Abstract
The TonB and TolA proteins are energy transducers that couple the ion electrochemical potential of the cytoplasmic membrane to support energy-dependent processes at the outer membrane of the gram-negative envelope. The transfer of energy to these transducers is facilitated by energy-harvesting complexes, which are heteromultimers of cytoplasmic membrane proteins with homologies to proton pump proteins of the flagellar motor. Although the cognate energy-harvesting complex best services each transducer, components of the complexes (for TonB, ExbB and ExbD; for TolA, TolQ and TolR) are sufficiently similar that each complex can imperfectly replace the other. Previous investigations of this molecular cross talk considered energy-harvesting complex components expressed from multicopy plasmids in strains in which the corresponding genes were interrupted by insertions, partially absent due to polarity, or missing due to a larger deletion. These questions were reexamined here using strains in which individual genes were removed by precise deletions and, where possible, components were expressed from single-copy genes with native promoters. By more closely approximating natural stoichiometries between components, this study provided insight into the roles of energy-harvesting complexes in both the energization and the stabilization of TonB. Further, the data suggest a distinct role for ExbD in the TonB energy transduction cycle.
The gram-negative bacterial envelope contains two distinct membranes, which are separated by an aqueous periplasmic space. A standard phospholipid bilayer forms the inner, cytoplasmic membrane (CM), a structure present in virtually all cells that provides a permeability barrier rich in proteins that generate and use ion gradients to support numerous essential processes. Conversely, the outer membrane (OM) is a unique structure, an asymmetric bilayer with an inner phospholipid face shielded by an outer leaflet bearing lipid-anchored anionic oligosaccharides. This charged surface hinders the passage of potentially harmful molecules soluble in typical membranes, while it allows diffusion of small hydrophilic nutrients via aqueous channels provided by porin proteins residing in the OM. Thus, the OM serves as a diffusion barrier, enabling gram-negative bacteria to exploit a variety of niches toxic to organisms lacking similar defenses. The major complication caused by this architecture is spatial separation of the OM from standard cellular energy sources; thus, energy-dependent OM processes must rely upon the transduction of energy from other sites.
The model system for the transfer of energy to the OM is the TonB system. In Escherichia coli and most other gram-negative species that have been examined, TonB couples the ion electrochemical gradient of the CM to drive the active transport of ferric siderophores, cobalamin, and potentially other ligands across the OM (for reviews, see references 44, 45, and 54). A second system, centered on the TolA protein, plays a major role in maintaining envelope integrity (37, 38), at least in part by supporting OM invagination during cell division (11), and it shares many characteristics with the TonB system. While a role in energy transduction has not been established as firmly for TolA as for TonB, the conformations of both proteins are responsive to changes in the CM electrochemical potential (5, 13, 35), and the proteins have membrane topologies consistent with such a role (17, 41, 46), with amino-terminal CM anchors, elongated central domains that are a suitable length to span the periplasmic space (9, 40), and carboxy-terminal domains that interact with OM components (4, 53). For these and additional reasons presented below, it is likely that both proteins function as energy transducers; however, the OM recipients of TonB- and TolA-transduced energy are sufficiently different that neither transducer is able to substitute for the other (3).
TonB and TolA interact with heteromultimeric protein complexes whose members share sequence homology with the MotA/MotB proton translocator complex of the bacterial flagellum (6, 29). Each energy transducer has a preferred energy-harvesting complex. For TonB, the complex consists of the proteins ExbB and ExbD, and for TolA, the complex consists of the proteins TolQ and TolR. The exact stoichiometry and arrangement of protein subunits in these energy-harvesting complexes remain unknown. For the ExbB/ExbD complex, protein stoichiometry (20) and in vivo chemical cross-linking studies (22) together have suggested that there is a large complex containing as many as four or five molecules of ExbD and 14 or 15 molecules of ExbB (18). Conversely, it has been suggested that the TolQ/TolR complex might consist of a pair of TolR molecules surrounded by four TolQ molecules, mimicking a structure proposed for flagellar motor components (6). While the details of the complex composition remain unresolved, the relationship between the complexes is evident, both in the conservation of primarily amino acid sequences among components (8, 49) and in membrane topology. ExbD and its paralogue TolR are both secured in the CM by a single amino-terminal signal anchor (24, 43), while ExbB and its paralogue TolQ are both anchored in the CM by three transmembrane domains and have a periplasmically exposed amino terminus, a relatively large cytoplasmic domain, a short periplasmic loop, and a cytoplasmically exposed carboxy terminus (25, 26, 52).
The cumulative evidence indicates that the ExbB/ExbD and TolQ/TolR energy-harvesting complexes couple the energy transducers TonB and TolA to the ion electrochemical potential of the CM. For both TonB and TolA, the presence of the preferred energy-harvesting complex is required for membrane energy-dependent conformational changes (13, 35) and, in the case of TolA, also for energy-dependent association with an OM protein (5). The interactions that drive these changes appear to be mediated through the amino-terminal signal anchors of the energy transducers, which share a conserved amino acid motif (28) containing several residues important for the function (12, 32, 34, 50). Mutations that disrupt this conserved face in TonB can be suppressed by second-site mutations that map to the energy-harvesting complex component ExbB (35, 36); similarly, the corresponding mutations in TolA can be suppressed by second-site mutations in TolQ (12).
While the energy transducers show a preference for their cognate energy-harvesting complexes, the preference is not absolute. This was first shown by the observation that mutations in exb genes resulted in a leaky tonB phenotype (8, 15), with the residual activity dependent upon the presence of a functional tolQ (and presumably tolR) gene (2). This molecular cross talk between energy-harvesting complexes is bidirectional, with the TolQ/TolR and ExbB/ExbD complexes each able to imperfectly replace the other (3).
This cross talk phenomenon suggests that these systems share a common mechanism of energy transfer. Interestingly, in the study mentioned above (3) TonB activities lower than the activities achieved by TolQ/TolR-mediated cross talk were evident in a strain carrying only the exbB and tolR genes. It is thus likely that ExbB and TolR can be assembled into an energy-harvesting complex; however, the low level of activity obtained suggests that the energy-harvesting complex that they form either is very inefficient, is formed only rarely, or is not stable. Activity was not detected in a strain carrying only the tolQ and exbD genes, indicating either that TolQ and ExbD do not assemble into complexes or that such complexes are not functional at levels detectable by the assays used. Regardless of the explanation, the lack of efficient energy transduction in strains encoding only one component of each energy-harvesting complex suggests that paired components of each energy-harvesting complexes have coevolved to the degree that the protein interactions within a complex are more stringent than those between the complex and the energy transducers. Consistent with this interpretation, Braun and Herrmann also found that overexpressed TonB, which is unstable unless ExbB is also overexpressed (10), could be stabilized by overexpression of TolQ, even though TolQ alone or in a potential complex with ExbD could not energize TonB (3).
The inefficiency of cross talk and the lack of interchangeability of energy-harvesting complex components suggest approaches for dissection and identification of the essential structural features that provide energy transduction. Specifically, we began to modify Tol system components by substituting the corresponding Exb regions, with the idea that this should identify motifs that are important for intracomplex interactions, as well as motifs that mediate transducer preference. Our initial approach was to assemble tol derivatives on plasmids and express their chimeric products from arabinose-regulated promoters at levels approximating those of their chromosomally encoded counterparts. As a baseline, we first revisited the question of potential mixed energy-harvesting complexes (3), pairing plasmids expressing individual energy-harvesting complex components under arabinose regulation in a strain carrying a large deletion encompassing the exb operon and an amber mutation in tolQ that is polar on tolR (KP1456) (Table 1). The data generated in these pilot experiments were difficult to interpret. First, we were unable to consistently express proteins at wild-type levels, and we found that overexpression of tol genes limited the ability of cells to form confluent lawns in colicin and phage susceptibility assays (data not shown). Second, the tolQ mutation used was clearly not completely polar on tolR (data not shown), a complicating factor also evident in the previous study (3). Finally, it was unclear whether the other genes affected by the exb deletion made phenotypic contributions.
TABLE 1.
E. coli strains and plasmids
| Strain or plasmida | Relevant genotype and/or phenotypeb | Reference |
|---|---|---|
| Strains | ||
| BW25113 | lacIqrrnBT14 ΔlacZwj16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 7 |
| W3110 | F-IN(rrnD-rrnE)1 | 23 |
| KP1456 | W3110 ΔexbBD tolQ(Am)37 | 18 |
| RA1003 | W3110 ΔexbBD::kan | 32 |
| RA1034 | W3110 ΔexbBD::kan ΔtolR | This study |
| RA1035 | W3110 ΔtolQR | This study |
| RA1044 | W3110 ΔexbB ΔtolR | This study |
| RA1045 | W3110 ΔtolQR ΔexbD | This study |
| RA1046 | W3110 ΔtolQR ΔexbB | This study |
| RA1051 | W3110 ΔexbBD::kan ΔtolQR | This study |
| Plasmids | ||
| pBAD24 | araBAD promoter, AraC, Ampr | 16 |
| pKP392 | pBAD24 araBAD-regulated exbB | 18 |
| pKP393 | pBAD24 araBAD-regulated exbD | This study |
| pRA001 | pBAD24 araBAD-regulated tolQ | This study |
| pRA002 | pBAD24 araBAD-regulated tolR | This study |
| pRA003 | pBAD24 araBAD-regulated tolQR | This study |
The identity of each plasmid construct generated in this study was confirmed by sequence determination.
The specific nature of each of the deletions involving exbB, exbD, tolQ, and tolR is as follows. The ΔexbBD deletion of KP1456 is a large, incompletely characterized deletion involving the exbBD operon and the adjacent metC gene, originally isolated by Guterman and Dann (15). The ΔexbBD::kan mutation involves the replacement of all but the initiation codon of exbB (codons 2 to 244) and its TAA termination codon, the six intervening nucleotides, and all of the codons of exbD (codons 1 to 141) by the kan gene from pACYC177, as previously described (32). The ΔexbB deletion removed exbB codons 1 to 244, the TAA termination codon, and the six additional nucleotides that occur prior to the exbD initiation codon. The ΔexbD deletion removed exbD codons 1 to 141 and the TAA termination codon. The ΔtolQR deletion removed tolQ codons 1 to 230, the TAA termination codon, the three nucleotides that occur prior to the tolR initiation codon, all 142 tolR codons, and the TAA termination codon. The ΔtolR deletion removed tolR codons 1 to 142 and the TAA termination codon.
Because some strains (such as the tolQ mutant described above) cause unwanted complications and because stoichiometry is an important consideration in these energy transduction systems (19, 31), our strategy was modified to, where possible, express the energy-harvesting complex proteins from single chromosomal copies under the control of their native promoters. To do this, we constructed a set of specific deletions in BW25113 with the λ red recombination technique (7), which were then paired in various combinations in a W3110 background by generalized transduction using P1vir (42), as summarized in Table 1. As described below, by evaluating individual components expressed from the chromosome, this strategy confirmed previous observations regarding the ability of plasmid-encoded products to support TonB-dependent energy transduction and yielded information concerning protein stability and interactions under more physiologically relevant conditions. Together, the observations provide insight into the role of individual components in TonB-dependent energy transduction not evident from plasmid-based studies.
Deletions of the exbB and exbD genes were verified by immunoblot analysis (as described below) of cell lysates, using ExbB-specific and ExbD-specific polyclonal antibodies (22). These analyses demonstrated not only that the specific gene product (ExbB or ExbD) was absent from the corresponding deletion strain but also that the deletions did not have polar effects (i.e., deletion of one member of the gene pair did not alter the expression of the other member) (data not shown). These observations were confirmed by colicin susceptibility assays (performed as described previously [31]) to determine loss of function, followed by complementation experiments in which individual deletion strains were transformed with either the ExbB-expressing plasmid pKP392 (18) or the ExbD-encoding plasmid pKP393 (constructed here by insertion of a PCR-amplified exbD gene from W3110 into pBAD24 [16] under control of the arabinose promoter). In each case, complementation with the appropriate plasmid restored wild-type levels of TonB activity as measured by susceptibility to colicins (data not shown).
Because specific antibodies were not available for proteins of the Tol system, an alternative approach was used to verify the deletions. First, the absence of the each gene or gene set was confirmed by PCR, using genomic DNA isolated from each strain and primers that flank the tolQRA gene cluster. The identities of resultant amplimers (and the deletions evident in the amplimers) were confirmed by restriction mapping (data not shown). The absence of Tol proteins was confirmed by colicin susceptibility assays to determine loss of function, followed by complementation experiments in which individual deletion strains were transformed with plasmids (constructed as described above for pKP393) carrying either tolQ (pRA001), tolR (pRA002), or tolQR (pRA003). While introduction of the tolQR-bearing plasmid restored TolA activity (as measured by susceptibility to colicins) with all tol gene deletions tested, complementation studies with individual genes indicated that deletion of tolQ was highly polar on tolR (data not shown). Thus, for strains in which we desired the absence of TolQ but the presence of TolR, it was necessary to provide TolR in trans from plasmid pRA002.
The TonB and TolA activities of strains expressing various combinations of energy-harvesting complex proteins were examined by spot titer assays (Table 2). Strain W3110, which contained functional copies of all four energy-harvesting complex genes, was used as the wild type and was sensitive to both the TolA-dependent agent colicin A (ColA) and the TonB-dependent agents colicin B (ColB) and bacteriophage φ80. Multiple-deletion strain RA1051, which lacked all four genes, served as the negative control and was fully resistant to each agent at the dilutions tested. When only TolQ and TolR were present (RA1003), the sensitivities to the TonB-dependent agents were reduced compared to the sensitivities of wild-type strain W3110 (2 or 3 fivefold dilutions less for ColB and 1 or 2 10-fold dilutions less for φ80, with the residual activity representing the cross talk contributions of TolQ and TolR). The susceptibility to the TolA-dependent agent ColA was similar to that of the wild-type strain. Because our tolQ deletion was polar on tolR, strain RA1034 (which expressed only TolQ) was complemented with the TolR-encoding plasmid pRA002 and compared with RA1003 to verify that functional TolR protein was made from the plasmid under these assay conditions. This pair was slightly more efficient at supporting cross talk and slightly less efficient at supporting the cognate TolA-dependent activity than RA1003, in which both TolQ and TolR were encoded in the chromosome. Thus, plasmid-derived TolR was certainly functional, but under the assay conditions used it was expressed at levels sufficiently different from the level of chromosomally encoded TolR to result in small differences in overall colicin susceptibility. When only ExbB and ExbD were present (RA1035), the sensitivities to TonB-dependent agents were similar to those of the wild-type strain, whereas the susceptibility to the TolA-dependent agent ColA was greatly decreased, with the residual activity representing the cross talk contribution of ExbB and ExbD. When only ExbB and TolR were present [RA1045(pRA002)], TonB activity was evident only with the φ80 spot titer assay, an assay that can detect levels of TonB lower than the levels that can be detected by the colicin spot titer assay (31). Conversely, no activity was detected by any assay when only TolQ and ExbD were present (RA1044) or when any of the energy-harvesting complex proteins were expressed alone [RA1045, RA1046, RA1034, and RA1051(pRA002)].
TABLE 2.
Susceptibility to group A and B colicins and phage φ80
| Straina | Phenotype | Susceptibility tob:
|
||
|---|---|---|---|---|
| ColA | ColB | φ80 | ||
| W3110(pBAD24) | ExbB+ ExbD+ TolQ+ TolR+ | 7, 7, 7 | 7, 7, 7 | 8, 8, 8 |
| RA1051(pBAD24) | ExbB− ExbD− TolQ− TolR− | R, R, R | R, R, R | R, R, R |
| RA1003(pBAD24) | ExbB− ExbD− TolQ+ TolR+ | 7, 7, 6 | 5, 5, 4 | 7, 6, 6 |
| RA1034(pRA002) | ExbB− ExbD− TolQ+ pRA002+ | 6, 6, 6 | 6, 6, 7 | 7, 7, 6 |
| RA1035(pBAD24) | ExbB+ ExbD+ TolQ− TolR− | R, R, 3 | 7, 8, 7 | 7, 8, 9 |
| RA1045(pRA002) | ExbB+ ExbD− TolQ− pRA002+ | R, R, R | R, R, R | 4, 4, 2 |
| RA1044(pBAD24) | ExbB− ExbD+ TolQ+ TolR− | R, R, R | R, R, R | R, R, R |
| RA1045(pBAD24) | ExbB+ ExbD− TolQ− TolR− | R, R, R | R, R, R | R, R, R |
| RA1046(pBAD24) | ExbB− ExbD+ TolQ− TolR− | R, R, R | R, R, R | R, R, R |
| RA1034(pBAD24) | ExbB− ExbD− TolQ+ TolR− | R, R, R | R, R, R | R, R, R |
| RA1051(pRA002) | ExbB− ExbD− TolQ− pRA002+ | R, R, R | R, R, R | R, R, R |
Most strains carried plasmid pBAD24; the exceptions were the strains which carried plasmid pRA002, which encodes TolR. Cells were grown at 37°C with aeration in LB broth supplemented with 100 μg ml−1 ampicillin to mid-exponential phase (an A550 of 0.4, as determined with a Spectronic 20 spectophotometer with a path length of 1.5 cm) and then plated in T-top medium supplemented with 100 μg ml−1 ampicillin and 0.01% (wt/vol) l-arabinose on similarly supplemented T-medium plates. Colicin and phage dilutions were added as 5-μl aliquots, and the plates were incubated at 37°C for 18 h and then scored for clearing.
The values are the highest numbers of dilutions (fivefold dilutions for colicins and 10-fold dilutions for φ80) of the agent that resulted in an evident zone of clearing in the cell lawn. R indicates resistance (i.e., no clearing) with the undiluted colicin or phage stock. The values for three platings are shown for each strain-agent pair.
The results described above are consistent with those obtained previously by Braun and Herrmann (3). In the study of these workers, energy-harvesting complex proteins were encoded in multicopy plasmids, and it is likely that there was an excess of energy-harvesting complexes relative to the amounts of energy transducers, which were chromosomally encoded. Here, with the necessary exception of providing TolR in trans in tolQ strains, energy-harvesting complex proteins were encoded by single-copy, chromosomal genes under regulation of native promoters. These conditions more closely approximate the normal stoichiometry of these systems, and thus, the present results strengthen the prior suggestion that cross talk between these systems, while potentially informative at the molecular and mechanistic levels, is probably not relevant to normal cell function. Indeed, when the ability of TonB to support iron transport was examined in these strains (Table 3), TolQ- and TolR-mediated cross talk provided barely detectable levels of [55Fe]ferrichrome uptake in the absence of ExbB and ExbD (RA1003) relative to the wild-type (W3110) and the ΔtolQR strain (RA1035) levels, while strains lacking either TolQ and ExbD [RA1045(pRA002)] or ExbB and TolR (RA1044) were indistinguishable from the strain lacking all four proteins (RA1051). The degree of cross talk activity detected in this assay was lower than the level detected by colicin susceptibility assays (Table 2). This was consistent with the ability of colicin assays to register levels of TonB lower than the levels that could be detected by transport assays (31). As expected, the steady-state levels of TonB varied between strains (Fig. 1); however, the differences in either the pattern or the intensity were not great enough to account for the differences in [55Fe]ferrichrome uptake observed (Table 3).
TABLE 3.
TonB-dependent ferrichrome transport
| Straina | Phenotype | [55Fe]ferrichrome uptakeb |
|---|---|---|
| W3110(pBAD24) | ExbB+ ExbD+ TolQ+ TolR+ | 1,982.5 ± 273 |
| RA1035(pBAD24) | ExbB+ ExbD+ TolQ− TolR− | 1,752.0 ± 33 |
| RA1003(pBAD24) | ExbB− ExbD− TolQ+ TolR+ | 11.3 ± 4.3 |
| RA1045(pRA002) | ExbB+ ExbD− TolQ− pRA002+ | −4.5 ± 2.1 |
| RA1044(pBAD24) | ExbB− ExbD+ TolQ+ TolR− | −6.7 ± 2.9 |
| RA1051(pBAD24) | ExbB− ExbD− TolQ− TolR− | −6.9 ± 6 |
Most strains carried plasmid pBAD24; the exception was the strain which carried plasmid pRA002, which encodes TolR. Cells were grown with aeration at 37°C to mid-exponential phase in T-broth supplemented with 100 μg ml−1 ampicillin and then centrifuged and suspended at a concentration of 2 × 108 CFU ml−1 in M9 salts containing 0.1 mM nitrilotriacetate. Transport was initiated by addition of 150 pmol [55Fe]ferrichrome as previously described (34).
Uptake is expressed in cpm 55Fe transported per 1 × 109 cells min−1. The values are means ± standard deviations of triplicate assays.
FIG. 1.
Steady-state levels of TonB protein in strains used for [55Fe]ferrichrome uptake. Aliquots (0.5 ml) of cells grown to mid-exponential phase (as described in Table 3, footnote a) were harvested just prior to the addition of [55Fe]ferrichrome, precipitated in 10% trichloroacetic acid at 4°C for 15 min, centrifuged, washed in 100 mM Tris-Cl (pH 8.0), suspended in 25 μl of Laemmli sample buffer, incubated at 98°C for 5 min, resolved on a sodium dodecyl sulfate-11% polyacrylamide gel (30), transferred to a polyvinylidene fluoride membrane, and probed using the TonB-specific monoclonal antibody 4F1 (33), with subsequent visualization by enhanced chemiluminescence as previously described (48). Following immunoblot analysis, membranes were stained for total protein with Coomassie blue and visually examined to confirm equivalent sample loading of all lanes. In this experiment, the wild-type (ExbB+ ExbD+ TolQ+ TolR+) sample was two- to threefold underloaded relative to the other samples. Strains are identified by their relevant phenotypes at the top, and the positions of molecular mass standards are indicated on the right. B, ExbB; D, ExbD; Q, TolQ; R, TolR.
In vivo chemical cross-linking produces a specific set of TonB-containing complexes, identifying proteins with which wild-type TonB engages in close physical interactions. The TonB-interactive proteins identified in this way include the energy-harvesting complex protein ExbB and the OM proteins Lpp and FepA (21, 47). The absence of the energy-harvesting complex proteins TolQ and TolR does not alter this pattern of TonB interaction, nor does the absence of ExbB and ExbD, except of course for the lack of a detectable interaction with ExbB itself (47). In the present study, the ability of TonB to form the cross-linked complexes in strains expressing partial or no energy-harvesting complex components was examined (Fig. 2). For each strain considered, in vivo cross-linking produced characteristic 180- and 43-kDa complexes containing TonB and the OM proteins FepA and Lpp, respectively. Indeed, the formation of these complexes appeared to be more efficient in the strains in which an ExbB/ExbD pair was not present, consistent with the observation that the majority of TonB partitions with the OM in fractionation studies of exbBD strains (39) and the observation that the TonB/FepA complex appears to represent the interaction between the OM receptor FepA and an inactive conformer of TonB (14). Cross-linked TonB complexes with ExbB did not form in strains lacking ExbB, nor was a TonB/TolQ complex evident for either of the ExbB− TolQ+ strains. Interestingly, neither ExbD nor its paralogue TolR appeared to be required for detectable interactions between TonB and ExbB to occur, as the TonB/ExbB complex was evident in all strains that expressed ExbB. This is consistent with the previous observation that the ability of a strain to form a cross-linked ExbB/TonB complex does not correlate with the ability to engage in TonB-dependent energy transduction. (34).
FIG. 2.
In vivo chemical cross-linking. Cells were grown in T-broth supplemented with 100 μg ml−1 ampicillin to mid-exponential phase, harvested in 1.0-ml aliquots, centrifuged, and suspended in 938 μl of 100 mM phosphate buffer (pH 6.8). Then 62 μl of 16% paraformaldehyde was added, and suspensions were incubated at room temperature for 15 min, centrifuged, and suspended in 25 μl of Laemmli sample buffer. Samples were incubated at 60°C for 5 min and then resolved on a sodium dodecyl sulfate-11% polyacrylamide gel, and a subsequent immunoblot analysis was performed as described in the legend to Fig. 1. Strains are identified by their relevant phenotypes at the top; the positions of molecular mass standards are indicated on the right, and the relative positions of monomeric TonB and the specific TonB-containing complexes are indicated on the left. B, ExbB; D, ExbD; Q, TolQ; R, TolR.
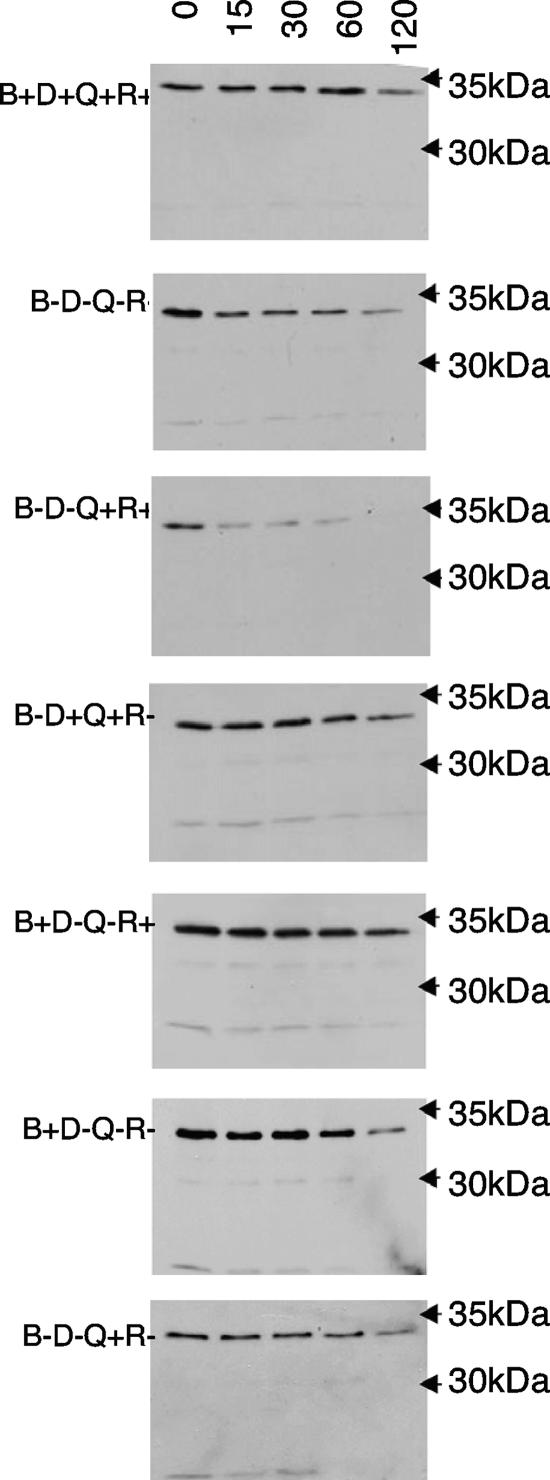
The lower apparent level of steady-state TonB in some of the strains used for [55Fe]ferrichrome transport (Fig. 1) suggested that TonB stability was different in different strains. The fact that the presence of energy-harvesting complexes has an effect on the stability of TonB is well known; chemical half-life studies have indicated that TonB is labile in exbBD strains (48), and when either ExbB or ExbD is absent, the steady-state half-life of TonB is approximately 30 min (1). Similarly, in the present study we found that relative to the wild-type strain (ExbB+ ExbD+ TolQ+ TolR+) (Fig. 3), the steady-state half-life of TonB was greatly reduced for both the strain lacking ExbB and ExbD (ExbB− ExbD− TolQ+ TolR+) and for the strain lacking all four energy-harvesting complex proteins (ExbB− ExbD− TolQ− TolR−). For strains encoding mixtures of energy-harvesting complex components (ExbB− ExbD+ TolQ+ TolR− or ExbB+ ExbD− TolQ− TolR+) and for strains expressing only either ExbB or TolQ, the steady-state half-life of TonB was similar to that in the wild-type strain (Fig. 3). For reasons that are not clear, the apparent stability of TonB in the strain lacking ExbB and TolR was not consistent with the smaller amount of steady-state TonB initially observed (Fig. 1).
FIG. 3.
Stability of TonB under steady-state conditions. All strains were grown to mid-exponential phase in LB broth supplemented with 100 μg ml−1 ampicillin. Chloramphenicol was then added to a final concentration of 100 μg ml−1 to halt protein synthesis. Samples were taken at 0, 15, 30, 60, and 120 min and precipitated in 10% (wt/vol) trichloroacetic acid. Samples were washed in 100 mM Tris-Cl (pH 8.0) and then suspended in 25 μl of Laemmli sample buffer, incubated at 98°C for 5 min, and resolved on sodium dodecyl sulfate-11% polyacrylamide gels, and a subsequent immunoblot analysis was performed as described in the legend to Fig. 1. Strains are identified by their relevant phenotypes on the left, sample times (in minutes) are indicated at the top, and the positions of molecular mass standards are indicated on the right. B, ExbB; D, ExbD; Q, TolQ; R, TolR.
How the proteins of the energy-harvesting complex influence the stability of TonB remains unresolved. Early studies showed that overexpressed TonB is unstable unless either ExbB or TolQ is concomitantly overexpressed, suggesting that these proteins might physically protect TonB (3, 10). Meanwhile, studies of TonB molecules lacking functional signal sequences (and therefore located in the cytoplasm) suggest a chaperone-like role for ExbB during either the synthesis or localization of TonB (27), and a later study suggested that the stability of TonB was dependent upon recruitment into functional energy-harvesting complexes, a process ostensibly mediated by ExbB and ExbD (1). Each of these scenarios implicitly involves the physical sequestration of TonB from endogenous proteinases but does not suggest how that sequestration might occur.
An alternative explanation is suggested by studies with second-site ExbB suppressors of TonB transmembrane domain mutants (35). In the absence of the suppressor, these TonB mutants are inactive and relatively stable; in the presence of the suppressor, they are active and very unstable, but only if a transportable ligand is present. Thus, instability in this case appears to involve a state that TonB normally achieves only following transfer of energy to an OM transporter.
The present results appear to bridge these two potential explanations. In the absence of either energy-harvesting complex, TonB was inactive and unstable. The presence of either ExbB or TolQ was sufficient to confer stability, but without supporting TonB activity. Similarly, strains encoding one component of each energy-harvesting complex (ExbB and TolR or TolQ and ExbD) proved to be unable to efficiently support TonB activity, but this did confer stability to TonB. Conversely, the TolQ/TolR energy-harvesting complex could support TonB activity but could not confer stability to TonB. Only in the presence of both ExbB and ExbD was TonB both active and stable. Together, these observations support the contention that the stabilization of TonB by energy-harvesting complex components is indirect, involving distinct TonB conformations.
The cumulative body of in vivo and in vitro data addressing TonB structure has led us to propose that the TonB carboxy-terminal domain can exist in either ordered or disordered conformations and that transitions between these conformations is regulated by the ExbB/ExbD complex (32, 45). Because it shares membrane topology with TonB, it has been speculated that ExbD might function as a chaperone to manage the disorder-order transitions of TonB that occur during the energy transduction cycle (51). The present data suggest that ExbD specifically mediates the transitions that return TonB to a stable conformation following energy transduction. Further, they raise the possibility that the greatly diminished levels of function evident when ExbB and TolR are paired might be due to incompatibilities between the periplasmic of domains TolR and TonB rather than between TolR and ExbB themselves.
Acknowledgments
The Postle laboratory provided strain KP1456, plasmids pKP392 and pKP393, monoclonal antibody 4F1, and the ExbB- and ExbD-specific polyclonal antisera. In addition to the reagents, we thank Kathleen Postle for many helpful conversations and for critical reading of the manuscript.
An award to R.L. from the National Science Foundation (grant MCB0315983) supported this research.
Footnotes
Published ahead of print on 26 October 2007.
REFERENCES
- 1.Ahmer, B. M., M. G. Thomas, R. A. Larsen, and K. Postle. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 1774742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun, V. 1989. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J. Bacteriol. 1716387-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and TolA-TolQ-TolR proteins. Mol. Microbiol. 8261-268. [DOI] [PubMed] [Google Scholar]
- 4.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 9610673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cascales, E., M. Gavioli, J. N. Sturgis, and R. Lloubes. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 38904-915. [DOI] [PubMed] [Google Scholar]
- 6.Cascales, E., R. Lloubes, and J. N. Sturgis. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42795-807. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eick-Helmerich, K., and V. Braun. 1989. Import of biopolymers into Escherichia coli: nucleotide sequences of exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J. Bacteriol. 1715117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, J. S., B. A. Levine, I. P. Trayer, C. J. Dorman, and C. F. Higgins. 1986. Sequence-imposed structural constraints in the TonB protein of E. coli. FEBS Lett. 208211-216. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, E., K. Günter, and V. Braun. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol. 1715127-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerding, M. A., Y. Ogata, N. D. Pecora, H. Niki, and P. A. J. de Boer. 2007. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 631008-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Germon, P., T. Clavel, A. Vianney, R. Portalier, and J.-C. Lazzaroni. 1998. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J. Bacteriol. 1806433-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Germon, P., M.-C. Ray, A. Vianney, and J.-C. Lazzaroni. 2001. Energy-dependent conformational change in the TolA protein of Escherichia coli involves its N-terminal domain, TolQ, and TolR. J. Bacteriol. 1834110-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, J., and K. Postle. 2005. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol. Microbiol. 55276-288. [DOI] [PubMed] [Google Scholar]
- 15.Guterman, S. K., and L. Dann. 1973. Excretion of enterochelin by exbA and exbB mutants of Escherichia coli. J. Bacteriol. 1141225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman, L. M., D., Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose P-BAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannavy, K., G. C. Barr, C. J. Dorman, J. Adamson, L. R. Mazengera, M. P. Gallagher, J. S. Evans, B. A. Levine, I. P. Trayer, and C. F. Higgins. 1990. TonB protein of Salmonella typhimurium: a model for signal transduction between membranes. J. Mol. Biol. 216897-910. [DOI] [PubMed] [Google Scholar]
- 18.Held, K. G., and K. Postle. 2002. ExbB and ExbD do not function independently in TonB-dependent energy transduction. J. Bacteriol. 1845170-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heller, K., R. J. Mann, and R. J. Kadner. 1985. Cloning and expression of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J. Bacteriol. 161896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgs, P. I., R. A. Larsen, and K. Postle. 2002. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 44271-281. [DOI] [PubMed] [Google Scholar]
- 21.Higgs, P. I., T. E. Letain, K. K. Merriam, N. S. Burke, H. Park, C. Kang, and K. Postle. 2002. TonB interacts with nonreceptor proteins in the outer membrane of Escherichia coli. J. Bacteriol. 1841640-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgs, P. I., P. S. Myers, and K. Postle. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 1806031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 787069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kampfenkel, K., and V. Braun. 1992. Membrane topology of the Escherichia coli ExbD protein. J. Bacteriol. 1745485-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kampfenkel, K., and V. Braun. 1993. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 2686050-6057. [PubMed] [Google Scholar]
- 26.Karlsson, M., K. Hannavy, and C. F. Higgins. 1993. A sequence-specific function for the N-terminal signal-like sequence of the TonB protein. Mol. Microbiol. 8378-388. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson, M., K. Hannavy, and C. F. Higgins. 1993. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8389-396. [DOI] [PubMed] [Google Scholar]
- 28.Koebnik, R. 1993. Microcorrespondence: the molecular interaction between components of the TonB-ExbBD-dependent and of the TolQRA-dependent bacterial uptake systems. Mol. Microbiol. 9219. [DOI] [PubMed] [Google Scholar]
- 29.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 4013041-13050. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277680-685. [DOI] [PubMed] [Google Scholar]
- 31.Larsen, R. A., G. J. Chen, and K. Postle. 2003. Performance of standard phenotypic assays for TonB activity, as evaluated by varying the level of functional, wild-type TonB. J. Bacteriol. 1854699-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen, R. A., G. Deckert, K. Kastead, S. Devanathan, K. L. Keller, and K. Postle. 2007. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J. Bacteriol. 1892825-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen, R. A., P. S. Myers, J. T. Skare, C. L. Seachord, R. P. Darveau, and K. Postle. 1996. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 1781363-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsen, R. A., and K. Postle. 2001. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 2768111-8117. [DOI] [PubMed] [Google Scholar]
- 35.Larsen, R. A., M. G. Thomas, and K. Postle. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 311809-1824. [DOI] [PubMed] [Google Scholar]
- 36.Larsen, R. A., M. G. Thomas, G. E. Wood, and K. Postle. 1994. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol. Microbiol. 13627-640. [DOI] [PubMed] [Google Scholar]
- 37.Lazzaroni, J.-C., J.-F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84391-397. [DOI] [PubMed] [Google Scholar]
- 38.Lazzaroni, J. C., P. Germon, M.-C. Ray, and A. Vianney. 1999. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177191-197. [DOI] [PubMed] [Google Scholar]
- 39.Letain, T. E., and K. Postle. 1997. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in gram-negative bacteria. Mol. Microbiol. 24271-283. [DOI] [PubMed] [Google Scholar]
- 40.Levengood, S. K., W. F. Beyer, Jr., and R. E. Webster. 1991. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc. Natl. Acad. Sci. USA 885939-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levengood, S. K., and R. E. Webster. 1989. Nucleotide sequences of the tolA and tolB genes and localization of their products, components of a multistep translocation system in Escherichia coli. J. Bacteriol. 1716600-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Muller, M. M., A. Vianney, J. C. Lazzaroni, R. E. Webster, and R. Portalier. 1993. Membrane topology of the Escherichia coli TolR protein required for cell envelope integrity. J. Bacteriol. 1756059-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49869-882. [DOI] [PubMed] [Google Scholar]
- 45.Postle, K., and R. A. Larsen. 2007. TonB dependent energy transduction between outer and cytoplasmic membranes. Biometals 20453-465. [DOI] [PubMed] [Google Scholar]
- 46.Roof, S. K., J. D. Allard, K. P. Bertrand, and K. Postle. 1991. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J. Bacteriol. 1735554-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skare, J. T., B. M. M. Ahmer, C. L. Seachord, R. P. Darveau, and K. Postle. 1993. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 26816302-16308. [PubMed] [Google Scholar]
- 48.Skare, J. T., and K. Postle. 1991. Evidence for a TonB-dependent energy transduction complex in Escherichia coli. Mol. Microbiol. 511000-11007. [DOI] [PubMed] [Google Scholar]
- 49.Sun, T.-P., and R. E. Webster. 1986. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J. Bacteriol. 165107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Traub, I., S. Gaisser, and V. Braun. 1993. Activity domains of the TonB protein. Mol. Microbiol. 8409-423. [DOI] [PubMed] [Google Scholar]
- 51.Vakharia-Rao, H., K. A. Kastead, M. I. Savenkova, C. M. Bulathsinghala, and K. Postle. 2007. Deletion and substitution analysis of the Escherichia coli TonB Q160 region. J. Bacteriol. 1894662-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vianney, A., T. M. Lewin, W. F. Beyer, Jr., J. C. Lazzaroni, R. C. Portalier, and R. E. Webster. 1994. Membrane topology and mutational analysis of the TolQ protein of Escherichia coli required for the uptake of macromolecules and cell envelope integrity. J. Bacteriol. 176822-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walburger, A., C. Lazdunski, and Y. Corda. 2002. The Tol/Pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol. Microbiol. 44695-708. [DOI] [PubMed] [Google Scholar]
- 54.Wiener, M. C. 2005. TonB-dependent outer membrane transport: going for Baroque? Curr. Opin. Struct. Biol. 15394-400. [DOI] [PubMed] [Google Scholar]