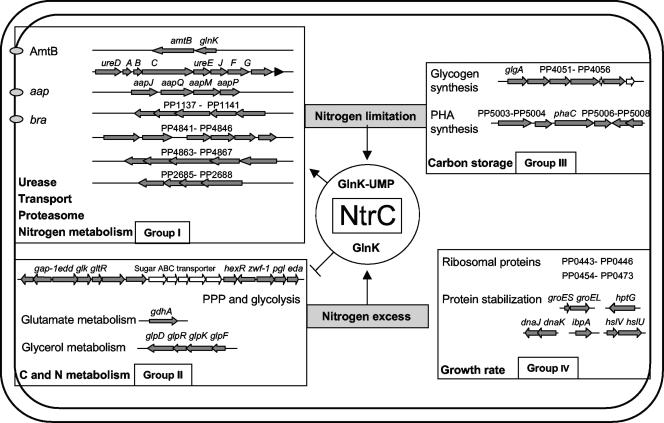
The expression of a number of genes, such as the genes coding for ribosomal proteins and chaperonins, including heat shock proteins, and even rpoS, apparently changed in an NtrC-dependent manner (group IV in Fig. 1; see Table S3 in the supplemental material). However, this change correlated with the growth rate (0.81 h−1 for the wild-type strain growing on ammonium plus serine, 0.59 h−1 for the wild-type strain growing on serine alone, and 0.18 h−1 for the ΔntrC mutant strain growing on serine) and was not consistent with nitrogen-regulated expression.
FIG. 1.
Classification of P. putida genes according to their responses to nitrogen and NtrC. Four groups of genes are indicated: group I, NtrC-activated genes; group II, NtrC-repressed genes; group III, NtrC-independent nitrogen-regulated genes; and group IV, genes responding to a differential growth rate.
The genes that were considered NtrC activated in response to nitrogen were the genes showing increased expression when organisms were growing in mid-exponential phase under nitrogen-limited conditions and in the wild-type strain compared to the ΔntrC mutant (group I in Fig. 1; see Table S3 in the supplemental material). We found 58 NtrC-activated open reading frames. These open reading frames included several operons involved in diverse systems for amino acid transport, including transport of amino acids in general (PP1297 to PP1300), branched-chain amino acids (PP1138 to PP1141, PP4841 to PP4845, and PP4863 to PP4867), or other amino acids, such as proline, arginine, or ornithine (PP0296 and PP1206), and for transport of three different putative porins (PP0046, PP0883, PP4465), as well as genes responsible for urea assimilation (PP2843 to PP2848) (see Table S3 in the supplemental material). P. putida KT2440 has quite broad transport capabilities, with approximately 370 membrane transport systems (5). It appears that a major response to nitrogen limitation is to activate transport systems to scavenge nitrogen-containing compounds that may be present in the medium. The high-affinity ammonium transporter, encoded by amtB (PP5233), which is presumed to be part of an operon with the upstream PII-encoding glnK gene, is NtrC activated in P. putida (see Table S3 in the supplemental material), as it is in enterobacteria (13, 33). Quantitative RT-PCR confirmation of amtB expression revealed that there was 62-fold induction in the presence of serine compared to the expression in the presence of ammonium plus serine for the wild-type strain and that there was a 76-fold change compared to the ΔntrC mutant. The fact that the change in the expression of glnK was much smaller than the change in the expression of amtB (Table 1) and the identification of a σ54-dependent promoter and two imperfect putative NtrC binding sites upstream of amtB provide an indication of the presence of an internal promoter upstream of amtB. We also detected NtrC-dependent activation of the glnA gene coding for glutamine synthetase. Quantitative RT-PCR data confirmed that the expression of glnA was lower in KT2442 in nitrogen-excess conditions (2.1-fold ± 0.07-fold) and in the ΔntrC mutant (3.7-fold ± 0.016-fold), thus indicating that there was NtrC activation. As it is in enteric bacteria, P. putida ntrC is downstream of glnA and is also induced by nitrogen limitation (3.3-fold as determined by DNA arrays and 8.1-fold ± 0.6-fold as determined by quantitative RT-PCR), and thus it presumably is part of an operon.
TABLE 1.
Verification of selected DNA microarray results by quantitative RT-PCR
| Gene | Ratio of expressiona
|
|||
|---|---|---|---|---|
| Nitrogen
|
NtrC
|
|||
| RT-PCRb | Microarray | RT-PCRb | Microarray | |
| amtB (PP5233) | 62.1 ± 35.9 | 25.4 | −75.8 ± 17.4 | −9.0 |
| glnK (PP5234) | 9.2 ± 3.2 | 5.7 | −4.9 ± 1.2 | −2.6 |
| gdhA (PP0675) | −2.9 ± 0.5 | −3.6 | 3.0 ± 1.1 | 2.5 |
| zwf-1 (PP1022) | −3.8 ± 1.8 | −6.6 | 3.1 ± 1.0 | 5.7 |
| gap-1 (PP1009) | −2.5 ± 0.43 | −2.9 | 3.6 ± 0.6 | 2.6 |
| glpF (PP1076) | −1.4 ± 0.1 | −1.7 | 7.5 ± 0.8 | 4.7 |
| phaC (PP5005) | 4.2 ± 1.3 | 2.3 | 1.2 ± 0.2 | 1.2 |
| GA1 (PP5008) | 4.7 ± 0.8 | 3.4 | 1.1 ± 0.1 | 1.1 |
| glgA (PP4050) | 4.6 ± 1.8 | 2.4 | 1.7 ± 0.5 | 1.7 |
| glgX (PP4055) | 32.2 ± 19.7 | 11 | −3.0 ± 1.3 | −2.4 |
Positive values for the ratio of expression in the nitrogen arrays indicated that the gene was upregulated in the presence of serine, and negative values indicated it was upregulated in the presence of ammonium plus serine. Positive values for the NtrC arrays indicated that the gene was upregulated in the mutant strain, and negative values indicated that it was upregulated in the wild-type strain. The same criteria were used for the quantitative RT-PCR data.
The quantitative RT-PCR values are the means ± standard deviations of at least three independent experiments.
The highest levels of induction were those of open reading frames corresponding to PP2685 to PP2688, which presumably constitute an operon (see Table S3 in the supplemental material). None of these genes are represented in the genomes of enteric bacteria, and their products have unknown functions. However, the PP2685 protein, which is conserved in other groups of bacteria, showed similarity to the beta subunit of the 20S proteasome of eukarya and archaea, which is the central enzymatic complex for nonlysosomal protein degradation in both the cytosol and the nucleus (11). In addition, the PP2686 protein showed high similarity to microbial homologues of transglutaminases, which have been proposed to be proteases (31). Induction of these genes suggests that an increase in protein turnover by proteolysis may be required to support the extensive changes in the protein profile of P. putida, particularly when these changes are in response to nitrogen limitation. This response appears to be specific for Pseudomonas since similar global analysis of E. coli did not show nitrogen regulation of genes coding for proteases (33).
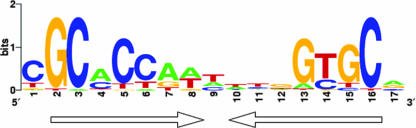
Analysis of P. putida intergenic regions using the E. coli NtrC consensus sequence led to identification of 120 potential sites in different operons, and the sites shown to be NtrC activated in the microarrays analysis were selected in order to determine a consensus sequence specific for P. putida (Fig. 2). Of 29 operons experimentally shown to be NtrC activated, 16 contained one or more NtrC binding sites, but we could not detect any NtrC binding sites in 13 of the operons, thus suggesting that they are not directly activated by NtrC. Attempts to find any conserved motif that could be recognized by an adapter in these promoters were unsuccessful.
FIG. 2.
Determination of the consensus NtrC binding site in P. putida (26). The consensus sequence was obtained by alignment of the promoter regions in the P. putida genome that contained the putative NtrC- consensus sequence of enterobacteria (TGCACCATAATGGTGCA) (7) and that were NtrC activated in the array experiments.
In E. coli, a subset of nine σ70-dependent operons that code for different transport systems for amino acids, other nitrogenated compounds, or porins are coordinately activated by nitrogen limitation through the adapter Nac (33). Two of these operons have homologues in P. putida and appear to be nitrogen regulated. These operons are codBA (PP3187 and PP3189; homologues of b0336 and b0337 in E. coli), which encodes a cytosine transporter and deaminase, and an operon involved in dipeptide transport (PP0885 to PP0878; homologues of E. coli b1243 to b1247 and b3540 to b3544). We observed 2.5-fold upregulation in the presence of serine of codA (PP3189), which is presumably part of an operon with codB, and 4- to 6-fold upregulation in the wild-type strain of PP0885 to PP0878. Both promoters are dependent on σ54, and one NtrC binding site is close to them. The other operons have no counterpart, or their putative counterparts are not nitrogen regulated in P. putida. Thus, although some P. putida operons may be indirectly regulated by NtrC, possibly in response to specific signals, the two comparable operons controlled by Nac in E. coli are directly activated by NtrC in P. putida.
In P. putida we detected a number of operons whose expression pattern indicates that there is repression by NtrC, since they exhibited increased expression in nitrogen-excess conditions and in the ΔntrC mutant (group II in Fig. 1; see Table S3 in the supplemental material). An example is gdhA (PP0675), encoding glutamate dehydrogenase. This enzyme may incorporate nitrogen through glutamate formation when a high concentration of ammonium is available. However, at low ammonium concentrations the reaction may proceed in the opposite direction, while ammonium is incorporated through the glutamine synthetase-glutamate synthase pathway (23). Strong repression of gdhA has been reported for Klebsiella pneumoniae and E. coli, which is exerted by Nac (3, 25). Interestingly, up to three potential NtrC binding sites have been found upstream of gdhA, further supporting the notion that NtrC may directly exert the regulatory control corresponding to that of Nac in E. coli, even when this control is negative.
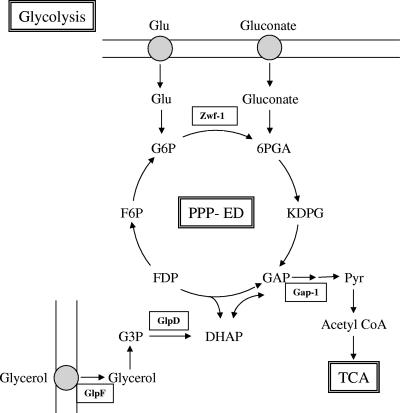
Two genes involved in sugar utilization, zwf-1 (PP1022) and gap-1 (PP1009), encoding glucose-6-phosphate 1-dehydrogenase and glyceraldehyde-3-phosphate 1-dehydrogenase, respectively, are also repressed by NtrC under low-nitrogen-availability conditions. KT2440 has an incomplete glycolytic pathway since it lacks 6-phosphofructokinase. Glucose is thus converted to glyceraldehyde-3-phosphate and pyruvate via the Entner-Doudoroff pathway, which is complete in this strain, in which 6-phosphogluconate is a key intermediate (6) (Fig. 3). In the P. putida NtrC mutant strain we also detected upregulation of the genes coding for a glycerol uptake facilitator protein (glpF [PP1076]) and a glycerol-3-phosphate dehydrogenase (glpD [PP1073]), which are involved in uptake of glycerol and its catabolism through the Entner-Doudoroff central pathway (4, 6, 18). We postulate that repression of glycerol uptake and metabolism by NtrC contribute to reduction of carbon fuelling in the tricarboxylic acid cycle when nitrogen is limiting growth. Repression of these genes provides a clear indication that NtrC may also control carbon catabolism in bacteria, particularly the major route of hexose utilization, probably to prevent an excess of catabolic flow under nitrogen-limiting conditions for growth.
FIG. 3.
Effect of nitrogen availability on expression of genes involved in carbon catabolism. Glycolysis, the pentose phosphate and Entner-Doudoroff pathways, and the glycerol uptake pathway are represented. Enzymes encoded by genes that are NtrC repressed are enclosed in boxes. Abbreviations: Zwf-1, glucose-6-phosphate 1-dehydrogenase; Gap-1, glyceraldehyde-3-phosphate 1-dehydrogenase; GlpD, glycerol-3-phosphate dehydrogenase; GlpF, glycerol uptake facilitator protein; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; FDP, fructose diphosphate; 6PGA, 6-phosphogluconate; KDPG, 2-keto-3-deoxy-6-phosphogluconate; GAP, glyceraldehyde-3-phosphate; PPP-ED, pentose phosphate and Entner-Doudoroff pathways; DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde-3-phosphate; CoA, coenzyme A; TCA, tricarboxylic acids.
Under nitrogen limitation conditions we also detected upregulation of a set of genes involved in the accumulation of carbon storage polymers, such as polyhydroxyalkanoates (PHAs) and glycogen. However, this regulation was mainly independent of NtrC (Fig. 1; see Table S3 in the supplemental material). Regulation of phaC (PP5005) encoding one of the two poly(3-hydroxyalkanoate) polymerases, PP5008 encoding the PHA granule-associated protein GA1 for PHA accumulation, and glgA (PP4050) and glgX (PP4055) for glycogen synthesis was confirmed by quantitative RT-PCR (Table 1). Most bacteria accumulate PHAs (12, 15) or glycogen (22) as carbon and energy storage material if excess amounts of a carbon source are provided or if another nutrient is limiting (nitrogen in our case but also sulfur, phosphate, iron, magnesium, potassium, or oxygen). This suggests that the regulatory system(s) controlling carbon storage polymers truly senses excess carbon relative to other nutrients rather than limitation of each nutrient, which is consistent with the NtrC-independent regulation of the genes in response to nitrogen availability.
In summary, we present here a global view of the P. putida transcriptional status in response to changing nitrogen availability conditions. Our data show that NtrC exerts the main control over transcription of nitrogen provision and carbon catabolism genes in response to nitrogen availability in P. putida.
Supplementary Material
Acknowledgments
We are grateful to all members of the Santero laboratory for their insights and helpful suggestions and to Maribel Ramírez, Guadalupe Martín, and Nuria Pérez for technical help. We thank Ildefonso Cases for his valuable help with NtrC consensus sequence determination.
This work was funded by grant GEN2001-4698-C05-04 and fellowship of the FPU program awarded to A.B.H. from the Spanish Ministry of Science and Technology. The institutional support from the Junta de Andalucia is acknowledged.
Footnotes
Published ahead of print on 26 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arcondeguy, T., R. Jack, and M. Merrick. 2001. P(II) signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 6580-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamini, Y., D. Drai, G. Elmer, N. Kafkafi, and I. Golani. 2001. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125279-284. [DOI] [PubMed] [Google Scholar]
- 3.Camarena, L., S. Poggio, N. Garcia, and A. Osorio. 1998. Transcriptional repression of gdhA in Escherichia coli is mediated by the Nac protein. FEMS Microbiol. Lett. 16751-56. [DOI] [PubMed] [Google Scholar]
- 4.Cuskey, S. M., and P. V. Phibbs, Jr. 1985. Chromosomal mapping of mutations affecting glycerol and glucose catabolism in Pseudomonas aeruginosa PAO. J. Bacteriol. 162872-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dos Santos, V. A., S. Heim, E. R. Moore, M. Stratz, and K. N. Timmis. 2004. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 61264-1286. [DOI] [PubMed] [Google Scholar]
- 6.Dos Santos, V. A., K. N. Timmis, B. Tummler, and C. Weinel. 2004. Genomic features of Pseudomonas putida strain KT2440, p. 77-112. In J. L. Ramos (ed.), Pseudomonas, vol. 1. Plenum Publishers, New York, NY. [Google Scholar]
- 7.Ferro-Luzzi Ames, G., and K. Nikaido. 1985. Nitrogen regulation in Salmonella typhimurium. Identification of an ntrC protein-binding site and definition of a consensus binding sequence. EMBO J. 4539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda, M. 1994. Degradation of recalcitrant synthetic chemical products: genetic engineering in pseudomonads. Bioprocess Technol. 19821-835. [PubMed] [Google Scholar]
- 9.Garcia-Gonzalez, V., F. Govantes, O. Porrua, and E. Santero. 2005. Regulation of the Pseudomonas sp. strain ADP cyanuric acid degradation operon. J. Bacteriol. 187155-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Gonzalez, V., F. Govantes, L. J. Shaw, R. G. Burns, and E. Santero. 2003. Nitrogen control of atrazine utilization in Pseudomonas sp. strain ADP. Appl. Environ Microbiol. 696987-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gille, C., A. Goede, C. Schloetelburg, R. Preissner, P. M. Kloetzel, U. B. Gobel, and C. Frommel. 2003. A comprehensive view on proteasomal sequences: implications for the evolution of the proteasome. J. Mol. Biol. 3261437-1448. [DOI] [PubMed] [Google Scholar]
- 12.Hankermeyer, C. R., and R. S. Tjeerdema. 1999. Polyhydroxybutyrate: plastic made and degraded by microorganisms. Rev. Environ. Contam. Toxicol. 1591-24. [DOI] [PubMed] [Google Scholar]
- 13.Javelle, A., E. Severi, J. Thornton, and M. Merrick. 2004. Ammonium sensing in Escherichia coli. Role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J. Biol. Chem. 2798530-8538. [DOI] [PubMed] [Google Scholar]
- 14.Jiménez, J. I., B. Miñambres, J. L. García, and E. Díaz. 2004. Genomic insights in the metabolism of aromatic compounds in Pseudomonas, p. 425-462. In J. L. Ramos (ed.), Pseudomonas, vol. 3. Plenum Publishers, New York, NY. [Google Scholar]
- 15.Kessler, B., and B. Witholt. 2001. Factors involved in the regulatory network of polyhydroxyalkanoate metabolism. J. Biotechnol. 8697-104. [DOI] [PubMed] [Google Scholar]
- 16.Kustu, S., E. Santero, J. Keener, D. Popham, and D. Weiss. 1989. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 53367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leigh, J. A., and J. A. Dodsworth. 2007. Nitrogen regulation in Bacteria and Archaea. Annu. Rev. Microbiol. 61349-377. [DOI] [PubMed] [Google Scholar]
- 18.Lessie, T. G., and P. V. Phibbs, Jr. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38359-388. [DOI] [PubMed] [Google Scholar]
- 19.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4799-808. [DOI] [PubMed] [Google Scholar]
- 21.Ninfa, A. J., and P. Jiang. 2005. PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 8168-173. [DOI] [PubMed] [Google Scholar]
- 22.Preiss, J. 1984. Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 38419-458. [DOI] [PubMed] [Google Scholar]
- 23.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57155-176. [DOI] [PubMed] [Google Scholar]
- 24.Rojo, F., D. H. Pieper, K. H. Engesser, H. J. Knackmuss, and K. N. Timmis. 1987. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science 2381395-1398. [DOI] [PubMed] [Google Scholar]
- 25.Rosario, C. J., and R. A. Bender. 2005. Importance of tetramer formation by the nitrogen assimilation control protein for strong repression of glutamate dehydrogenase formation in Klebsiella pneumoniae. J. Bacteriol. 1878291-8299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 186097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timmis, K. N., R. J. Steffan, and R. Unterman. 1994. Designing microorganisms for the treatment of toxic wastes. Annu. Rev. Microbiol. 48525-557. [DOI] [PubMed] [Google Scholar]
- 28.van Beilen, J. B., and B. Witholt. 2004. Alkane degradation by pseudomonads, p. 397-424. In J. L. Ramos (ed.), Pseudomonas, vol. 3. Plenum Publishers, New York, NY. [Google Scholar]
- 29.Walsh, U. F., J. P. Morrissey, and F. O'Gara. 2001. Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr. Opin. Biotechnol. 12289-295. [DOI] [PubMed] [Google Scholar]
- 30.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yee, V. C., L. C. Pedersen, I. Le Trong, P. D. Bishop, R. E. Stenkamp, and D. C. Teller. 1994. Three-dimensional structure of a transglutaminase: human blood coagulation factor XIII. Proc. Natl. Acad. Sci. USA 917296-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuste, L., A. B. Hervas, I. Canosa, R. Tobes, J. I. Jimenez, J. Nogales, M. M. Perez-Perez, E. Santero, E. Diaz, J. L. Ramos, V. de Lorenzo, and F. Rojo. 2006. Growth phase-dependent expression of the Pseudomonas putida KT2440 transcriptional machinery analysed with a genome-wide DNA microarray. Environ. Microbiol. 8165-177. [DOI] [PubMed] [Google Scholar]
- 33.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 9714674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.