Abstract
Rhodobacter sphaeroides 2.4.1 is a facultative photosynthetic anaerobe that grows by anoxygenic photosynthesis under anaerobic-light conditions. Changes in energy generation pathways under photosynthetic and aerobic respiratory conditions are primarily controlled by oxygen tensions. In this study, we performed time series microarray analyses to investigate transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration. Major changes in gene expression profiles occurred in the initial 15 min after the shift from anaerobic-light to aerobic-dark conditions, with changes continuing to occur up to 4 hours postshift. Those genes whose expression levels changed significantly during the time series were grouped into three major classes by clustering analysis. Class I contained genes, such as that for the aa3 cytochrome oxidase, whose expression levels increased after the shift. Class II contained genes, such as those for the photosynthetic apparatus and Calvin cycle enzymes, whose expression levels decreased after the shift. Class III contained genes whose expression levels temporarily increased during the time series. Many genes for metabolism and transport of carbohydrates or lipids were significantly induced early during the transition, suggesting that those endogenous compounds were initially utilized as carbon sources. Oxidation of those compounds might also be required for maintenance of redox homeostasis after exposure to oxygen. Genes for the repair of protein and sulfur groups and uptake of ferric iron were temporarily upregulated soon after the shift, suggesting they were involved in a response to oxidative stress. The flagellar-biosynthesis genes were expressed in a hierarchical manner at 15 to 60 min after the shift. Numerous transporters were induced at various time points, suggesting that the cellular composition went through significant changes during the transition from anaerobic photosynthesis to aerobic respiration. Analyses of these data make it clear that numerous regulatory activities come into play during the transition from one homeostatic state to another.
One of the challenges of present genomic research in microbiology is to move from our understanding of microbial systems obtained through studies of steady-state-grown cultures to the fundamental property of any microbial system, namely, adaptation, moving from one environmental state to a new set of environmental parameters. Rhodobacter sphaeroides 2.4.1 is a facultative photosynthetic (PS) bacterium that has a remarkable ability to grow under a variety of environmental conditions. It grows by aerobic respiration in the presence of oxygen. It also grows anaerobically by anoxygenic photosynthesis in the presence of light or by anaerobic respiration in the presence of alternative electron acceptors, such as dimethyl sulfoxide, under anaerobic-dark conditions. R. sphaeroides synthesizes an intracytoplasmic-membrane (ICM) system when grown anaerobically in the light or dark (40). The ICM houses all of the membrane components necessary for the light reactions of photosynthesis. It contains three PS protein-pigment complexes, comprised of the reaction center and two light-harvesting complexes, B875 (LHI) and B800-B850 (LHII). Oxygen tension is a major determinant controlling the formation of the ICM and other PS components (58, 83, 84). The expression of the PS genes (photosynthesis) is mainly regulated by the interplay of three transcriptional regulatory systems, PrrBA, AppA-PpsR, and FnrL (24, 27, 44, 64, 82). PrrBA, designated RegBA in Rhodobacter capsulatus (23), is a two-component regulatory system consisting of the membrane-bound sensor kinase PrrB and the response regulator PrrA (24, 44). PrrB is proposed to respond to oxygen tension by indirectly monitoring electron flow through the cbb3 cytochrome c oxidase (41, 57). The AppA-PpsR antirepressor-repressor system is reported to be responsive to the redox status of the quinone pool (10, 58). AppA also mediates a blue-light regulation of PS gene expression (10, 50). FnrL is a homologue of FNR (fumarate nitrate reduction regulation protein) of Escherichia coli, which contains an iron-sulfur cluster and senses intracellular oxygen tension directly via the redox state of the bound iron-sulfur cluster (75). A fourth system, TspO, appears to modulate the rate of gene expression as cells undergo induction of PS gene expression (78).
A large number of research studies have been published which deal with the transcriptional regulation of PS gene expression controlled by these transcription factors. In addition to those research studies, recent transcriptome and proteome analyses of cells grown under steady-state aerobic-dark, anaerobic-light, and anaerobic-dark conditions have provided a wealth of information pertaining to the genes and proteins regulated by oxygen tension and light intensity in cells growing in the steady state (13, 61, 71). However, there is little information concerning the dynamics of gene expression during adaptation to changing environmental conditions. In this study, we focus on the dynamic change in the transcriptome profile when growth conditions are shifted from anaerobic-light to aerobic-dark, i.e., PS to respiratory metabolism. We seek to address the question as to whether known regulatory systems can account for changes in gene expression profiles. The results presented here show that, in addition to the expected changes in the expression of the genes for energy and carbon metabolism, a number of genes are temporarily upregulated during the transition process. In addition to the genes specifically expressed under steady-state growth conditions, e.g., respiratory or PS, transiently expressed genes may be necessary for the versatile growth ability of R. sphaeroides as cells achieve homeostasis under new growth conditions.
MATERIALS AND METHODS
Growth conditions.
R. sphaeroides 2.4.1 was grown in Sistrom's minimal medium A containing succinate as a carbon source at 30°C (16). The culture was continuously bubbled with a gas mixture of 95% N2 and 5% CO2 and illuminated at a light intensity of 10 W/m2 for anaerobic PS conditions. The gas mixture was 69% N2, 30% O2, and 1% CO2, and the culture was not illuminated for aerobic conditions.
RNA extraction.
R. sphaeroides 2.4.1 was grown in 800 ml of medium in 1-liter glass bottles under anaerobic PS conditions. When the optical density at 600 nm (OD600) reached approximately 0.2 to 0.3, the light was turned off and the composition of the gas mixture was abruptly changed for aerobic growth conditions. RNA was isolated from a 50-ml aliquot of the culture prior to the shift to aerobic conditions (0 min) and at 15 min, 30 min, 1 h, 2 h, and 4 h after the shift. The final OD600 at 4 h was approximately 0.3 to 0.4, representing not quite a culture doubling. The experiment was performed in quadruplicate independent cultures. RNA was extracted by a modified hot-phenol method as described previously (71).
Microarray experiments and GeneChip data analyses.
The R. sphaeroides 2.4.1 GeneChip was custom designed and manufactured by Affymetrix Inc. (Santa Clara, CA) (61). A quadruplicate series of RNA samples at the six time points during the transition from anaerobic-light to aerobic-dark conditions was used for the analyses. cDNA synthesis, fragmentation, and labeling were performed according to the instructions designed for the Pseudomonas aeruginosa genome array by Affymetrix and methods described previously (71). cDNA was synthesized from 10 μg of total RNA with Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). RQ1 DNase I (Promega, Madison, WI) was used for fragmentation of the cDNA. The Enzo BioArray terminal labeling kit (Affymetrix) was used for 3′ labeling of the fragmented cDNA to an approximate size of 50 to 200 bases. Target hybridization, staining, and scanning were performed by using a GeneChip Hybridization Oven 640, a Fluidics Station 400, and the Agilent GeneArray scanner under the control of the Affymetrix Microarray Suite 5.0 (MAS5). The scanned images were analyzed using the MAS5 program to obtain signal values, detection calls (present/absent/marginal), and P values. The relative change (n-fold) and hierarchical clustering of the hybridization intensities of the experimental probe sets were calculated using dChip software (46). The filtering criterion between the group means was a 1.5-fold change using the 90% confidence boundary for 0-fold change, which was calculated using the standard errors of the mean from quadruplicate experiments. For hierarchical analysis, genes significantly up- or downregulated during the transition were filtered by the following two criteria: the variation across samples (after pooling of replicate arrays) was 0.50 < standard deviation/mean < 1,000, and the present call percentages in the array were ≥20%. The criterion used for filtering the gene is described in the dChip manual (http://biosun1.harvard.edu/complab/dchip/filter_gene.htm). A subset of 483 probe sets was obtained with the above criteria and used for hierarchical clustering analysis. The Pearson correlation coefficient value (r value) was calculated using the Microsoft Excel program.
Quantitative analysis of carotenoid, bacteriochlorophyll, and spectral complexes.
Photopigments were extracted with acetone-methanol (7:2 [vol/vol]) from cell pellets by vigorous vortexing. After centrifugation at 13,000 rpm for 15 min, the supernatant was used for quantitation of carotenoid and bacteriochlorophyll as described previously (16).
For analysis of spectral complexes, the harvested cells were resuspended in ICM buffer (10 mM potassium phosphate and 1 mM EDTA, pH 7.2) and disrupted by passage through a French pressure cell at 100 MPa. Cell extracts were obtained following centrifugation at 13,000 rpm for 15 min to remove cell debris. Spectra were recorded with a UV 1601PC spectrophotometer (Shimadzu Corp., Columbia, MD). The B800-B850 and B875 complex levels were determined by the method described previously (52) from the spectral data.
The protein concentrations of cell extracts were determined with the bicinchoninic acid protein assay kit (Pierce, Rockford, IL) with bovine serum albumin as a standard.
RESULTS
Overview of microarray analysis.
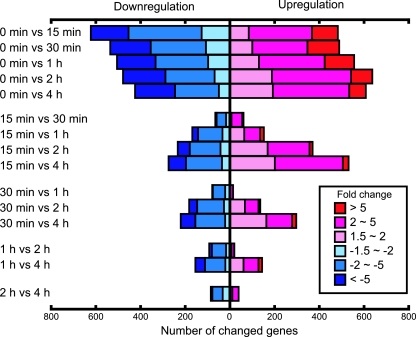
The global gene expression profiles obtained for R. sphaeroides 2.4.1 at six time points during the transition from PS to aerobic conditions were analyzed by using the R. sphaeroides microarray GeneChip (61). The percentages of open reading frames (ORFs) whose expression was detected as “present” by the MAS5 program were 64.0% ± 2.1%, 60.6% ± 3.6%, 62.1% ± 2.3%, 63.6% ± 1.2%, 62.9% ± 2.9%, and 62.1% ± 4.6% at 0-min, 15-min, 30-min, 1-h, 2-h, and 4-h time points, respectively. The data are means ± standard deviations from the series of independent experiments performed in quadruplicate. The number of expressed ORFs appeared to undergo a transient decrease soon after the shift, but the differences were not statistically significant. The Pearson correlation coefficient value (r value) obtained by comparison of the quadruplicate data sets at each time point, which indicates the experimental reproducibility between samples, was between 0.965 and 0.996. The r values obtained by comparison between different time points are shown in Table 1. The values observed between zero min and the other time points were substantially lower than those of the other combinations of time points, and the discrepant values decreased over time, indicating that major changes in gene expression patterns occurred in the initial 15 min postshift but that changes continued to occur even up to 4 h, although these changes were very few.
TABLE 1.
Pearson correlation coefficients between the data sets
| Time | Correlation ata:
|
||||
|---|---|---|---|---|---|
| 15 min | 30 min | 1 h | 2 h | 4 h | |
| 0 min | 0.802 ± 0.018 | 0.790 ± 0.008 | 0.784 ± 0.015 | 0.718 ± 0.006 | 0.696 ± 0.012 |
| 15 min | 0.975 ± 0.003 | 0.936 ± 0.008 | 0.856 ± 0.013 | 0.812 ± 0.021 | |
| 30 min | 0.970 ± 0.006 | 0.893 ± 0.013 | 0.843 ± 0.018 | ||
| 1 h | 0.955 ± 0.005 | 0.913 ± 0.015 | |||
| 2 h | 0.976 ± 0.008 | ||||
The Pearson correlation coefficient values were obtained by comparison of the 5,306 probe sets to one another within the six data sets for each experiment. The values are means ± standard deviations from the quadruplicate series of experiments at each time point.
The numbers of genes/ORFs that showed more than a 1.5-fold change relative to one of the other time points with 90% confidence boundaries were evaluated for all combinations of time points and are depicted in Fig. 1. Large numbers of the genes/ORFs were either upregulated or downregulated compared to the 0-min time point. Expression levels of ∼9.1% (483) or ∼11.7% (621) of the genes/ORFs either increased or decreased within the initial 15 min after the shift, respectively, i.e., over 20% of the genome. The number of upregulated genes relative to the zero time point remained high for both 2 h and 4 h, and we could see that over time, significant numbers of genes increased in expression over the earlier time point. On the other hand, the number of downregulated genes continued to decrease at the later time points, although there were many such genes relative to the zero time point. A total of 37.8% of the genes/ORFs were more than 1.5-fold up- or downregulated within 4 h following the transition.
FIG. 1.
Gene expression changes during the transition from PS to aerobic conditions. The number of genes showing increased and decreased expression and their relative changes are shown. The relative changes for the hybridization intensities of 5,306 probe sets were calculated using dChip. The filtering criterion between the group means was a 1.5-fold change using the 90% confidence boundary for 0-fold change, which was calculated using the standard errors of the mean from independent quadruplicate experiments.
Hierarchical clustering analysis.
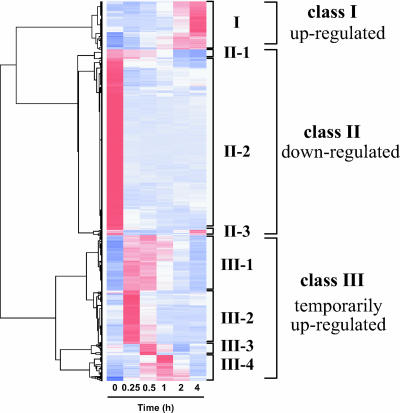
The hierarchical tree consisting of 483 probe sets, which were filtered by the criteria indicated in Materials and Methods, showed three major nodes (I, II, and III), and the genes/ORFs can be classified according to the branching of the tree (Fig. 2). The probe sets used for the hierarchical analysis and their GeneChip data can be found in the supplemental material (see Table S1 in the supplemental material). Classes I and II contained those genes whose expression levels increased or decreased, respectively, after the shift from PS to aerobic conditions. Class III contained genes whose expression levels were transiently increased during the transition. The overall gene expression pattern also suggests that the cells went through three physiologic phases during adaptation to aerobic conditions after the shift, i.e., the early phase (∼30 min), middle phase (30 min to 2 h), and late phase (2 h on). A detailed description of each class follows.
FIG. 2.
Changes in gene expression profiles after transition from PS to aerobic conditions. The hierarchical tree cluster was generated using the dChip program with a subset of 483 genes, which were filtered by the following criteria: variation across samples (after replicate arrays were pooled) was 0.50 < standard deviation/mean < 1,000, and present call percentages in the array were ≥20%. Significantly up- and downregulated genes are shown in red and blue, respectively. The change level (n-fold) is indicated by the color intensity.
(i) Class I.
Class I contained the late-phase genes whose expression levels gradually increased after the shift and showed the highest expression levels at 2 to 4 h. These genes are suggested to be specific for aerobic growth. The hemF gene (AffyChip designation RSP0682), encoding an oxygen-dependent coproporphyrinogen III oxidase, which is involved in aerobic heme biosynthesis (85), was 8.0-fold upregulated at 4 h. Subunit II of the aa3 cytochrome oxidase (RSP1829) was in this class, showing an 11.0-fold upregulation. The other subunits of the aa3 oxidase (RSP1826 and RSP1877) also showed an expression pattern similar to that of subunit II and were 8.2- and 8.4-fold upregulated, respectively, although they were not identified by the filtering criteria. A gene cluster encoding glycerol kinase and glycerol-3-phosphate dehydrogenase and a putative ABC glycerol transporter (RSP2661-RSP2670) was 3.4- to 23.5-fold upregulated. The genes for an ABC glycerol-3-phosphate transporter (RSP3289 and RSP3290) were also in this class (9.7- and 9.5-fold upregulated, respectively). These results indicate that glycerol metabolism was active at later stages after the shift. The structural and accessory genes for periplasmic nitrate reductase (RSP4112-RSP4118), which might be involved in nitrate assimilation, were 4.6- to 15.4-fold upregulated, despite the fact that ammonium was used as a nitrogen source. Putative diguanylate cyclases with GGDEF domains (RSP0406 and RSP3401) were 7.3- and 23.3-fold upregulated, respectively. These gene products might control the intracellular level of a ubiquitous bacterial second-messenger molecule, cyclic di-GMP, which is known to regulate various cellular processes, such as cell surface adhesiveness, biofilm formation, and motility (18, 72). Other genes in this class that are significantly upregulated encode a cold shock protein (RSP2346; 8.5-fold), a putative calcium-binding hemolysin protein (RSP3539; 22.3-fold), and a putative outer membrane adhesin protein (RSP3540; 24.3-fold).
(ii) Class II.
Class II contained genes whose expression levels decreased after the shift. These genes are likely to be specific for anaerobic growth. Most of these genes overlap with the “class I” genes reported by Pappas et al., whose expression was upregulated under both anoxic-dark and anoxic-light conditions (61). Class II can be divided into three subclasses (II-1, II-2, and II-3). The expression level of the genes in class II-1 gradually decreased over time following the shift, with those genes in class II-2 showing a rapid decrease after the shift. Expression of the class II-3 genes was only temporarily downregulated during the transition to aerobic growth.
Class II-1 contained only pufBA (RSP0258; represented by one probe set) and pucBA (RSP0314; represented by one probe set) as genes with known functions. pufBA and pucBA encode proteins of the B875 light-harvesting complex (LHI) and the B800-B850 light-harvesting complex (LHII), respectively. They were 11.6- and 26.3-fold downregulated, respectively, at 4 h compared to 0 min. The regulation of these light-harvesting complex protein genes is different from those of the other PS genes, because most of the other PS genes were categorized as belonging to class II-2 (see below).
Class II-2 was the largest group among the different subclasses. All genes in the large PS gene cluster (RSP0255-RSP0292), except pufBA (RSP0258; class II-1) and ppsR (RSP0282), were in this subclass, although some genes were not identified by using the filtering criteria. The genes of unknown function (RSP0293-RSP0295), which are located in the vicinity of the large PS gene cluster, also showed the same class II-2 expression pattern, suggesting a relationship to PS gene expression, which remains to be tested. Other photosynthesis-related genes that showed the class II-2 expression pattern were cycA (RSP0296), encoding cytochrome c2, which is an electron carrier for the PS reaction center (20); pucC (RSP0315), which is involved in LHII assembly; osp (RSP0869), which is required for optimal synthesis of spectral complexes, as well as for optimal growth under dark-dimethyl sulfoxide conditions (59); blrB (RSP1261), a blue-light receptor of the BLUF family; prrA (RSP1518), a response regulator involved in oxygen regulation of the photosynthesis genes (24); the second LHII genes pucBA2 (RSP1556-RSP1557) (87); and appA (RSP1565), an antirepressor of PpsR (25). dxsA (RSP0254), which is located downstream of the puf genes, is also in this class. dxsA encodes deoxy-d-xylulose 5-phosphate synthase, which is involved in the biosynthesis pathway of isoprenoids and is therefore required for biosynthesis of bacteriochlorophylls and carotenoids (30). dxs (RSP1134), encoding an isoenzyme of deoxy-d-xylulose 5-phosphate synthase, and dxr (RSP2709), encoding deoxyxylulose 5-phosphate reductoisomerase, which is another enzyme of the isoprenoid biosynthesis pathway, also showed a similar expression pattern. The genes encoding enzymes for biosynthesis of protoporphyrin IX, a precursor of bacteriochlorophyll, such as hemA (RSP2984), hemB (RSP2848), hemC (RSP0679), and hemE (RSP0680), are in this class. In particular, hemN (RSP0317) and hemZ (RSP0699), encoding oxygen-independent coproporphyrinogen III oxidases, were significantly (40.9- and 27.1-fold, respectively) downregulated at 15 min. Both of these genes are known to be upregulated by FnrL under anaerobic conditions. It is evident that hemF encodes an aerobic coproporphyrinogen III oxidase, while hemN and hemZ are critical for anaerobic growth. Thus, we predict that hemF is not regulated by either FnrL, PrrA, or PpsR.
A gene cluster (RSP0100-RSP0112) encoding one of two sets of multicomponent NADH-ubiquinone oxidoredctases was 4.3- to 12.0-fold downregulated after the shift and remained low during the transition. The plasmid-encoded putative F0F1-type ATPase genes (RSP3929-RSP3936) were 2.8- to 5.5-fold downregulated at 15 min. This result is inconsistent with the previous report by Pappas et al. (61), in which this gene cluster was upregulated under anaerobic-dark conditions but unchanged under anaerobic-light conditions compared to the aerobic-dark conditions. fbcQ (RSP2687), encoding subunit IV of the bc1 complex, and qxtA (RSP3212), encoding subunit I of the quinol oxidase, were 6.8- and 4.5-fold downregulated, respectively. The genes of the other subunits of these respiratory complexes also showed similar downregulation of their expression patterns. Data presented elsewhere indicate that many of these genes are positively regulated by PrrA (38).
R. sphaeroides has two sets of cbb gene clusters for the Calvin-Benson-Bassham (CBB) cycle enzymes (RSP1278-RSP1285 and RSP3266-RSP3271), including the genes for the form I and form II ribulose-1,5-bisphosphate carboxylase/oxygenases (RubisCOs), respectively. Both cbb gene clusters were significantly (5.7- to 80.1-fold and 26.6- to 303.7-fold) downregulated at 15 min and almost absent after that. The CBB cycle is utilized not only for autotrophic CO2 fixation, but also as an electron sink for maintaining cellular redox balance (31), and is apparently not required for heterotrophic growth as an electron sink in the presence of oxygen. These genes are known to be regulated, at least in part, by PrrA.
Some proteases (RSP0465, RSP2045, RSP3496, and RSP4072), putative restriction endonucleases (RSP2729 and RSP3361), and the DnaK suppressor protein (RSP0166) were found to be members of this class, although their specific functions under PS conditions are unclear. The gene for a universal stress protein (RSP0697), which is located between fnrL and the ccoN operon encoding the cbb3 cytochrome oxidase, was 12.4-fold downregulated at 15 min. Transcription of the RSP0697 gene is probably under the control of FnrL, because there are two FNR-binding motifs in the promoter region between ccoN and the RSP0697 gene (82). RSP0697 is similar to PA4352 of Pseudomonas aeruginosa, which has recently been shown to be essential for survival under anaerobic energy stress conditions (9, 73).
A putative fatty acid desaturase gene (RSP1467), which was highly expressed under anaerobic-light conditions, was significantly (18.8-fold) downregulated after the shift and placed in this subclass. Specific expression of the RSP1467 gene under PS growth conditions was also observed as a result of the proteomic analysis in PS-grown cells (13). Although the physiological function of RSP1467 is not clear, these results seem to contradict the idea that the amount of saturated fatty acids decreases when the growth conditions are changed from anaerobic-light to aerobic-dark (76). This may reflect differences in the fatty acid biosynthetic pathway under anaerobic and aerobic conditions. Another possibility is that the need for fatty acids decreases as the cellular abundance of the total membranes decrease. Altogether, these data reveal numerous gene products which may be parts of the PrrA, PpsR, and FnrL regulons.
Class II-3 genes were temporarily downregulated after the shift from PS to aerobic conditions and showed their lowest expression level at 15 to 30 min. Only some genes, such as bfr (RSP3342), encoding bacterioferritin, and cbbR (RSP1286), encoding a regulator for expression of RubisCOs, were identified in this subclass. However, many genes, especially regulatory genes, such as ppsR (RSP0282) and fnrL (RSP0698), also showed the class III-3 expression pattern. They failed to be identified using the filtering criteria because their expression levels and rates of change were low. Because some of these genes are known to be regulated by FnrL and PrrA, it may be that these changes are important early in the transitions, but other regulatory elements may be involved after the shift.
(iii) Class III.
Class III contains genes whose expression levels temporarily increased during the transition. This class can be divided into four subclasses (III-1, III-2, III-3, and III-4) (Fig. 2). The expression levels of the genes in class III-1 quickly increased after the shift, remained high for 30 to 60 min, and gradually decreased thereafter, often to their preshift levels. Many genes for metabolism or transport of carbohydrates or lipids were characteristically found in this subclass (see Table S1 in the supplemental material). The most highly upregulated (201.2-fold) gene was RSP2364, encoding l-fuculose phosphate aldolase. This gene is clustered with genes (RSP2363-RSP2372) encoding putative proteins for an ABC sugar (ribose) transporter, sugar kinase, 3-oxoacyl-(acyl-carrier protein) reductase, and aldehyde dehydrogenase. All members of this cluster were in this subclass. Interestingly, another putative l-fuculose phosphate aldolase gene (RSP0476), which is in the putative PrrA regulon (48), was significantly downregulated after the shift. These observations suggest that PrrA is not a player in the regulation of class III-1 genes, at least directly. A gene cluster (RSP0480-RSP0488) for several glucuronate metabolic enzymes and a tripartite ATP-independent (TRAP)-type transporter was 7.7- to 62.0-fold upregulated at 15 to 30 min. A gene cluster (RSP1602-RSP1605) for a similar TRAP-type transporter also showed a similar expression pattern. These gene clusters are preceded by the genes encoding similar transcriptional regulators belonging to the GntR family (RSP2362, RSP0489, and RSP1606), suggesting that these transcriptional regulators may be responsible for the class III-1 expression pattern observed. This observation remains to be tested.
A gene cluster (RSP0431-RSP0443) containing the suf genes, which are involved in the assembly and stability of iron-sulfur clusters (7), showed a transient upregulation at 15 to 60 min. This result indicates that the de novo synthesis and/or repair of iron-sulfur proteins is required after exposure to oxygen, which might be expected. Genes encoding a TonB-ExbB-ExbD complex (RSP0920-RSP0922) and two ABC Fe3+ siderophore transporters (RSP2913 and RSP3079), which are essential for ferric iron uptake (5), and a gene cluster (RSP1543-RSP1548) encoding bacterioferritin and related proteins, which might be involved in iron storage, were also found in this subclass, suggesting that ferric iron is stringently required after the shift from anaerobic to aerobic conditions.
The class III-2 genes were immediately upregulated after the shift and showed expression peaks at 15 min, but their expression levels significantly decreased soon after that. These gene products might be involved in a “stress-like” response to the abrupt exposure to oxygen. Heat shock proteins, chaperones, and proteases, such as HSP20 type (RSP1016 and RSP1572), HSP70 type (RSP1172-RSP1173), heat shock proteosome HslVU (RSP1531-RSP1532), Lon protease (RSP2806), and other types (RSP0554, RSP1408, RSP1742, and RSP2710), were 2.6- to 17.3-fold upregulated at 15 min. Two isozymes of peptide methionine sulfoxide reductases (RSP0559 and RSP2617), which counter methionine oxidation, were 2.7- and 2.1-fold upregulated. One of two heat shock sigma factors (RpoH1; RSP2410) (29) was also in this class. These results suggest that exposure to oxygen caused misfolding and oxidative damage of proteins, which may be a reflection of a general stress response. Since the earliest time point after the shift was 15 min, it is possible that some of these genes, and even others not observed, were expressed at even higher levels and that we were witnessing their expression on the downward side of this trend.
Genes for the biosynthesis of methionine (RSP2172 and RSP3346-RSP3347) and an ABC methionine uptake transporter (RSP0130-RSP0132) were more than fourfold upregulated. The cysteine synthase gene (RSP1109) was also in this class. Oxidative stress has harmful effects on thiol groups of reactive cysteine residues in proteins, including cobalamin-independent methionine synthase, MetE, which causes methionine auxotrophy in the case of E. coli (33, 45). The transient upregulation of the genes for the biosynthesis and uptake of methionine in E. coli after exposure to oxygen was also reported recently (62). These results indicate that cells are subjected to temporary deprivation of sulfur-containing amino acids during the transition from anaerobic to aerobic conditions. Gene clusters encoding sulfide reductase (RSP1940-RSP1944), an ABC sulfate transporter (RSP3696-RSP3699), and sulfurtransferase (RSP2738) also showed high expression levels at 15 min postshift. Thus, the metabolism of sulfur compounds seems to be generally upregulated after exposure to oxygen.
A gene cluster (RSP2196-RSP2200) that might be involved in fatty acid metabolism showed very high expression levels (25.1-to 51.6-fold) immediately following the shift, followed by near-preshift expression levels. A gene cluster encoding lipolytic enzyme and a possible ABC efflux transporter (RSP3601-RSP3603) was upregulated 3.0- to 4.7-fold but dropped thereafter. From these results, together with the results of class III-1, lipid metabolism seems to be generally activated at earlier stages after exposure to oxygen. Genes for an ABC-type divalent-cation transporter (RSP0904-RSP0908) and a putative sodium/calcium exchanger (RSP2638) were upregulated 7.6- to 26.8-fold at 15 min, suggesting that the ion composition in the cells significantly changed after the shift from anaerobic to aerobic conditions. Such results could also be symptomatic of a transient stress response. What regulators might be involved in these responses remains to be determined.
Class III-3 includes genes that were temporarily upregulated during the transition, with an expression peak at 30 min after the shift. The genes in this class are exclusively involved in biogenesis of the flagellum, and most of them are in the large flagellar-gene cluster (RSP0032-RSP0083). One of four σ54 factors, RpoN2 (RSP0068), which regulates flagellar-gene expression (67), is also in this class. Regulation of total flagellar-gene expression is discussed below.
Class III-4 includes the middle-phase-specific genes, which were transiently expressed at 1 h or had a broad expression peak at around 1 h after the shift. Many transporter genes are characteristically found in this class. The genes related to siderophore transporters or receptors (RSP1437-RSP1440, RSP3056, RSP3221-RSP3224, and RSP3678) were 5.2- to 122.8-fold upregulated at 1 h after the shift, indicating that these gene products were involved in iron uptake during the middle phase of the experiment. A gene cluster (RSP2273-RSP2279) encoding an ABC branched-chain amino acid transporter, putative anaerobic phenylacetate coenzyme A (CoA) ligase, and long-chain acyl-CoA synthetase was 13.1- to 37.2-fold upregulated. The gene for methylcrotonyl-CoA carboxylase (RSP2509), which is probably involved in leucine metabolism, was highly (65.6-fold) upregulated at 1 h after the shift. The gene for a putative fumarylacetoacetate hydrolase (RSP3169), which is probably involved in tyrosine metabolism, was also in this class. These results suggest that metabolism of some aliphatic and aromatic amino acids was active at this stage. Genes for other transporters, such as ABC transporters (RSP3081 and RSP3167-RSP-3168) and the TRAP-type C4 dicarboxylate transport system (RSP3371-RSP3372), were also in this class.
We are well aware that the mere presence or lack of a particular RNA species does not readily imply that the protein products behave similarly. However, where data are available, there is a remarkably good correlation between the two. Similarly, these studies do not take into account mRNA stability.
DISCUSSION
Because of the extensive display of information presented in this analysis, we discuss the integration of these results extensively below rather than in Results. We summarize these discussions in “Conclusions.”
Switching of energy metabolism after the shift from anaerobic-light to aerobic-dark conditions.
The energy conservation system of R. sphaeroides abruptly switches from photosynthesis to aerobic respiration following the shift from anaerobic-light to aerobic-dark conditions. Most of the PS-related genes showed rapid downregulation after the shift and were categorized as falling into class II-2. This class includes the genes for a reaction center (puhA, pufL, and pufM), the bch genes for bacteriochlorophyll biosynthesis, the crt genes for carotenoid biosynthesis, the gene involved in assembly of the spectral complex (pufQ), the genes for putative spectral-complex-specific assembly factors (RSP0290, RSP0292, RSP0293, and RSP0295, which correspond to lhaA, puhB, puhC, and puhE, respectively, of R. capsulatus) (1-3, 80, 81), and genes for the biosynthesis of isoprenoids, which are the precursors for bacteriochlorophyll, carotenoids, and quinols. On the other hand, the pufBA and pucBA genes for the light-harvesting complex proteins of B875 and B800-B850, respectively, were more slowly downregulated and were categorized in class II-1 (Fig. 3A). Two transcripts with different half-lives have been identified for both pufBA and pucBA (43, 88). The small 0.5-kb pufBA-specific transcript and the 0.5-kb pucBA-specific transcript have long half-lives of 20 and 20.5 min, respectively. The large 2.6-kb transcript of pufBALMX and the 2.3-kb transcript of pucBAC have half-lives of 9 min and less than 5 min, respectively (21, 39, 43, 88). The observed expression of pufBA and pucBA at 15 min and later time points is probably attributable to the 0.5-kb transcripts, because the expression of pufLMX and pucC on the large transcripts decreased quickly after the shift (Fig. 3A). Thus, the slow response of pufBA and pucBA might be at least partly due to the long turnover of the small transcripts of pufBA and pucBA. These results have important physiological implications. If the cells abruptly return to PS growth conditions, any new bacteriochlorophyll synthesized will be immediately sequestered by the potentially abundant apoproteins of the light-harvesting complexes and thus will not pose a potential danger to the cell.
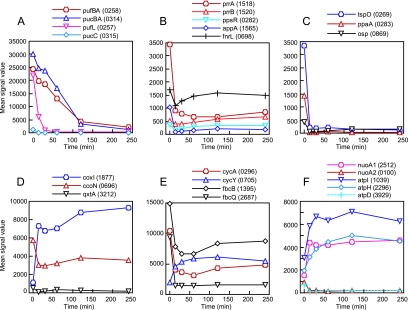
FIG. 3.
Changes in expression of the genes for energy metabolism during the transition from anaerobic photosynthesis to aerobic respiration. (A) The PS genes for light-harvesting complexes. (B) The genes for major oxygen-responsive regulators. (C) The genes for minor regulators for PS gene expression. (D) The genes for terminal oxidases for aerobic respiration. (E) The genes for cytochrome c and cytochrome bc1 complex. (F) The genes for NADH dehydrogenases and F0F1 ATPases. One representative gene was chosen for multisubunit enzymes or multigene operons. RSP numbers of the genes are indicated in parentheses.
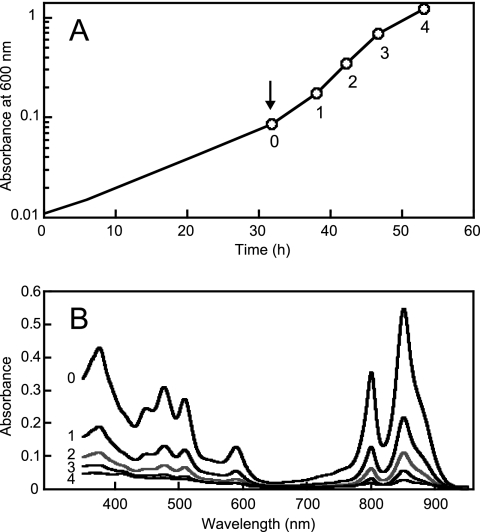
The expression of the genes for the PS reaction center and the genes for the biosynthesis of bacteriochlorophyll and carotenoids was immediately shut off after exposure to oxygen. However, the light-harvesting complexes, bacteriochlorophyll, and carotenoids did not disappear immediately after the shift but decreased by approximately half at each culture doubling (Fig. 4 and Table 2). These results indicate that the PS apparatus housed in the ICM vesicles was generally diluted out as a function of cell division rather than being actively degraded under aerobic growth conditions. Failure to degrade the PS apparatus quickly may be advantageous for the survival of R. sphaeroides in natural environments, where growth conditions fluctuate between anaerobic and aerobic and/or light and dark. Thus, the imposition of PS growth conditions finds these cells with an active PS membrane system, as discussed further below.
FIG. 4.
Effects of the shift in growth conditions from anaerobic-light to aerobic-dark on the levels of spectral complexes. (A) Growth profile of R. sphaeroides 2.4.1. After the strain was grown anaerobically by sparging it with 95% N2-5% CO2 in the light (10 W/m2) to an OD600 of 0.086, the growth conditions were shifted to aerobic-dark by sparging with 69% N2-30% O2-1% CO2. The time point when the growth conditions were shifted is indicated by an arrow and a number zero. The numbers one to four indicate the first, second, third, and fourth culture-doubling times after the shift, respectively. (B) Absorption spectra of crude cell extracts obtained from the cells during the transition from photosynthesis to aerobic respiration. Samples were prepared just before the shift (0) and at each doubling time (1 to 4) after the shift. Equal protein concentrations (0.17 μg/ml) were used for the measurement.
TABLE 2.
Effects of the shift in growth conditions from anaerobic-light to aerobic-dark on the concentrations of spectral complexes and photopigmentsa
| Doublingb | Amt of spectral complexes (nmol/mg of protein)c
|
Amt of photopigments (μg/mg of protein)c
|
||
|---|---|---|---|---|
| B875 | B800-B850 | BChl | CRT | |
| 0 | 5.32 (100) | 9.21 (100) | 29.9 (100) | 6.20 (100) |
| 1 | 2.18 (41.0) | 3.66 (39.7) | 11.6 (38.8) | 2.89 (46.6) |
| 2 | 1.09 (20.5) | 1.90 (20.6) | 5.14 (17.2) | 1.52 (24.5) |
| 3 | 0.52 (9.83) | 0.97 (10.5) | 1.94 (6.49) | 0.73 (11.8) |
| 4 | 0.19 (3.53) | 0.38 (4.15) | 0.56 (1.87) | 0.40 (6.45) |
After R. sphaeroides 2.4.1 was grown anaerobically by sparging it with 95% N2-5% CO2 in the light (10 W/m2) to an OD600 of 0.086, the growth conditions were shifted to aerobic-dark by sparging with 69% N2-30% O2-1% CO2.
Samples were taken just before the shift (0) and at the first, second, third, and fourth doubling times after the shift (1 to 4) as indicated in the legend to Fig. 4.
The values in parentheses are percentages relative to the initial values before the shift. BChl, bacteriochlorophyll; CRT, carotenoids.
Expression of the PS genes is primarily regulated by three major transcriptional regulators, PrrBA, FnrL, and PpsR (24, 25, 44, 64, 82). Expression of these regulators during the transition is shown in Fig. 3B. PrrBA is a two-component global regulator that controls the expression of the genes not only for photosynthesis, but also for CO2 fixation, N2 fixation, and H2 metabolism (22), as well as numerous other functions (J. M. Eraso and S. Kaplan, unpublished data). PrrA (RSP1518) is a response regulator, and PrrB (RSP1520) is a membrane-bound histidine kinase. The expression levels of PrrA decreased immediately after the shift and remained low during the transition, and it was placed in class II-2. This result appears to contradict previous results (71), in which the expression level of prrA did not significantly change between the anaerobic-light and aerobic-dark conditions. Because the expression of prrA started to increase at 4 h, the expression level could be recovered when the cells reach aerobic steady-state growth. This result also indicates that the prrA gene itself is subject to anaerobic regulation. Recent studies have shown that the expression of prrA is dependent on the AppA-PpsR antirepressor/repressor system (53), although this cannot provide for the return of prrA expression under steady-state aerobic growth. The expression of appA is dependent on PrrA (38). Such an interplay between the AppA-PpsR and PrrBA systems is likely to be important for global gene regulation responding to oxygen and light, as suggested previously (32, 34). The expression level of prrB decreased slightly over time after the shift and then increased over time, but the change was not significant, indicating that prrA and prrB are regulated differently.
FnrL (RSP0698) is a homologue of the oxygen-sensing regulator FNR of E. coli (82). The expression level of fnrL was transiently downregulated at 15 min postshift but soon recovered and remained high throughout the transition (Fig. 3B). FNR of E. coli is also constitutively expressed irrespective of the oxygen concentration (35). Recent studies have shown that FNR-dependent genes rapidly respond to the shift from aerobic to microaerobic conditions (63). The constitutive expression of FnrL may provide an advantage for the rapid response to environmental oxygen fluctuations. The immediate downregulation of fnrL should have an effect on such genes as bchE, the cco operon, and hemN or hemZ, because FnrL is important in their expression.
PpsR (RSP0282) is a repressor of PS pigment synthesis (64), acting on the bch and crt genes, to mention only a few. The expression pattern of ppsR was similar to that of fnrL, although the level of expression was always low (Fig. 3B). AppA (RSP1565), the antirepressor of PpsR (26), was significantly downregulated at 15 min postshift and remained low during the transition (Fig. 3B). Thus, the PpsR-dependent repression of PS genes after exposure to oxygen was effectively increased due to the lowered expression levels of AppA. The other gene regulators involved in PS gene expression, tspO (RSP0269) (77), ppaA (RSP0283) (28), and osp (RP0869) (59), also showed a class II-2 expression pattern (Fig. 3C). The transcriptional values observed for tspO accurately reflect the steady-state expression levels observed for this gene (79). What is significant is the rapidity with which tspO transcription shuts down after the shift. Since TspO is involved in the export of excess porphyrin from the cell under PS conditions, maintenance of this activity could constitute a potential problem when cells are placed under aerobic growth conditions.
R. sphaeroides possesses a highly branched aerobic respiratory chain (47). The multiple genes for each respiratory complex were differently regulated during the transition. Cytochrome c oxidases of the aa3 type, encoded by the cox genes (RSP1877, RSP1826, and RSP1829), and the cbb3 type, encoded by the cco genes (RSP0693-RSP0696), are important under high- and low-oxygen conditions, respectively. The cbb3 oxidase was downregulated and the aa3 oxidase was upregulated soon after the shift (Fig. 3D), which took place in a high (30%) oxygen concentration after the shift. The kinetic values observed for the ccoNOQP operon is in keeping with our knowledge of the relative steady-state abundance of the cbb3 complex. We have seen (54) that the cbb3 complex is approximately 2.0- to 3.0-fold more abundant under PS conditions than under conditions of high aeration. The expression level of the Qxt quinol oxidase (RSP3210-RSP3212) was low and downregulated at 15 min. The second quinol oxidase, Qox (RSP3097-RSP3098) (55), and another putative cytochrome oxidase (RSP0117-RSP0118), which were found by genome analyses, were not expressed under the tested conditions (data not shown). These two oxidases, as has been previously reported, have never been shown to be expressed. The cco genes have been shown to be regulated by both PrrA and FnrL, which themselves are downregulated after the shift.
Two cytochromes c, soluble cytochrome c2 and membrane-bound cytochrome cy, encoded by cycA (RSP0296) and cycY (RSP0705), respectively, are known to act as electron donors for both the aa3 and cbb3 oxidases (17). While cytochrome c2 is also required for PS growth and cytochrome cy is not functional in PS electron transport (12, 20, 56), cycA was significantly downregulated soon after the shift and, conversely, cycY was upregulated after the shift (Fig. 3E). These results were consistent with those of previous studies (13, 61, 71) and fit the physiological functions ascribed to these cytochromes. Although cycA was downregulated after the shift, its expression level was comparable to that of cycY at 4 h, suggesting that both cytochromes were functioning under aerobic conditions. The subunits of cytochrome bc1 complex, encoded by fbcCBF (RSP1394-RSP1396) and fbcQ (RSP2687), were downregulated after the shift (Fig. 3E). The rapidity with which fbcQ transcription falls off is to be contrasted with the more moderate decreases observed for fbcB, suggesting that the Q subunit is not essential for the aerobic functioning of the bc1 complex. The result is consistent with previous studies showing that fbcQ is not expressed under highly aerated conditions and is unnecessary for aerobic growth (14). These observations reveal an underlying interesting regulatory pattern. Further, it is important to point out that the fbcQ gene maps 1.84 Mb away from the main fbc locus on chromosome I and that the Q subunit of the bc1 complex has been found only in R. sphaeroides. Interestingly, the expression patterns of fbcCBF, cycA, and cco genes were very similar, suggesting the coordinate regulation and function of the cytochrome bc1 complex, cytochrome c2, and cbb3 cytochrome oxidase. Since the requirement for these gene products is substantially increased under PS conditions, these results were not unexpected. In line with these findings, it is noteworthy that the expression of hemA (RSP2984) also drops significantly after the shift, reflecting the decreased need for porphyrins.
The genes for NADH dehydrogenase (RSP2512-RSP2531) were significantly upregulated soon after the shift and remained high during the transition (Fig. 3F). On the other hand, another gene cluster for a putative NADH dehydrogenase (RSP0100-RSP0112) was downregulated after the shift, as reported previously (61, 71). Two chromosomal gene clusters for the F0F1-type ATPase (RSP1035-RSP1039 and RSP2296-RSP2300) were upregulated during the transition (Fig. 3F). The expression levels at 4 h, although still higher than at earlier times after the shift, are expected to decrease over time because their expression levels are reported to be unchanged between steady-state aerobic-dark and anaerobic-light conditions (61). On the other hand, the expression level of a third putative F0F1-type ATPase (RSP3929-RSP3936), which is encoded on plasmid A, was low and downregulated soon after the shift (Fig. 3F).
Changes in carbon metabolism during the transition.
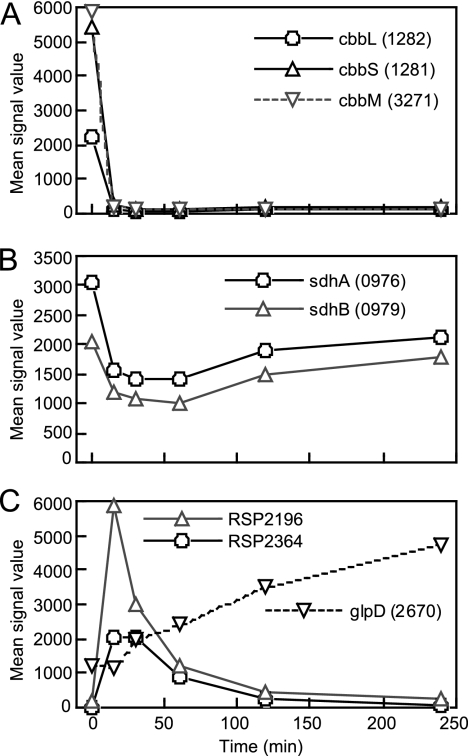
R. sphaeroides utilizes CO2 as a primary carbon source via the CBB cycle under anaerobic PS conditions. The CBB cycle is also used as an electron sink for the maintenance of the redox poise of the cells under PS conditions (31). Transcription of both the cbbLS and cbbM genes, encoding high-affinity and low-affinity RubisCO enzymes, respectively, which catalyze CO2 assimilation, was shut off immediately after the shift from anaerobic-light to aerobic-dark conditions (class II-2), indicating that CO2 was no longer utilized as a primary carbon source under aerobic conditions (Fig. 5A). Because the growth medium contained only succinate as a carbon source, aerobic growth is expected to be supported by succinate. Some TRAP-type transporters, which may be involved in the transport of C4-dicarboxylates, were significantly upregulated after exposure to oxygen. They were placed in class III-1 (RSP0484-RSP0487 and RSP1602-RSP1605) or III-4 (RSP3370-RSP3372). Another, similar transporter was also expressed at high level and shown to possess a class I-type expression pattern (RSP0910-RSP0912). At least one of these transporters might be responsible for the uptake of succinate. However, the expression of succinate dehydrogenase (sdh; RSP0974-RSP0979) was temporarily downregulated after the shift to aerobic conditions. The expression levels of the sdh genes at 4 h were lower than those before the shift (Fig. 5B), suggesting succinate was also utilized under PS conditions before the shift, as indicated previously (13). It is used partly as a source of reducing power for the assimilation of CO2.
FIG. 5.
Changes in expression of genes for CO2 fixation and carbon metabolism during the transition from anaerobic photosynthesis to aerobic respiration. (A) The genes for form I (cbbLS) and form II (cbbM) RubisCOs. (B) The genes for succinate dehydrogenase. (C) Representative genes for carbohydrate (RSP2196), lipid (RSP2364), and glycerol (glpD) metabolism that show typical temporal expression patterns. RSP numbers of the genes are indicated in parentheses.
Unexpectedly, metabolic enzymes and transporters for glucuronates and other carbohydrates were significantly upregulated at early stages during the transition (Fig. 5C). It is likely that carbohydrates accumulated during PS growth were mobilized and initially utilized as carbon sources after the shift to aerobic conditions. Oxidation of those carbohydrates might be advantageous for the maintenance of cellular redox homeostasis after exposure to oxygen. It is also likely that the cells become more dependent upon exogenous carbohydrates after the shift.
Metabolic and transporter genes for lipids were also temporarily upregulated after the shift to aerobic conditions. The genes for glycerol metabolism and transport were upregulated at a later time (Fig. 5C). These results suggest that some lipids were degraded soon after the transition and the derived glycerol was degraded later during the transition. The number of ICM vesicles is known to decrease under aerobic growth conditions (40), but it is not certain whether the lipids derived from the ICM are degraded for reutilization as carbon sources. As mentioned above, the amount of PS pigments housed in the ICM vesicles decreased by approximately half at each cell generation, suggesting that the ICM components were not actively degraded (Fig. 4 and Table 2). Also, the genes for lipid metabolism that were transiently upregulated were downregulated during a later phase of the transition, when more than half of the PS pigments still remained (Fig. 4 and 5). Since the ICM of R. sphaeroides has been shown to alter its lipid composition during steady-state PS growth, the subsequent degradation of some lipid materials would not have as adverse an effect upon the stability of the spectral complexes, and the lipid could serve as a readily available carbon source (37).
Iron homeostasis during the transition.
The aerobic upregulation of iron transport genes usually occurs in bacteria (5, 8). Iron is present in the soluble ferrous form under anaerobic conditions, but it is oxidized to the insoluble ferric form in the presence of oxygen. Therefore, solubilization by ferric chelators, called siderophores, is necessary for the uptake of iron in order to synthesize iron-containing enzymes and components for aerobic metabolism and respiration. The ferrous iron transport system FeoB (RSP1818), which can import ferrous iron directly, was significantly downregulated soon after the shift (class II-2), suggesting that ferrous iron was quickly oxidized and was no longer available after exposure to 30% oxygen. The TonB-ExbB-ExbD protein complex (RSP0920-RSP0922) and ABC-type transporters (RSP2913 and RSP3079), both of which are required for siderophore-mediated ferric iron uptake (5), showed a significant temporary upregulation at early times (15 to 30 min) during the transition (class III-1). As mentioned above, de novo synthesis and repair of iron-sulfur cluster proteins seemed to be active at early times after the shift. This observation might also lead to the temporary high iron demand after the shift. Such responses have been observed in many bacteria, including R. sphaeroides, under oxidative-stress conditions (8, 62, 86). Other genes for a siderophore-mediated ferric iron transport system (RSP1437-RSP1440, RSP3056, RSP3221-RSP3224, and RSP3678) were also found in class III-4. They were transiently upregulated at midphase (30 min to 2 h). Thus, R. sphaeroides appears to utilize different uptake systems at early and middle phases following the shift. The reason is not certain, but it is probably related to the minimization of the toxic effect of iron by controlling uptake efficiency.
The gene for one of the bacterioferritins (RSP1546), which is clustered with some genes for other putative iron-related functions (RSP1543-RSP1548), was temporarily upregulated early postshift. Bacterioferritin is a heme-containing iron storage protein and is believed to function as an intracellular iron supplier under iron starvation conditions (5). Bacterioferritin also functions to limit the amount of iron available to prevent generation of the highly toxic hydroxyl radical. Interestingly, the gene for another bacterioferritin (RSP3342), which was highly expressed during PS growth, showed an opposite expression pattern, that is, significant temporal downregulation after the shift (class II-3). These results indicate that R. sphaeroides utilizes different bacterioferritins under anaerobic and aerobic conditions.
Oxidative-stress response.
The shift from anaerobic photosynthesis to aerobic conditions would be expected to induce oxidative stress. The genes for mediating an oxidative-stress response are expected to be placed in class III-1 or class III-2, i.e., they are significantly upregulated soon after the shift. Actually, the genes for some heat shock protein families and proteases, many of which are also reported to be responsible for the H2O2-induced oxidative stress, showed a sharp expression peak at 15 min (class III-2) (86). Reactive sulfur sites, such as thiol groups and iron-sulfur clusters, are sensitive to oxidative stress. Genes for the biosynthesis of methionine or cysteine, the repair of oxidized methionine residues, and the uptake of sulfate were all upregulated at 15 min and were placed in class III-2. These results suggest that the sulfur-containing amino acid residues are damaged and stringently required soon after the shift, but only for a short period. On the other hand, genes for assembly and repair of iron-sulfur clusters and ferric iron uptake were upregulated soon after the shift and remained at high levels for 30 to 60 min (class III-1), suggesting that iron-sulfur clusters were subjected to oxidative damage for a relatively long period compared to the thiol groups.
An operon of two genes, rpoE (RSP1092) and chrR (RSP1093), which encode one of the alternative RNA polymerase sigma factors of the extracytoplasmic function (ECF) family and its anti-sigma factor, respectively, was significantly (∼4.4-fold) upregulated at 15 to 30 min (Fig. 6A). It has been reported that the activity of RpoE increases when cells are exposed to the highly reactive oxygen species singlet oxygen, (1O2) and that RpoE-dependent gene expression is required for protecting cells from 1O2 when the cells contain low levels of carotenoids (6), although at 15 to 30 min postshift, carotenoid levels are high. The expression of RpoE is also reported to be upregulated by blue-light irradiation or exposure to H2O2 (11). 1O2 is known to be produced by energy transfer from excited-state triplet chlorophyll to oxygen (42). Probably, the abrupt exposure of the PS cells to 30% oxygen caused the production of 1O2 and the RpoE-dependent transcriptional response was required for adaptation to the aerobic conditions.
FIG. 6.
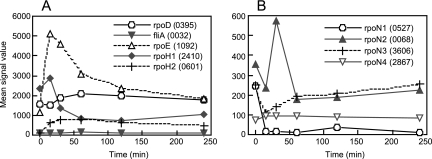
Changes in expression of the RNA polymerase sigma factor genes during the transition from anaerobic photosynthesis to aerobic respiration. (A) The family of sigma 70. (B) The family of sigma 54. RSP numbers of the genes are indicated in parentheses.
Differential expression of multiple RNA polymerase sigma factors.
R. sphaeroides has 17 genes for RNA polymerase sigma factors, which include one σ70 type (rpoD), one σ28 type (fliA), two heat shock response σ32 types (rpoH1 and rpoH2), and four RpoN (σ54) types (rpoN1-rpoN4). Another nine genes are predicted to encode members of the ECF family (47). The gene for housekeeping σ70 factor (RSP0395) was slightly upregulated at the middle phase but constitutively expressed during the transition (Fig. 6A). The fliA gene (RSP0032) is required for flagellar biosynthesis, and its regulation is discussed below. One of the ECF sigma factor genes, rpoE (RSP1092), which is responsive to 1O2, as mentioned above, was significantly upregulated after the shift (Fig. 6A). The expression levels of the other ECF sigma genes remained low or were not significantly changed during the transition, suggesting that they have no significant function under the tested conditions (data not shown).
Two heat shock sigma factor genes, rpoH1 (RSP2410) and rpoH2 (RSP0601), were upregulated at 15 min. While rpoH1 was significantly downregulated after 30 min, rpoH2 remained induced during the transition, though the expression level was lower than that of rpoH1 (Fig. 6A). It has been suggested that RpoH1 and RpoH2 regulate different yet overlapping regulons in response to heat shock and other stresses (29). Because the expression level of rpoH1 before the shift was significantly higher than that of rpoH2, RpoH1 might play a significant role under anaerobic PS conditions. On the other hand, the expression level of rpoH2 under PS conditions was low. It has been reported that the expression of rpoH2 is dependent on RpoE and is therefore controlled by 1O2 (6). These results suggest that RpoH2 specifically works under oxidative-stress conditions in the presence of oxygen.
R. sphaeroides has four different σ54-type sigma factors, which are not functionally interchangeable (65). rpoN1 (RSP0527) is clustered with the nif genes for nitrogen fixation. Although disruption of rpoN1 of R. sphaeroides strain HR has no effect on diazotrophic growth, the rpoN1 mutant of strain WS8 shows a severe defect in diazotrophic growth, and therefore, the physiological function of RpoN1 is predicted to be involved in controlling the expression of the nitrogen fixation genes (51, 65). rpoN1 was expressed under PS conditions but significantly downregulated to absent after the shift to aerobic conditions (Fig. 6B). Because nitrogen fixation specifically occurs under low-oxygen or anaerobic conditions, the expression pattern of rpoN1 is consistent with its predicted function. rpoN2 (RSP0068) is clustered with the flagellar genes and is required for motility (65). The expression of rpoN2 was transiently upregulated at 30 min after the shift, as is the case for most of the flagellar genes, as discussed below (Fig. 6B and 7). rpoN3 (RSP3606) showed transient downregulation at 15 to 30 min after the shift (Fig. 6B). A recent report has shown that RpoN3 regulates one of the chemosensory operons, cheOp3, which clusters with the flagellar genes (49). The function of rpoN4 (RSP2867) is not certain at present, but it has been shown by real-time reverse transcription-PCR that rpoN4 is transcribed under photoheterotrophic conditions (65). The expression level of rpoN4 remained low during the transition in our microarray data, suggesting that RpoN4 was not functional under the growth conditions of this study (Fig. 6B).
FIG. 7.
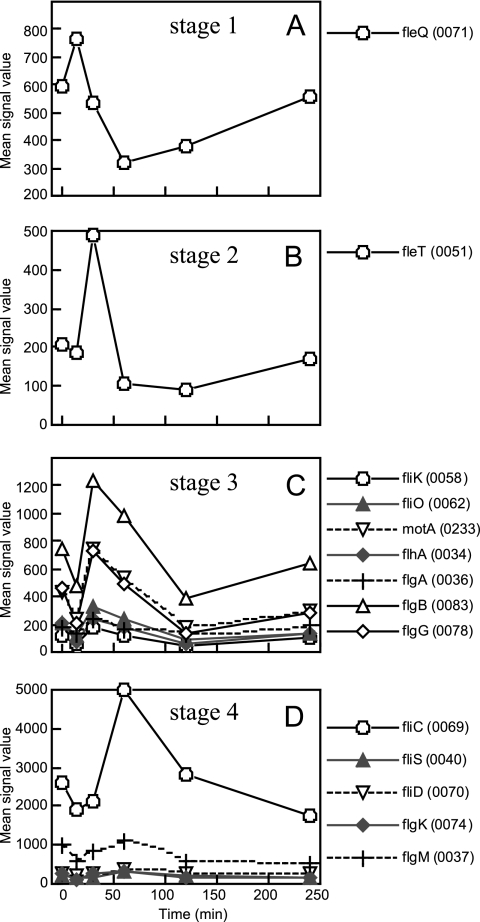
Four-tiered hierarchical expression of the flagellar genes during the transition from anaerobic photosynthesis to aerobic respiration. The genes of classes I to IV of the flagellar transcriptional hierarchy are shown in stages 1 (A) to 4 (D), respectively. Initial genes were chosen as representatives of putative multigene operons. RSP numbers of the genes are indicated in parentheses.
Hierarchical expression of the flagellar genes during the transition.
Biogenesis of the bacterial flagellum requires more than 40 genes. The expression of those genes is coordinately regulated transcriptionally and posttranslationally by a hierarchical regulatory network for the efficient and accurate assembly of the flagellum. The flagellar genes have been placed in three or four classes that correspond to the temporal requirement during flagellum assembly or the hierarchy of regulation (4, 15). R. sphaeroides has a four-tiered (“classes I to IV”) transcriptional hierarchy (66) that is similar to that of Vibrio cholerae or P. aeruginosa (19, 69). To avoid confusion in the names of the classifications, we will call “classes I to IV” of the flagellar hierarchy “stages 1 to 4” here. The stage 1 gene is the master regulator gene fleQ (RSP0071), which encodes a σ54-dependent activator and is required for expression of all flagellar genes. fleQ was marginally (1.3-fold) upregulated at 15 min after the shift (Fig. 7A). The stage 2 genes are members of the fleT-fliEFGHIJ operon (RSP0051-RSP0057), which encodes another σ54-dependent regulator, FleT; an MS ring; a C ring; and proteins involved in assembly and export of the flagellar components. The genes for most of the other basal-body components belong to stage 3 and are transcribed from the fliK (RSP0058), fliO (RSP0062), motA (RSP0233), flhA (RSP0034), flgA (RSP0036), flgB (RSP0083), and flgG (RSP0078) promoters. All promoters for the stage 2 and 3 genes are σ54 dependent, and one of four σ54 factors, RpoN2 (RSP0068), specifically regulates these promoters. The fleT promoter of stage 2 also requires FleQ, and the promoters of stage 3 require both FleQ and FleT. The microarray data showed that the genes of stages 2 and 3 had expression peaks at 30 min after the shift (Fig. 7B and C). While the stage 2 genes were significantly downregulated at 1 h, the stage 3 genes were highly expressed at 1 h, indicating that the expression peak time of the stage 3 genes was later than that of the stage 2 genes. The significant downregulation of the stage 2 genes at 1 h was probably due to the self-repression of the fleT promoter by FleT (66). The rpoN2 gene showed the same expression pattern as the stage 2 genes mentioned above (Fig. 6B). The stage 4 genes, which are expressed at the last stage of flagellar biogenesis, are transcribed from the fliC (RSP0069), fliS (RSP0040), fliD (RSP0070), flgK (RSP0074), and flgM (RSP0037) promoters. Their expression is dependent on the σ28 factor FliA (RSP0032). The stage 4 genes were transiently upregulated, with a peak at 1 h after the shift (Fig. 7D). The fliA gene also showed upregulation at 1 h, though its expression level was low compared to those of the other sigma factor genes (Fig. 6A). The expression levels at 30 min and 1 h were almost equal, suggesting that the expression peak time of fliA was earlier than those of the stage 4 genes and somewhere between 30 min and 1 h. Regulation of the biogenesis of the flagellum is believed to be coupled with the cell cycle (36, 60, 70, 74). Our results from the time series microarray experiment during the transition from anaerobic-photosynthesis to aerobic-respiration conditions clearly demonstrate the four-tiered hierarchical expression of the flagellar genes. The results also suggest that the cell cycle of R. sphaeroides was synchronized after the exposure to oxygen, and that was probably because of the temporary cessation of growth soon after the shift.
R. sphaeroides also has another complete set of flagellar genes (RSP1302-RSP1336, RSP2220, and RSP3878-RSP3881) (47, 68). These second flagellar genes seemed to be nonfunctional under the tested conditions, because the expression levels of the genes were low throughout the transition.
Conclusions.
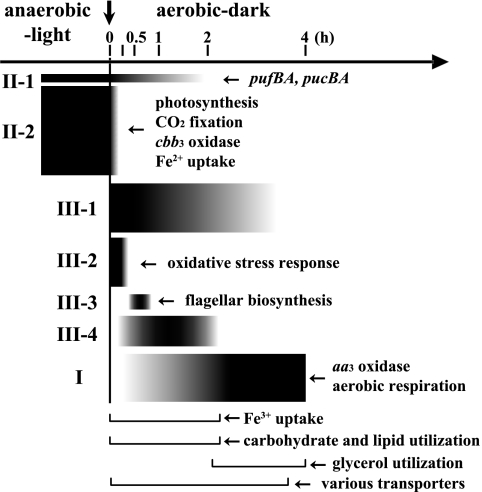
Although it would be easy to focus our attention on any one of the numerous changes depicted under categories I, II, and III, the more important matters of concern center about the underlying mechanisms that permit R. sphaeroides to sense, evaluate, and respond to those perturbations accompanying the transition from one steady state to another. For example, what regulatory systems are involved, and how do these systems integrate the totality of the environmental changes into physiological adjustments involving diverse metabolic parameters to new metabolic alignments. We believe that the data profiles presented here can lead to the formulation of testable hypotheses. In this study, we have revealed that more than one-third of the genes/ORFs of R. sphaeroides are significantly (>1.5-fold) up- or downregulated after a shift in growth conditions from anaerobic photosynthesis to aerobic respiration. The gene expression patterns during the transition state are somewhat arbitrarily categorized into several classes, as described schematically in Fig. 8 for ease of presentation and discussion, although the patterns are quite reproducible. The transcriptome and proteome profiles of steady-state PS and aerobic conditions have been reported previously (13, 61, 71). As predicted from those previous studies, genes such as those for photosynthesis, carbon dioxide fixation, and the high-affinity cbb3 cytochrome oxidase were downregulated and genes for the low-affinity aa3 cytochrome oxidase were upregulated after the shift. The transcriptional responses of those genes were generally very quick. The results here also demonstrate that a large number of genes that had not been previously identified by any earlier steady-state analyses are transiently expressed during the transition. Those genes are expected to be specifically involved in the adaptation of PS cells to the aerobic environment, rather than to steady-state aerobic growth. In addition to some protein repair chaperones, the genes for biosynthesis and repair of sulfur-containing amino acids, thiol group, or iron-sulfur clusters were significantly upregulated soon after the shift, suggesting that they are both necessary for aerobic growth and counteract oxidative stress. Uptake of ferric iron is also important for adaptation to aerobic conditions. Unexpectedly, the most significantly upregulated genes during the transition were metabolic and transport genes for carbohydrates or lipids. Probably, those compounds were accumulated during PS growth and were initially utilized as carbon sources soon after the shift. Interestingly, the ordered expression of the flagellar genes according to the proposed transcriptional hierarchy was observed at 15 to 60 min. The results suggest that there was a cell cycle synchronization after the shift from anaerobic-light to aerobic-dark conditions. In addition to those genes with known or predictable functions, many genes with unknown functions were found to be transiently expressed. Further detailed analyses of those genes would provide novel information about the mechanism and strategy utilized by R. sphaeroides for adaptation and survival in changing environments. Finally, we have suggested the importance of known regulatory elements during this transition, and we have also provided evidence for the activities of new regulatory elements, which remain to be determined.
FIG. 8.
Schematic view of the changes in gene expression during the transition from anaerobic photosynthesis to aerobic respiration. Classification of the gene expression patterns was based on the hierarchical tree shown in Fig. 2. The thicknesses of the black bars indicate the degree of change in gene expression. The down-pointing arrow indicates the time when the growth conditions were shifted.
Supplementary Material
Acknowledgments
This work was supported by UPHS grant GM15590. H.A. was financially supported by MEXT of Japan during his stay in Houston.
We are grateful to Jesus Eraso (UTHSC-Houston) for useful discussions and advice.
Footnotes
Published ahead of print on 26 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aklujkar, M., R. C. Prince, and J. T. Beatty. 2005. The puhE gene of Rhodobacter capsulatus is needed for optimal transition from aerobic to photosynthetic growth and encodes a putative negative modulator of bacteriochlorophyll production. Arch. Biochem. Biophys. 437186-198. [DOI] [PubMed] [Google Scholar]
- 2.Aklujkar, M., R. C. Prince, and J. T. Beatty. 2005. The PuhB protein of Rhodobacter capsulatus functions in photosynthetic reaction center assembly with a secondary effect on light-harvesting complex 1. J. Bacteriol. 1871334-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aklujkar, M., R. C. Prince, and J. T. Beatty. 2006. The photosynthetic deficiency due to puhC gene deletion in Rhodobacter capsulatus suggests a PuhC protein-dependent process of RC/LH1/PufX complex reorganization. Arch. Biochem. Biophys. 45459-71. [DOI] [PubMed] [Google Scholar]
- 4.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5160-165. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, S. C., A. K. Robinson, and F. Rodríguez-Quiñones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27215-237. [DOI] [PubMed] [Google Scholar]
- 6.Anthony, J. R., K. L. Warczak, and T. J. Donohue. 2005. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc. Natl. Acad. Sci. USA 1026502-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barras, F., L. Loiseau, and B. Py. 2005. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv. Microb. Physiol. 5041-101. [DOI] [PubMed] [Google Scholar]
- 8.Beller, H. R., T. E. Letain, A. Chakicherla, S. R. Kane, T. C. Legler, and M. A. Coleman. 2006. Whole-genome transcriptional analysis of chemolithoautotrophic thiosulfate oxidation by Thiobacillus denitrificans under aerobic versus denitrifying conditions. J. Bacteriol. 1887005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boes, N., K. Schreiber, E. Härtig, L. Jaensch, and M. Schobert. 2006. The Pseudomonas aeruginosa universal stress protein PA4352 is essential for surviving anaerobic energy stress. J. Bacteriol. 1886529-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 45827-836. [DOI] [PubMed] [Google Scholar]
- 11.Braatsch, S., O. V. Moskvin, G. Klug, and M. Gomelsky. 2004. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J. Bacteriol. 1867726-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandner, J. P., A. G. McEwan, S. Kaplan, and T. J. Donohue. 1989. Expression of the Rhodobacter sphaeroides cytochrome c2 structural gene. J. Bacteriol. 171360-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callister, S. J., C. D. Nicora, X. Zeng, J. H. Roh, M. A. Dominguez, C. L. Tavano, M. E. Monroe, S. Kaplan, T. J. Donohue, R. D. Smith, and M. S. Lipton. 2006. Comparison of aerobic and photosynthetic Rhodobacter sphaeroides 2.4.1 proteomes. J. Microbiol. Methods 67424-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, Y. R., S. Usui, C. A. Yu, and L. Yu. 1994. Role of subunit IV in the cytochrome b-c1 complex from Rhodobacter sphaeroides. Biochemistry 3310207-10214. [DOI] [PubMed] [Google Scholar]
- 15.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen-Bazire, G., W. R. Sistrom, and R. Y. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Physiol. 4925-68. [DOI] [PubMed] [Google Scholar]
- 17.Daldal, F., S. Mandaci, C. Winterstein, H. Myllykallio, K. Duyck, and D. Zannoni. 2001. Mobile cytochrome c2 and membrane-anchored cytochrome cy are both efficient electron donors to the cbb3- and aa3-type cytochrome c oxidases during respiratory growth of Rhodobacter sphaeroides. J. Bacteriol. 1832013-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 1502497-2502. [DOI] [PubMed] [Google Scholar]
- 19.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50809-824. [DOI] [PubMed] [Google Scholar]
- 20.Donohue, T. J., A. G. McEwan, S. Van Doren, A. R. Crofts, and S. Kaplan. 1988. Phenotypic and genetic characterization of cytochrome c2 deficient mutants of Rhodobacter sphaeroides. Biochemistry 271918-1925. [DOI] [PubMed] [Google Scholar]
- 21.Donohue, T. J., P. J. Kiley, and S. Kaplan. 1988. The puf operon region of Rhodobacter sphaeroides. Photosynth. Res. 1939-61. [DOI] [PubMed] [Google Scholar]
- 22.Dubbs, J. M., and F. R. Tabita. 2004. Regulators of nonsulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol. Rev. 28353-376. [DOI] [PubMed] [Google Scholar]
- 23.Elsen, S., L. R. Swem, D. L. Swem, and C. E. Bauer. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eraso, J. M., and S. Kaplan. 1994. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 17632-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomelsky, M., and S. Kaplan. 1995. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J. Bacteriol. 1771634-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomelsky, M., and S. Kaplan. 1995. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1774609-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomelsky, M., and S. Kaplan. 1997. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 179128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomelsky, L., J. Sram, O. V. Moskvin, I. M. Horne, H. N. Dodd, J. M. Pemberton, A. G. McEwan, S. Kaplan, and M. Gomelsky. 2003. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology 149377-388. [DOI] [PubMed] [Google Scholar]
- 29.Green, H. A., and T. J. Donohue. 2006. Activity of Rhodobacter sphaeroides RpoHII, a second member of the heat shock sigma factor family. J. Bacteriol. 1885712-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahn, F. M., L. M. Eubanks, C. A. Testa, B. S. Blagg, J. A. Baker, and C. D. Poulter. 2001. 1-Deoxy-d-xylulose 5-phosphate synthase, the gene product of open reading frame (ORF) 2816 and ORF 2895 in Rhodobacter capsulatus. J. Bacteriol. 1831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallenbeck, P. L., R. Lerchen, P. Hessler, and S. Kaplan. 1990. Roles of CfxA, CfxB, and external electron acceptors in regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase expression in Rhodobacter sphaeroides. J. Bacteriol. 1721736-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Happ, H. N., S. Braatsch, V. Broschek, L. Osterloh, and G. Klug. 2005. Light-dependent regulation of photosynthesis genes in Rhodobacter sphaeroides 2.4.1 is coordinately controlled by photosynthetic electron transport via the PrrBA two-component system and the photoreceptor AppA. Mol. Microbiol. 58903-914. [DOI] [PubMed] [Google Scholar]
- 33.Hondorp, E. R., and R. G. Matthews. 2004. Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jäger, A., S. Braatsch, K. Haberzettl, S. Metz, L. Osterloh, Y. Han, and G. Klug. 2007. The AppA and PpsR proteins from Rhodobacter sphaeroides can establish a redox-dependent signal chain but fail to transmit blue-light signals in other bacteria. J. Bacteriol. 1892274-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones, H. M., and R. P. Gunsalus. 1987. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J. Bacteriol. 1693340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalir, S., J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M. G. Surette, and U. Alon. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 2922080-2083. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan, S., B. D. Cain, T. J. Donohue, W. D. Shepherd, and G. S. Yen. 1983. Biosynthesis of the photosynthetic membranes of Rhodopseudomonas sphaeroides. J. Cell. Biochem. 2215-29. [DOI] [PubMed] [Google Scholar]
- 38.Kaplan, S., J. Eraso, and J. H. Roh. 2005. Interacting regulatory networks in the facultative photosynthetic bacterium, Rhodobacter sphaeroides 2.4.1. Biochem. Soc. Trans. 3351-55. [DOI] [PubMed] [Google Scholar]
- 39.Kiley, P. J., and S. Kaplan. 1987. Cloning, DNA sequence, and expression of the Rhodobacter sphaeroides light-harvesting B800-850-α and B800-850-β genes. J. Bacteriol. 1693268-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiley, P. J., and S. Kaplan. 1988. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol. Rev. 5250-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, Y. J., I. J. Ko, J. M. Lee, H. Y. Kang, Y. M. Kim, S. Kaplan, and J. I. Oh. 2007. Dominant role of the cbb3 oxidase in the regulation of photosynthesis gene expression through the PrrBA system in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1895617-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krieger-Liszkay, A. 2005. Singlet oxygen production in photosynthesis. J. Exp. Bot. 56337-346. [DOI] [PubMed] [Google Scholar]
- 43.Lee, J. K., P. J. Kiley, and S. Kaplan. 1989. Posttranscriptional control of puc operon expression of B800-850 light-harvesting complex formation in Rhodobacter sphaeroides. J. Bacteriol. 1713391-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, J. K., and S. Kaplan. 1992. cis-acting regulatory elements involved in oxygen and light control of puc operon transcription in Rhodobacter sphaeroides. J. Bacteriol. 1741146-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leichert, L. I., and U. Jakob. 2004. Protein thiol modifications visualized in vivo. PLoS Biol. 2e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 9831-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mackenzie, C., M. Choudhary, F. W. Larimer, P. F. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. J. Donohue, J. P. Hosler, J. E. Newman, J. P. Shapleigh, R. E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 7019-41. [DOI] [PubMed] [Google Scholar]
- 48.Mao, L., C. Mackenzie, J. H. Roh, J. M. Eraso, S. Kaplan, and H. Resat. 2005. Combining microarray and genomic data to predict DNA binding motifs. Microbiology 1513197-3213. [DOI] [PubMed] [Google Scholar]
- 49.Martin, A. C., M. Gould, E. Byles, M. A. Roberts, and J. P. Armitage. 2006. Two chemosensory operons of Rhodobacter sphaeroides are regulated independently by sigma 28 and sigma 54. J. Bacteriol. 1887932-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masuda, S., and C. E. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110613-623. [DOI] [PubMed] [Google Scholar]
- 51.Meijer, W. G., and F. R. Tabita. 1992. Isolation and characterization of the nifUSVW-rpoN gene cluster from Rhodobacter sphaeroides. J. Bacteriol. 1743855-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meinhardt, S. W., P. J. Kiley, S. Kaplan, A. R. Crofts, and S. Harayama. 1985. Characterization of light-harvesting mutants of Rhodopseudomonas sphaeroides. I. Measurement of the efficiency of energy transfer from light-harvesting complexes to the reaction center. Arch. Biochem. Biophys. 236130-139. [DOI] [PubMed] [Google Scholar]
- 53.Moskvin, O. V., L. Gomelsky, and M. Gomelsky. 2005. Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as a master regulator of photosystem development. J. Bacteriol. 1872148-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mouncey, N. J., and S. Kaplan. 1998. Oxygen regulation of the ccoN gene encoding a component of the cbb3 oxidase in Rhodobacter sphaeroides 2.4.1T: involvement of the FnrL protein. J. Bacteriol. 1802228-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mouncey, N. J., E. Gak, M. Choudhary, J. I. Oh, and S. Kaplan. 2000. Respiratory pathways of Rhodobacter sphaeroides 2.4.1T: identification and characterization of genes encoding quinol oxidases. FEMS Microbiol. Lett. 192205-210. [DOI] [PubMed] [Google Scholar]
- 56.Myllykallio, H., D. Zannoni, and F. Daldal. 1999. The membrane-attached electron carrier cytochrome cy from Rhodobacter sphaeroides is functional in respiratory but not in photosynthetic electron transfer. Proc. Natl. Acad. Sci. USA 964348-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh, J. I., and S. Kaplan. 2000. Redox signaling: globalization of gene expression. EMBO J. 194237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oh, J. I., and S. Kaplan. 2001. Generalized approach to the regulation and integration of gene expression. Mol. Microbiol. 391116-1123. [DOI] [PubMed] [Google Scholar]
- 59.Oh, J. I., I. J. Ko, and S. Kaplan. 2003. Digging deeper: uncovering genetic loci which modulate photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. Microbiology 149949-960. [DOI] [PubMed] [Google Scholar]
- 60.Østerås, M., and U. Jenal. 2000. Regulatory circuits in Caulobacter. Curr. Opin. Microbiol. 3171-176. [DOI] [PubMed] [Google Scholar]
- 61.Pappas, C. T., J. Sram, O. V. Moskvin, P. S. Ivanov, R. C. Mackenzie, M. Choudhary, M. L. Land, F. W. Larimer, S. Kaplan, and M. Gomelsky. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 1864748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Partridge, J. D., C. Scott, Y. Tang, R. K. Poole, and J. Green. 2006. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J. Biol. Chem. 28127806-27815. [DOI] [PubMed] [Google Scholar]
- 63.Partridge, J. D., G. Sanguinetti, D. P. Dibden, R. E. Roberts, R. K. Poole, and J. Green. 2007. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J. Biol. Chem. 28211230-11237. [DOI] [PubMed] [Google Scholar]
- 64.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 1762869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2002. The four different σ54 factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol. Microbiol. 4675-85. [DOI] [PubMed] [Google Scholar]
- 66.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2005. The flagellar hierarchy of Rhodobacter sphaeroides is controlled by the concerted action of two enhancer-binding proteins. Mol. Microbiol. 58969-983. [DOI] [PubMed] [Google Scholar]
- 67.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2006. Transcriptional specificity of RpoN1 and RpoN2 involves differential recognition of the promoter sequences and specific interaction with the cognate activator proteins. J. Biol. Chem. 28127205-27215. [DOI] [PubMed] [Google Scholar]
- 68.Poggio, S., C. Abreu-Goodger, S. Fabela, A. Osorio, G. Dreyfus, P. Vinuesa, and L. Camarena. 2007. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J. Bacteriol. 1893208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 391595-1609. [DOI] [PubMed] [Google Scholar]
- 70.Prüß, B. M., and P. Matsumura. 1997. Cell cycle regulation of flagellar genes. J. Bacteriol. 1795602-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roh, J. H., W. E. Smith, and S. Kaplan. 2004. Effects of oxygen and light intensity on transcriptome expression in Rhodobacter sphaeroides 2.4.1. Redox active gene expression profile. J. Biol. Chem. 2799146-9155. [DOI] [PubMed] [Google Scholar]
- 72.Römling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57629-639. [DOI] [PubMed] [Google Scholar]
- 73.Schreiber, K., N. Boes, M. Eschbach, L. Jaensch, J. Wehland, T. Bjarnsholt, M. Givskov, M. Hentzer, and M. Schobert. 2006. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J. Bacteriol. 188659-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shapiro, L., and R. Losick. 2000. Dynamic spatial regulation in the bacterial cell. Cell 10089-98. [DOI] [PubMed] [Google Scholar]
- 75.Unden, G., and J. Schirawski. 1997. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Mol. Microbiol. 25205-210. [DOI] [PubMed] [Google Scholar]
- 76.Wood, B. J. B., B. W. Nichols, and A. T. James. 1965. The lipids and fatty acid metabolism of photosynthetic bacteria. Biochim. Biophys. Acta 106261-273. [DOI] [PubMed] [Google Scholar]
- 77.Yeliseev, A. A., and S. Kaplan. 1995. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 27021167-21175. [DOI] [PubMed] [Google Scholar]
- 78.Yeliseev, A. A., and S. Kaplan. 1999. A novel mechanism for the regulation of photosynthesis gene expression by the TspO outer membrane protein of Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 27421234-21243. [DOI] [PubMed] [Google Scholar]
- 79.Yeliseev, A. A., and S. Kaplan. 2000. TspO of Rhodobacter sphaeroides. A structural and functional model for the mammalian peripheral benzodiazepine receptor. J. Biol. Chem. 2755657-5667. [DOI] [PubMed] [Google Scholar]
- 80.Young, C. S., and J. T. Beatty. 1998. Topological model of the Rhodobacter capsulatus light-harvesting complex I assembly protein LhaA (previously known as ORF1696). J. Bacteriol. 1804742-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Young, C. S., R. C. Reyes, and J. T. Beatty. 1998. Genetic complementation and kinetic analyses of Rhodobacter capsulatus ORF1696 mutants indicate that the ORF1696 protein enhances assembly of the light-harvesting I complex. J. Bacteriol. 1801759-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeilstra-Ryalls, J. H., and S. Kaplan. 1995. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene. J. Bacteriol. 1776422-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zeilstra-Ryalls, J., M. Gomelsky, J. M. Eraso, A. Yeliseev, J. O'Gara, and S. Kaplan. 1998. Control of photosystem formation in Rhodobacter sphaeroides. J. Bacteriol. 1802801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeilstra-Ryalls, J. H., and S. Kaplan. 2004. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell Mol. Life Sci. 61417-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zeilstra-Ryalls, J. H., and K. L. Schornberg. 2006. Analysis of hemF gene function and expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 188801-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zeller, T., O. V. Moskvin, K. Li, G. Klug, and M. Gomelsky. 2005. Transcriptome and physiological responses to hydrogen peroxide of the facultatively phototrophic bacterium Rhodobacter sphaeroides. J. Bacteriol. 1877232-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeng, X., M. Choudhary, and S. Kaplan. 2003. A second and unusual pucBA operon of Rhodobacter sphaeroides 2.4.1: genetics and function of the encoded polypeptides. J. Bacteriol. 1856171-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu, Y. S., P. J. Kiley, T. J. Donohue, and S. Kaplan. 1986. Origin of the mRNA stoichiometry of the puf operon in Rhodobacter sphaeroides. J. Biol. Chem. 26110366-10374. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.