Abstract
Biofilms are structured communities characterized by distinctive gene expression patterns and profound physiological changes compared to those of planktonic cultures. Here, we show that many gram-negative bacterial biofilms secrete high levels of a small-molecular-weight compound, which inhibits the growth of only Escherichia coli K-12 and a rare few other natural isolates. We demonstrate both genetically and biochemically that this molecule is the amino acid valine, and we provide evidence that valine production within biofilms results from metabolic changes occurring within high-density biofilm communities when carbon sources are not limiting. This finding identifies a natural environment in which bacteria can encounter high amounts of valine, and we propose that in-biofilm valine secretion may be the long-sought reason for widespread but unexplained valine resistance found in most enterobacteria. Our results experimentally validate the postulated production of metabolites that is characteristic of the conditions associated with some biofilm environments. The identification of such molecules may lead to new approaches for biofilm monitoring and control.
Biofilms are matrix-encased bacterial communities that develop on most surfaces and often constitute a reservoir of bacterial pathogens. Detrimental biofilms are difficult to eradicate due to a characteristic tolerance to biocides. They cause serious health and economic problems when they form on medical and industrial devices (11, 19, 24). While biofilm-associated antibiotic tolerance is considered a major physiological trait that distinguishes free cells from surface-attached cells, biofilms are also characterized by gene expression patterns that are distinctive compared to those of planktonic cultures (4, 35, 40, 41, 58). These changes are likely to result from modifications of growth conditions within biofilms, and they have been proposed to correspond to ill-understood responses triggered by the biofilm lifestyle (3, 26).
Recently, studies conducted with simplified, mixed communities composed of two bacterial species revealed that biofilm bacteria express competitive or cooperative behavior that does not take place within classical planktonic cultures (7, 22). These results suggested that biofilm-associated weaponry could contribute to the dynamics of bacterial biofilm populations. However, thus far, the production of molecules involved in antagonistic bacterial relationships within biofilms has been poorly investigated.
Here we show that the continuous-flow biofilms formed by many Escherichia coli strains and other gram-negative bacteria accumulate high levels of a small-molecular-weight compound with inhibitory activity against E. coli K-12 strains. We demonstrate both genetically and biochemically that this compound is the amino acid valine. We provide evidence that valine secretion within biofilms could be the cause of the long-known but unexplained widespread valine resistance observed for most enterobacteria. Beyond the biological significance of in-biofilm valine secretion, our results experimentally support the hypothesis that metabolites can be secreted more particularly under biofilm conditions, a finding that opens up novel research alleys for the development of markers with which to monitor biofilm formation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are listed in Table 3. Bacteria were grown at 37°C in 0.4% glucose-minimal M63B1 medium (M63B1glu) (31), unless specified otherwise, supplemented with the appropriate antibiotics, as follows: kanamycin (Km; 50 μg ml−1), chloramphenicol (Cm; 25 μg ml−1), ampicillin (Amp; 100 μg/ml), apramycin (Apr; 30 μg ml−1), or zeocin (Zeo; 50 μg ml−1). Nonessential- and essential-amino-acid solutions were purchased from Invitrogen. The nonessential-amino-acid solution contained l-alanine, l-asparagine, l-aspartic acid, l-glutamic acid, glycine, l-proline, and l-serine at a concentration of 50 μg ml−1 each. The essential-amino-acid solution contained l-arginine·HCl, l-cysteine, l-histidine·HCl·H2O, l-isoleucine, l-leucine, l-lysine·HCl, l-methionine, l-phenylalanine, l-threonine, l-tryptophan, l-tyrosine, and l-valine at a concentration of 50 μg ml−1 each. Single-amino-acid solutions were prepared in distilled water at 2,500 μg ml−1 each and used at a final concentration of 50 μg ml−1 each. For growth curve experiments or for supplementation in agar plates, filter-sterilized supernatant (20% [vol/vol]) was added to the M63B1glu. For complementation experiments, E. coli strain CFT073 genes sspA and tufA were PCR amplified using specific primers (see Table S1 at www.pasteur.fr/recherche/unites/Ggb/supmat.html) and cloned into plasmid pZE12 under the control of a synthetic plac promoter (29), leading to the plasmids pSspA and pTufA. Plasmids pSM11 and pMM100 (lacIq) were used for the overexpression of ppGpp. One millimolar isopropyl-β-d-thiogalactopyranoside (IPTG) was used for the activation of the plac promoters.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| MG1655 | Commensal E. coli K-12 containing a frameshift mutation in ilvG | Laboratory collection |
| MG1655ΔlivH | MG1655ΔlivH::km Kmr | This study |
| MG1655F′ | MG1655 F′tet-ΔtraD::apra plasmid, Aprr, Tetr | 54 |
| KS272 | Commensal E. coli K-12 | Laboratory collection |
| BL21 | Commensal E. coli K-12 | Laboratory collection |
| MG1655λattgfp-amp | MG1655 with a gfp gene inserted at the λatt site; Ampr | 13 |
| MG1655λattgfp-Km | MG1655 with a gfp gene inserted at the λatt site; Kmr | 13 |
| MG1655ilvG+ | MG1655 with a hybrid active ilvG gene of K-12 and CFT073 | This study |
| CFT073 | Uropathogenic isolate | Laboratory collection |
| CFT073ΔkpsD | CFT073 ΔkpsD::kan; Kmr | 54 |
| CFT073ΔmchB | CFT073 ΔmchB::kan; Kmr | This study |
| CFT073ΔsoxR | CFT073 ΔsoxR::kan; Kmr | This study |
| CFT073ΔoxyR | CFT073 ΔoxyR::kan; Kmr | This study |
| CFT073ΔcpxR | CFT073 ΔcpxR::kan; Kmr | This study |
| CFT073ΔrpoS | CFT073 ΔrpoS::kan; Kmr | This study |
| CFT073Δhns | CFT073 Δhns::kan; Kmr | This study |
| CFT073ΔrelA | CFT073 ΔrelA::cm; Cmr | This study |
| CFT073ΔrelA-ΔspoT | CFT073 ΔrelA::cm ΔspoT::zeo; Cmr Zeor | This study |
| CFT073 pRelA* | Strain with plasmids pMM100 and pSM11 | This study |
| E. coli 195 | Commensal isolate belonging to phylogenetic group B1, obtained from a Pyrenees farm sheep | 44; also see Table S2c |
| E. coli 057 | Commensal isolate belonging to phylogenetic group B1.341 from a Pyrenean chamois | 44; also see Table S2 |
| 1092a | Commensal isolate from a healthy patient | C. Le Bouguenec |
| 1091a | Commensal isolate from a healthy patient | C. Le Bouguenec |
| 1102a | Commensal isolate from a healthy patient | C. Le Bouguenec |
| 1103a | Commensal isolate from a healthy patient | C. Le Bouguenec |
| 1110a | Commensal isolate from a healthy patient | C. Le Bouguenec |
| 1125a | Commensal isolate from a healthy patient | C. Le Bouguenec |
| 536a | Uropathogenic isolate (O6:K15:H31) | J. Hacker |
| J96a | Uropathogenic isolate (O4:K6) | J. Hacker |
| E. coli collection ECORb | 33; also see Table S2 | |
| E. coli collection | 341 commensal isolates constituted from several animal populations | 44; also see Table S2 |
| E. coli collection | Commensal isolates constituted from human populations | 15; also see Table S2 |
| UPEC collection | Uropathogenic clinical isolates U1-3, U6-7, U9-11, U13-20 | 54; also see Table S2 |
| Klebsiella spp. | ||
| K. oxytoca U12 | Clinical isolate | C. Forestier |
| K. oxytoca U10 | Clinical isolate | C. Forestier |
| K. oxytoca U33 | Clinical isolate | C. Forestier |
| K. pneumoniae U112 | Clinical isolate | C. Forestier |
| K. pneumoniae U22 | Clinical isolate | C. Forestier |
| K. pneumoniae U12 | Clinical isolate | C. Forestier |
| K. pneumoniae U21 | Clinical isolate | C. Forestier |
| Enterobacter | ||
| E. cloacae U28 | Clinical isolate | C. Forestier |
| E. cloacae U4 | Clinical isolate | C. Forestier |
| Serratia marcescens U11 | Clinical isolate | C. Forestier |
| Salmonella enterica serovar Enteritidis | ||
| 3934 | Clinical isolate | 46 |
| 921 | Clinical isolate | 46 |
| Pseudomonas aeruginosa | ||
| PAK | Laboratory strain | 8 |
| KK02 | Clinical isolate | S. de Bentmann |
| KK14 | Clinical isolate | S. de Bentmann |
| KK11 | Clinical isolate | S. de Bentmann |
| Plasmids | ||
| pMM100 | Derivative of pACYC184 that expresses the LacI repressor | 34 |
| pSM11 | pSMll contains 455 codons from the E. coli relA gene placed under the control of the lac promoter; promoter induction leads to increased ppGpp production in the cell | 42 |
| pZE12 | Used as a cloning vector; Ampr ColE1 ori | 29 |
| pSspA | CFT073 sspA placed under the control of the synthetic lacP promoter on the pZE12 vector; Ampr ColE1 ori | This study |
| pTufA | CFT073 tufA placed under the control of the synthetic lacP promoter on the pZE12 vector; Ampr ColE1 ori | This study |
Tested for growth inhibition activity of in-biofilm supernatant on an MG1655 overlayer (Table 1).
ECOR, Escherichia coli Reference Collection (33). Isolates 1 to 4, 7 to 12, and 14 to 19 were used; see Table S2 in the supplemental material.
In the supplemental material.
DNA manipulations and genetic techniques.
DNA techniques were performed as described by Sambrook et al. (39). Oligonucleotides were purchased from Sigma, France. DNA sequencing was performed using MWG services. Mutants were constructed using a three-step PCR procedure described previously (see reference 28 and http://www.pasteur.fr/recherche/unites/Ggb/matmet.html).
To obtain a CFT073ΔrelA ΔspoT mutant growing on minimal medium, a spontaneous prototroph was selected. In a relA spoT double mutant, the absence of (p)ppGpp prevents growth on minimal medium. This auxotrophy can be suppressed by spontaneous mutations in RNA polymerase subunits (9). Such a spontaneous mutant (presumably an rpoB mutant) was selected by plating a relA spoT double mutant previously grown in LB broth on M63B1glu. To construct the MG1655ilvG+ mutant, a 950-bp PCR fragment amplified from the ilvG gene of CFT073 was recombined with the ilvG gene of MG1655. This enabled the correction of the frameshift mutation present in the ilvG gene of MG1655 mariner and led to the mutant strain MG1655ilvG+.
Transposon mutagenesis.
Mariner transposon mutagenesis of E. coli CFT073 and MG1655F′ was performed as described previously (12). To select MG1655 mutants resistant to the CFT073 inhibitor, 10,000 transposon mutants were tested for their ability to grow on plates of M63B1glu supplemented with the CFT073 supernatant (10% [vol/vol]). The mutants that were isolated were transduced to MG1655F′ to verify that their resistance was due to the transposon insertion only. To screen for CFT073 mutants able to secrete valine in a planktonic culture, 10,000 transposon mutants were grown for 24 h in M63B1glu at 37°C in 96-well microtiter plates. The inhibitory effect was analyzed by spotting 5 μl of culture on an overlayer of strain MG1655 cells. Transposon insertion sites were determined as described previously (12). Homology searches were performed using BLAST 2.0. Deletion mutants were generated as described elsewhere (www.pasteur.fr/recherche/unites/Ggb/3SPCRprotocol.html) using perimers described in Table S1 (www.pasteur.fr/recherche/unites/Ggb/supmat.html).
Purification and toxicity test of the supernatants. (i) Planktonic supernatant.
Bacteria cultured to an optical density of 1 at 600 nm (OD600) (1 ml at an OD of 1) for 48 h in M63B1glu were centrifuged for 30 min at 5,000 rpm at 4°C. The supernatant was filtered through a 0.2-μm filter.
(ii) Biofilm supernatant.
Bacterial biofilms were obtained using a continuous-flow system (60-ml microfermentors) as described previously (see reference 16 and www.pasteur.fr/recherche/unites/Ggb/biofilmfermenter.html). Briefly, inoculation was performed by dipping the microfermentor's removable spatula into a culture containing 108 bacteria/ml for 2 min and subsequently reintroducing it into the microfermentor. The medium was pumped through the microfermentors at a constant rate (0.75 ml/min). After 48 h, the biofilm formed was rapidly resuspended in 10 ml of M63B1 medium (without glucose) by vortexing for 30 s, the equivalent of an OD600 of 1 of the bacterial resuspension was centrifuged, and the supernatant was filtered through a 0.2-μm filter. For supernatants purified from colonies grown overnight on M63B1glu plates at 37°C, a sufficient number of colonies was picked from the plate and resuspended in 1 ml of M63B1 medium to obtain a minimum concentration of an OD600 of 1. Bacteria were centrifuged, and the supernatant was filtered through a 0.2-μm filter. For supernatants purified from static biofilms, the biofilms were grown in M63B1glu in microtiter plates at 37°C. After 48 h of growth, the cultures in the microtiter plates were rinsed briefly with M63B1 medium, and biofilms were resuspended in M63B1 medium to reach an OD of 1 and treated as described above.
(iii) Growth inhibition test.
The inhibitory effect of filter-sterilized supernatants was evaluated by spotting 8 μl of the supernatant on an overlayer of MG1655 cells on M63B1glu plates. Plates were incubated overnight at 37°C before pictures were taken. When specified, the filtered supernatant was mixed at a 1:1 ratio with other solutions (LB, amino acids, etc.), and 8 μl of the mixed solution was spotted onto an overlayer of MG1655 cells on M63B1glu plates.
Molecular characterization of the inhibitory compound.
The filter-sterilized supernatant was concentrated using a Vivaspin 2 column with a 10,000-Da cutoff (Sartorius). The eluting fraction was further fractionated with a Vivaspin 2 column with a 5,000-Da cutoff (Sartorius). Each concentrated fraction was washed twice with sterilized water before being assayed for inhibition on an overlayer of MG1655 cells on M63B1glu plates.
Role of the stress response in valine production analysis.
CFT073 cells were cultured for 48 h at 37°C in M63B1glu supplemented with 0.05, 0.1, 0.5, and 1 mM paraquat or in M63B1glu whose pH had been decreased to pH 5 by the addition of HCl. Supernatants from these cultures were purified, and their toxicity levels were tested on an overlayer of MG1655 cells on M63B1glu plates, as described above.
Amino acid composition analysis.
The supernatants were prepared as described above. Amino acid composition was determined by total hydrolysis by F. Baleux at the Unité de Chimie Organique of the Institut Pasteur.
Competitive growth experiment.
For competition experiments, we used strains labeled with an antibiotic marker inserted at the chromosomal λatt site, using a three-step PCR procedure. Cultures were grown overnight in LB medium at 37°C and diluted to an OD of 1 with fresh LB medium. Diluted cultures of MG1655attλ-amp and MG1655ilvG+/attλ-Km or of MG1655attλ-Km and MG1655ilvG+/attλ-amp were mixed at a 1:1 ratio and grown at 37°C for 24 h. A sample of the mixed culture was taken at time zero and at 24 h, serially diluted, and plated on LB, LB plus Km, and LB plus Amp plates, in order to estimate the CFU of the two strains in the mixed culture over time.
Selection of valine-resistant mutants under valine selection pressure.
Planktonic cultures of MG1655F′, CFT073ΔmchB ΔkpsD, and KS272 were grown overnight in M63B1glu at 37°C. Cultures were diluted to an OD of 1 with M63B1glu and mixed at a 1:1 ratio. The mixed cultures were grown under planktonic conditions at 37°C for 24 h and simultaneously used to inoculate microfermentors, as described above, to allow the formation of mixed biofilms grown for 24 h at 37°C. Serial dilutions of the mixed planktonic and biofilm cultures were plated onto M63B1glu plus Apr with or without 20% CFT073 filtered biofilm supernatant. The frequency of the appearance of valine-resistant mutants was measured by determining the ratio of clones resistant to both Apr and CFT073 in-biofilm supernatant to the total number of Aprr clones.
RESULTS
Inhibitory molecules are secreted within E. coli CFT073 biofilms.
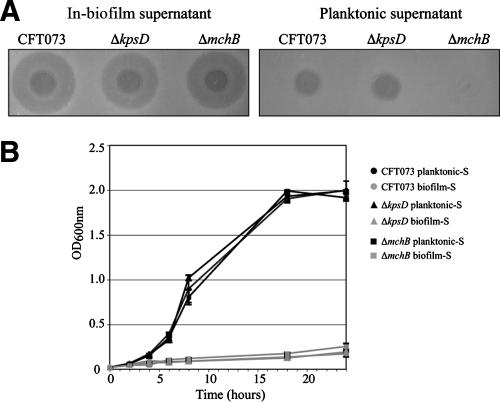
We previously showed that the uropathogenic E. coli (UPEC) isolate CFT073 and most extraintestinal pathogenic E. coli isolates release capsular polysaccharides that inhibit bacterial adhesion and could contribute to competitive interactions within bacterial communities encountered during the UPEC colonization process (54). We hypothesized that the secretion of bacterial interference molecules could occur within bacterial biofilms. To test this, 48-h-old biofilms formed in a continuous-flow biofilm microfermentor, using CFT073 or the stronger former of biofilm, the capsule-deficient mutant CFT073ΔkpsD (54), were resuspended in minimal medium and filter sterilized. The resulting biofilm-derived supernatants (hereafter referred to as in-biofilm supernatants), as well as the supernatants of equivalent 48-h-old planktonic cultures (planktonic supernatants) of these strains, were spotted onto an overlayer of the E. coli K-12 MG1655 strain on M63B1glu plates. Interestingly, both the CFT073 and the CFT073ΔkpsD in-biofilm supernatants produced a large growth inhibition halo that was not observed for the planktonic supernatants (Fig. 1A). MG1655 planktonic growth was also severely impaired in the presence of the CFT073 and the CFT073ΔkpsD in-biofilm supernatants (Fig. 1B). Bacteria incubated for 10 h in the presence of a CFT073 in-biofilm supernatant resumed growth after dilution in fresh medium and displayed a wild-type level of sensitivity to in-biofilm supernatants, indicative of bacteriostatic rather than bactericidal activity at the concentration used (data not shown). The supernatants extracted either from resuspended 48-h biofilms formed in microtiter plates or from resuspended colonies formed on agar plates did not inhibit MG1655 growth. These results indicate that growth-inhibiting molecules are produced and accumulate within mature CFT073 biofilms formed under continuous-flow conditions.
FIG. 1.
Bacteriotoxic effect of the CFT073 in-biofilm supernatant. (A) Growth inhibition activity of CFT073, CFT073ΔkpsD, and CFT073ΔmchB biofilm supernatants on an overlayer of E. coli K-12 cells. (B) Growth curves of MG1655 in the presence of 20% CFT073, CFT073ΔkpsD, and CFT073ΔmchB planktonic supernatant or in-biofilm supernatant. Cultures were performed in M63B1glu. Experiments were performed in duplicate. Error bars represent standard deviations of the means.
The inhibitor secreted within CFT073 biofilms is not microcin H47.
Attempts to determine the size of the inhibitory molecule using cutoff size-differential filters showed that fractions smaller than 5,000 Da still retained activity (data not shown). This small size suggested that the inhibitory molecule could be a microcin, a small peptide with bacteriostatic or bactericidal activity produced by many enterobacteria (17). Under certain environmental conditions, CFT073 was shown to secrete microcin H47, which inhibits the growth of E. coli and several phylogenetically related enterobacterial strains (25, 45). Growth inhibition associated with CFT073 in-biofilm supernatants could therefore be due to the overproduction of microcin H47 under biofilm conditions. To test this hypothesis, we constructed a mutation in mchB, one of four chromosomal genes involved in microcin synthesis (mchABCD) in CFT073 and tested the activity of its in-biofilm supernatant on an MG1655 overlayer. As shown in Fig. 1A, H47 microcin is responsible only for the slight growth inhibition displayed by CFT073 planktonic supernatants, while supernatants from the ΔmchB mutant biofilms still produced a large growth inhibition halo and impaired MG1655 liquid growth (Fig. 1B). These results indicate that an inhibitor distinct from microcin H47 is produced within the CFT073 biofilms.
Genetic and biochemical evidence for high levels of valine in the CFT073 biofilm matrix.
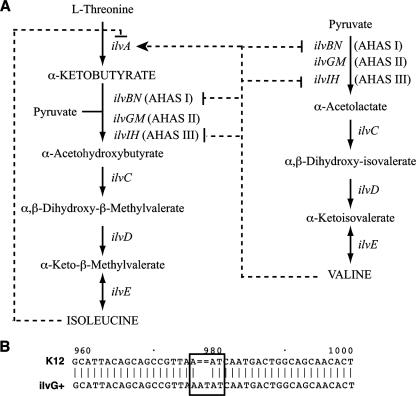
To determine the nature of the inhibitory molecule produced within CFT073 biofilms, we reasoned that E. coli MG1655 mutants resistant to the molecule might provide insight into its entry pathway or cellular target. We performed mariner transposon mutagenesis in the sensitive strain MG1655 and plated the mutants onto M63B1glu agar plates supplemented with filter-sterilized in-biofilm supernatant. One resistant transposon mutant carried a mutation in the livH gene, encoding a membrane component of the high-affinity branched-chain amino acid transport system (for the import of monomeric isoleucine, leucine, and valine). The allelic exchange deletion of livH from MG1655 also led to a high level of resistance to CFT073 in-biofilm supernatants, confirming that the inhibitor studied used the LivH transporter to enter the cell (Fig. 2A). Interestingly, the inhibition of E. coli K-12 growth by exogenous valine is a well-known phenomenon (49). It is due to the loss in E. coli K-12 of one of the three heterodimeric acetohydroxy acid synthase (AHAS) isozymes. While feedback from AHAS I and III is inhibited by valine, the valine-insensitive isozyme AHAS II (ilvGM) is inactivated in E. coli K-12 by a frameshift mutation in ilvG (14) (Fig. 3B). Valine inhibition of the growth of K-12ilvG is due to accumulation of the AHAS substrate α-ketobutyrate, a toxic inhibitor of the glucose phosphotransferase system (38). This accumulation results both from the valine stimulation of threonine deaminase (ilvA) production, which catalyzes α-ketobutyrate formation, and from the valine inhibition of the remaining valine-sensitive AHAS I and AHAS III isozymes, required for isoleucine synthesis (Fig. 3A). Valine inhibition is reversed by isoleucine, which inhibits IlvA activity, thus blocking the accumulation of α-ketobutyrate caused by valine (Fig. 3A) (38).
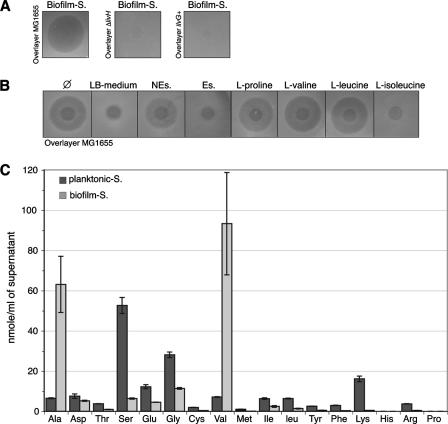
FIG. 2.
A high level of valine is present in biofilm supernatant. (A) Inhibitory activity of the CFT073 in-biofilm supernatant (Biofilm-S) on overlays of MG1655, MG1655ΔlivH, and MG1655ilvG+ cells. (B) Inhibitory activity of CFT073 in-biofilm supernatant supplemented at a 1:1 ratio with LB medium, nonessential amino acids (NEs; which includes l-alanine; see Materials and Methods), essential amino acids (Es), or the amino acid l-proline, l-valine, l-leucine, or l-isoleucine (all amino acids at a concentration of 50 μg ml−1 each) on an overlayer of MG1655 cells. LB and Es solutions contained l-isoleucine. Ø means that the CFT073 in-biofilm supernatant was not supplemented with exogenous amino acids. (C) The amino acid composition of CFT073 supernatants purified from planktonic culture (Planktonic-S) and biofilm (Biofilm-S). Experiments were performed in triplicate. Error bars represent standard deviations of the means.
FIG. 3.
Isoleucine and valine biosynthesis pathways. (A) ilvBN, ilvGM and ilvIH code for the heterodimeric acetohydroxy acid synthase isozymes AHAS I, AHAS II, and AHAS III, respectively. AHAS I and AHAS III are inhibited in the presence of high amounts of valine, whereas isozyme AHAS II is valine insensitive. Regulations by valine and isoleucine are shown by dashed lines terminated by an arrow for activation or by a vertical line for inhibition. (B) In E. coli K-12, AHAS II is inactivated by a frameshift mutation in ilvG. ilvG sequences of E. coli K-12 and MG1655ilvG+ are aligned. The numbers above indicate base numbers from the ATG translation start according to the ilvG+ sequence.
To determine whether E. coli K-12 growth inhibition by CFT073 in-biofilm supernatants reflected valine secretion, we first checked the effect of l-isoleucine on CFT073 in-biofilm supernatant activity. The addition of l-isoleucine to a CFT073 in-biofilm supernatant alleviated its inhibition of E. coli K-12 MG1655, whereas the addition of other amino acids had no effect (Fig. 2B). Moreover, no growth inhibition was observed for rich LB medium plates (data not shown). We then used the CFT073 ilvG+ sequence to correct the ilvG frameshift mutation in MG1655 (MG1655ilvG+) (see Fig. 3B) and tested the sensitivity of the valine-resistant MG1655ilvG+ to CFT073 in-biofilm supernatant. While MG1655 (ilvG) growth is severely inhibited on minimal medium plates by a CFT073 in-biofilm supernatant, strain MG1655ilvG+ displayed a high level of resistance (Fig. 2A). Finally, analysis of the amino acid composition of a CFT073 in-biofilm supernatant showed that it indeed contains much higher levels of l-valine as well as l-alanine than a CFT073 planktonic supernatant (Fig. 2C). The concentration of valine found in the CFT073 biofilm supernatant is about 12 μg/ml. Consistently, whereas l-alanine has no inhibitory activity (data not shown and Fig. 2B), the addition of exogenous l-valine at a concentration of 15 μg/ml to the planktonic (inactive) supernatant of CFT073 reproduced the inhibitory effect of the CFT073 in-biofilm supernatant on an overlayer of MG1655 cells and not on an overlayer of the ΔlivH mutant cells (see Fig. S1 in the supplemental material). Taken together, these results demonstrate that the growth inhibition of E. coli K-12 by CFT073 in-biofilm supernatants is due to accumulation of monomeric valine.
Many E. coli isolates and other bacteria secrete valine in biofilms.
In order to investigate the occurrence of valine production under biofilm conditions by E. coli isolates and other bacteria, the growth inhibition activities of in-biofilm supernatants of 44 E. coli isolates of different origins and supernatants of Klebsiella pneumoniae, K. oxytoca, Enterobacter cloacae, Serratia marcescens, Salmonella enterica serovar Enteritidis, and Pseudomonas aeruginosa were tested with an MG1655 overlayer. As shown in Table 1, 24 out of 44 (55%) E. coli in-biofilm supernatants, as well as those from K. pneumoniae, E. cloacae, Salmonella serovar Enteritidis, and P. aeruginosa, produced a growth inhibition halo. This inhibition was likely to be due to valine secretion, since inhibition was alleviated by isoleucine and the tested in-biofilm supernatants had no effect on the valine-resistant mutant MG1655ilvG+ (Table 1). These results therefore indicate that the secretion of valine within the biofilm matrix is not restricted to CFT073 but is a widespread phenotype among natural E. coli isolates, as well as other gram-negative bacteria.
TABLE 1.
Occurrence of growth inhibition activity of in-biofilm supernatant of different gram-negative bacteria on E. coli MG1655
| Organism | No. of strains tested | Biofilm-S strains inducing inhibitiona
|
|
|---|---|---|---|
| No. of strains | % of strains tested | ||
| E. colib | 44 | 24 | 55 |
| K. oxytoca | 3 | 0 | 0 |
| K. pneumoniae | 4 | 3 | 75 |
| E. cloacae | 2 | 1 | 50 |
| S. marcescens | 1 | 0 | 0 |
| Salmonella serovar Enteritidis | 2 | 2 | 100 |
| P. aeruginosa | 4 | 4 | 100 |
Number and percentage of the strains tested whose in-biofilm supernatants (Biofilm-S) induced the appearance of a growth inhibition halo on an overlayer of MG1655 cells on M63B1glu plates and for which the inhibition was alleviated by the addition of isoleucine.
The E. coli strains tested for growth inhibition activities of their in-biofilm supernatants are specified in Table 3.
E. coli stress responses do not contribute to valine production within biofilm.
Mature biofilms constitute a heterogeneous stationary phase-like environment in which the induction of several stress responses has been demonstrated (4, 58). To investigate the potential link between bacterial stress responses and valine secretion, we first analyzed the level of valine secretion by CFT073 grown planktonically when bacteria were exposed to acid or oxidative stresses. Neither the use of different levels of superoxide radicals generated by a range of paraquat concentrations (up to 1 mM) nor acidic pH conditions (pH 5) induced valine secretion by CFT073 planktonic cells (data not shown). We then studied the role of several major regulators of E. coli stress or signaling responses, including H-NS, RpoS (stationary phase), OxyR, SoxR (oxidative stress), CpxR (envelope stress), and LuxS (quorum sensing). None of the mutants lacking one of these regulators displayed altered valine production in filter-sterilized planktonic or in-biofilm supernatants (data not shown). These results showed that none of the tested stresses or stress response regulators is involved in valine secretion within biofilms.
Valine secretion within biofilms is not due to the stringent response.
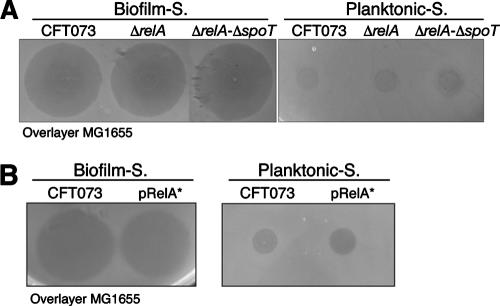
The stringent response, a complex cellular adaptation to starvation for amino acids and sources of carbon, nitrogen, and phosphate, has been shown to be important for E. coli biofilm formation under slow-growth or nutrient-limited conditions (2, 9). The bacterial stringent response is essentially mediated by two proteins, RelA and SpoT, which control the intracellular level of ppGpp. This nucleotide acts as a general starvation signal leading simultaneously to down-regulation of nucleic acid and protein synthesis and to up-regulation of protein degradation and amino acid synthesis. Whereas both RelA and SpoT convert GTP to pppGpp (which is further converted to ppGpp), only SpoT mediates pppGpp and ppGpp hydrolysis (9). To explore the potential relationship between in-biofilm valine secretion and the stringent response, we first tested the phenotypic consequences of the absence of ppGpp and introduced either a single relA or a relA spoT double mutation into CFT073. In their planktonic supernatants, neither the ΔrelA mutant nor the ΔrelA ΔspoT mutant produced valine, and their in-biofilm supernatants still displayed valine-associated inhibition (Fig. 4A).
FIG. 4.
Effect of the stringent response on valine-related inhibition. Valine-related inhibition on an MG1655 overlayer of (A) the in-biofilm (Biofilm-S) and planktonic (Planktonic-S) supernatants of the CFT073ΔrelA and CFT073ΔrelA ΔspoT mutants and (B) the supernatants of CFT073 cells overproducing ppGpp (pRelA*) and nonoverproducing cells, purified from biofilm (Biofilm-S) or planktonic culture (Planktonic-S).
We then tested the consequence of an increased level of ppGpp by introducing into CFT073 a plasmid-borne plac-controlled allele of relA (pRelA*), which has been shown to lead to starvation-independent ppGpp production (9). We did not observe any valine-related inhibition by the planktonic supernatant of the CFT073(pRelA*) strain, and the presence of the plasmid did not prevent valine secretion in biofilm (Fig. 4B). These results suggest that an increased ppGpp level does not lead to biofilm-independent valine secretion nor does it inhibit in-biofilm valine secretion. Hence, no correlation between in-biofilm valine secretion and the stringent response could be demonstrated.
Screening for CFT073 mutants secreting valine in planktonic cultures.
We sought to investigate further the genetic basis for valine secretion within CFT073 biofilms and performed mariner transposon mutagenesis with CFT073. However, due to the practical difficulty of screening for the loss of growth inhibition activity of in-biofilm supernatants extracted from a large number of mutants grown in microfermentors, we instead screened approximately 10,000 mariner transposon mutants for biofilm-independent inhibitory activity when grown as colonies on an overlayer of E. coli MG1655. We identified eight mutants that produced a larger growth inhibition halo than the wild-type CFT073. These mutants corresponded to a transposon insertion at eight different loci (Table 2). Two of the mutations were in genes involved in the branched-chain amino acid biosynthesis pathway (ilvA and ilvY). Most of the mutants (ilvA, ilvY, nadB, nadC, C0746, and aceE mutants) had a reduced growth rate on minimal glucose plates compared to that of wild-type CFT073 (data not shown), although two mutants, the tufA and sspA mutants, displayed wild-type levels of growth. The stringent starvation protein A (SspA) was previously described as an RNA polymerase-associated activator for the lytic development of phage P1, which also has global regulatory functions and is essential for stationary phase-induced acid tolerance in E. coli (20, 21). tufA is one of two unlinked genes (with tufB) coding for the translation elongation factor Tu, which promotes GTP-dependent binding of aminoacyl-tRNA to the A site of ribosomes during protein synthesis. Like sspA, tufA may play an important regulatory role in the bacterial response to nutrient deprivation (57, 59).
TABLE 2.
E. coli CFT073 transposon mutants producing valine under planktonic conditions
| COG classificationa | Protein | CFT073 gene no. | BLAST description |
|---|---|---|---|
| Amino acid biosynthesis genes | IlvA | C4694 | Threonine dehydratase biosynthesis |
| IlvY | C4695 | Transcriptional activator protein | |
| Growth-rate-dependent regulatory genes | SspA | C3981 | Stringent starvation protein |
| TufA | C4111 | Elongation factor Tu | |
| Other | NadB | C3098 | l-Aspartate oxidase |
| NadC | C0128 | Nicotine-nucleotide pyrophosphorylase | |
| H protein | C0746 | Hypothetical protein | |
| AceE | C0142 | Pyruvate dehydrogenase |
COG, cluster of orthologous groups.
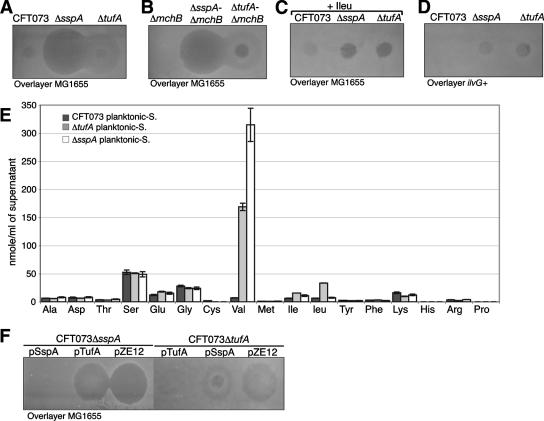
In order to confirm the role of these mutations in MG1655 growth inhibition, we performed nonpolar allelic replacement of the sspA and tufA genes in E. coli CFT073. The planktonic supernatant of the two mutants displayed the same inhibitory phenotype as those of the corresponding transposon mutants (Fig. 5A). Furthermore, the MG1655 growth inhibition activity of the CFT073ΔsspA and CFT073ΔtufA planktonic supernatants was independent of mchB; it was abolished by the addition of isoleucine and was inefficient against MG1655ilvG+ (Fig. 5B to D). Finally, the two mutants' planktonic supernatants contained high levels of l-valine (Fig. 5E). We then cloned sspA and tufA under the control of the plac promoter, leading to plasmids pSspA and pTufA, respectively. As expected, no valine-associated inhibition was observed in the planktonic supernatants of the mutants CFT073ΔsspA and CFT073ΔtufA complemented by pSspA and pTufA, respectively (Fig. 5F). These results therefore demonstrated that we had identified mutations in strain CFT073 leading to valine secretion under planktonic growth conditions.
FIG. 5.
Phenotypic analysis of the planktonic valine-secreting mutants. Valine-related inhibition by supernatants purified from planktonic cultures of CFT073, CFT073ΔsspA, and CFT073ΔtufA (A) and CFT073ΔmchB, CFT073ΔsspA ΔmchB, and CFT073ΔtufA ΔmchB (B) on an overlayer of MG1655 cells. (C) Effect of l-isoleucine addition to the CFT073ΔsspA and the CFT073ΔtufA planktonic supernatant activity on an overlayer of MG1655 cells. (D) Inhibitory activity of the CFT073ΔsspA and the CFT073ΔtufA planktonic supernatants on an overlayer of MG1655ilvG+. (E) Analysis of the amino acid composition of the CFT073ΔsspA and the CFT073ΔtufA planktonic supernatants. Experiments were performed in triplicate. Error bars represent standard deviations of the means. (F) Complementation of the CFT073ΔsspA and CFT073ΔtufA mutants with pSspA, pTufA, and the empty plasmid pZE12 as the control.
Although an ilvG strain outcompetes an ilvG+ strain under planktonic conditions, valine-sensitive E. coli isolates are rare in natural populations.
Valine-sensitive strains occur at very low frequencies among natural E. coli isolates, according to an early study (30). In agreement with that study, we identified only two valine-sensitive strains (E. coli 195 and E. coli 057) among 552 commensal E. coli isolates of extremely varied origins (Table 3). The sequences of their ilvG alleles indicated that at least one of these strains (E. coli 057) did indeed carry ilvG (see Fig. S2 at http://www.pasteur.fr/recherche/unites/Ggb/supmat.html). Hence, although it is sometimes considered a laboratory allele, an inactive ilvG gene may give bacteria a selective advantage under certain environmental conditions. To test whether the presence of a wild-type ilvG+ allele is associated with a detectable fitness loss, we used the corrected valine-resistant MG1655ilvG+ strain to compare the growth of isogenic MG1655ilvG with that of the ilvG+ strain under planktonic conditions. Growth curves indicated a clear yield defect of MG1655ilvG+ strains in LB medium but not in M63B1glu (Fig. 6A and data not shown). These results suggest that, under some growth conditions, the expression of ilvG may constitute a genetic burden that is alleviated in ilvG strains. To further investigate the potential consequences of this fitness loss, we performed competition experiments in which a 1:1 ratio of tagged isogenic MG1655ilvG+ and MG1655ilvG strains were mixed under planktonic culture conditions in LB medium. After 24 h of growth, plating assays revealed a competitive advantage for the ilvG strain, with a 1.5:1 ratio of ilvG to ilvG+ cells (Fig. 6B).
FIG. 6.
Inactivation of ilvG confers an advantageous phenotype on E. coli K-12. (A) Growth curves of MG1655 and MG1655ilvG+ in LB medium at 37°C. (B) Competition experiments: MG1655λattgfp-Km (MG1655) and MG1655λattgfp-amp (MG1655ilvG+) cells grown overnight and diluted to an OD of 1 with fresh LB medium were mixed at a 1:1 ratio and grown in LB at 37°C. The population composition was estimated by serial dilutions and plating in order to estimate the CFU of the two strains in the mixed culture. Results are shown as the percentages of colonies of MG1655 and MG1655ilvG+ in the mixed culture at 24 h after mixing. Experiments were performed in triplicate. Error bars represent standard deviations of the means.
This suggests that, although a valine-sensitive ilvG E. coli strain may have a selective advantage over an ilvG+ strain in LB broth, valine-sensitive ilvG frameshift mutants are not favored over valine-resistant E. coli in most natural ecological niches.
In-biofilm valine secretion selects valine resistance in mixed E. coli biofilms.
Valine-resistant mutants with mutations that correct the translational reading frame of the ilvG gene can be readily isolated among mutants growing when a moderate concentration of valine (1 μg/ml or more) is present in isoleucine-free growth medium. This genetic switch from ilvG (valine-sensitive) to ilvG+ (valine-resistant) E. coli K-12 strains is one of the best-documented examples of activation of an otherwise inactive (“cryptic”) pseudogene (18). The high frequency of valine-resistant strains among natural E. coli isolates found in early studies led to the hypothesis that in unidentified valine-rich environments, valine resistance might be an advantageous phenotype. Our finding that many E. coli and other bacterial isolates naturally secrete high amounts of valine within biofilms led us to hypothesize that this widespread valine-rich environment may be the selection pressure favoring genetic maintenance of the ilvG+ genotype.
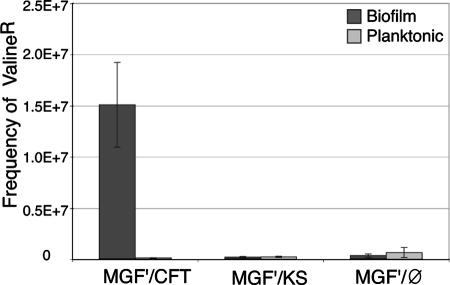
To test this hypothesis, we compared the frequency of valine-resistant derivatives produced in mixed biofilm or planktonic cultures in which valine-sensitive E. coli MG1655 F′, which does not produce in-biofilm valine, was mixed 1:1 with the valine-producing and valine-resistant strain CFT073ΔmchB ΔkpsD. As shown in Fig. 7, after 24 h, 15-fold more valine-resistant derivatives were isolated from mixed biofilm cultures than from mixed planktonic cultures. In contrast, the frequency of valine-resistant derivatives in a mixed biofilm formed with two E. coli K-12 strains that do not secrete valine (KS272 and MG1655 F′) was similar to that obtained under planktonic conditions with E. coli MG1655 F′ and CFT073ΔmchB ΔkpsD (Fig. 7). These results thus indicate that the secretion of valine within bacterial biofilms increases the frequency of valine-resistant derivatives in the valine-sensitive MG1655 F′ population. In-biofilm valine secretion is therefore a plausible selective force leading to the persistence of valine-resistant strains in natural E. coli populations living mostly in mixed biofilm-like environments where valine can be produced by many different bacteria.
FIG. 7.
Frequency of MG1655 valine-resistant derivative mutants. MG1655F′ valine-resistant derivatives were isolated from mixed cultures of MG1655F′ and the valine-producing strain CFT073ΔmchBΔkpsD (MGF′/CFT), MG1655F′ and the non-valine-producing strain KS272 (MGF′/KS) or MG1655F′ alone (MGF′/Ø) grown in biofilm or in planktonic culture. Results are expressed as the frequency of valine-resistant mutants of MG1655 isolated after 24 h of growth of the mixed culture. Experiments were performed in triplicate. Error bars represent standard deviations of the means.
DISCUSSION
Over 60 years ago, Tatum noted that in the absence of isoleucine, valine inhibits the growth of E. coli K-12 (49). The cause of this toxicity is now well understood (see Fig. 3), and amino acid antagonisms have been instrumental in the study of the branched-chain amino acid biosynthetic pathways and have led to the elucidation of several important biological concepts such as feedback inhibition and allostery (10, 14, 38, 52). However, we still know little about the actual biological or ecological relevance of these antagonisms.
In this study, we observed that many E. coli strains, irrespective of their specific phylogenetic groups, have the ability to produce valine when grown as continuous-flow biofilms in glucose-rich minimal medium but not after incubation in batch cultures of the same medium where carbon sources are rapidly used and depleted. Although valine is a bacteriostatic inhibitor of E. coli K-12 derivatives and a few natural isolates, it does not inhibit most of the 552 E. coli strains of extremely varied origin tested in this study. This narrow range of target strains therefore strongly suggests that valine is unlikely to be a biofilm bacteriotoxin. Nevertheless, valine could play several other specific roles, such as that of protective osmolyte or a role signaling (32, 37, 43, 48). However, none of the tested stress or adaptive response regulators affected in-biofilm valine production, and mutants overexpressing valine (sspA or tufA) or impaired in valine acquisition (livH) displayed a wild-type biofilm phenotype (data not shown). Moreover, the lack of effect of exogenously added in-biofilm supernatant on biofilm growth of several valine-resistant strains (see Table S2 at www.pasteur.fr/recherche/unites/Ggb/supmat.html) led us to favor the hypothesis that in-biofilm valine secretion is rather a metabolic adaptation taking place within the mature biofilm formed under our test conditions.
Increased valine concentrations due to an imbalance in amino acid biosynthetic pathways have been reported to result from acid stress or from the addition of exogenous homocysteine to the medium (50, 51, 53). Valine secretion under several unusual growth conditions, such as high-cell-density planktonic E. coli cultures (with ODs of up to 50) used in recombinant protein production biotechnology, has also been described previously (1). These conditions are reminiscent of those within biofilms, raising the possibility that in-biofilm valine production may be a consequence of complex alterations of metabolic pools within biofilms. Webb reported previously that the growth inhibition of Klebsiella pneumoniae by aminopterin led to the production of valine and l-alanine as a metabolic consequence of pyruvate accumulation (55, 56). Aminopterin is an inhibitor of dihydrofolate reductase and thus of tetrahydrofolate, a precursor of all one-carbon units. Under aerobic conditions, the central metabolite pyruvate leads to acetyl-coenzyme A (acetyl-CoA) and the citric acid cycle, while under anaerobic conditions, in addition to forming acetyl-CoA, it is the precursor of most fermentation products. Because pyruvate is a precursor of valine (the condensation of two pyruvates leads to one valine) and l-alanine (by transamination), Webb's studies suggest that in the absence of a limited source of carbon, reduced growth or oxygen rarefaction may slow down the citric acid (Kreb's) cycle and consequently lead to an increased drain of pyruvate toward biosynthesis of l-alanine and l-valine, both of which are increased in the CFT073 in-biofilm supernatant (55, 56).
In an attempt to investigate the genetic basis for in-biofilm valine production, we screened for mutants of CFT073 that were able to secrete valine under nonbiofilm conditions. We found eight mutants that secreted valine in their planktonic supernatant (Table 2). Two of the mutations affect the valine biosynthetic pathway genes ilvA and ilvY, which could cause an increase in the pyruvate intracellular pool by reducing its incorporation into α-acetohydroxybutyrate by the lack of α-ketobutyrate (the ilvA mutation) or by the reduction of conversion of α-acetohydroxybutyrate into α-β-keto-β-methylvalerate (IlvY is a transcriptional activator of ilvC) (Fig. 3). One mutation (aceE) affected pyruvate dehydrogenase, the inactivation of which has been recently shown to cause pyruvate accumulation and valine secretion (5, 6). The sspA and tufA mutations affect the protein synthesis rate, and the amino acid pool utilization may be also associated with a reduced pyruvate utilization. Mutations leading to valine secretion in planktonic cultures do not cause alanine secretion and, sensu stricto, may not be fully correlated with events leading to in-biofilm valine secretion. However, a transcriptome analysis performed under similar experimental conditions with E. coli K-12 showed consistent biofilm-induced modification of the expression of genes involved in carbohydrate and energy metabolism, including enzymes of the tricarboxylic acid cycle such as fumarate deshydrogenase, malate deshydrogenase, or succinyl-CoA synthetase (4). Our data therefore strongly suggest that in-biofilm valine production reflects physiological adjustments of bacteria to continuous-flow biofilm conditions such as low oxygen and reduced growth rate when carbon sources are not limiting. These physiological conditions would lead to an excess of pyruvate, dissimilated through relaxed synthesis and secretion of valine and alanine. Consistent with this hypothesis, biofilm supernatants from cultures grown in minimal medium containing glycerol or low glucose (0.05%), where pyruvate is not expected to accumulate, did not display valine-related inhibitory activity (data not shown). Although the determination of the metabolic bases of in-biofilm valine production in the UPEC strain CFT073 will require further study, we have demonstrated that this property is widespread among gram-negative bacteria. These bacteria are commonly found in environments that share some of the structural characteristics of multispecific biofilms (e.g., intestinal flora and lung), and we therefore sought to investigate the consequence of in-biofilm valine secretion, in particular with respect to the occurrence of valine resistance. Early studies showed that valine-resistant E. coli K-12 mutants were readily isolated on minimal medium in the presence of valine. The activation of the ilvG pseudogene of E. coli K-12 now even constitutes a classical example of cryptic gene activation. The relative fitness of bacteria with the cryptic or the active gene will depend on the environmental conditions. In the case of the ilvG mutant, it has been pointed out that a functional AHAS II isozyme (ilvG+ strains) is not required under most growth conditions, since isozymes AHAS I and III have adequate biosynthetic capacity (see Fig. 3). Previous studies suggested that the ilvG+ allele could constitute a genetic burden, possibly due to the wasteful overexpression of downstream genes (ilvEDA) when wild-type ilvG is expressed (27, 30). We have shown here that the characteristics of the ilvG+ mutant indeed negatively affect the E. coli MG1655 growth yield and fitness compared to that of MG1655 (ilvG), although we confirmed that the occurrence of valine-sensitive strains and ilvG isolates is rare. Hence, while one of the potential beneficial consequences of naturally occurring frame shifts in the ilvG gene may be to alleviate the detrimental effect of ilvG expression under certain conditions, selection pressure has been hypothesized as the explanation for the prevalence of ilvG+ valine-resistant strains in nature.
While it was possibly acquired under laboratory conditions, the E. coli K-12 frameshift mutation in ilvG raised the question of the long-sought rationale for valine resistance within natural E. coli isolates. We demonstrate that mixing valine-resistant and valine-sensitive strains led to the accumulation of valine-resistant mutants when the valine-sensitive cells were grown in a mixed biofilm with a valine secretor. These results suggest that in-biofilm valine secretion may be the most relevant environmental condition, leading to the persistence of a valine-resistant phenotype over that of valine sensitivity in E. coli strains, thus providing a plausible explanation for the widespread occurrence of valine-resistant bacterial strains.
Global transcriptome analyses comparing surface-attached planktonic culture conditions revealed that the biofilm lifestyle triggers extensive modifications in gene expression that are proposed to correspond to profound physiological changes (3, 26, 40). However, with the exceptions of increased antibiotic tolerance and cyclic di-GMP-mediated regulation of motility and cell surface adhesins, very few bacterial functions have yet been characterized as restricted to the biofilm lifestyle (23, 24). Here, we identified valine secretion as a metabolic response associated with mature biofilms formed under conditions in which carbon sources are not severely limited. Valine secretion is not strictly restricted to biofilm environments, as it is occasionally produced in some industrial planktonic fermentation bioprocesses; however, these conditions are highly artificial (1). By contrast, high-cell-density biofilms formed under conditions where carbon sources are renewed by environmental flow correspond to a number of natural settings. Valine can then accumulate, since it is locally trapped within this reduced-diffusion environment composed of a high-cell-density biomass and the biofilm matrix (1, 36, 47).
In conclusion, this study validates the postulated existence of molecules whose secretion is associated with the biofilm environment. Further characterization of such metabolic signatures may allow the development of new markers with which to monitor or control the biofilm lifestyle.
Supplementary Material
Acknowledgments
We thank C. Forestier, E. Denamur, S. de Bentzman, and A. Filloux for providing strains used in this study. We are grateful to C. Beloin, I. Matic, B. Le Quéré, L. Ferrieres, E. Denamur, P. Delepelaire, G. Cohen, A. Ullmann, and A. Danchin for helpful discussions and critical reading of the manuscript.
J.V. was a Marie Curie fellow. S.D.R. was supported by Sanofi-Pasteur.
This work was supported by grants from the Institut Pasteur, CNRS, the Network of Excellence EuroPathoGenomics (LSHB-CT-2005-512061), and the Fondation BNP PARIBAS.
Footnotes
Published ahead of print on 2 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andersen, D. C., J. Swartz, T. Ryll, N. Lin, and B. Snedecor. 2001. Metabolic oscillations in an E. coli fermentation. Biotechnol. Bioeng. 75212-218. [DOI] [PubMed] [Google Scholar]
- 2.Balzer, G. J., and R. J. McLean. 2002. The stringent response genes relA and spoT are important for Escherichia coil biofilms under slow-growth conditions. Can. J. Microbiol. 48675-680. [DOI] [PubMed] [Google Scholar]
- 3.Beloin, C., and J. M. Ghigo. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 1316-19. [DOI] [PubMed] [Google Scholar]
- 4.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51659-674. [DOI] [PubMed] [Google Scholar]
- 5.Blombach, B., M. E. Schreiner, J. Holatko, T. Bartek, M. Oldiges, and B. J. Eikmanns. 2007. l-Valine production with pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum. Appl. Environ. Microbiol. 732079-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blombach, B., M. E. Schreiner, M. Moch, M. Oldiges, and B. J. Eikmanns. 2007. Effect of pyruvate dehydrogenase complex deficiency on L-lysine production with Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 76615-623. [DOI] [PubMed] [Google Scholar]
- 7.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 10116630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley, D. E. 1974. Adsorption of bacteriophages specific for Pseudomonas aeruginosa R factors RP1 and R1822. Biochem. Biophys. Res. Commun. 57893-900. [DOI] [PubMed] [Google Scholar]
- 9.Cashel, M., D. R. Gentry, V. J. Hernandz, and D. Vinella. 1996. The stringent response, p. 1458-1496. In R. Curtiss III, A. Böck, J. L. Ingraham, J. B. Kaper, F. C. Neidhardt, M. Riley, and C. L. Squires (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 10.Changeux, J. P. 1964. On the allosteric properties of biosynthetic L-threonine deaminase. 3. Interpretation of the inhibitory effect of L-isoleucine: steric hindrance or allosteric effect. Bull. Soc. Chim. Biol. (Paris) 461151-1173. (In French.) [PubMed] [Google Scholar]
- 11.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41435-464. [DOI] [PubMed] [Google Scholar]
- 12.Da Re, S., and J. M. Ghigo. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 1883073-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Da Re, S., B. Le Quere, J. M. Ghigo, and C. Beloin. 2007. Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Appl. Environ. Microbiol. 733391-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Felice, M., M. Levinthal, M. Iaccarino, and J. Guardiola. 1979. Growth inhibition as a consequence of antagonism between related amino acids: effect of valine in Escherichia coli K-12. Microbiol. Rev. 4342-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escobar-Paramo, P., A. Le Menac'h, T. Le Gall, C. Amorin, S. Gouriou, B. Picard, D. Skurnik, and E. Denamur. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 81975-1984. [DOI] [PubMed] [Google Scholar]
- 16.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412442-445. [DOI] [PubMed] [Google Scholar]
- 17.Gillor, O., B. C. Kirkup, and M. A. Riley. 2004. Colicins and microcins: the next generation antimicrobials. Adv. Appl. Microbiol. 54129-146. [DOI] [PubMed] [Google Scholar]
- 18.Hall, B. G., S. Yokoyama, and D. H. Calhoun. 1983. Role of cryptic genes in microbial evolution. Mol. Biol. Evol. 1109-124. [DOI] [PubMed] [Google Scholar]
- 19.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 295-108. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, A. M., H. Lehnherr, X. Wang, V. Mobley, and D. J. Jin. 2003. Escherichia coli SspA is a transcription activator for bacteriophage P1 late genes. Mol. Microbiol. 481621-1631. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, A. M., Y. Qiu, N. Yeh, F. R. Blattner, T. Durfee, and D. J. Jin. 2005. SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol. Microbiol. 56719-734. [DOI] [PubMed] [Google Scholar]
- 22.Hansen, S. K., P. B. Rainey, J. A. Haagensen, and S. Molin. 2007. Evolution of species interactions in a biofilm community. Nature 445533-536. [DOI] [PubMed] [Google Scholar]
- 23.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40385-407. [DOI] [PubMed] [Google Scholar]
- 24.Kolter, R., and E. P. Greenberg. 2006. Microbial sciences: the superficial life of microbes. Nature 441300-302. [DOI] [PubMed] [Google Scholar]
- 25.Laviña, M., C. Gaggero, and F. Moreno. 1990. Microcin H47, a chromosome-encoded microcin antibiotic of Escherichia coli. J. Bacteriol. 1726585-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazazzera, B. A. 2005. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Curr. Opin. Microbiol. 8222-227. [DOI] [PubMed] [Google Scholar]
- 27.Leavitt, R. I., and H. E. Umbarger. 1962. Isoleucine and valine metabolism in Escherichia coli. XI. Valine inhibition of the growth of Escherichia coli strain K-12. J. Bacteriol. 83624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lesic, B., S. Bach, J. M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 521337-1348. [DOI] [PubMed] [Google Scholar]
- 29.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 251203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manten, A., and D. Rowley. 1953. Genetic analysis of valine inhibition in the K-12 strain of Bacterium coli. J. Gen. Microbiol. 9226-233. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Monds, R. D., and G. A. O'Toole. Metabolites as intracellular signals for regulation of community level traits. In B. Bassler and S. Winans (ed.), Chemical communication among microbes, in press. ASM Press, Washington, DC.
- 33.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Page, A. L., H. Ohayon, P. J. Sansonetti, and C. Parsot. 1999. The secreted IpaB and IpaC invasins and their cytoplasmic chaperone IpgC are required for intercellular dissemination of Shigella flexneri. Cell. Microbiol. 1183-193. [DOI] [PubMed] [Google Scholar]
- 35.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 1815993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redfield, R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10365-370. [DOI] [PubMed] [Google Scholar]
- 37.Roesch, P. L., P. Redford, S. Batchelet, R. L. Moritz, S. Pellett, B. J. Haugen, F. R. Blattner, and R. A. Welch. 2003. Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. Mol. Microbiol. 4955-67. [DOI] [PubMed] [Google Scholar]
- 38.Salmon, K. A., C.-R. Yang, and G. W. Hatfield. 28 February 2006, posting date. Chapter 3.6.1.5, Biosynthesis and regulation of the branched-chain amino acids. In R. Curtiss III, A. Böck, J. L. Ingraham, J. B. Kaper, F. C. Neidhardt, M. Riley, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Sauer, K. 2003. The genomics and proteomics of biofilm formation. Genome Biol. 4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48253-267. [DOI] [PubMed] [Google Scholar]
- 42.Schreiber, G., S. Metzger, E. Aizenman, S. Roza, M. Cashel, and G. Glaser. 1991. Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 2663760-3767. [PubMed] [Google Scholar]
- 43.Shahjee, H. M., K. Banerjee, and F. Ahmad. 2002. Comparative analysis of naturally occurring L-amino acid osmolytes and their D-isomers on protection of Escherichia coli against environmental stresses. J. Biosci. 27515-520. [DOI] [PubMed] [Google Scholar]
- 44.Skurnik, D., R. Ruimy, A. Andremont, C. Amorin, P. Rouquet, B. Picard, and E. Denamur. 2006. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 571215-1219. [DOI] [PubMed] [Google Scholar]
- 45.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 726373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43793-808. [DOI] [PubMed] [Google Scholar]
- 47.Starkey, M., A. K. Gray, S. I. Chang, and M. Parsek. 2004. A sticky business: the extracellular polymeric substance matrix of bacterial biofilms, p. 478. In M. Ghannoum and G. A. O'Toole (ed.), Microbial biofilms, vol. 336. ASM Press, Washington, DC. [Google Scholar]
- 48.Taneja, S., and F. Ahmad. 1994. Increased thermal stability of proteins in the presence of amino acids. Biochem. J. 303147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tatum, E. L. 1946. Induced biochemical mutations in bacteria. Cold Spring Harbor Symp. Quant. Biol. 11278-284. [Google Scholar]
- 50.Tuite, N. L., K. R. Fraser, and C. P. O'Byrne. 2005. Homocysteine toxicity in Escherichia coli is caused by a perturbation of branched-chain amino acid biosynthesis. J. Bacteriol. 1874362-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tweeddale, H., L. Notley-McRobb, and T. Ferenci. 1999. Assessing the effect of reactive oxygen species on Escherichia coli using a metabolome approach. Redox Rep. 4237-241. [DOI] [PubMed] [Google Scholar]
- 52.Umbarger, H. E. 1992. The origin of a useful concept: feedback inhibition. Protein Sci. 11392-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uzan, M., and A. Danchin. 1978. Correlation between the serine sensitivity and the derepressibility of the ilv genes in Escherichia coli relA-mutants. Mol. Gen. Genet. 16521-30. [DOI] [PubMed] [Google Scholar]
- 54.Valle, J., S. Da Re, N. Henry, T. Fontaine, D. Balestrino, P. Latour-Lambert, and J. M. Ghigo. 2006. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. USA 10312558-12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webb, M. 1958. Aminopterin inhibition in Aerobacter aerogenes: alanine and valine accumulation during the inhibition and their utilization on recovery. Biochem. J. 70472-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webb, M. 1968. Pyruvate accumulation in growth-inhibited cultures of Aerobacter aerogenes. Biochem. J. 106375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weijland, A., K. Harmark, R. H. Cool, P. H. Anborgh, and A. Parmeggiani. 1992. Elongation factor Tu: a molecular switch in protein biosynthesis. Mol. Microbiol. 6683-688. [DOI] [PubMed] [Google Scholar]
- 58.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413860-864. [DOI] [PubMed] [Google Scholar]
- 59.Young, C. C., and R. W. Bernlohr. 1991. Elongation factor Tu is methylated in response to nutrient deprivation in Escherichia coli. J. Bacteriol. 1733096-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.