Abstract
PY100 is a lytic bacteriophage with a broad host range within the genus Yersinia. The phage forms plaques on strains of the three human pathogenic species Yersinia enterocolitica, Y. pseudotuberculosis, and Y. pestis at 37°C. PY100 was isolated from farm manure and intended to be used in phage therapy trials. PY100 has an icosahedral capsid containing double-stranded DNA and a contractile tail. The genome consists of 50,291 bp and is predicted to contain 93 open reading frames (ORFs). PY100 gene products were found to be homologous to the capsid proteins and proteins involved in DNA metabolism of the enterobacterial phage T1; PY100 tail proteins possess homologies to putative tail proteins of phage AaΦ23 of Actinobacillus actinomycetemcomitans. In a proteome analysis of virion particles, 15 proteins of the head and tail structures were identified by mass spectrometry. The putative gene product of ORF2 of PY100 shows significant homology to the gene 3 product (small terminase subunit) of Salmonella phage P22 that is involved in packaging of the concatemeric phage DNA. The packaging mechanism of PY100 was analyzed by hybridization and sequence analysis of DNA isolated from virion particles. Newly replicated PY100 DNA is cut initially at a pac recognition site, which is located in the coding region of ORF2.
Bacteriophages were discovered independently by F. Twort in 1915 and F. d'Herelle in 1917. In the first decades after their discovery, phages were used to combat bacterial infections, first in animals and later in humans (46, 47). The use of phages for the treatment of bacterial infections was abandoned in Western countries with the advent of antibiotics, largely since many of the results of early phage therapies had been ambiguous. In the 1940s in the United States scientists began using phages in basic genetic studies, and the findings and results of those experiments formed the basis of molecular biology (48).
In the genus Yersinia, phages have been used for typing, and phage sets have been worked out for typing Yersinia enterocolitica (6, 24). The genomes of two lytic yersiniophages have been determined; both phages show a close relationship to the Escherichia coli phages T3 and T7 and possess short noncontractile tails. Phage ΦYeO3-12 is specific for Y. enterocolitica serotype O3 (31), and phage ΦA1122 has been used for typing of Y. pestis (17). Their genome sizes are 37,555 and 39,600 bp, respectively. Recently, the lytic yersiniophage ΦR1-37 was described; this phage has a broader host range within Y. enterocolitica and possesses a contractile tail, however, the genome size of ΦR1-37 is estimated to be 270 kb (25). In our laboratory the temperate yersiniophage PY54 with a genome size of 46,339 bp is studied because of its replication modus as a linear prophage (20, 21). The host range of PY54 is restricted to Y. enterocolitica strains serotypes O5 and O5,27.
Reports on phage therapy experiments with yersiniophages are rare in the literature; however, one remarkable historic report is from the experiments of d'Herelle, who reported the successful treatment of four plague patients with lytic phages (13, 4). The renewed interest in phage therapy experiments due to the increase in antibiotic resistance of several pathogenic bacteria prompted us to search for Yersinia phages that lyse their hosts at 37°C. The aim of these experiments was to study the possibility to reduce or eradicate enteropathogenic Y. enterocolitica in infected pigs, which are the main reservoir for human food-borne infections (16). Applications of phages against bacterial pathogens in the food production chain are intended to reduce zoonotic bacteria in domestic animals or to use them as biocontrol agents for food preservation (18, 22).
We isolated the phage PY100 from the manure of a pig farm in Germany. PY100 was found by its ability to form large clear plaques on susceptible Y. enterocolitica strains at 37°C. We investigated, in an animal model for enteropathogenic Y. enterocolitica, whether PY100 or the yersiniophage ΦYeO3-12 (30) was able to inhibit colonization of the guts of mice by strains of Y. enterocolitica biotype 4, serotype O3 (43), which causes most of the human cases of yersiniosis in Europe.
Our studies of the PY100 biology revealed a very broad host range of the phage in the genus Yersinia, which included also the two other human pathogenic species Y. pseudotuberculosis and Y. pestis. In all cases, PY100 formed large plaques at 37°C on the bacterial lawns and may be of interest for treating infections with these pathogens. Especially, since Y. pestis is classified as a potential biowarfare or bioterror agent, phage therapy may be considered as an approach to counter such a threat (4, 17). We report here on the host range, burst size, genome sequence, proteomic characteristics, and packaging mechanism of this phage.
MATERIALS AND METHODS
Bacterial strains.
Yersinia strains and other enterobacterial strains were obtained from the Robert Koch Institute collection (27, 44) and the Institute Pasteur, Paris, France. Y. pestis strains KIM (12) and EV76 (38) were tested in the biosafety level III laboratory of the Robert Koch Institute. Other investigated strains used were Y. enterocolitica 8081 (serotype O:8) (49), Y. enterocolitica 6471/76 (serotype O:3) (42), and Y. pseudotuberculosis YPIII (7).
Isolation of PY100, propagation, and plaque assay.
Manure was centrifuged twice at 10,000 × g, and the supernatant was passed through a 0.45-μm-pore-size filter. Serial dilutions of the supernatant were made in SM buffer (5.8 g of NaCl per liter, 2.0 g of MgSO4·7H2O per liter, 50 mM Tris-HCl [pH 7.5]) (39). Portions (20 μl) of the dilutions were spotted onto a lawn of the indicator strain Y. enterocolitica 13169 (44). PY100 was purified by repeated single plaque isolation.
To determine the titer of PY100 preparations, 0.1-ml portions of the phage dilutions were mixed with 0.1 ml of the overnight culture of strain 13169, followed by incubation for 15 min. After the addition of 3 ml of 48°C warm soft agar medium (Luria-Bertani [LB] medium with 10 mM CaCl2, 10 mM MgSO4, and 0.6% agar), the mixture was poured on LB solid medium and incubated overnight at 37°C.
The host range of PY100 was determined by spotting 20 μl of phage suspensions containing approximately 108, 106, or 103 PFU ml−1 (determined on strain 13169) on lawns of test bacteria, followed by incubation overnight at 37°C. The overlays were prepared with 0.1 ml overnight cultures grown in LB broth that were mixed with 3 ml of LB soft agar (see above). Each strain was tested three times, except for Y. pestis strains that were tested only twice. One-step growth curves were carried out with mid-exponential-phase cultures of strain 13169 with 5 × 106 PFU of PY100. Samples were taken in intervals of 5 min.
Electron microscopy.
Phages were isolated in CsCl step gradients as described earlier (45). Transmission electron microscopy was performed with a Philips 400 electron microscope. Negative staining of phage preparations were performed with 1% uranyl acetate or 1% phosphotungstic acid (pH 6.8) (44).
Cloning and nucleotide sequence determination.
A random or “shotgun” library containing 1.4-kb DNA fragments was constructed using a previously described fast nebulization method (35). The vector pCR4Blunt-TOPO (Invitrogen) was used according to the manufacturer's recommendations. Plasmid DNA was prepared by using the QIAprep 96 Turbo BioRobot kit (Qiagen) on a BioRobot 8000 (Qiagen). Cycle sequencing reactions from plasmid inserts were performed by using an ABI Prism BigDye terminator cycle sequencing kit (Applied Biosystems) on a GeneAmp PCR System 9700 (Applied Biosystems). Sequence reactions were analyzed on an ABI Prism 3700 DNA Analyzer (Applied Biosystems). Sequencing was continued until an eightfold coverage of sequence data was reached. Assembly of sequences was performed by using the SeqMan module of the Lasergene software (DNASTAR, Inc., Madison, WI). Persisting gaps were closed by primer walking on genomic DNA.
Open reading frame (ORF) prediction and initial analysis was performed with GLIMMER (HUSAR package at http://genius.embnet.dkfz-heidelberg.de/menu/w2h/w2hdkfz/). In addition, the Find ORF feature of SeqEdit and Coding Prediction-Borodovsky method of GeneQuest (DNASTAR) was used to visually scan the sequence for potential genes (cutoff, 90 bp). Putative translated proteins were scanned for homologues using BLASTP and Psi-BLAST (2, 3) at http://www.ncbi.nlm.nih.gov. A tRNA search was performed with tRNAscan (http://www.genetics.wustl.edu/eddy/tRNAscan-SE) (28). A terminator search was performed with the tool TERMINATOR search (HUSAR package). Further analyses were carried out with the software package of MacVector (version 7.0; Oxford Molecular Group).
Nucleotide sequence accession number.
The nucleotide sequence of the PY100 genome was submitted to EMBL data library under accession number AM076770.
SDS-PAGE and mass spectrometric analysis of PY100 proteins.
The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system as described by Laemmli (26) was used. Samples were suspended in loading buffer (Bio-Rad, Munich, Germany), boiled for 10 min, and electrophoresed on a 12% (wt/vol) polyacrylamide gel at 20 mA at 8°C. Proteins were visualized by using Coomassie brilliant blue R-250 (Bio-Rad, Munich, Germany) staining. For further analysis the bands of interest were excised, and in-gel digestion was performed according to a standard procedure. For peptide sequence determination, liquid chromatography-mass spectrometry (LC-MS) measurements were performed using a Qstar XL hybrid mass spectrometer (Applied Biosystems, Foster City, CA) coupled to an Ultimate Nano-HPLC system (LC Packings, Amsterdam, The Netherlands) by using a nano-electrospray source. Tandem MS spectra were acquired during high-pressure liquid chromatography run using the data-dependent acquisition abilities of the Analyst software package (Applied Biosystems). Peptide sequences were assigned manually using Bioanalyst software (Applied Biosystems), and peptide sequences were aligned to the phage genome using MacVector (Accelrys, Cambridge, United Kingdom).
Analysis of pac fragments.
Virion DNA was digested with BglII and run on an 0.8% agarose gel. The pac fragment was cut out and digested with DraI. The resulting DNA fragments were ligated with pLitmus38 which had been digested with EcoRV to have blunt ends. The ligation mixture was introduced into E. coli DH5α, and AmpR transformants were selected. Plasmid DNA was prepared and hybridized to a PCR probe (345 bp) generated with the primers PY100-5 (5′-CAAGTAGCGATGGTTCTATGAGTCC-3′) and PY100-6 (5′-CTTGCTTTGCGTCTTCCAGTG-3′). The probe was labeled with fluorescein, and hybridization was carried out according to standard procedures (39). Hybridizing plasmids were sequenced.
RESULTS AND DISCUSSION
Isolation and host range of PY100.
PY100 was isolated from pig manure collected on a farm in Germany by single plaque purification on Y. enterocolitica 13169 (43). Since the phage formed clear large plaques at 37°C, it was chosen for further experiments. We found that the incubation temperature during the development of the bacterial lawn influenced the number of plaques significantly. Plaque titers of the same PY100 preparation were lower in bacterial lawns after incubation at 30°C or 20°C compared to 37°C, suggesting that phage infections are more efficient at 37°C.
To determine the host range of the phage a number of bacterial strains were tested (Table 1) . The host range of the phage was very broad within the genus Yersinia and included strains from the three human pathogenic species Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica. Among the susceptible Yersinia strains are Y. enterocolitica 8081 (serotype O:8), Y. enterocolitica 6471/76 (serotype O:3), Y. pseudotuberculosis YPIII, and Y. pestis KIM. In addition, we tested the phage on other species belonging to the Enterobacteriaceae: E. coli (12 strains) Shigella sp. (9 strains), Salmonella enterica (10 strains), Proteus sp. (3 strains), Enterobacter cloacae (1 strain), Citrobacter freundii (2 strains), and Serratia marcescens (1 strain). We did not detect any plaque formation under the described conditions, suggesting that the host range of the phage is restricted to Yersinia strains.
TABLE 1.
Host range of PY100a
| Organism | Source | Strain properties | No. of strains tested | No. of PY100-sensitive strains |
|---|---|---|---|---|
| Y. enterocolitica | RKI, IP | Serogroup O:3 | 10 | 9 |
| Y. enterocolitica | RKI, IP | Serogroup O:5,27 | 5 | 5 |
| Y. enterocolitica | RKI, IP | Serogroup O:9 | 9 | 8 |
| Y. enterocolitica | RKI, IP | Serogroup O:8 | 6 | 4 |
| Y. enterocolitica | RKI, IP | Biogroup 1A, various serogroups, NT | 20 | 15 |
| Y. pseudotuberculosis | RKI, IP | Serogroups I to VII | 10 | 10 |
| Y. pestis | KIM, EV76 | 2 | 2 | |
| Y. intermedia | RKI, IP | Various serogroups, NT | 10 | 6 |
| Y. kristensenii | RKI, IP | Various serogroups, NT | 10 | 6 |
| Y. frederiksenii | RKI, IP | Various serogroups, NT | 10 | 4 |
| Y. mollaretii | RKI, IP | Various serogroups, NT | 10 | 3 |
| Y. rohdei | RKI | 1 | 0 |
Tests were performed by spotting 20 μl of phage suspensions containing approximately 108, 106, or 103 PFU/ml on lawns of test bacteria (100 μl of overnight culture mixed with 3 ml of LB soft agar). Plaques were determined after overnight incubation at 37°C. Abbreviations: RKI, strain collection of the Robert Koch Institute; IP, strains from the Institute Pasteur, Paris, France. NT, strains not typeable with standard sera.
Morphology of phage.
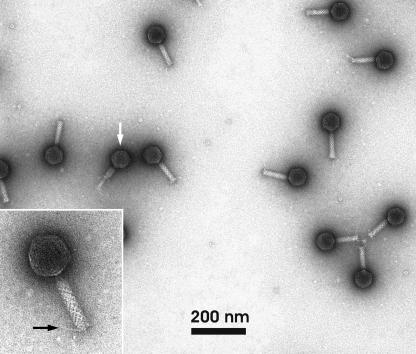
Electron micrographs revealed that PY100 virions possess an icosahedral capsid and a contractile tail with fibers (Fig. 1). The diameter of the capsid is approximately 70 nm, and the length of the tail approximately 80 nm. The capsid contains double-stranded DNA with a length of 50,291 bp. Based on these properties the phage belongs to the family of Myoviridae (1).
FIG. 1.
Electron micrograph of CsCl-purified phage PY100 particles. The white arrow indicates phage with a contracted tail; the black arrow (inset) indicates tail fiber.
Propagation of PY100 and burst size.
Occasionally, plaques of PY100 on bacterial lawns appeared to be turbid. Therefore, we isolated bacteria from these plaques and investigated whether the bacteria might have become lysogenic. After repeated streaking on agar plates, single colonies were picked and grown in liquid culture. Phage induction was carried out by mitomycin C addition in the early logarithmic phase. However, we never succeeded in isolating viable phages from such liquid cultures, and bacteria from these cultures tested for phage susceptibility always allowed plaque formation. Thus, we conclude that PY100 does not integrate into the chromosome and replicates only via the lytic cycle.
To estimate the burst size of the phage, single growth experiments were carried out with strain Y. enterocolitica 13169 (30). The lytic cycle lasts approximately 30 min, and the burst size deduced from these experiments was estimated to be about 120 phages per infected cell at 37°C (data not shown).
Genome analysis of PY100.
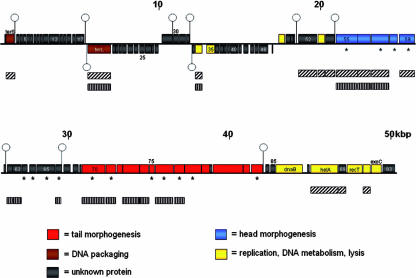
The complete nucleotide sequence of the PY100 genome (GenBank accession AM076770) was determined by sequencing random DNA clones of virion DNA fragments as described in Materials and Methods. The sequence was assembled into a single sequence of 50,291 bp in length, which is predicted to code for 93 genes (Fig. 2). To define the left end of the circular permutated genome, the order of the genes was arranged commencing with the small terminase gene. This arrangement facilitates the comparison with genomic maps of other phages with the gene order “terminase - head - tail” (37). A description of annotated genes is summarized in Table 2; no tRNA genes were found in the PY100 genome.
FIG. 2.
Genetic map of the PY100 genome. Colored boxes above the black line represent ORFs of the positive strand; colored boxes below the black line represent ORFs of the negative strand. Colors indicate functional assignments. Rho-independent terminators are indicated by hairpins (above the sequence, terminators on the positive strand; below the sequence, terminators on the negative strand). Asterisks indicates ORFs whose gene products were detected by MS. Filled boxes with diagonal and vertical lines indicate regions of homology to the genome of phage T1 and phage AaΦ23, respectively.
TABLE 2.
Selected genes of phage PY100a
| ORF | Gene | Start-stop | Protein size (aa)b | Potential function (gene) | Significant matches (source) |
|---|---|---|---|---|---|
| 2 | ORF2 | 491-1045 | 184 | Terminase small subunit (terS) | Terminase small subunit (enterobacterium phage P22) |
| 18 | ORF18 | 6909-5440 | 489 | Terminase large subunit (terL) | Putative terminase large subunit TerL (bacteriophage AaΦ23) |
| 34 | ORF34 | 12756-12109 | 215 | Dam-methylase (dam) | Phage DNA methyltransferase (Sodalis glossinidius strain) (enterobacterium phage T1) |
| 35 | ORF35 | 13367-12804 | 187 | Single-stranded DNA binding (ssb) | Single-stranded DNA-binding protein (Vibrio sp. strain MED222) |
| 48 | ORF48 | 17422-17838 | 127 | Endolysine | Bacteriophage P7-related protein (Yersinia pseudotuberculosis IP 32953) |
| 52 | ORF52 | 18620-19810 | 396 | Hypothetical protein T1p16 (enterobacterium phage T1) | |
| 53 | ORF53 | 19854-20312 | 152 | Polynucleotide kinase/phosphatase | Putative polynucleotide kinase/phosphatase (enterobacterium phage T1) |
| 54 | ORF54 | 20235-20912 | 195 | Hypothetical protein T1p62 (enterobacterium phage) | |
| 55 | ORF55 | 20979-22340 | 453 | Portal protein | Hypothetical protein (XF1571 Xylella fastidiosa 9a5c) (T1p52 enterobacterium phage T1) |
| 56 | ORF56 | 22300-23133 | 277 | Head protein | Putative bacteriophage protein (Acinetobacter sp. strain ADP1) (T1p51 enterobacterium phage T1) |
| 57 | ORF57 | 23145-24305 | 386 | Head protein | Hypothetical protein (lin1728 Listeria innocua Clip11262) (T1p50 enterobacterium phage T1) |
| 58 | ORF58 | 24308-24796 | 162 | Hypothetical protein lin1727 (Listeria innocua Clip11262) | |
| 59 | ORF59 | 24886-25893 | 335 | Head protein | Hypothetical protein (CGSHi22121_00927) (Haemophilus influenzae 22.1-21) (T1p47 enterobacterium phage T1) |
| 63 | ORF63 | 27035-27403 | 122 | ||
| 64 | ORF64 | 27400-27930 | 176 | ||
| 65 | ORF65 | 27975-29114 | 379 | Bacteriophage Felix 01 | |
| 66 | ORF66 | 29118-29546 | 142 | Hypothetical protein PD1187 (Xylella fastidiosa Temecula1) | |
| 70 | ORF70 | 30763-32271 | 502 | Tail tape measure protein | Phage-related protein (Xylella fastidiosa Ann-1) |
| 71 | ORF71 | 32271-32918 | 215 | Putative bacteriophage protein (Ralstonia eutropha JMP134) | |
| 73 | ORF73 | 33313-34380 | 355 | Phage-related protein (Xylella fastidiosa Dixon) | |
| 74 | ORF74 | 34358-34951 | 197 | Baseplate protein | Conserved hypothetical protein (Xylella fastidiosa Dixon) |
| 75 | ORF75 | 34948-35304 | 118 | Hypothetical protein CGSHi22121_00997 (Haemophilus influenzae 22.1-21) | |
| 76 | ORF76 | 35315-36490 | 391 | COG3299: uncharacterized homolog of phage Mu protein gp47 (Magnetospirillum magnetotacticum MS-1) | |
| 77 | ORF77 | 36493-37158 | 221 | Hypothetical protein plu2875 (Photorhabdus luminescens subsp. laumondii TTO1) | |
| 78 | ORF78 | 37151-38206 | 351 | Tail fiber protein | Hypothetical protein (Ecol5_01002866 Escherichia coli 53638) |
| 79 | ORF79 | 38179-38736 | 185 | Tail fiber assembly protein | Putative bacteriophage protein (Yersinia pseudotuberculosis IP 32953) |
| 81 | ORF81 | 38887-40809 | 640 | Tail fiber protein | Tail fiber protein (Yersinia phage PY54) |
| 82 | ORF82 | 40794-41225 | 143 | Tail fiber assembly protein | Tail fiber assembly protein (Yersinia phage PY54) |
| 83 | ORF83 | 41268-42035 | 255 | Tail protein | gp37 long tail fiber, distal subunit (enterobacterium phage T4) |
| 86 | ORF86 | 42787-44475 | 562 | Replicative DNA helicase (dnaB) | Hypothetical protein PBPRB1439 (Photobacterium profundum SS9) |
| 88 | ORF88 | 44964-46682 | 572 | NTP-dependent helicase (helA) | Putative ATP-dependent helicase (enterobacterium phage T1) |
| 89 | ORF89 | 46682-47185 | 167 | Hypothetical protein T1p21 (enterobacterium phage T1) | |
| 90 | ORF90 | 47215-48213 | 332 | DNA recombination protein (recT) | Phage-related DNA recombination protein (Bordetella bronchiseptica RB50) |
| 91 | ORF91 | 48214-48702 | 162 | HNH endonuclease (endC) | Bacteriophage Felix 01 |
| 92 | ORF92 | 48702-49400 | 232 | Exonuclease | Exonuclease (Escherichia coli O157:H7 strain Sakai) |
ORFs whose gene products were detected by MS are indicated in boldface type.
aa, amino acids.
The left side of the genome (nucleotides 1 to 20,000) contains mostly smaller ORFs. Homology to known proteins was found only for a few gene products. ORF2 encodes a putative protein with significant similarity to the P22 gene 3 product (small terminase) of Salmonella phage P22 (33, 50). The putative gene product of ORF18, located on the minus strand, shows similarity to the large terminase subunits of a number of phages. The arrangement of ORF2 and ORF18 is unusual since most terminase genes are adjacent and located on the same DNA strand (11). In PY100 both putative terminase genes are followed by a transcription terminator. A number of small ORFs, whose putative gene products have no similarity to known proteins, are located between ORF2 and ORF18. The next ORFs coding for gene products with significant homology to known proteins are ORF34 and ORF35 on the minus strand. Their products are similar to DNA methyltransferases and single-stranded DNA-binding proteins, respectively. The putative gene product of ORF48 may be an endolysin. Although few functional assignments could be made for genes of the left side of the phage genome, the arrangement of rho-independent terminators (indicated by hairpins, see Fig. 2) support the ORF analysis. Table 3 displays the sequences of potential terminators.
TABLE 3.
Potential rho-independent terminators in the PY100 genome
| Position | Direction | ORF no.a | Sequenceb |
|---|---|---|---|
| 1055 | + | 2 | AAAGCCCTTGACATTGTTCAGGGGCTTTTCTATTATATGCGC |
| 5407 | + | 17 | GCCGGATCGGGTTTCCCTTTTCCGGCTTTTCTTTTATCCCCAC |
| 5433 | − | 18 | AGCCGGAAAAGGGAAACCCGATCCGGCTCTTTTTTTTGTT |
| 10843 | + | 29 | AAGCGCCCTTAATTGGGCGCTTTTACTATCAACTT |
| 11922 | + | 32 | AAGGGGCCATATGGCCCCTTTCTTTTGTCTATTCAACTG |
| 11941 | − | 33 | AAGGGGCCATATGGCCCCTTAATCGTTTTTAGCGCTG |
| 18502 | + | 51 | TGCCCCGCTCCGGCGGGGTATTTTTTTATCTGAAA |
| 20929 | + | 54 | CCCGTAAGCTATAATAGTTTACGGGCTTATTTATTTTTCGAG |
| 26094 | + | 60 | GGGGAGAGGCTACGGCCTTTCCCCTTTTTTATATGGAGGC |
| 29554 | + | 66 | AAAAGGGGTTGACTTCGGTCAATCCCTTTTCTATTATGCGC |
| 42038 | + | 83 | AAAGGGGCTTCGGCCCCTTTATTTTTTTATAAA |
That is, the preceding ORF.
Underlining indicates dyad symmetries that potentially yield stable hairpin structures in the RNA transcript.
The putative gene products of ORF52, ORF53, and ORF54 display similarity to the three enterobacterial phage T1 proteins T1p16, T1p64, and T1p62, respectively. Protein T1p64 may be a polynucleotide kinase or phosphatase (37), while for the protein homologs to the ORF52 and ORF54 gene products no functions are known.
ORF55 to ORF83, spanning the region from nucleotides 20900 to 42300, encode the structural proteins of the virion particle and are also discussed with the results of the proteomic investigation (see below). ORF55 may encode a portal protein and gene products of ORF56, and ORF57 and ORF59 are related to different head proteins of other phages. Protein homologs to the ORF61, ORF62, and ORF66 gene products exist in phage AaΦ23 of Actinobacillus actinomycetemcomitans, although no function of these gene products is known (36). ORF70 is related to phage tail tape measure proteins, and ORF74, ORF78, ORF79, ORF81, ORF82, and ORF83 encode proteins related to different tail proteins, such as baseplate proteins, tail fiber proteins, and the tail fiber assembly proteins of other phages (Table 2).
The right side of the genome from ORF84 to ORF93 contains some genes that encode proteins that are known to play a role in DNA metabolism, including synthesis, degradation, and recombination. ORF86 encodes a protein with homology to the replicative helicase RepA of gram-negative plasmids belonging to the DnaB-like helicase family. These proteins unwind double-stranded DNA into single-stranded intermediates fueled by nucleoside triphosphate hydrolysis (29). The putative protein encoded by ORF88 shows high homology to the T1p22 helicase, another protein involved in DNA unwinding and synthesis (37). Interestingly, the putative gene product of ORF89 is similar to the putative T1p21 protein, for which no function is known thus far. ORF90 encodes a DNA recombinase protein, and ORF91 encodes an endonuclease related to the T1p63 gene product of phage T1. The latter protein is a zinc-dependent HNH homing endonuclease, whose function in phages is unknown. It has been proposed that these endonuclease genes in phage genomes can be considered as analogous to insertion or transposon elements in bacterial genomes (37). ORF92 encodes an exonuclease that may be involved in host DNA degradation.
Proteomic analysis of virion particles.
The bioinformatic analysis of the PY100 genome suggested that the region from ORF55 to ORF83 harbors most of the genes encoding the proteins building the mature virion particle. From Coomassie blue-stained two-dimensional gels, one protein encoded by ORF59 could be identified, which was isolated from several spots, indicating that it might be modified posttranslationally (data not shown). We therefore decided to analyze one-dimensional SDS gels, to cut out all visible bands and perform an MS analysis after trypsin digestion. The LC-MS analysis of eight protein bands visualized by Coomassie staining (Fig. 3, bands A to H) led to the identification of 15 structural proteins of the phage PY100. The sequence coverage and the number of identified peptides are given in Table 4. We were able to identify up to three proteins in one band by using the LC-MS system with integrated tandem MS capabilities. In some cases, peptides confirming the N terminus of the PY100 proteins were found (see Table 4).
FIG. 3.

SDS-PAGE analysis of proteins from PY100 virions (different protein concentrations were loaded from left to right). Capital letters indicate protein bands excised for MS analyses.
TABLE 4.
PY100 virion proteins detected by MS
| Banda | Observed avg mass (kDa) | ORF | Theoretical avg mass (kDa) | Amino acids | No. of peptides found | Sequence coverage (%) | Putative functionb | N-terminal peptidec |
|---|---|---|---|---|---|---|---|---|
| A | 51 | 70 | 53.9 | 502 | 7 | 15.1 | Tail-length tape measure protein | |
| B | 48 | 55 | 50.0 | 453 | 7 | 16.6 | Portal protein | + |
| 48/40 | 76 | 42.7 | 391 | 4 | 13.0 | - | ||
| C | 40 | 65 | 42.6 | 379 | 3 | 5.3 | - | |
| D | 38 | 78 | 37.7 | 351 | 6 | 21.1 | Tail fiber | |
| E | 35 | 59 | 37.2 | 335 | 15 | 30.7 | Head protein | |
| 35 | 83 | 27.7 | 255 | 7 | 19.2 | Tail fiber | ||
| F | 28 | 77 | 24.8 | 221 | 7 | 23.9 | - | + |
| 28 | 71 | 23.7 | 215 | 4 | 19.0 | - | + | |
| G | 19 | 57 | 42.6 | 386 | 3 | 7.4 | Head protein | |
| 19 | 64 | 19.8 | 176 | 8 | 44.3 | - | ||
| 19 | 58 | 17.2 | 162 | 8 | 38.8 | - | + | |
| H | 16 | 66 | 15.2 | 142 | 7 | 32.4 | - | |
| 16 | 63 | 14.2 | 122 | 3 | 22.1 | - | ||
| 16 | 75 | 13.1 | 118 | 1 | 9.3 | - |
See Fig. 3.
-, The annotation was “phage related, unknown function.”
+, Peptides were found corresponding to the deduced N terminus.
Based on homology to known phage proteins, the detected PY100 proteins of ORF55, ORF57, ORF58, and ORF59 form the capsid. The head protein encoded by ORF59 is the highly abundant protein seen in band E (Fig. 3). The proteomic approach proves that some ORFs are indeed protein coding sequences, although the proteins have no functionally assigned homologs in the database, e.g., proteins of ORF63, ORF64, and ORF65 (see Table 3). A homolog of the ORF66 protein with unknown function is encoded in the genome of phage AaΦ23. The ORFs downstream (ORF70 to ORF83) encode the tail proteins with its various structures, such as tail length tape measure (ORF70 protein), fibers, base plate, etc. (see Table 2). In most cases, the observed molecular masses correspond well to the theoretical molecular size deduced from the corresponding ORF. However, ORF57 would encode a protein of 366 amino acids with a size of 43.3 kDa, while peptides of the ORF57 gene product were found in a protein band with an observed mass of 19 kDa. Since the detected peptides are from the N-terminal part of the protein (amino acid residues 21 to 28, 36 to 44, and 161 to 172), no obvious explanation for this observation can be given. It is possible that the C-terminal part of the protein is proteolytically cut off during assembly of the phage particle, as is described for some phage proteins, e.g., gp5 protein of phage T4 (52).
Relationship of PY100 to other bacteriophages and prophage sequences.
BLASTN searches with the PY100 sequence against the database (nucleotide collection [nr/nt]; http://www.ncbi.nlm.nih.gov/BLAST/) yielded only short sequences (<50 bp) of similarity. All functional assignments were made on the basis of translated ORFs against the database (BLASTX). Thus, the PY100 genome harbors several gene products with homology to proteins of the enterobacterial phage T1 (37). Gene products of ORFs showing homology to the T1 proteins are the capsid proteins and proteins involved in replication and DNA packaging (Fig. 2). Phage T1 is a Siphoviridae, a phage with a flexible, noncontractile tail, whereas PY100 has a contractile tail. Another phage possessing several putative gene products with homology to the PY100 is the temperate bacteriophage AaΦ23 of A. actinomycetemcomitans, which has a contractile tail like PY100. The homologous gene products of phage AaΦ23 are putative tail and head proteins (36).
The BLASTX search of the PY100 ORFs revealed some genomic regions in other gram-negative bacteria that encode putative gene products with significant homology to the PY100 structural head and tail proteins. These genomic regions are prophage remnants, for example, two genomic regions in the S. enterica strain CT18. Thirteen putative gene products of the region from STY1058 to STY1073 and eleven gene products of the region from STY2038 to STY2015 share homology with some of the proteins from ORF55 to ORF77 of PY100 and have a similar gene order (32). Other putative gene products of prophage regions with homologies to the head and tail PY100 proteins are found in Xylella fastidiosa strain 9a5c (region from genes XF1571 to XF1595 and region from genes XF1676 to XF1706, accession no. NC_002488 [41]). In Photorhabdus luminescens subsp. laumondii TTO1, several genes of phage origin encode putative proteins with homology to PY100 proteins (accession no. NC_005126 [14]).
Packaging mechanism of PY100.
In an attempt to elucidate whether cos sites were present in the PY100 genome, DNA of the phage was ligated and digestions with selected restriction enzymes were carried out. When the restriction patterns of unligated and ligated phage DNA obtained with the same enzyme were compared, no fragments arising from ligation of the cos sites were detectable.
The analysis of the genomic sequence indicated a gene product (encoded by ORF2) with significant similarity to the product of gene 3 of the Salmonella phage P22 (33, 50). This protein is part of the terminase complex of phage P22 and initiates the packaging of phage DNA by cutting the concatemeric P22 DNA close to or at the pac site. The pac site is 22 bp long with a consensus sequence of 12 bp (AAGATTTATCTG) and lies within the coding region of the P22 gene 3. After the initial endonucleolytic cut, packaging of DNA proceeds in one direction and sequentially DNA from the concatemer is packaged by a headful mechanism (9, 51). As a consequence of the packaging process starting at a pac site, a restriction fragment with reduced abundance is found in agarose gels after digestion of virion DNA (“pac fragment”) with suitable restriction enzymes (23). The pac fragment is also visualized by hybridization when using a hybridization probe consisting of sequences downstream from the pac site.
In an experiment analogous to that used to examine P22, we performed restriction enzyme analysis of the PY100 virion DNA and hybridization studies. Analysis of the restriction patterns of the PY100 DNA revealed the presence of the expected restriction fragments plus an additional fragment with a lower intensity. Since the pac site of P22 is located within the coding region of gene 3, we deduced primers for a 348-bp hybridization probe from the PY100 sequence (primers PY100-5 and PY100-6) spanning the 3′ region of ORF2 and its downstream region (see Fig. 5). Hybridization studies with a labeled PCR probe revealed two positive bands in each digest (Fig. 4). One of these hybridizing bands corresponded to the band with lower abundance and results from initiation of DNA packaging at the pac site (“pac fragment”). The second band was the expected restriction fragment derived from the genomic sequence containing the probe sequence (Fig. 5).
FIG. 5.
Generation of pac fragments. (A) Cutting of concatemeric DNA starting from the pac site (○), with sequential cuts according to the headful mechanism. (B) Partial genomic map of PY100 showing the position of the putative pac site (boxed nucleotides) and selected restriction sites of enzymes generating the pac fragments (see also Fig. 4). The black box indicates the PCR probe (primers PY100-5 and PY100-6) that was used for hybridization.
FIG. 4.
(A) Detection of pac fragment. The left panel shows a 0.8% agarose gel of restriction of PY100 genomic DNA with BglII (lane 1), SalI (lane 2), Eco147I (lane 3), and PvuI (lane 4). Lane M contains lambda DNA digested with Eco130I. The right panel shows a Southern blot with PCR probe generated with the primers PY100-5 and PY100-6 (see Fig. 5). (B) Sizes of restriction fragments containing ORF2 and pac fragments (in base pairs). The positions of restriction sites are given according to PY100 numbering. Asterisks indicate sites at the 3′ end of pac fragments. Enzymes SalI and Eco1471 have only one recognition site in the PY100 genome.
To determine the position of the pac site in the PY100 sequence more precisely, an alignment of the PY100 ORF2 DNA sequence and the gene 3 sequence of P22 was performed (data not shown). The overall similarity of the two coding regions is 39% (four gaps introduced). A nucleotide sequence identical to the pac consensus sequence of P22 (51) is not present in the coding region of ORF2. Allowing mismatches, two sequences can be found in which 8 of 12 bp are identical to the pac consensus sequence of P22. One of these sequences (AAttTTTAcCaG) is at a similar position in the coding sequence of PY100 ORF2 (approximately the middle of the coding region, positions 821 to 833 of the genomic sequence) as the pac site in gene 3 of P22 (position 269; length of coding sequence, 489 bp) and may serve as a recognition site for the terminase complex (Fig. 5).
Since another short nucleotide sequence has also been proposed to function as a secondary pac site for the P22 gp3 protein (10, 34), we performed an additional search. The putative secondary pac site of P22 was assumed to consist of nine nucleotides and is located 110 bp downstream of the consensus pac site in the gene 3 coding sequence. We also found a similar sequence (seven out of nine identical) in the corresponding region of ORF2 of PY100 (160 bp downstream of the pac site; data not shown). The functionality of this secondary pac site, however, was questioned later (51).
In P22 the pac site serves as a packaging recognition signal for the terminase complex. The initial cleavage of the concatemeric DNA, however, takes place in six “end” regions that span 120 bp within the coding region of gene 3. The end regions are separated by approximately 20 bp or multiples of 20 bp, and individual cuts might be spread over up to 3 to 5 bp (9). The pac consensus recognition site lies between end regions 3 and 4.
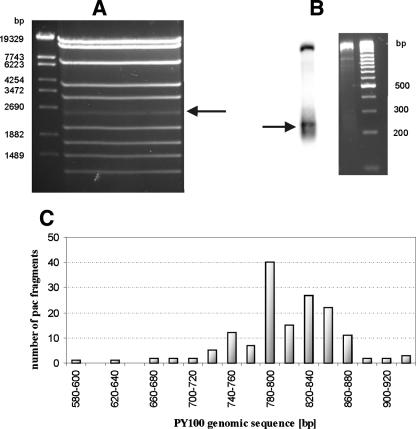
To determine the initial cleavage site of the terminase reaction in the PY100 genome, the pac fragment obtained after BglII digestion of virion DNA was cut out from agarose gels (Fig. 6A). After digestion with DraI and hybridization with the PCR probe obtained with the primers PY100-5 and PY100-6, the hybridization signal indicated a variable length of the pac fragment of PY100 (Fig. 6B). The fragment was cloned into a standard vector, and a number of recombinant plasmids were sequenced. In total, the molecular ends formed at the packaging initiation site of 154 pac fragments were determined. Figure 6C shows the distribution of the cleavage sites along the genomic sequence of PY100. All sites are within the coding region of ORF2 (small terminase homolog), and most of the cuts (87%) lie within 140 bp (from positions 740 to 880). The distribution of the cleavage sites does not show any obvious clustering, nor does the analysis of the nucleotides at the sites reveal any significant structures. The putative pac site at positions 821 to 833 lies in the middle of the cleavage sites region, as is also the case for the pac site of P22 (5, 10).
FIG. 6.
Determination of the position of end sites. (A) BglII digest of PY100 DNA. The pac fragment was excised (arrow). (B) Digest of the BglII pac fragment with DraI and a Southern blot with probe (see Fig. 5B). The arrow indicates fragments with end sites. (C) Distribution of initial cuts within the coding region of ORF2 (positions are according to PY100 sequence numbering).
PY100 for genetic studies of Yersinia?
The similarity of the packaging mechanism of PY100 and P22 suggests that PY100 might be able to perform transduction in Yersinia. Bacteriophage P22 was the first phage shown to be able to perform generalized transduction (53). During the assembly of progeny particles host DNA is “incorrectly” recognized for packaging by the P22 gene 3 protein in ca. 2% (15). The particles thus formed can inject a contiguous fragment of host DNA into a susceptible bacterium. For this reason, P22 can move genes from one bacterium to another and is used in genetic studies of S. enterica serovar Typhimurium Given its wide host range, PY100 could be a useful genetic tool for Yersinia.
If PY100 should be used for genetic experiments, a high plating efficiency resulting in comparable titers on different recipient strains would be preferable. It is known that phage particles growing well and efficiently on one strain of bacteria are often unable to do so in other strains of the same bacterial species due to restriction modification systems. We investigated the plating efficiency of PY100 preparations on 11 Y. enterocolitica strains belonging to the four most important pathogenic serotypes (O3, O5,27, O8, and O9; Table 5). From each strain phage lysates were prepared and tested on the donor and on all other strains, yielding 121 (11 × 11) phage titer experiments. We observed no restriction of phage growth between the O3 and O5,27 strains. One O8 strain (ATCC 9610) and two O9 strains restricted phage growth to some extent; however, in total only in 4 cases (out of 121) did lysates not result in plaque formation. These experiments indicate that PY100 has a high plating efficiency, which is a prerequisite for its use as genetic tool.
TABLE 5.
Efficiency of plaque formation of PY100 in Y. enterocoliticaa
| Recipient strain | Efficiency of plaque formation of PY100 phage lysate (host strain) for serotype:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| O3, strain:
|
O5,27, strain:
|
O9, strain:
|
O8, strain:
|
||||||||
| 2* | 16* | 738/84* | 83/88 | 44 | 97 | 9610* | 8081 | 8 | 36 | 38 | |
| 2* | 100% | + | + | + | + | + | + | + | + | + | + |
| 16* | + | 100% | + | + | + | + | + | + | + | + | + |
| 738/84* | + | + | 100% | + | + | + | + | + | + | + | + |
| 83/88* | + | + | + | 100% | + | + | + | + | + | + | + |
| 44* | + | + | + | + | 100% | + | + | + | + | + | + |
| 97* | + | + | + | + | +/- | 100% | + | + | + | + | + |
| 9610† | Low | - | Low | Low | - | +/- | 100% | Low | +/- | +/− | +/- |
| 8081 | + | + | + | + | + | + | + | 100% | + | + | + |
| 8* | + | + | + | + | +/- | + | + | + | 100% | + | +/- |
| 36* | +/- | Low | + | +/- | - | +/- | + | +/- | +/- | 100% | +/- |
| 38* | +/- | Low | + | +/- | - | +/- | + | + | +/- | +/- | 100% |
PY100 preparations derived from 11 host strains belonging to the main pathogenic serotypes O3, O5,27, O8, and O9 were prepared. Efficiency was determined as follows. The PFU titer of lysate titered on the same strain was set to 100% (100% values). All PY100 titers on host strains were greater than 107 PFU ml−1; only PY100 preparations derived from strains 36 and 38 were 5 × 105 and 3 × 105, respectively. Key: +, approximately the same PFU on the recipient strain as on the host strain; +/−, 1 to 10% PFU on the recipient strain compared to the PFU on the host strain; Low, <1% PFU on the recipient strain compared to the PFU on the host strain; −, no PFU. *, Y. enterocolitica RKI strain collection; †, Y. enterocolitica ATCC 9610.
Conclusions.
PY100 is a lytic phage with a broad host range in Yersinia. The genome sequence revealed that several gene products possess homologies to proteins of the lambdoid phages T1 and aaphi23. According to the modular theory of phage evolution, phages are built from interchangeable units that can be multigenic and fulfill homologous functions but may lack any sequence similarity (8). From this point of view, PY100 possesses some modules that are known from other phages, especially the units that encode the structural proteins of the head and tail. Some of these structural proteins were also identified in a proteomic analysis.
However, surprisingly, a significant portion of ORFs located on the left arm of the PY100 genome encode gene products for which at present no functional assignment can be made. ORFs encoding known integrases and repressor/antirepressor proteins were not identified, indicating that PY100 does not lysogenize its hosts and confirming the experimental observations. This is a good precondition if one considers antimicrobial strategies such as phage therapy using PY100. The ability of PY100 to lyse susceptible strains at 37°C is remarkable, since most described yersiniophages are propagated at temperatures below 30°C (6, 24, 25, 30).
We applied PY100 phage preparations by the oral route in a mouse model with enteropathogenic Y. enterocolitica O3. These trials, however, did not yield satisfying results since it was not possible to prevent the colonization of the gut by Y. enterocolitica (43). Nevertheless, PY100 may still be an interesting tool for antimicrobial strategies, since the efficacy of phage treatments depends heavily on the route (oral, intramuscular, aerosol inhalation, etc.) and/or the frequency of the phage application (4). Optimal conditions for PY100 applications may have yet to be found. Antimicrobial strategies using lytic phages against Y. pestis is still in consideration, especially if antibiotic-resistant strains of this human pathogen would be used as a bioterror agent.
The genome sequence analysis suggested that PY100 uses a headful packaging strategy starting at a pac recognition site in the concatemeric DNA formed by rolling-circle replication. This mechanism could be confirmed and a clustering of cuts around a putative pac site within ORF2 was found. Based on the similarity to the packaging strategy of Salmonella phage P22, it seems possible that PY100 can be used for transduction. Lytic Yersinia phages from environmental sources have been shown to perform generalized transduction of small plasmids (19). Plasmid transduction with phage P22 was also reported either by homologous recombination between phage DNA and plasmid DNA that harbored P22 sequences (cointegrate formation), or when a pac-like sequence was present on a plasmid that can be packaged in a headful mechanism from concatemeric DNA (40). It seems feasible to develop PY100 phage derivatives that can be used for transduction.
Acknowledgments
We thank M. Özel and G. Holland, Robert Koch Institute, for the electron micrographs and S. Beck, University of Edinburgh, for support in delivering the PY100 sequence to the EBI database. We are grateful to S. Hertwig for critical reading of the manuscript.
Footnotes
Published ahead of print on 26 October 2007.
REFERENCES
- 1.Ackermann, H.-W. 2006. Classification of bacteriophages, p. 8-17. In R. Calender (ed.), The bacteriophages, vol. 2. Oxford University Press, New York, NY. [Google Scholar]
- 2.Altschul, S. F., and D. J. Lipman. 1990. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. USA 875509-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anisimov, A. P., and K. K. Amoako. 2006. Treatment of plague: promising alternatives to antibiotics. J. Med. Microbiol. 551461-1475. [DOI] [PubMed] [Google Scholar]
- 5.Backhaus, H. 1985. DNA packaging initiation of Salmonella bacteriophage P22: determination of cut sites within the DNA sequence coding for gene 3. J. Virol. 55458-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergan, T., and J. R. Norris. 1978. Bacteriophage typing of Yersinia enterocolitica. Methods Microbiol. 1225-36. [Google Scholar]
- 7.Bolin, I., L. Norlander, and H. Wolf-Watz. 1982. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect. Immun. 37506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brüssow, H., and F. Desiere. 2006. Evolution of tailed phages: insights from comparative phage genomics, p. 26-37. In R. Calender (ed.), The bacteriophages, vol. 2. Oxford University Press, New York, NY. [Google Scholar]
- 9.Casjens, S., and W. M. Huang. 1982. Initiation of sequential packaging of bacteriophage P22 DNA. J. Mol. Biol. 157287-298. [DOI] [PubMed] [Google Scholar]
- 10.Casjens, S., W. M. Huang, M. Hayden, and R. Parr. 1987. Initiation of bacteriophage P22 DNA packaging series: analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J. Mol. Biol. 194411-422. [DOI] [PubMed] [Google Scholar]
- 11.Casjens, S. R., E. B. Gilcrease, D. A. Winn-Stapley, P. Schicklmaier, H. Schmieger, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2005. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 1871091-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 1844601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieudonne, A., and R. Otto. 1931. Pest, p. 179-303. In W. Kolle (ed.), Handbuch der pathogenen Mikroorganismen, 3. Auflage. Urban & Fischer, Jena, Germany.
- 14.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, et al. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 211307-1313. [DOI] [PubMed] [Google Scholar]
- 15.Ebel-Tsipsis, J., D. Botstein, and M. Fox. 1972. Generalized transduction by phage P22 in Salmonella typhimurium. J. Mol. Biol. 71433-448. [DOI] [PubMed] [Google Scholar]
- 16.Fredriksson-Ahomaa, M., A. Stolle, and H. Korkeala. 2006. Molecular epidemiology of Yersinia enterocolitica infections. FEMS Immunol. Med. Microbiol. 47315-329. [DOI] [PubMed] [Google Scholar]
- 17.Garcia, E., J. M. Elliott, E. Ramanculov, P. S. Chain, M. C. Chu, and I. J. Molineux. 2003. The genome sequence of Yersinia pestis bacteriophage phiA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J. Bacteriol. 1855248-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greer, G. G. 2005. Bacteriophage control of foodborne bacteria. J. Food Prot. 681102-1111. [DOI] [PubMed] [Google Scholar]
- 19.Hertwig, S., A. Popp, B. Freytag, R. Lurz, and B. Appel. 1999. Generalized transduction of small Yersinia enterocolitica plasmids. Appl. Environ. Microbiol. 653862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hertwig, S., I. Klein, V. Schmidt, S. Beck, J. A. Hammerl, and B. Appel. 2003. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 331605-622. [DOI] [PubMed] [Google Scholar]
- 21.Hertwig, S., I. Klein, R. Lurz, E. Lanka, and B. Appel. 2003. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 48989-1003. [DOI] [PubMed] [Google Scholar]
- 22.Hudson, J. A., C. Billington, G. Carey-Smith, and G. Greening. 2005. Bacteriophages as biocontrol agents in food. J. Food Prot. 68426-437. [DOI] [PubMed] [Google Scholar]
- 23.Jackson, E. N., D. A. Jackson, and R. J. Deans. 1978. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 118365-388. [DOI] [PubMed] [Google Scholar]
- 24.Kawaoka, Y., T. Mitani, K. Otsuki, and M. Tsubokura. 1987. Isolation and use of eight phages for typing Yersinia enterocolitica O3. J. Med. Microbiol. 23349-352. [DOI] [PubMed] [Google Scholar]
- 25.Kiljunen, S., K. Hakala, E. Pinta, S. Huttunen, P. Pluta, A. Gador, H. Lonnberg, and M. Skurnik. 2005. Yersiniophage phiR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology 1514093-4102. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lewin, A., E. Strauch, S. Hertwig, B. Hoffmann, H. Nattermann, and B. Appel. 1996. Comparison of plasmids of strains of Yersinia enterocolitica biovar 1A with the virulence plasmid of a pathogenic Y. enterocolitica strain. Zentbl. Bakteriol. 28552-63. [DOI] [PubMed] [Google Scholar]
- 28.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niedenzu, T., D. Roleke, G. Bains, E. Scherzinger, and W. Saenger. 2001. Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol. 306479-487. [DOI] [PubMed] [Google Scholar]
- 30.Pajunen, M., S. Kiljunen, and M. Skurnik. 2000. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 1825114-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pajunen, M. I., S. J. Kiljunen, M. E. Soderholm, and M. Skurnik. 2001. Complete genomic sequence of the lytic bacteriophage phiYeO3-12 of Yersinia enterocolitica serotype O:3. J. Bacteriol. 1831928-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhill, J., G. Dougan, K. D. James, et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413848-852. [DOI] [PubMed] [Google Scholar]
- 33.Pedulla, M. L., M. E. Ford, T. Karthikeyan, J. M. Houtz, R. W. Hendrix, G. F. Hatfull, A. R. Poteete, E. B. Gilcrease, D. A. Winn-Stapley, and S. R. Casjens. 2003. Corrected sequence of the bacteriophage p22 genome. J. Bacteriol. 1851475-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petri, J. B., C. Schmidt, and H. Schmieger. 1992. Sequence comparison among DNA fragments from different sources with pac site function for the packaging apparatus of Salmonella phage P22. Intervirology 33103-108. [DOI] [PubMed] [Google Scholar]
- 35.Pohl, T. M., and E. Maier. 1995. Sequencing 500 kb of yeast DNA using a GATC 1500 direct blotting electrophoresis system. BioTechniques 19482-486. [PubMed] [Google Scholar]
- 36.Resch, G., E. M. Kulik, F. S. Dietrich, and J. Meyer. 2004. Complete genomic nucleotide sequence of the temperate bacteriophage AaΦ23 of Actinobacillus actinomycetemcomitans. J. Bacteriol. 1865523-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, M. D., N. L. Martin, and A. M. Kropinski. 2004. The genome and proteome of coliphage T1. Virology 318245-266. [DOI] [PubMed] [Google Scholar]
- 38.Russell, P., S. M. Eley, S. E. Hibbs, R. J. Manchee, A. J. Stagg, and R. W. Titball. 1995. A comparison of plague vaccine, USP, and EV76 vaccine induced protection against Yersinia pestis in a murine model. Vaccine 131551-1556. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Schmidt, C., and H. Schmieger. 1984. Selective transduction of recombinant plasmids with cloned pac sites by Salmonella phage P22. Mol. Gen. Genet. 196123-128. [DOI] [PubMed] [Google Scholar]
- 41.Simpson, A. J., et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406151-157. [DOI] [PubMed] [Google Scholar]
- 42.Skurnik, M. 1984. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Appl. Bacteriol. 56355-363. [DOI] [PubMed] [Google Scholar]
- 43.Skurnik, M., and E. Strauch. 2006. Phage therapy: facts and fiction. Int. J. Med. Microbiol. 2965-14. [DOI] [PubMed] [Google Scholar]
- 44.Strauch, E., H. Kaspar, C. Schaudinn, P. Dersch, K. Madela, C. Gewinner, S. Hertwig, J. Wecke, and B. Appel. 2001. Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl. Environ. Microbiol. 675634-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauch, E., R. Lurz, and L. Beutin. 2001. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect. Immun. 697588-7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sulakvelidze, A., Z. Alavidze, and J. G. J. Morris. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Summers, W. C. 2001. Bacteriophage therapy. Annu. Rev. Microbiol. 55437-451. [DOI] [PubMed] [Google Scholar]
- 48.Summers, W. C. 2006. Phage and early development of molecular biology, p. 3-8. In R. Calender (ed.), The bacteriophages, vol. 2. Oxford University Press, New York, NY. [Google Scholar]
- 49.Thomson, N. R., S. Howard, B. W. Wren, M. T. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall. K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G.Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vander, B. C., and A. M. Kropinski. 2000. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 1826472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, H., L. Sampson, R. Parr, and S. Casjens. 2002. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 451631-1646. [DOI] [PubMed] [Google Scholar]
- 52.Ye, N., and N. Nemoto. 2004. Processing of the tail lysozyme (gp5) of bacteriophage T4. J. Bacteriol. 1866335-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zinder, N., and J. Lederberg. 1952. Genetic exchange in Salmonella. J. Bacteriol. 64679-699. [DOI] [PMC free article] [PubMed] [Google Scholar]