Abstract
The PilE pilin subunit protein of Neisseria gonorrhoeae undergoes unique covalent modifications with phosphoethanolamine (PE) and phosphocholine (PC). The pilin phospho-form transferase A (PptA) protein, required for these modifications, shows sequence relatedness with and architectural similarities to lipopolysaccharide PE transferases. Here, we used regulated expression and mutagenesis as means to better define the relationships between PptA structure and function, as well as to probe the mechanisms by which other factors impact the system. We show here that pptA expression is coupled at the level of transcription to its distal gene, murF, in a division/cell wall gene operon and that PptA can act in a dose-dependent fashion in PilE phospho-form modification. Molecular modeling and site-directed mutagenesis provided the first direct evidence that PptA is a member of the alkaline phosphatase superfamily of metalloenzymes with similar metal-binding sites and conserved structural folds. Through phylogenetic analyses and sequence alignments, these conclusions were extended to include the lipopolysaccharide PE transferases, including members of the disparate Lpt6 subfamily, and the MdoB family of phosphoglycerol transferases. Each of these enzymes thus likely acts as a phospholipid head group transferase whose catalytic mechanism involves a trans-esterification step generating a protein-phospho-form ester intermediate. Coexpression of PptA with PilE in Pseudomonas aeruginosa resulted in high levels of PE modification but was not sufficient for PC modification. This and other findings show that PptA-associated PC modification is governed by as-yet-undefined ancillary factors unique to N. gonorrhoeae.
Zwitterionic phospho-forms are increasingly being recognized as influential substituents of pathogenic and commensal microbe cell surfaces. Phosphoethanolamine (PE) modification of lipopolysaccharide (LPS) has been documented in a number of important pathogens (3, 13, 23, 38, 57) and has been implicated in resistance to cationic microbial peptides (24, 53) and recognition by complement component C4b (34). Phosphocholine (PC) modification of LPS is documented in Haemophilus influenzae and commensal Neisseria species (25, 42), while in Streptococcus pneumoniae, PC is found covalently linked to teichoic/lipoteichoic acid, as well as to some capsular polysaccharides (55). Activities ascribed to the PC moiety found in LPS, teichoic/lipoteichoic acids, and complex oligosaccharides include enhancing epithelial and endothelial cell adherence through binding to the receptor for platelet-activating factor (7, 41, 51) and acting as a ligand for C-reactive protein (41, 60) and PC-recognizing antibodies (4). The PilE protein subunit of the Neisseria gonorrhoeae type IV pilus (Tfp) colonization factor can be uniquely modified by the covalent addition of PE, as well as PC, to select serine residues (1, 14). This situation thus contrasts with all other examples noted, in which PE and PC are exclusively linked through carbohydrate moieties.
Given their increasing recognition as biologically active substituents, heightened attention has been focused on characterizing biosynthetic pathways involved in zwitterionic phospho-form modifications. Phospholipid headgroups represent abundant and conveniently localized donor sources for such phospho-form modifications. The Escherichia coli EptB protein (formerly called YhjW) has been shown to be directly responsible for modifying the inner core of LPS with PE, using phosphatidylethanolamine as a precursor (35). Most gram-negative species possess multiple genes whose products show structural homology to EptB. They include the Salmonella enterica gene cptA (53) and pmrC (24), as well as lptA (6) and the Lpt3 (26) and Lpt6 (70) genes from Neisseria meningitidis. Although phosphatidylcholine is found in significant amounts in diverse groups of bacteria, there are as yet no known instances in which its head group serves as a precursor. Rather, PC modifications of both LPS and teichoic/lipoteichoic acids involve a CDP-choline-type pathway utilizing enzymes encoded by the lic genes (61, 71) that requires exogenous choline or choline-containing compounds as precursors. Although present in commensal Neisseria species, the lic genes are absent in both N. gonorrhoeae and N. meningitidis. We recently identified PptA as a putative pilin phospho-form transferase required for modification of N. gonorrhoeae PilE with both PE and PC (1). The orthologous N. meningitidis PptA protein was implicated early on in PC modification of pilin, as high-frequency frame-shifting events within pptA correlated with phase (on-off) variation of the PilE PC epitope (58). PptA shares multiple structural features with E. coli EptB and related proteins implicated in LPS PE modification, which are all grouped together in the so-called YhjW/YjdB/YijP family, which comprises a subfamily of the larger alkaline phosphatase superfamily. The members of this superfamily have conserved core structures and active-site residues, which has led to the proposal that these enzymes involve catalytic cycles of phosphorylation, sulfatation, or phosphonation of conserved Ser/Cys/Thr residues (12). It has also been suggested that these enzymes have the same reaction scheme as was originally proposed for E. coli alkaline phosphatase (AlkP) (21).
The structural relatedness of PptA with EptB and other LPS PE transferases likely utilizing phosphatidylethanolamine as a donor strongly suggests similar modes of action. However, phosphatidylcholine has been documented only once in N. gonorrhoeae (49), while a more recent study of gonococcal phospholipids using fast atom bombardment-mass spectrometry (MS) and gas liquid chromatography-MS technologies failed to detect its presence (33). Two bacterial pathways for phosphatidylcholine synthesis have been characterized in bacteria: one in which endogenous phosphatidylethanolamine undergoes methylation by phospholipid N-methyltransferases and another in which it is synthesized via direct condensation of exogenous choline with CDP-diacylglyceride mediated by the activity of phosphatidylcholine synthase (46). To date, no phospholipid N-methyltransferases or phosphatidylcholine synthase orthologs are readily identifiable within neisserial genome sequences.
An additional confounding factor in pilin phospho-form modification involves the pilin-like PilV protein. Pilin PC modification has been seen only in pilV null mutants, a background that is also associated with PE hypermodification while PilV overexpression is associated with PE hypomodification (1). The mechanisms by which PilV impacts each of these aspects of Tfp biology remain enigmatic.
The goal of this study was to characterize pptA in regard to its genetic organization and to probe the structure-function relationships of PptA, as well as to gain insight into the relationships between PE and PC modifications mediated by PptA.
MATERIALS AND METHODS
Bacterial strains, vectors, and culture conditions.
The bacterial strains used in this study are described in Table 1. N. gonorrhoeae strains were grown on conventional GC medium as described previously (9) unless otherwise described as grown on defined, choline-free medium (30). E. coli DH5α or HB101 was used for plasmid propagation and cloning experiments and was grown on Luria-Bertani (LB) medium (37). The antibiotics used for selection of transformants and transconjugants were at the following concentrations: in N. gonorrhoeae, chloramphenicol, 10 μg/ml, and erythromycin, 8 μg/ml; in E. coli, chloramphenicol, 30 μg/ml; erythromycin, 300 μg/ml; kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; tetracycline, 15 μg/ml; and streptomycin, 100 μg/ml; in Pseudomonas aeruginosa PAK, carbenicillin, 1,000 μg/ml, and kanamycin, 1,000 μg/ml. For growth of P. aeruginosa PAK, the concentrations of carbenicillin and kanamycin were reduced to 300 μg/ml. Isolation and purification of plasmid DNA were performed by using QIAprepSpin Miniprep columns (no. 27106) according to the manufacturer's specifications (Qiagen, Chatsworth, CA). The nucleotide sequences of all clones and constructs described were determined from plasmid DNA or directly from PCR products derived from N. gonorrhoeae mutant strains at GATC Biotech AG (Konstanz, Germany).
TABLE 1.
Strains used in this study
| Strain | Parental strain | Relevant genotype | Reference |
|---|---|---|---|
| N. gonorrhoeae | |||
| VD300a | MS11 | 22 | |
| N400 | VD300 | recA6b | 56 |
| GV1 | N400 | pilV(Fs) | 65 |
| GE68 | N400 | pilE(S68A) | 1 |
| GE68V | GE68 | pilE(S68A) pilV(Fs) | 1 |
| KS1 | GV1 | pilV(Fs) pptA::Tn5 | This study |
| KS2 | GV1 | pilV(Fs) pptA::Tn9 | This study |
| KS3 | GV1 | pilV(Fs) pptA::Tn11 | This study |
| KS4 | GV1 | pilV(Fs) pptA::Tn22 | This study |
| KS5 | N400 | pptA::Tn5 | This study |
| KS6 | N400 | pptA::Tn9 | This study |
| KS7 | N400 | pptA::Tn11 | This study |
| KS8 | N400 | pptA::Tn22 | This study |
| KS9 | N400 | pptA::kan | 44 |
| KS10 | GV1 | pilV(Fs) pptA::kan | This study |
| GD2 | N400 | pptALS1::kan | 44 |
| GD3 | GV1 | pilV(Fs) pptALS1::kan | 1 |
| KS11 | KS10 | pilV(Fs) pptA::kan iga::pptA(Ind) | This study |
| KS12 | KS5 | pptA::Tn5 iga::pptA(Ind) | This study |
| KS13 | KS1 | pilV(Fs) pptA::Tn5 iga::pptA(Ind) | This study |
| KS14 | KS11 | pilV(Fs) pptA::kan iga::pptA(Ind)::Tn7 | This study |
| KS15 | KS11 | pilV(Fs) pptA::kan iga::pptA(Ind)::Tn23 | This study |
| KS16 | GV1 | pilV(Fs) pptA(S241A) | This study |
| KS17 | GV1 | pilV(Fs) pptA(ΔS241) | This study |
| KS18 | GV1 | pilV(Fs) pptA(G239A) | This study |
| KS19 | GV1 | pilV(Fs) pptA(H374A) | This study |
| KS20 | GV1 | pilV(Fs) pptA(G377A) | This study |
| KS21 | GV1 | pilV(Fs) pptA(S378A) | This study |
| KS22 | GV1 | pilV(Fs) pptA(H379A) | This study |
| KS23 | GV1 | pilV(Fs) pptA(S434A) | This study |
| KS24 | GV1 | pilV(Fs) pptA(D435A) | This study |
| KS25 | GV1 | pilV(Fs) pptA(H436A) | This study |
| KS26 | GV1 | pilV(Fs) iga::pptA | This study |
| KS27 | KS10 | pilV(Fs) pptA::kan iga::pptA | This study |
| KS28 | N400 | iga::pptA(Ind) | This study |
| KS29 | KS9 | pptA::kan iga::pptA(Ind) | This study |
| KS30 | GE68 | pilE(S68A) iga::pptA(Ind) | This study |
| KS31 | GE68V | pilV(Fs) pilE(S68A) iga::pptA(Ind) | This study |
| P. aeruginosa | |||
| PAKΔpilA | pilA | 18 | |
| KS32 | PAKΔpilA | pilA pilT | This study |
| KS33 | KS32 | pilA pilT pMMB67EH pilE | This study |
| KS34 | KS33 | pilA pilT pMMB67EH pilE pJT19 pptA | This study |
| KS35 | KS32 | pilA pilT pMMB67EH pilE(S68A) | This study |
| KS36 | KS32 | pilA pilT pilY1 | This study |
| KS37 | KS36 | pilA pilT pilY1 pMMB67EH pilE | This study |
| KS38 | KS37 | pilA pilT pilY1 pMMB67EH pilE pJT19 pptA | This study |
| KS39 | KS32 | pilA pilT pilB | This study |
| KS40 | KS39 | pilA pilT pilB pMMB67EH pilE | This study |
| KS41 | KS40 | pilA pilT pilB pMMB67EH pilE pJT19 pptA | This study |
| E. coli | |||
| S17.1 | RP4 2-Tc::Mu-Km::Tn7 pro res mod+ | 43 | |
| KS42 | DH5α | pMMB67EH pilE | This study |
| KS43 | KS42 | pMMB67EH pilE pJT19 pptA | This study |
VD300 is an Opa− derivative of MS11.
recA6 is an IPTG-inducible allele of recA.
Construction of an N. gonorrhoeae pptA null mutant and strains carrying site-specific mutations.
The pptA::kan allele carried on plasmid pLS3 (44) was introduced into the wt strain N400 and into GV1 [pilV(Fs) (a null allele of pilV with a frameshift mutation at the G−1 codon (65)], by transformation and selection for kanamycin-resistant transformants, to generate the pptA null mutants designated KS9 (pptA::kan) and KS10 [pilV(Fs) pptA::kan]. The strains carrying the pptALS1::kan allele carried on plasmid pLS1 (44) have been previously described (1).
The pptA gene was amplified from genomic DNA from the N. gonorrhoeae strain N400 with the primers pptA5′ and pptA3′. The resulting PCR fragment was digested with restriction endonucleases cutting at EcoRI and BamHI (see Table S1 in the supplemental material) and cloned into the polylinker of the plasmid pUP6 (67), yielding plasmid pUP6 pptA. Point mutations in pptA were made by using a modified version of the PCR-based technique termed splicing by overlap extension (SOE). The SOE PCR products were subcloned into pUP6 pptA, and new restriction sites created by the point mutation were used for screening. For the point mutations listed in Table S1 in the supplemental material, the two PCR products for each site-specific mutation were subjected to SOE using the flanking primers pptA5′1150 and pptA3′1927 and cut with BsteEII and NsiI (521 bp). The resulting fragment was then ligated with the NsiI/NheI (2,009-bp)- and the BstEII/NheI (2,509-bp)-cut fragments of pUP6 pptA. For pptA(G239A), the upstream region was amplified using primers G239A5′ and pptA3′, whereas the downstream primers were pptA5′ and G239A3′. The two PCR products were spliced together using the flanking primers pptA5′747 and pptA3′. The resulting PCR fragment was cut with XmnI and BstEII and ligated with the XmnI/NheI- and the BstEII /NheI-cut fragments of pUP6 pptA, resulting in pUP6 pptA(G239A). The G239A mutation created a unique DdeI restriction site. The following point mutations were made by subcloning SOE PCR products into pUP6 pptA(G239A) and screening for loss of the DdeI restriction site: pUP6 pptA(S241A) was made using PCR products from S241A5′ plus pptA3′ and pptA5′ plus S241A3′; pUP6 pptA(Δ241A) was made using PCR products from ΔS241_5′ plus pptA3′ and pptA5′ plus ΔS241_3′.
All resulting plasmids were introduced into strain GV1 [pilV(Fs)] for further analysis. pUP6 pptA(804) (see below) was cut with the restriction endonucleases AdhI and NotI, and the fragment was transformed into KS25 and KS22 in order to rescue the phenotype of the pptA(H436) and pptA(H379) point mutations.
Expression of pptA from an inducible promoter.
To create an isopropylthiogalactoside (IPTG)-regulatable pptA construct, the NotI-blunted fragment of pVD300 recA6 carrying lacIq, the tandem lac operator promoter sequences tacOP and UV5OP, and the tetM gene (39) was inserted into the HpaI site of pUP6 pptA. The resulting plasmid was cut with XhoI and BamHI, and the 3′ recessed ends were filled in using Klenow (New England BioLabs Inc.). The resulting fragment containing lacIOP pptA was inserted into the SacI site (filled in using Klenow) of p2/16/1 (67), resulting in p2/16/1 pptA(Ind).
Transposon mutagenesis of the pptA locus.
The two target plasmids, pUP6 pptA(1315) and pUP6 pptA(804), were constructed by amplifying the pptA gene in overlapping fragments from N. gonorrhoeae strain N400. The first 894 bp of pptA, along with 421 bp upstream of the gene, were amplified using the primers pptA5′ and pptA1315_3′. The terminal 1,327 bp of pptA, along with 227 bp downstream of the gene, were amplified using primers dca804_5′ and pptA3′. The resulting PCR fragments were digested with unique flanking EcoRI and BamHI sites (see Table S1 in the supplemental material) and cloned into the polylinker of the plasmid pUP6, yielding plasmids pUP6 pptA(1315) and pUP6 pptA(804), respectively. Transposon mutagenesis was performed on pUP6 pptA(1315) and pUP6 pptA(804) as described previously (40). Transposon insertions were isolated, sequenced, and transformed into the gonococcal strain N400 as previously described (9).
P. aeruginosa strains and construction of mutants.
The P. aeruginosa mutants used in this study were derived from the wt strain PAK (52). Construction of the pilA null allele was previously described (18), as were constructs for the pilT and pilB mutations (50). The pilY1 null allele was assembled by removing an internal fragment of coding sequence (amino acids 7 to 1094) from the targeted gene by SOE PCR to generate a nonpolar in-frame deletion. Specific primer sequences are available upon request. Deletion alleles were cloned into the suicide vector pEXGmGW (68) and introduced onto the chromosome of strain PAK as described previously (16). All deletions were confirmed by PCR and sequencing of the relevant chromosomal regions.
Conjugation of plasmids into P. aeruginosa PAK.
E. coli S17.1 (Table 1) was used as a donor for conjugation of plasmids into P. aeruginosa PAK strains. Donor and recipient strains were grown in LB broth at 37°C to an optical density at 600 nm (OD600) of ∼0.4 to 0.8, and 1 ml of each was mixed in a centrifuge tube and pelleted. The cells were resuspended in 100 μl LB medium and deposited as a drop on LB agar overnight at 37°C. The resulting growth was diluted in 6 ml of LB medium, and 10× to 1,000× dilutions were plated onto pseudomonas isolation agar (Difco) with the appropriate antibiotics.
Expression of PilE and PptA in P. aeruginosa PAK and E. coli.
To express gonococcal PilE in E. coli and P. aeruginosa, a Bsu36I/SmaI-blunted fragment from pPilE2 (1) was ligated into pMMB67EH (10), cut with SmaI, and transformed into E. coli DH5α and S17.1, respectively. The plasmid in S17.1 was transferred to KS32 by conjugation. To express gonococcal PptA in E. coli and KS32, an NdeI restriction site was fused to the ATG start codon of pptA by SOE PCR, using pUP6 pptA as a template. The upstream region was amplified using primers pUP6_5′ and pptA_NdeI_3′, whereas the downstream primers were pptA_NdeI_5′ and pptA3′1000. The two PCR products were subjected to SOE together, using flanking primers pUP6_5′ and pptA3′1000. The resulting PCR fragment was digested with restriction endonucleases recognizing the unique EcoRI and BsgI sites and subcloned into pUP6 pptA cut with the same restriction enzymes, yielding plasmid pUP6 pptAM1NdeI, which was then cut at the unique NdeI and BamHI restriction sites. The resulting fragment was cloned into the polylinker of pJT19 (64), yielding plasmid pJT19 pptA. This plasmid was transformed into E. coli DH5α and S17.1, respectively, using S17.1 to transfer the plasmid into KS32 by conjugation. For induction of PilE and PptA in E. coli and KS32, the bacteria were grown to an OD660 of 0.8 and induced with 0.4 mM IPTG and 2 mM m-toluate, respectively, for 2 hours. The cells were spun at 4,000 × g for 20 min, and the cell pellets were used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and MS (see below).
Immunofluorescence microscopy, SDS-PAGE, and immunoblotting.
For immunofluorescence microscopy, immobilization of bacteria was performed by incubating 100 μl of gonococcal cells at an OD660 of 0.1 for 1 h at 37°C in GC medium on poly-l-lysine-coated glass coverslips and fixing them for 1 h with 2% formaldehyde in phosphate-buffered saline. After being washed three times for 5 min each time in phosphate-buffered saline, the gonococci were labeled using a 1:10 dilution of MicroTrak Neisseria gonorrhoeae Culture Conformation Test according to the manufacturer's specifications (Trinity Biotech PIc) and mounted in the supplied mounting fluid before being viewed in a Nikon Eclipse C400 fluorescence microscope.
The procedures for SDS-PAGE and immunoblotting have been described previously (9). PC-decorated proteins were detected by using a 1:1,000 dilution of the monoclonal antibody (MAb) TEPC-15 (Sigma) and alkaline phosphatase-conjugated goat anti-mouse immunoglobulin A (Sigma) (14). PilE, PilV, and PptA were detected by immunoblotting of whole-cell lysates, using rabbit polyclonal antibodies and alkaline phosphatase-coupled goat anti-rabbit antibodies (Tago Inc.). PilV-specific sera have been described previously (65). PilE-specific antisera were generated against isolated pili from N. gonorrhoeae strain N400 (Invitrogen [lot 904]). PptA-specific sera were generated and affinity purified against recombinant His-tagged protein (Agrisera AB, Sweden). The recombinant His-tagged protein encompassed amino acids 230 to 548 of PptA and was amplified from N. gonorrhoeae strain N400 with primers pptA:sulfatase5′ and pptA:sulfatase3′. The resulting PCR fragment was digested using restriction endonucleases cutting at the unique, flanking BamHI and HindIII sites (see Table S1 in the supplemental material) and cloned into the polylinker of the plasmid pQE30 (Qiagen), resulting in the plasmid pQE30 pptA, which was transformed into E. coli strain M15[pREP4]. For protein purification, 6 liters of LB medium was inoculated with M15[pREP4] pQE30 pptA and was first incubated for 4 h to approximately an OD600 of 1.3 before induction with 1 mM IPTG and further incubation for 2 h to approximately an OD600 of 1.9. The cells were harvested at 4,000 × g for 20 min and frozen in liquid nitrogen. The frozen cells were lysed using an X-press French press (AB BIOX, Sweden) and dissolved in 100 ml 20 mM NaH2PO4-20 mM Na2HPO4 with 5 mg DNase I added. The suspension was agitated for 10 min, and urea was directly added to a final concentration of 8 M and stirred until it completely dissolved. Cellular debris was removed by centrifugation at 20,000 rpm in a JA25-50 rotor for 90 min, and the supernatant was filtered through a 0.22-μm filter (Millipore). The filtered solution was applied to a 5-ml Hitrap Chelating Histrap column mounted on an ÁKTApurifier FPLC (Amersham Biosciences). The column was washed twice with wash buffer 1 (20 mM NaH2PO4, 20 mM Na2HPO4, 8 M urea) and wash buffer 2 (20 mM NaH2PO4, 20 mM Na2HPO4, 8 M urea, 10 mM imidazole). The protein was then removed from the column by using an elution buffer (20 mM NaH2PO4, 20 mM Na2HPO4, 8 M urea, 0.5 M imidazol). The fractions containing the recombinant PptA protein were concentrated through an Amicon Ultra 12 MWCO 30,000 centrifugal filter (Millipore) and applied to a Tricorn Superdex high-performance column (Superdex 200 10/300 GL; Amersham Biosciences) with baseline separation. The fractions with PptA were concentrated through a Centricon 10 centrifugal filter (Millipore) for 3 h at 5,000 × g and diluted in 20 mM Tris, pH 7.4, 4 M urea.
Quantitative real-time RT-PCR.
RNA was isolated from N. gonorrhoeae using Trizol (Invitrogen) according to the manufacturer's instructions. The RNA was purified using an RNeasy minikit (Qiagen) with the RNase-free DNase set (Qiagen) according to the manufacturer's instructions. Ten nanograms of RNA was reverse transcribed and used as a template for quantitative real-time reverse transcription (RT)-PCR on a Light Cycler (Roche) in the same reaction, using a Quantitect Sybr green RT-PCR kit (Roche). The cycler conditions were the same for all reactions; 57°C was used as the annealing temperature, and fluorescence was read at 81°C. The primers used as the loading control were tbpA5′ and tbpA3′. Transcription of murF and pptA was carried out using the primer pairs murF5′-murF3′ and pptA5′1469- pptA3′1706, respectively. Negative controls without reverse transcriptase were performed in parallel.
Sample preparation and infusional MS analysis of intact PilE.
Pilus purification was carried out as described previously (66), and preparations were further processed using a methanol/chloroform precipitation procedure (1, 62). All data were acquired on a quadrupole time-of-flight mass spectrometer (Q-Tof micro; Micromass, Manchester, United Kingdom) equipped with the standard Z-spray electrospray ionization (ESI) source as described previously (1).
RESULTS
Genetic characterization of pptA.
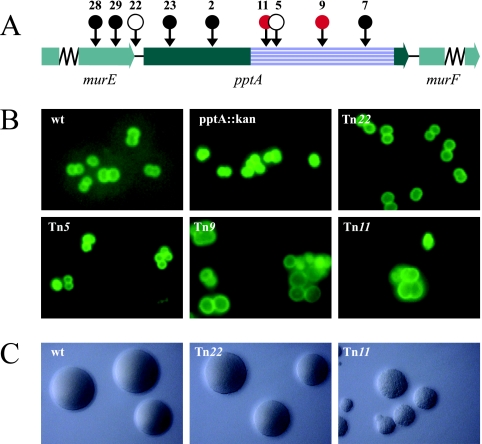
pptA maps upstream of murF, whose product is required for synthesis of UDP- MurNAc pentapeptide, a precursor essential to peptidoglycan synthesis (45). When the genetic organization of this locus was examined by transposon mutagenesis, only four of the nine insertions constructed in E. coli were recoverable in strain N400 (Fig. 1A). Moreover, transposon insertion mutants 9 and 11 uniquely displayed aberrant cell septation and “giant” cell morphology phenotypes that were paralleled by unusual colony morphologies and poor growth properties (Fig. 1B and C). Steady-state levels of murF mRNA were significantly reduced in the Tn9 and Tn11 backgrounds, slightly less diminished in the Tn22 mutant, and surprisingly elevated in the Tn5 background (Fig. 2A). Unlike the kanamycin resistance gene cassette insertion mutant (in which the level of murF mRNA was indistinguishable from the wt background), all four of the transposon insertion mutants displayed abnormal levels of murF mRNA.
FIG. 1.
Mapping and phenotypic characterization of pptA-linked transposon insertion mutants. (A) Physical map of the gonococcal pptA locus with transposon insertions indicated by circles. •, transposon insertions that could not be recovered in N. gonorrhoeae; ○, insertions with wt growth phenotype. The red circles represent transposon insertions that resulted in aberrant cell septation and coccal morphology. The precise sites of transposon insertion are shown in Fig. S1 in the supplemental material. The striated box defines the Pfam PF00884 sulfatase domain of PptA. (B) Cell septations and coccal morphologies of transposon mutants detected by immunofluorescence microscopy. Gonococci were detected using fluorescein-labeled MAbs (green). (C) Gonococcal colonies on conventional medium showing the different growth phenotypes of transposon mutants. The strains used were wt (N400), pptA::kan (KS9), Tn22 (pptA::Tn22; KS8), Tn5 (pptA::Tn5; KS5), Tn9 (pptA::Tn9; KS6), and Tn11 (pptA::Tn11; KS7).
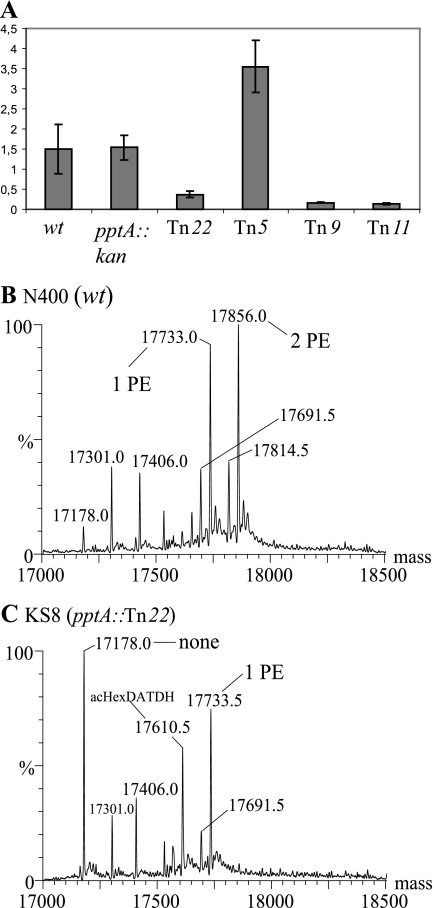
FIG. 2.
PptA-linked transposon insertion mutations perturb murF expression and PptA activity. (A) Real-time RT-PCR analysis of murF mRNA levels in mutants carrying transposon insertions. Amplification of tbpA (5) served as an internal reference and control. Negative controls were performed without reverse transcriptase treatment (not shown). The values are means ± standard errors of the mean; n = 3. The strains used were wt (N400), pptA::kan (KS9), Tn22 (pptA::Tn22; KS8), Tn5 (pptA::Tn5; KS5), Tn9 (pptA::Tn9; KS6), and Tn11 (pptA::Tn11; KS7). The Tn22 mutation reduces PilE phospho-form modification. (B and C) Intact mass analysis of PilE N400 (wt) (B) and KS8 (pptA::Tn22) (C). Species labeled as bearing PE/PC modifications also carry the hexose-2,4-diacetamido-2,4,6-trideoxyhexose (HexDATDH) dissacharide in its acetylated form. Peaks in the MS spectra that are not labeled are listed in Table S3 in the supplemental material.
PilE phospho-form modification was abolished in backgrounds in which transposon insertions disrupted the pptA open reading frame (ORF) (Tn5, -9, and -11) (data not shown) and reduced in that carrying the insertion upstream of the pptA ORF (Tn22) (Fig. 2B and C). These findings were consistent with our earlier work documenting the requirement for PptA in phospho-form modification (1). However, complementation there employed an ectopic, intact pptA gene copy (encompassing the pptA ORF and 420 bp of upstream sequence) that failed to restore phospho-form modification to levels seen in the wt background. Based on a real-time RT-PCR assay, complementation with the ectopic pptA gene copy restored pptA mRNA detection in a pptA::kan background, but not to levels seen in the wt background (Fig. 3). These studies were carried out in a pilV null background that enhanced PptA-mediated modification and led to PC decoration in order to facilitate phospho-form analyses and quantification. It is important to note, however, that pptA mRNA levels in a pilV background were not significantly altered from those seen in a wt background (Fig. 3, lane 2).
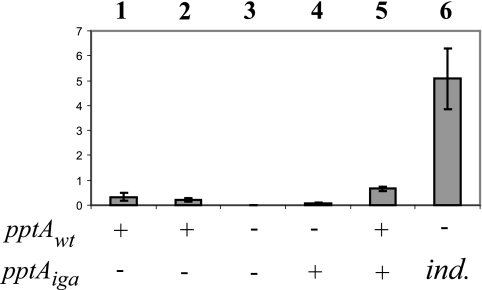
FIG. 3.
Relative pptA mRNA levels in defined isogenic backgrounds. Real-time RT-PCR amplification of tbpA mRNA (5) served as an internal reference/control. Negative controls included reactions performed without reverse transcriptase; no products were detected under these conditions (not shown). The values are means ± standard errors of the mean; n = 3. Bars: 1, wt (N400); 2, pilV(Fs) (GV1); 3, pilV(Fs) pptA::kan (KS10); 4, pilV(Fs) pptA::kan iga::pptA (KS27); 5, pilV(Fs) iga::pptA (KS26); 6, pilV(Fs) pptA::kan iga::pptA(Ind) (KS11) induced with 0.25 mM IPTG. pptAwt and pptAiga refer to pptA allele status, with the gene at either the endogenous locus (wt) or the ectopic locus (in iga). ind., ectopic, inducible pptA in its derepressed state.
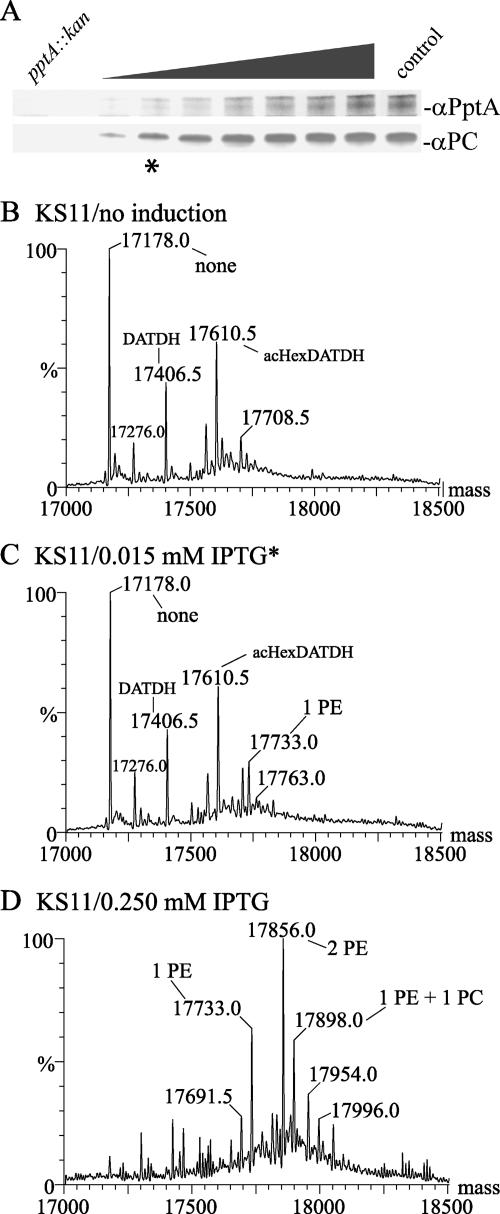
To provide higher levels of pptA for complementation, an inducible pptA allele carrying a tandem lac operator-promoter sequence was placed in the iga locus. In its repressed state, no mRNA was detected (data not shown), while when maximally derepressed, mRNA levels 17-fold higher than those seen for the wt strain were observed (Fig. 3, lane 6). Using a pptA::kan pilV background (KS11), a clear correlation was observed between levels of inducer, PptA protein, and the PilE PC epitope (as assessed by reactivity with the PC epitope recognizing TEPC-15 MAb) to a point where PC epitope levels plateaued while PptA levels increased (Fig. 4A). MS analyses of intact PilE protein from purified Tfp confirmed that increasing pptA expression was paralleled by concomitant increases in PE and PC site occupancy (Fig. 4B to D). Despite the high levels of pptA mRNA seen when maximally derepressed, detection of the pilin PC epitope remained dependent on the absence of PilV and was not seen under any conditions in the presence of the wt pilV allele (data not shown).
FIG. 4.
PilE PC modification levels parallel regulated expression of PptA. (A) Lane 1 (from left), KS9 (pptA::kan); lane 2, KS11 without IPTG [pilV(Fs pptA::kan iga::pptA(Ind)]. Lanes 3 to 9, KS11 with IPTG in the following concentrations: lane 3, 0.01 mM; lane 4, 0.015 mM; lane 5, 0.02 mM; lane 6, 0.03 mM; lane 7, 0.04 mM; lane 8, 0.05 mM; lane 9, 0.1 mM; lane 10, control (GV1). (B to D) Deconvoluted molecular weight spectra from intact PilE ESI MS analyses in strains KS11 without IPTG [pilV(Fs) pptA::kan iga::pptA(Ind)] (B), KS11 with IPTG (0.015 mM) (C), and KS11 with IPTG (0.250 mM) (D). Species labeled as bearing PE/PC modifications also carry the hexose-2,4-diacetamido-2,4,6-trideoxyhexose (HexDATDH) dissacharide in its acetylated (ac) form. A complete list of m/z forms and corresponding species of PilE can be found in Table S3 in the supplemental material. Asterisks indicate samples in which pptA was induced with 0.015 mM IPTG (A and C).
While transformants bearing Tn7 and Tn23 were not recoverable in a wt background, they could be established in a pptA diploid background (data not shown), and in all these instances, the insertions mapped to the ectopic gene copy. Together, these results are consistent with the hypotheses that (i) pptA and murF are part of a single transcriptional unit whose promoter maps at least 400 bp upstream of the pptA ORF, (ii) the growth defects seen in some pptA transposon mutants are the result of perturbed cell wall metabolism ensuing from reduced levels of MurF and are not directly related to altered PptA expression, and (iii) the pilV null allele does not lead to increased pptA transcription.
Pilin PC modification does not require exogenous choline.
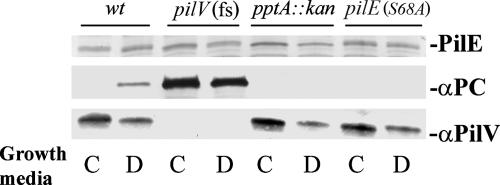
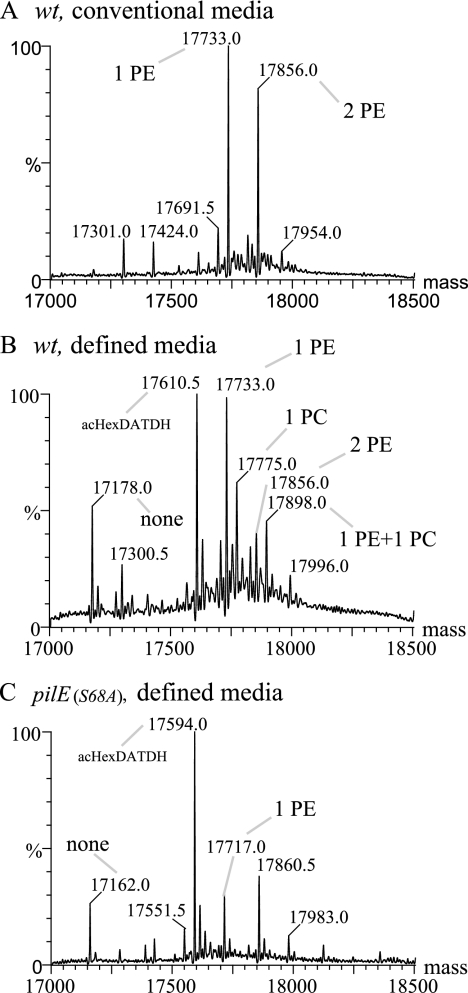
Both the phosphatidylcholine synthase- and lic gene-based pathways require exogenous choline as a precursor (46). Although neither pathway has been documented in pathogenic Neisseria species, we nonetheless tested if the pilin PC modification occurred in the absence of exogenous choline by growing the bacteria on chemically defined, choline-free medium (30). As seen by pilin reactivity with the TEPC-15 MAb for a pilV strain, PC modification occurred independently of an exogenous choline source (Fig. 5). Surprisingly, PC modification of pilin was seen for the first time in a wt background when propagated in defined, choline-free medium. Moreover, PC modification was PptA dependent under these conditions and was not detectable when a strain expressing PilE carrying an alanine substitution in place of serine at position 68 (the major residue for phospho-form modification) was used. To examine pilin phospho-form modifications occurring under these growth conditions in more detail, MS analysis of intact pilin was performed. In contrast to the sample derived from growth in conventional medium, one of the major species detected corresponded to PilE bearing the modified disaccharide and no phospho-form (Fig. 6A and B). Along with signals corresponding to glycosylated PilE carrying either one or two PE moieties, species corresponding to PilE bearing the modified disaccharide and a single PC and PilE bearing the modified disaccharide, one PE, and one PC were found in this sample. The signals for PilE derived from the pilE(S68A) strain grown in choline-free medium were not notably different from those observed previously for that mutant propagated in conventional medium (Fig. 6C). Given the unique finding of PC modification in a wt background, the status of PilV expression was assessed. In all backgrounds, PilV levels in cells cultured in defined, choline-free medium were reduced two- to threefold relative to levels seen in samples from conventional medium (Fig. 5, bottom). These findings demonstrate that the PilE PC moiety can be endogenously synthesized and corroborated earlier data showing that PC modification is associated with diminished PilV expression.
FIG. 5.
PilE PC modification occurs in a wt background when propagated in defined, choline-free medium. (Top) Immunoblot of whole-cell lysates using polyclonal antibodies specific for PilE. (Middle) MAb TEPC-15 specific for PC. (Bottom) Polyclonal antibodies specific for PilV. N. gonorrhoeae strains were grown either on solid conventional medium (C) or on solid defined, choline-free medium (D). Lanes: 1 and 2, wt (N400); 3 and 4, pilV(Fs) (GV1); 5 and 6, pptA::kan (KS9); 7 and 8, pilE(S68A) (GE68).
FIG. 6.
MS analysis of intact PilE from N. gonorrhoeae strains grown in defined, choline-free medium. (A) Wt (N400) grown on solid conventional medium. (B) Wt (N400) grown on solid defined, choline-free medium. (C) pilE(S68A) (GE68) grown on solid defined, choline-free medium. The species labeled as bearing PE/PC modifications also carry the hexose-2,4-diacetamido-2,4,6-trideoxyhexose dissacharide (HexDATDH) in its acetylated (ac) form. The peaks in the MS spectra that are not labeled are listed in Table S3 in the supplemental material.
Comparative modeling of the putative catalytic domain of PptA.
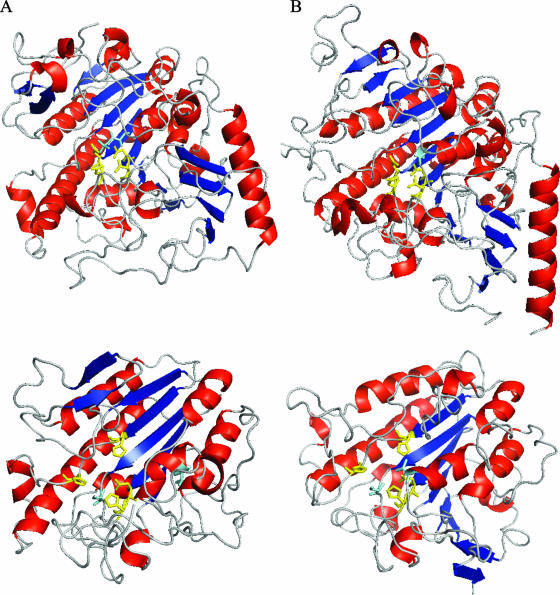
PptA shares basic structural features with LPS PE transferases, including an N-terminal contiguous region containing five consecutive transmembrane domains and a large sulfatase domain of the Pfam PF00884 family. Based on topological studies of PmrC, the sulfatase domain is predicted to be oriented in the periplasm (24). Included within the PF00884 domain family are the catalytic sites of members of the alkaline phosphatase superfamily of metalloenzymes as defined on the basis of biochemical and X-ray structural studies (2, 21, 36). We structurally modeled the PptA sulfatase domain to assess the similarities of its potential catalytic core to known protein structures. Although modeling with the CPHmodels and SwissModel programs failed to give significant hits, two models of significance were found using the ModWeb Modeler server (32) (Fig. 7A and B). These high-quality models derived from the human arylsulfatases ArsA and N-acetylgalactosamine-4-sulfatase both predicted prototypic α/β topology consisting of a central core of β-strands circumferentially flanked by α- helices. Moreover, putative metal-binding residues predicted previously (11, 12) (H374, H379, D435, and H436) were well colocalized, and there was remarkable conservation in their spatial positioning relative to the active-site structures of other alkaline phosphatase superfamily members. While not precisely superimposable on the structures of the other enzymes, the results strongly suggest that PptA possesses the same highly conserved catalytic core structure and therefore acts through a similar catalytic mechanism. If true, a nucleophile would likely be present in the vicinity of the metal coordination residues, and here, the hydroxyl of S241 is a good candidate. It is important to specify here that despite their overall congruence, the two models differ, with the ArsA-based model being comprised of six central β-strands while the N-acetylgalactosamine-4-sulfatase-based model has five central β-strands. This discrepancy influences the positioning of another putative active-site nucleophile (T278), which coordinates only with the other residues of the putative catalytic domain in the model based on ArsA.
FIG. 7.
Comparative modeling of the catalytic domain of PptA. (A) (Top) Crystallographic structure of human N-acetylgalactosamine-4-sulfatase (residues 43 to 533; Protein Data Bank [PDB] no. 1fsu). (Bottom) Three-dimensional structure of the sulfatase domain of PptA (residues 230 to 540) based on comparative modeling with 1fsu (sequence identity, 15%; E value, 1e-16; model score, 1.00). (B) (Top) Crystallographic structure of human arylsulfatase A (residues 19 to 503; PDB no. 1auk). (Bottom) Three-dimensional structure of the sulfatase domain of PptA (residues 230 to 547) based on comparative modeling with 1auk (sequence identity, 14%; E-value, 6e-42; model score, 0.90). Color coding of the protein backbone is as follows: red, α-helices; blue, β-strands; gray, loop regions. Color coding for residues is as follows: yellow, residues involved in metal ion coordination (human N-acetylgalactosamine-4-sulfatase, D53, D54, D300, and N301; arylsulfatase A, D29, D30, D281, and N282); cyan, active-site residues (human N-acetylgalactosamine-4-sulfatase, C91; arylsulfatase A, C69). Yellow, analogous residues in PptA, i.e., putative residues involved in metal ion coordination (H374, H379, D435, and H436); cyan, putative active-site residues (S241 and T278).
Site-directed mutagenesis of putative active-site residues in PptA.
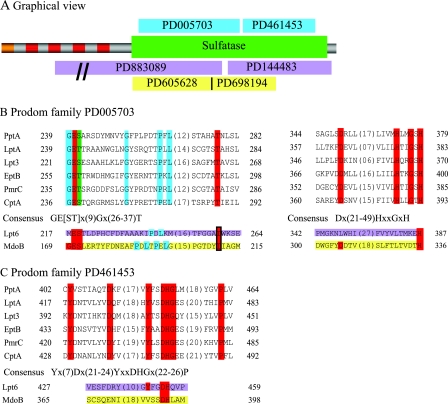
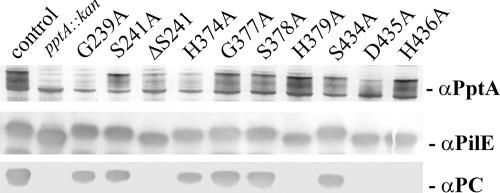
The modeled structures of the PptA active site and the inferred mechanism of activity predicted that a number of amino acid residues were likely to be of significant importance to enzyme catalysis. We also assessed the degrees of conservation of these candidate residues within the LPS PE transferase family (previously described as the YhjW/YjdB/YijP family), as found in Prodom entries PD005703 and PD461453, which encompass S241, H374, H379, D435, and H436 (Fig. 8). PptA, all known LPS PE transferases (excluding those related to Lpt6-type enzymes), and over 100 other uncharacterized proteins possess the PD005703 defined domain. All of these proteins carry readily identifiable, highly conserved residues corresponding to S241 (serine or threonine) and conserved residues corresponding to H374 and H379. Likewise, PptA and all known LPS PE transferases (excluding those related to Lpt6-type enzymes) and over 100 other uncharacterized proteins possess the PD461453 defined domain, including conserved residues corresponding to D435 and H436. Other absolutely conserved residues are seen in proteins bearing the PD005703 and PD461453 defined domains, and very often, these map adjacent to the residues noted above. To test the relevance of the PptA models and the significance of putative active-site residues, alanine substitution mutations for those residues modeled as being in the active site, along with certain conserved flanking residues, were constructed and introduced into a pilV null background. This background facilitated screening of the mutants for PptA function by virtue of PilE reactivity with the TEPC-15 MAb recognizing the PC epitope and the reduction in PilE mobility associated with phospho-form modification. As shown in Fig. 9, three of the nine pptA missense mutations (H379, D435, and H436) resulted in the pptA null phenotypes seen as loss of reactivity with the TEPC-15 MAb and increased PilE mobility in SDS-PAGE. MS analysis of intact PilE confirmed the absence of PC and PE modified forms (see Fig. S2 in the supplemental material and data not shown). In contrast, mutation of neighboring residues (G377, S378, and S434) had no observed effect on these PptA phenotypes. Mutants with alanine substitutions at either S241 or H374 retained TEPC-15 MAb reactivity and retarded PilE migration (Fig. 9). However, MS analyses of intact pilin from these two backgrounds were consistent with phospho-form hypomodification and showed pilin subunits exclusively modified with a single PC moiety (see Fig. S2 in the supplemental material). Finally, deleting residue S241 resulted in a pptA null phenotype, as seen by MS analysis of intact pilin and reaction to TEPC-15 MAb, although expression of PptA was normal (Fig. 9 and data not shown). The residues shown here to impact on PptA activity are illustrated in the context of the modeled active-site structures based on arylsulfatase A and N-acetylgalactosamine-4-sulfatase (see Fig. S3 in the supplemental material).
FIG. 8.
Alignment of sulfatase domains from the PptA/LPS PE transferase protein family. (A) Graphical overview of the domain structure of PptA. Color codes: orange, signal peptide (residues 1 to 33) with a hidden transmembrane region (residues 13 to 32); red, transmembrane regions (residues 42 to 61, 68 to 90, 122 to 144, and 156 to 175); green, sulfatase domain PF00884 (residues 230 to 516); blue, Prodom families PD005703 and PD461453; pink, Prodom families PD883089 and PD144483 (found in Lpt6); yellow, Prodom families PD605628 and PD698194 (found in MdoB). The domain structures are similar for all the proteins belonging to Prodom families PD005703 and PD461453. (B) Juxtaposed alignment of the sulfatase domain-containing proteins from Prodom family PD005703. Color codes: red, absolutely conserved residues; green, either S or T; blue, residues conserved only between characterized proteins. Lpt6 is part of the Prodom families PD883089 and PD144483, as indicated by the pink coloring of the residues. MdoB is part of the Prodom families PD605628 and PD698194, as indicated by the yellow coloring of the residues. (C) Alignment of characterized proteins from Prodom family PD461453. The color coding is the same as in panel B. The aligned proteins are as follows: PptA, Q9RMJ3 NEIGO; LptA, Q7DD94 NEIMB; Lpt3, Q9JXJ7 NEIMB; EptB, Q38J80 ECOLI (formerly YhjW); PmrC, YjdB SALTY; CptA, Q7CPC0 SALTY; Lpt6, Q9JWE8 NEIMA; MdoB, P39401 (OPGB) ECOLI.
FIG. 9.
PptA activities in structure-based, site-directed PptA mutants. (Top) Immunoblot of whole-cell lysates using polyclonal antibodies specific for PptA. (Middle) Polyclonal antibodies specific for PilE. (Bottom) MAb TEPC-15 specific for PC. Lanes: control, pilV(Fs) (GV1); pptA::kan, pilV(Fs) pptA::kan (KS10); G239A, G239A (KS18); S241A, S241A (KS16); ΔS241, ΔS241 (KS17); H374A, H374A (KS19); G377A, G377A (KS20); S378A, S378A (KS21); H379A, H379A (KS22); S434A, S434A (KS23); D435A, D435A (KS24); H436A, H436A (KS25). As controls for all mutants, immunoblotting was used to ensure that reduced or absent phospho-form modification did not result from decreased PptA stability.
Potential implications for the Lpt6 subfamily of LPS PE transferases and MdoB.
PE modifications at position 6 of inner core heptose II of LPS in H. influenzae and some strains of N. meningitidis involve a unique family of putative transferases termed Lpt6 (19, 70). Despite their high degree of identity to one another, H. influenzae and N. meningitidis Lpt6 were reported to have an extremely limited degree of relatedness to PptA and the other putative LPS PE transferases. This situation is mirrored in the fact that although the Lpt6 proteins share the sulfatase domain of the Pfam PF00884 family and analogous transmembrane domains of other LPS PE transferases, they lack the PD005703 and PD461453 defined domains (Fig. 8). A previous sequence comparison of the two Lpt6 proteins against Lpt3 and LptA aligned only 3 residues common to all four (corresponding to Y432, D435, and H436 of PptA) out of the total of 14 that are conserved in all proteins with the PD005703 and PD461453 defined domains (70). Another work concluded that Lpt6 “had no significant amino acid homology with the Lpt3 family proteins” (19). Rather, Lpt6 proteins are characterized by the PD883089 and PD144483 entries that nonetheless map within their Pfam PF00884 domains. By juxtaposing the PD005703 and PD883089 domains in reference to the common PF00884 sulfatase domain, it becomes obvious that the Lpt6 proteins have well-conserved, similarly positioned residues corresponding to those identified in the PptA/LPS PE transferases (Fig. 8). For example, 33 of 34 proteins possessing the PD144483 domain have a histidine residue juxtaposable to PptA H379, and all 34 have the DH residues (equivalent to PptA D435 H436) 60 to 70 residues distal to it. Parenthetically, it is intriguing to note that over half of the PD144483 domain-bearing proteins are from gram-positive organisms. This finding may give some insight into the evolutionary origins of Lpt6 PE transferases.
The same analysis used for Lpt6 was applied to MdoB, a member of the periplasmically localized phosphoglycerol transferase family that catalyzes the addition of phosphoglycerol to membrane-derived oligosaccharides from phosphatidylglycerol (17). This protein and its orthologues also possess the Pfam PF00884 domain but are uniquely defined by the PD605628 and PD698194 domains. By juxtaposing these domains in reference to the shared sulfatase domain, MdoB and related proteins have conserved, similarly positioned residues corresponding to those identified in the PptA/LPS PE transferases. Here, 29 of 33 proteins possessing the PD698194 domain have a histidine residue juxtaposable to PptA H379, while 13 of 33 have the DH residues (equivalent to PptA D435 H436) 60 to 70 residues distal to it. The latter value is misleadingly low due to the fact that the DH signature is at the very end of the domain, and manual inspection shows that over half of the PD698194 domain-bearing proteins actually have similarly positioned DH residues. The striking conservation of putative active-site residues for all these likely phospholipid head group transferases strongly suggests that each involves the same reaction mechanisms defined for better-characterized members of the phospho-/sulfo-coordinating metalloenzymes.
PilE phospho-form modification in heterologous host species.
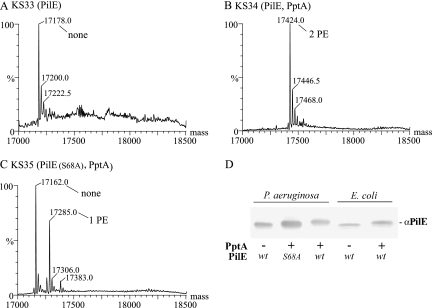
As another approach to probing the relationship between PE and PC PilE modifications, we expressed both N. gonorrhoeae PptA and PilE in P. aeruginosa. Unlike the case in pathogenic Neisseria species, phosphatidylcholine is an established phospholipid constituent in P. aeruginosa, with a well-characterized phosphatidylcholine synthase biosynthetic pathway (63). Here, the relaxed specificity of the P. aeruginosa pilus assembly machinery made it possible to carry out MS analyses of intact PilE from purified Tfp. In the presence of PptA, the sole species observed in the mass spectrum of intact PilE corresponded to the mature protein modified with two PE moieties, while in its absence, a single signal reflecting unmodified PilE was found (Fig. 10A and B). Coexpression of PptA and PilE with an alanine substitution at S68 showed two signals corresponding to unmodified PilE and PilE with a single PE attached (Fig. 10C). Save for the absence of glycan-related species, these findings were remarkably similar to those seen for this altered PilE derived from N. gonorrhoeae. Moreover, given the stoichiometry of PE site occupancy in the wt and S68 mutant PilE evidenced by the signals seen from P. aeruginosa-derived samples, the heterologous system recapitulates the hierarchical nature of PE modification seen in N. gonorrhoeae, with modification at S68 significantly increasing the likelihood that a second site will be modified (1). The hierarchical nature of PilE PE modification is thus an intrinsic property of the substrate and enzyme.
FIG. 10.
PptA-mediated PilE modification in P. aeruginosa and E. coli. (A) MS analysis of intact PilE expressed in P. aeruginosa. (B) MS analysis of intact PilE coexpressed with PptA in P. aeruginosa. (C) Intact mass analysis of a PilE(S68A) mutant coexpressed with PptA in P. aeruginosa. (D) Immunoblot of whole-cell lysates using polyclonal antibodies specific for PilE. Lanes 1 to 3 (from left), P. aeruginosa pilA pilT expressing N. gonorrhoeae proteins: lane 1, PilE; lane 2, PilE(S68A); lane 3, PilE and PptA. Lanes 4 and 5, E. coli DH5α expressing the proteins: lane 4, PilE; lane 5, PilE and PptA. Peaks in the MS spectra that are not labeled are listed in Table S3 in the supplemental material.
To confirm that the PilE species seen in the top-down MS analysis reflected the major forms expressed in P. aeruginosa, immunoblotting of whole-cell lysates was performed. As seen in Fig. 10D, migration of wt PilE expressed in conjunction with PptA was clearly retarded relative to its unmodified form, while the mobility of the S68 mutant PilE was intermediate between those of unmodified and modified forms of wt PilE. The mobilities of wt PilE coexpressed with and without PptA in E. coli were also examined by immunoblotting, and again, a significant PptA-dependent reduction in mobility was seen. Finally, these samples were also probed with the PC epitope recognizing TEPC-15 MAb with negative results (data not shown). Thus, PptA alone is sufficient for pilin modification with PE in P. aeruginosa and appears to have a similar activity when expressed in E. coli. However, there was no evidence for PilE PC modification in P. aeruginosa under the same conditions, despite the availability of the potential precursor phosphatidylcholine, as well as functional pathways for phosphatidylcholine biosynthesis.
DISCUSSION
Accumulating evidence suggests that the decoration of microbial surfaces with PE and PC moieties is a surprisingly common and important factor in host-parasite interactions. However, the biosynthetic pathways and enzymes that generate the modified PE and PC molecules presented on microbial surfaces are poorly understood. In this study, we addressed several outstanding questions regarding PptA and pilin phospho-form modification biology. Lacking in vitro assays for pilin phospho-form modification activity, we focused on genetic characterization of pptA as a means to probe the relationships between PptA expression, structure, and function, as well as the mechanisms by which other factors impact the system. In addition to confirming the role of PptA in pilin phospho-form modification, phenotypes associated with transposon mutations mapping upstream and in its ORF show that pptA expression is tightly coupled to that of murF and that MurF is undoubtedly an essential protein. Taken together with the low levels of pptA mRNA and PptA activity imparted by an ectopic allele containing 420 bp 5′ of the ORF, murE, pptA, and murF are likely part of a single operon whose promoter precedes mraZ (28). This finding is interesting because pptA expression in N. meningitidis has been shown to undergo phase variation due to frameshifting within a monotonous stretch of nucleotides (58). Such a pptA regulatory system is one of the few, if not the only, genetic control mechanisms compatible with its polycistronic organization that would not disturb distal murF expression. Based on the unperturbed murF expression seen for the pptA::kan allele, this background was used to show a dose-dependent relationship between PptA and pilin phospho-form modification and full stoichiometric restoration of phospho-form site occupancy. It is important to note here that although PC substitution may be an indirect readout of PptA activity (see below), assays to detect the PC epitope are more sensitive and quantitative for monitoring pilin modification at low to moderate PptA levels than MS detection of the PE substituent.
Galperin et al. and Galperin and Jedrzejas (11, 12), first noted the pattern of sequence similarities between phosphopentomutases, cofactor-independent phosphoglycerate mutases, alkaline phosphatases, and sulfatases defining the alkaline phosphatase superfamily. Encompassed within the catalytic core structures for each are similarly located serine and threonine (and cysteine in sulfatases) residues shown to form the covalent intermediates and residues implicated as metal-binding and coordinating sites. Included in that early analysis were EptB, PmrC, CptA, and orthologous proteins from H. influenzae, but at that time, their functions were not known. The relevance of those findings to establishing structural and catalytic similarities between PptA and the other alkaline phosphatase superfamily members was first demonstrated here by molecular modeling, with high-quality models derived using human lysosomal sulfatases predicting a striking colocalization of putative active-site and metal-coordinating residues. The significance of these findings was further validated by mutagenesis with individual substitution mutations of three residues implicated in metal coordination abolishing PptA activity, whereas that of a fourth such residue diminished activity. Alanine substitution at a highly conserved serine potentially corresponding to the catalytic residue (S241) reduced PptA activity. While this result might imply that S241 is not the site of formation of a covalent intermediate, the loss of the phosphorylatable hydroxyl group nucleophile might be compensated for by a water molecule, as shown for catalytic residue S102 substitution mutants of AlkP (48). The idea that active-site architecture (together with an appropriate nucleophile source) might contribute to catalysis is further attested to by the complete lack of activity seen for the SΔ241 mutant. Nonetheless, at this point, we cannot rule out the importance of a highly conserved T278 as an active-site residue. Potentially highly conserved residues synonymous with those implicated in PptA catalysis are readily identifiable within the Prodom PD005703 and PD461453 domains defining the LPS PE transferase family members, the Prodom PD883089 and PD144483 domains in the Lpt6 LPS PE transferase family members, and even the Prodom PD605628 and PD698194 domains defining MdoB phosphoglycerol transferase family members. Accordingly, the conserved structure and modular organization of these enzymes clearly attest to common sites of action and conserved reaction mechanisms.
The bulk of evidence supports the hypothesis that PptA is a PE transferase that utilizes the phospholipid head group of phosphatidylethanolamine as a donor. To date however, utilization of phosphatidylethanolamine as a precursor for PE has been demonstrated only for EptB (35), N. gonorrhoeae Lpt3 (31), and modifications of membrane-derived oligosaccharides (17). The enzyme responsible for the last activity is unknown, although the enzyme MdoB (which catalyzes the addition of phosphoglycerol derived from phosphatidylglycerol to membrane-derived oligosarcharides) has been ruled out (29). Assuming phosphatidylethanolamine is the precursor to PptA-mediated modifications, the question of what the precursor for PC is remains a perplexing problem. We previously proposed two plausible scenarios, one in which PptA might utilize phosphatidylcholine as a head group donor and another in which PptA would decorate PilE with PE that would be modified in situ into PC by the action of PE methyltransferase(s) (1). The phosphatidylcholine donor model is challenged by a number of observations. First, the presence of phosphatidylcholine in N. gonorrhoeae remains unsubstantiated. Second, the activities and active-site architecture of alkaline phosphatase superfamily enzymes are quite specific, and the ability of PptA to accommodate PE, as well as PC, with its additional trimethylammonium group, would likely require a high degree of relaxed specificity. Third, PC modification is not seen, even in the phosphatidylcholine possessing P. aeruginosa background despite high levels of PptA-mediated PE site occupancy. Finally, well-characterized structural modules and domains implicated in PC recognition or binding, such as those seen in PC esterase (15), CRP (54), phospholipase C (27), and pneumococcal choline binding proteins (8), are not readily recognizable in PptA (data not shown). An alternative model invoking methylation of PE bound to pilin is also problematic, as this would undoubtedly have to occur in the periplasm and the methyl donor S-adenosylmethionine is unlikely to be available at this site. Nonetheless, we have targeted potential N. gonorrhoeae PE methyltransferase homologs identified using the criteria detailed previously (46) for mutagenesis. However, the results are equivocal, since in some cases it has not been possible to recover null mutants, while in those cases where knockouts were viable, PC modification was not altered. Further evaluation of the alternative models would be dramatically facilitated by high-resolution structural analysis of the PptA sulfatase domain. Likewise, the availability of the PE pilin modification system in P. aeruginosa and E. coli should provide unique opportunities for both biochemical and genetic screens for PC-generating activities.
The relationships between PE and PC modifications are further complicated by the influence of the PilV pilin-like protein. Both PE hypermodification and PC modification are associated with pilV null backgrounds, and conversely, phospho-form hypomodification has been demonstrated in pilV-overexpressing backgrounds. However, levels of pptA mRNA are not discernibly elevated in a pilV null background, and overexpression of pptA does not in and of itself lead to PC modification in either N. gonorrhoeae or P. aeruginosa. Conversely, PC modification is seen even at low levels of pptA expression, provided that PilV is not present. Therefore, PilV does not impact on the system by modulating levels of PptA but rather may influence PptA activity or PC donor availability. It is interesting to note here that there is evidence in related periplasmic, phospholipid head group transferase systems that enzyme activity can be dramatically influenced by environmental signals. For example, the activity of E. coli EptB requires elevated levels of Ca2+ in a heptose-deficient mutant (35), PE modification of N. meningitidis lipid A by LptA is reported to be significantly influenced by growth conditions (6), and the activity of MdoB phosphoglycerol transferase is regulated directly by osmolarity (20). Even in the case of the seemingly unrelated system of lic1-mediated PC modification of LPS in H. influenzae, it has been suggested that levels of PC modification are influenced by growth conditions in a manner that cannot be solely accounted for by alterations in lic1 mRNA levels (69). It is therefore not without precedent that PptA activity might be altered by environmental conditions, although how PilV might be involved here is open to speculation.
Despite the findings made here, a considerable number of facets of the systems remain to be explored. Is PptA dedicated solely to pilin modification, or are other proteins modified? This is a particularly daunting task, as protein PE modifications are only recognizable through MS analyses. Likewise, do other members of the PptA/LPS PE transferase family possess protein-modifying activity? And what structural features, domains, and residues dictate the seemingly exclusive propensities for most family members to target LPS while PptA modifies pilin? Finally, there remains a serious gap in our knowledge of the enzymes directly engaged in PC modification of molecules elaborated on bacterial cell surfaces. In the case of PC modifications of LPS and teichoic acid associated with lic genes, the role of LicD as a PC transferase is inferred from genetic data indicating that licD allele status influences the sites of PC modifications in the target moieties (59, 71). Furthermore, the enzymes involved in the incorporation of PC into certain pneumococcal capsular polysaccharides are undefined (47). Formally, then, the enzymes that carry out the committed step of PC modification have yet to be unambiguously identified in any of these systems.
Supplementary Material
Acknowledgments
Work done at the University of Oslo and CMBN is supported by a Storforsk (“big research”) grant from the Research Council of Norway (RCN) and the Consortium for Advanced Microbial Sciences and Technologies (CAMST) national technology platform funded through the Functional Genomics (FUGE) program of the RCN and EMBIO (the steering board for research in molecular biology, biotechnology, and bioinformatics at the University of Oslo).
We are indebted to Svein Valla (NTNU, Trondheim, Norway), as well as William Shafer (Emory University School of Medicine, Atlanta, GA), for gifts of plasmids and strains.
Footnotes
Published ahead of print on 19 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aas, F. E., W. Egge-Jacobsen, H. C. Winther-Larsen, C. Løvold, P. G. Hitchen, A. Dell, and M. Koomey. 2006. Neisseria gonorrhoeae type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J. Biol. Chem. 28127712-27723. [DOI] [PubMed] [Google Scholar]
- 2.Bond, C. S., P. R. Clements, S. J. Ashby, C. A. Collyer, S. J. Harrop, J. J. Hopwood, and J. M. Guss. 1997. Structure of a human lysosomal sulfatase. Structure 5277-289. [DOI] [PubMed] [Google Scholar]
- 3.Brabetz, W., S. Muller-Loennies, O. Holst, and H. Brade. 1997. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in an Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur. J. Biochem. 247716-724. [DOI] [PubMed] [Google Scholar]
- 4.Briles, D. E., C. Forman, and M. Crain. 1992. Mouse antibody to phosphocholine can protect mice from infection with mouse-virulent human isolates of Streptococcus pneumoniae. Infect. Immun. 601957-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelissen, C. N., G. D. Biswas, J. Tsai, D. K. Paruchuri, S. A. Thompson, and P. F. Sparling. 1992. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to TonB-dependent outer membrane receptors. J. Bacteriol. 1745788-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, A. D., J. C. Wright, J. Li, D. W. Hood, E. R. Moxon, and J. C. Richards. 2003. Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. J. Bacteriol. 1853270-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cundell, D. R., N. P. Gerard, C. Gerard, I. Idanpaan-Heikkila, and E. I. Tuomanen. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377435-438. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Tornero, C., R. Lopez, E. Garcia, G. Gimenez-Gallego, and A. Romero. 2001. A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol. 81020-1024. [DOI] [PubMed] [Google Scholar]
- 9.Freitag, N. E., H. S. Seifert, and M. Koomey. 1995. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 16575-586. [DOI] [PubMed] [Google Scholar]
- 10.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48119-131. [DOI] [PubMed] [Google Scholar]
- 11.Galperin, M. Y., A. Bairoch, and E. V. Koonin. 1998. A superfamily of metalloenzymes unifies phosphopentomutase and cofactor-independent phosphoglycerate mutase with alkaline phosphatases and sulfatases. Protein Sci. 71829-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galperin, M. Y., and M. J. Jedrzejas. 2001. Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins 45318-324. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, B. W., J. W. Webb, R. Yamasaki, S. J. Fisher, A. L. Burlingame, R. E. Mandrell, H. Schneider, and J. M. Griffiss. 1989. Structure and heterogeneity of the oligosaccharides from the lipopolysaccharides of a pyocin-resistant Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 8617-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegge, F. T., P. G. Hitchen, F. E. Aas, H. Kristiansen, C. Løvold, W. Egge-Jacobsen, M. Panico, W. Y. Leong, V. Bull, M. Virji, H. R. Morris, A. Dell, and M. Koomey. 2004. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc. Natl. Acad. Sci. USA 10110798-10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermoso, J. A., L. Lagartera, A. Gonzalez, M. Stelter, P. Garcia, M. Martinez-Ripoll, J. L. Garcia, and M. Menendez. 2005. Insights into pneumococcal pathogenesis from the crystal structure of the modular teichoic acid phosphorylcholine esterase Pce. Nat. Struct. Mol. Biol. 12533-538. [DOI] [PubMed] [Google Scholar]
- 16.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 21277-86. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, B. J., and E. P. Kennedy. 1983. The biosynthesis of membrane-derived oligosaccharides. A membrane-bound phosphoglycerol transferase. J. Biol. Chem. 2582394-2398. [PubMed] [Google Scholar]
- 18.Kagami, Y., M. Ratliff, M. Surber, A. Martinez, and D. N. Nunn. 1998. Type II protein secretion by Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin-like component XcpT by the cytoplasmic component XcpR. Mol. Microbiol. 27221-233. [DOI] [PubMed] [Google Scholar]
- 19.Kahler, C. M., A. Datta, Y. L. Tzeng, R. W. Carlson, and D. S. Stephens. 2005. Inner core assembly and structure of the lipooligosaccharide of Neisseria meningitidis: capacity of strain NMB to express all known immunotype epitopes. Glycobiology 15409-419. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy, E. P. 1982. Osmotic regulation and the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. Proc. Natl. Acad. Sci. USA 791092-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, E. E., and H. W. Wyckoff. 1991. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J. Mol. Biol. 218449-464. [DOI] [PubMed] [Google Scholar]
- 22.Koomey, J. M., and S. Falkow. 1987. Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J. Bacteriol. 169790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulshin, V. A., U. Zahringer, B. Lindner, C. E. Frasch, C. M. Tsai, B. A. Dmitriev, and E. T. Rietschel. 1992. Structural characterization of the lipid A component of pathogenic Neisseria meningitidis. J. Bacteriol. 1741793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, H., F. F. Hsu, J. Turk, and E. A. Groisman. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 1864124-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lysenko, E., J. C. Richards, A. D. Cox, A. Stewart, A. Martin, M. Kapoor, and J. N. Weiser. 2000. The position of phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae affects binding and sensitivity to C-reactive protein-mediated killing. Mol. Microbiol. 35234-245. [DOI] [PubMed] [Google Scholar]
- 26.Mackinnon, F. G., A. D. Cox, J. S. Plested, C. M. Tang, K. Makepeace, P. A. Coull, J. C. Wright, R. Chalmers, D. W. Hood, J. C. Richards, and E. R. Moxon. 2002. Identification of a gene (lpt-3) required for the addition of phosphoethanolamine to the lipopolysaccharide inner core of Neisseria meningitidis and its role in mediating susceptibility to bactericidal killing and opsonophagocytosis. Mol. Microbiol. 43931-943. [DOI] [PubMed] [Google Scholar]
- 27.Martin, S. F., B. C. Follows, P. J. Hergenrother, and B. K. Trotter. 2000. The choline binding site of phospholipase C (Bacillus cereus): insights into substrate specificity. Biochemistry 393410-3415. [DOI] [PubMed] [Google Scholar]
- 28.Mengin-Lecreulx, D., J. Ayala, A. Bouhss, J. van Heijenoort, C. Parquet, and H. Hara. 1998. Contribution of the Pmra promoter to expression of genes in the Escherichia coli mra cluster of cell envelope biosynthesis and cell division genes. J. Bacteriol. 1804406-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, K. J., and E. P. Kennedy. 1987. Transfer of phosphoethanolamine residues from phosphatidylethanolamine to the membrane-derived oligosaccharides of Escherichia coli. J. Bacteriol. 169682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morse, S. A., and L. Bartenstein. 1980. Purine metabolism in Neisseria gonorrhoeae: the requirement for hypoxanthine. Can. J. Microbiol. 2613-20. [DOI] [PubMed] [Google Scholar]
- 31.O'Connor, E. T., A. Piekarowicz, K. V. Swanson, J. M. Griffiss, and D. C. Stein. 2006. Biochemical analysis of Lpt3, a protein responsible for phosphoethanolamine addition to lipooligosaccharide of pathogenic Neisseria. J. Bacteriol. 1881039-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pieper, U., N. Eswar, H. Braberg, M. S. Madhusudhan, F. P. Davis, A. C. Stuart, N. Mirkovic, A. Rossi, M. A. Marti-Renom, A. Fiser, B. Webb, D. Greenblatt, C. C. Huang, T. E. Ferrin, and A. Sali. 2004. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 32D217-D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahman, M. M., V. S. Kolli, C. M. Kahler, G. Shih, D. S. Stephens, and R. W. Carlson. 2000. The membrane phospholipids of Neisseria meningitidis and Neisseria gonorrhoeae as characterized by fast atom bombardment mass spectrometry. Microbiology 1461901-1911. [DOI] [PubMed] [Google Scholar]
- 34.Ram, S., A. D. Cox, J. C. Wright, U. Vogel, S. Getzlaff, R. Boden, J. Li, J. S. Plested, S. Meri, S. Gulati, D. C. Stein, J. C. Richards, E. R. Moxon, and P. A. Rice. 2003. Neisserial lipooligosaccharide is a target for complement component C4b. Inner core phosphoethanolamine residues define C4b linkage specificity. J. Biol. Chem. 27850853-50862. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds, C. M., S. R. Kalb, R. J. Cotter, and C. R. Raetz. 2005. A phosphoethanolamine transferase specific for the outer 3-deoxy-d-manno- octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J. Biol. Chem. 28021202-21211. [DOI] [PubMed] [Google Scholar]
- 36.Rigden, D. J., E. Lamani, L. V. Mello, J. E. Littlejohn, and M. J. Jedrzejas. 2003. Insights into the catalytic mechanism of cofactor-independent phosphoglycerate mutase from X-ray crystallography, simulated dynamics and molecular modeling. J. Mol. Biol. 328909-920. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schweda, E. K., O. E. Hegedus, S. Borrelli, A. A. Lindberg, J. N. Weiser, D. J. Maskell, and E. R. Moxon. 1993. Structural studies of the saccharide part of the cell envelope lipopolysaccharide from Haemophilus influenzae strain AH1-3 (lic3+). Carbohydr. Res. 246319-330. [DOI] [PubMed] [Google Scholar]
- 39.Seifert, H. S. 1997. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 188215-220. [DOI] [PubMed] [Google Scholar]
- 40.Seifert, H. S., R. S. Ajioka, D. Paruchuri, F. Heffron, and M. So. 1990. Shuttle mutagenesis of Neisseria gonorrhoeae: pilin null mutations lower DNA transformation competence. J. Bacteriol. 17240-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serino, L., and M. Virji. 2002. Genetic and functional analysis of the phosphorylcholine moiety of commensal Neisseria lipopolysaccharide. Mol. Microbiol. 43437-448. [DOI] [PubMed] [Google Scholar]
- 42.Serino, L., and M. Virji. 2000. Phosphorylcholine decoration of lipopolysaccharide differentiates commensal Neisseriae from pathogenic strains: identification of licA-type genes in commensal Neisseriae. Mol. Microbiol. 351550-1559. [DOI] [PubMed] [Google Scholar]
- 43.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 44.Snyder, L. A., N. J. Saunders, and W. M. Shafer. 2001. A putatively phase variable gene (dca) required for natural competence in Neisseria gonorrhoeae but not Neisseria meningitidis is located within the division cell wall (dcw) gene cluster. J. Bacteriol. 1831233-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobral, R. G., A. M. Ludovice, H. de Lencastre, and A. Tomasz. 2006. Role of murF in cell wall biosynthesis: isolation and characterization of a murF conditional mutant of Staphylococcus aureus. J. Bacteriol. 1882543-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sohlenkamp, C., I. M. Lopez-Lara, and O. Geiger. 2003. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 42115-162. [DOI] [PubMed] [Google Scholar]
- 47.Sorensen, U. B., R. Agger, J. Bennedsen, and J. Henrichsen. 1984. Phosphorylcholine determinants in six pneumococcal capsular polysaccharides detected by monoclonal antibody. Infect. Immun. 43876-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stec, B., M. J. Hehir, C. Brennan, M. Nolte, and E. R. Kantrowitz. 1998. Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102. J. Mol. Biol. 277647-662. [DOI] [PubMed] [Google Scholar]
- 49.Sud, I. J., and D. S. Feingold. 1975. Phospholipids and fatty acids of Neisseria gonorrhoeae. J. Bacteriol. 124713-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sundin, C., M. C. Wolfgang, S. Lory, A. Forsberg, and E. Frithz-Lindsten. 2002. Type IV pili are not specifically required for contact dependent translocation of exoenzymes by Pseudomonas aeruginosa. Microb. Pathog. 33265-277. [DOI] [PubMed] [Google Scholar]
- 51.Swords, W. E., M. R. Ketterer, J. Shao, C. A. Campbell, J. N. Weiser, and M. A. Apicella. 2001. Binding of the non-typeable Haemophilus influenzae lipooligosaccharide to the PAF receptor initiates host cell signalling. Cell Microbiol. 3525-536. [DOI] [PubMed] [Google Scholar]
- 52.Takeya, K., and K. Amako. 1966. A rod-shaped Pseudomonas phage. Virology 28163-165. [DOI] [PubMed] [Google Scholar]
- 53.Tamayo, R., B. Choudhury, A. Septer, M. Merighi, R. Carlson, and J. S. Gunn. 2005. Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar Typhimurium lipopolysaccharide core. J. Bacteriol. 1873391-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, D., M. B. Pepys, and S. P. Wood. 1999. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure 7169-177. [DOI] [PubMed] [Google Scholar]
- 55.Tomasz, A. 1981. Surface components of Streptococcus pneumoniae. Rev. Infect. Dis. 3190-211. [DOI] [PubMed] [Google Scholar]
- 56.Tonjum, T., N. E. Freitag, E. Namork, and M. Koomey. 1995. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol. Microbiol. 16451-464. [DOI] [PubMed] [Google Scholar]
- 57.Vaara, M., T. Vaara, M. Jensen, I. Helander, M. Nurminen, E. T. Rietschel, and P. H. Makela. 1981. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 129145-149. [DOI] [PubMed] [Google Scholar]
- 58.Warren, M. J., and M. P. Jennings. 2003. Identification and characterization of pptA: a gene involved in the phase-variable expression of phosphorylcholine on pili of Neisseria meningitidis. Infect. Immun. 716892-6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiser, J. N., J. M. Love, and E. R. Moxon. 1989. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell 59657-665. [DOI] [PubMed] [Google Scholar]
- 60.Weiser, J. N., N. Pan, K. L. McGowan, D. Musher, A. Martin, and J. Richards. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187 631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiser, J. N., M. Shchepetov, and S. T. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138141-143. [DOI] [PubMed] [Google Scholar]
- 63.Wilderman, P. J., A. I. Vasil, W. E. Martin, R. C. Murphy, and M. L. Vasil. 2002. Pseudomonas aeruginosa synthesizes phosphatidylcholine by use of the phosphatidylcholine synthase pathway. J. Bacteriol. 1844792-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winther-Larsen, H. C., J. M. Blatny, B. Valand, T. Brautaset, and S. Valla. 2000. Pm promoter expression mutants and their use in broad-host-range RK2 plasmid vectors. Metab. Eng. 292-103. [DOI] [PubMed] [Google Scholar]
- 65.Winther-Larsen, H. C., F. T. Hegge, M. Wolfgang, S. F. Hayes, J. P. van Putten, and M. Koomey. 2001. Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc. Natl. Acad. Sci. USA 9815276-15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hebert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29321-330. [DOI] [PubMed] [Google Scholar]
- 67.Wolfgang, M., J. P. van Putten, S. F. Hayes, D. Dorward, and M. Koomey. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 196408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4253-263. [DOI] [PubMed] [Google Scholar]
- 69.Wong, S. M., and B. J. Akerley. 2005. Environmental and genetic regulation of the phosphorylcholine epitope of Haemophilus influenzae lipooligosaccharide. Mol. Microbiol. 55724-738. [DOI] [PubMed] [Google Scholar]
- 70.Wright, J. C., D. W. Hood, G. A. Randle, K. Makepeace, A. D. Cox, J. Li, R. Chalmers, J. C. Richards, and E. R. Moxon. 2004. lpt6, a gene required for addition of phosphoethanolamine to inner-core lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae. J. Bacteriol. 1866970-6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, J. R., I. Idanpaan-Heikkila, W. Fischer, and E. I. Tuomanen. 1999. Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 311477-1488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.