Abstract
Magnetospirillum gryphiswaldense and related magnetotactic bacteria form magnetosomes, which are membrane-enclosed organelles containing crystals of magnetite (Fe3O4) that cause the cells to orient in magnetic fields. The characteristic sizes, morphologies, and patterns of alignment of magnetite crystals are controlled by vesicles formed of the magnetosome membrane (MM), which contains a number of specific proteins whose precise roles in magnetosome formation have remained largely elusive. Here, we report on a functional analysis of the small hydrophobic MamGFDC proteins, which altogether account for nearly 35% of all proteins associated with the MM. Although their high levels of abundance and conservation among magnetotactic bacteria had suggested a major role in magnetosome formation, we found that the MamGFDC proteins are not essential for biomineralization, as the deletion of neither mamC, encoding the most abundant magnetosome protein, nor the entire mamGFDC operon abolished the formation of magnetite crystals. However, cells lacking mamGFDC produced crystals that were only 75% of the wild-type size and were less regular than wild-type crystals with respect to morphology and chain-like organization. The inhibition of crystal formation could not be eliminated by increased iron concentrations. The growth of mutant crystals apparently was not spatially constrained by the sizes of MM vesicles, as cells lacking mamGFDC formed vesicles with sizes and shapes nearly identical to those formed by wild-type cells. However, the formation of wild-type-size magnetite crystals could be gradually restored by in-trans complementation with one, two, and three genes of the mamGFDC operon, regardless of the combination, whereas the expression of all four genes resulted in crystals exceeding the wild-type size. Our data suggest that the MamGFDC proteins have partially redundant functions and, in a cumulative manner, control the growth of magnetite crystals by an as-yet-unknown mechanism.
The ability of magnetotactic bacteria (MTB) to orient in the earth's magnetic field is based on specific organelles, the magnetosomes, which are membrane-enveloped crystals of a magnetic mineral that are arranged in chain-like structures within the cell (5). Magnetosome crystals display a variety of species-specific shapes and sizes, which for most MTB are between 35 and 120 nm (3, 4). In MTB of the genus Magnetospirillum, cubo-octahedral nanocrystals of the magnetic mineral magnetite (Fe3O4) are synthesized within magnetosome membrane (MM) vesicles, which are roughly spherical and are formed by invagination from the cytoplasmic membrane prior to magnetite biomineralization (2, 15, 16). The MM is a phospholipid bilayer with a distinctive biochemical composition (8, 29, 35). In Magnetospirillum gryphiswaldense, a specific set of 20 MM proteins (MMPs) was identified by biochemical and proteomic approaches (9, 10, 22), and these proteins were speculated to be involved in magnetosome biomineralization (29). However, the individual functions of MMPs have remained largely unknown, and only a few magnetosome proteins have been characterized experimentally with respect to their roles in magnetosome formation. For example, the tetratricopeptide repeat protein MamA, although not essential for magnetosome formation, was speculated to activate magnetosome vesicles for magnetite biomineralization by an unknown mechanism (16). The Mms6 protein is an MMP that in vitro exhibits iron-binding activity and affects crystal morphology in crystallization assays (1, 21). Another example is the acidic MamJ protein, which is involved in the assembly and stabilization of magnetosome chains (25), probably by aligning the individual particles along a cytoskeletal magnetosome filament that is likely formed by the actin-like MamK protein (15, 20, 26). These studies revealed that magnetosome formation is a complex process with strict control over MM vesicle differentiation and formation, iron transport, and the nucleation and growth of magnetite crystals, as well as their assembly into chain-like structures. In order for crystals to function effectively in magnetic orientation, crystal sizes must be controlled precisely within the narrow magnetic single-domain range, as the magnetic properties of magnetite nanocrystals change drastically with particle dimensions (3, 7). The isolation of spontaneous M. gryphiswaldense mutants that produce small and aberrantly shaped particles compared to the wild type (14, 37) indicated that crystal dimensions are under genetic control. Clearly, the growth of magnetite crystals has to be regulated to generate the species-specific shapes and sizes of particles. However, it is unknown how this regulation is achieved at the structural and genetic levels.
The four small MMPs MamG, MamF, MamD, and MamC together account for approximately 35% of all magnetosome-associated polypeptides in the MM of M. gryphiswaldense and are encoded by the mamGFDC operon within the conserved magnetosome island of M. gryphiswaldense and other magnetospirilla (9, 22, 37). Transcriptional analyses revealed that these genes are transcribed from a single promoter and are constitutively expressed (28, 39). With the exception of mamG, which is a Magnetospirillum-specific gene with no orthologs in other MTB, the mamD, mamF, and mamC genes are part of the MTB-specific set of 28 signature genes revealed by a comparative genomics approach (22). This means that these genes are specifically associated with the magnetotactic phenotype and occur in all MTB analyzed so far but do not have detectable homologs in any nonmagnetic organism. The 12.4-kDa MamC protein, which is the most abundant MMP, was shown to be resistant against solubilization by weak detergents and is tightly associated with the MM (10), probably owing to its two predicted transmembrane segments. Immunogold staining has shown that in M. magnetotacticum, MamC localizes exclusively in the MM (36). The second most abundant protein identified in MM preparations from M. gryphiswaldense is the 12.3-kDa MamF protein, which contains three predicted transmembrane segments and tends to form stable oligomers even in the presence of sodium dodecyl sulfate (9). The hydrophobic proteins MamD (30.2 kDa) and MamG (7.7 kDa) are partially identical and share a conspicuous motif containing a Leu-Gly-dipeptide repeat, which is reminiscent of repetitive sequences found in self-aggregating framework proteins (6, 34, 41). Based on their abundance in the MM, their exclusive occurrence in MTB, and their high level of conservation, MamC and the further gene products of the mamGFDC operon have been suggested to play a key role in magnetite crystal formation (29, 36, 40). However, until the present study, their specific functions in magnetosome formation have remained completely elusive.
In this study, we analyzed the functions of the MamGFDC proteins in magnetosome formation by targeted mutagenesis utilizing the Cre-loxP system (18). By the analysis of mutants lacking either mamC or the entire mamGFDC operon, we showed that, unexpectedly, neither MamC nor MamG, MamF, and MamC are essential for magnetite biomineralization. However, by complementation analysis, we further demonstrated that the sizes of magnetosome crystals are controlled by the cumulative action of the mamGFDC proteins.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
In this study, liquid cultures of M. gryphiswaldense strain R3/S1 (32) were grown in modified flask standard medium (FSM) (12). Colonies of M. gryphiswaldense were obtained on activated charcoal agar medium (ACAM) that was incubated microaerobically at 28°C (32). Growth experiments were carried out under microoxic conditions in 1-liter flasks containing 100 ml of low- or high-iron-content medium. Low-iron medium (LIM) is essentially FSM lacking yeast extract and ferric citrate, whereas for high-iron medium, ferric citrate was added to LIM to a concentration of 500 μM. To grow magnetite-free cells (with no magnetic response [Cmag]), M. gryphiswaldense strains were passaged for three successive transfers in LIM. Optical densities and Cmag values of M. gryphiswaldense cultures were measured turbidimetrically at 565 nm with immotile cells inactivated by the addition of formaldehyde (Fluka, Switzerland) to a final concentration of 0.074% prior to the measurement (26, 30). Magnetosomes were isolated as described previously (9) from cultures grown under microoxic conditions. For conjugation experiments, Escherichia coli strain S17-1 (33) was used as a donor and cultivated as previously described (23).
DNA techniques.
DNA isolation, digestion, ligation, and transformation essentially followed standard methods (23). Primers and plasmids used in this study are listed in Table 1 and Table S1 in the supplemental material. PCR products and vector inserts were sequenced using BigDye Terminator version 3.1 chemistry (Applied Biosystems, Darmstadt, Germany) on an ABI 3700 capillary sequencer. Sequence data were analyzed with Lasergene 6 (DNAStar Inc., Madison, WI) and MacVector 7.0 (Oxford Molecular Ltd., Oxford, United Kingdom) programs.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| M. gryphiswaldense strains | ||
| MSR-1 R3/S1 | Rifr Smr spontaneous mutant | 31 |
| ΔC::Kan | ΔmamC::Knr | This study |
| ΔC | ΔmamC | This study |
| ΔC_C | ΔC(pAS35) | This study |
| ΔGFDC | ΔmamGFDC | This study |
| ΔGFDC_MCS2 | ΔGFDC(pBBR1MCS-2) | This study |
| ΔGFDC_GFDC | ΔGFDC(pAS31) | This study |
| ΔGFDC_G | ΔGFDC(pAS32) | This study |
| ΔGFDC_F | ΔGFDC(pAS33) | This study |
| ΔGFDC_D | ΔGFDC(pAS34) | This study |
| ΔGFDC_C | ΔGFDC(pAS35) | This study |
| ΔGFDC_GD | ΔGFDC(pAS36) | This study |
| ΔGFDC_GC | ΔGFDC(pAS37) | This study |
| ΔGFDC_FD | ΔGFDC(pAS38) | This study |
| ΔGFDC_FC | ΔGFDC(pAS39) | This study |
| ΔGFDC_DC | ΔGFDC(pAS40) | This study |
| ΔGFDC_GFD | ΔGFDC(pAS41) | This study |
| ΔGFDC_GFC | ΔGFDC(pAS42) | This study |
| ΔGFDC_GDC | ΔGFDC(pAS43) | This study |
| ΔGFDC_FDC | ΔGFDC(pAS44) | This study |
| Plasmids | ||
| pBBR1MCS-2 | KnrlacZa | 17 |
| pK19mobsacB | KnrsacB (modified from Bacillus subtilis) lacZ | 24 |
| pCM184 | Apr Knr | 18 |
| pCM157 | Tcr | 18 |
| pDC2 | pK19mobsacB with upstream and downstream mamGFDC cluster-flanking sequences | This study |
| pAG3 | pCM184 with upstream mamC-flanking sequence between MunI and NdeI | This study |
| pAG4 | pAG3 with downstream mamC-flanking sequence between ApaI and SacI | This study |
| pAS100 | pSP72 with 2.941-kb construct consisting of the 2,077-bp mamGFDC operon and 705-bp upstream and 159-bp downstream sequences between XhoI and SacI restriction sites | This study |
| pAS101 | pAS100 cut with NaeI and Eco47III and self-ligated | This study |
| pAS102 | pAS100 cut with PvuII and PsiI and self-ligated | This study |
| pAS103 | pAS100 cut with NruI and BfrBI, blunted, and self-ligated | This study |
| pAS104 | pAS100 cut with EcoRI and self-ligated | This study |
| pAS105 | pAS100 cut with NaeI and PsiI and self-ligated | This study |
| pAS106 | pAS100 cut with NaeI and BamHI, blunted, and self-ligated | This study |
| pAS107 | pAS100 cut with PvuII and BamHI, blunted, and self-ligated | This study |
| pAS109 | pAS105 cut with EcoRI and self-ligated | This study |
| pAS110 | pAS100 cut with PvuII and EcoRI, blunted, and self-ligated | This study |
| pAS111 | pAS101 cut with NruI and EcoRI, blunted, and self-ligated | This study |
| pAS112 | pAS104 cut with PvuII and PsiI and self-ligated | This study |
| pAS113 | pAS104 cut with NaeI and Eco47III and self-ligated | This study |
| pAS114 | pAS101 cut with NruI and BamHI, blunted, and self-ligated | This study |
| pAS31 | pBBR1MCS-2 with 2,941-bp XhoI-SacI fragment of pAS100, for mamGFDC expression | This study |
| pAS32 | pBBR1MCS-2 with 1,014-bp XhoI-SacI fragment from pAS110, for mamG expression | This study |
| pAS33 | pBBR1MCS-2 with 1,229-bp XhoI-SacI fragment from pAS111, for mamF expression | This study |
| pAS34 | pBBR1MCS-2 with 1,826-bp XhoI-SacI fragment from pAS109, for mamD expression | This study |
| pAS35 | pBBR1MCS-2 with 1,538-bp XhoI-SacI fragment from pAS106, for mamC expression | This study |
| pAS36 | pBBR1MCS-2 with 2,104-bp XhoI-SacI fragment from pAS112, for mamGD expression | This study |
| pAS37 | pBBR1MCS-2 with 1,819-bp XhoI-SacI fragment from pAS107, for mamGC expression | This study |
| pAS38 | pBBR1MCS-2 with 2,165-bp XhoI-SacI fragment from pAS113, for mamFD expression | This study |
| pAS39 | pBBR1MCS-2 with 2,038-bp XhoI-SacI fragment from pAS114, for mamFC expression | This study |
| pAS40 | pBBR1MCS-2 with 2,375-bp XhoI-SacI fragment from pAS105, for mamDC expression | This study |
| pAS41 | pBBR1MCS-2 with 2,401-kb XhoI-SacI fragment from pAS104, for mamGFD expression | This study |
| pAS42 | pBBR1MCS-2 with 2,265-bp XhoI-SacI fragment from pAS103, for mamGFC expression | This study |
| pAS43 | pBBR1MCS-2 with 2,668-kb XhoI-SacI fragment from pAS102, for mamGDC expression | This study |
| pAS44 | pBBR1MCS-2 with 2,722-kb XhoI-SacI fragment from pAS101, for mamFDC expression | This study |
Generation of ΔmamGFDC and ΔmamC mutant strains.
An M. gryphiswaldense mutant lacking the mamGFDC cluster was generated using plasmid pDC2. For the construction of pDC2, we amplified 670 bp of the sequence upstream of mamGFDC, including the ATG start codon of mamG, with primer pair G/EcoRI-for and G/PstI_rev and 810 bp of the sequence downstream of mamGFDC, including the TGA stop codon of mamC, with primer pair C/PstI-for and C/XbaI_rev. Both amplification products were fused by three-fragment ligation between the EcoRI and XbaI sites of plasmid pK19mobsacB to produce pDC2. Plasmid pDC2 was introduced into M. gryphiswaldense R3/S1 by conjugation from E. coli S17-1, and clones that had chromosomally integrated pDC2 were selected on kanamycin (Kan)-containing ACAM. As we failed to obtain double-crossover mutants by sucrose selection due to unstable sacB expression, 300 randomly selected colonies were replica-plated onto ACAM (with and without Kan), and Southern blotting analysis of three clones that showed sensitivity to Kan confirmed the deletion of the mamGFDC operon. One mutant clone, designated ΔGFDC, was selected for further studies.
For the generation of a mamC mutant, we alternatively used the broad-host-range Cre-loxP antibiotic marker recycling system described by Marx and Lidstrom (18) in order to test its usability for M. gryphiswaldense. As most mam and mms genes are arranged in polycistronic operons, mutagenesis strategies require the construction of unmarked in-frame deletions, the generation of which in MTB has remained notoriously cumbersome due to difficulties in enforcing multiple double-crossover events. We found this system to provide an advantage over the conventional technique, and the exchange of the targeted locus with a selective marker allows selection against revertant growth. In addition, marker recycling by the site-specific Cre recombinase may enable the generation of strains bearing multiple genetic modifications with only a single selectable marker gene.
Searches for lox sites (34 bp composed of a short core sequence between two inverted repeats) in a draft version of the M. gryphiswaldense genome sequence identified no site identical to the characteristic lox sequence which might have been targeted by the Cre recombinase. The Cre recombinase of bacteriophage P1 catalyzes site-specific recombination between lox sites and, in particular, the in vivo excision of DNA regions flanked by codirectional loxP recognition sites (19). Cre expression from plasmid pCM157 (18) in M. gryphiswaldense was verified by reverse transcription-PCR. Cells expressing Cre did not show any apparent change in growth or magnetosome biomineralization, suggesting that Cre does not catalyze recombination between sequence sites inherent in the chromosome of M. gryphiswaldense. For the mamC deletion construct, the regions immediately flanking mamC were PCR amplified using the following primer pairs: AGmamCu_f/MunI and AGmamCu_r/NdeI-2 for the upstream region and AGmamCDd_f/ApaI and AGmamCd_r/SacI for the downstream region. A fragment of 1,822 bp upstream of mamC was introduced between the MunI and NdeI sites of pCM184 (18), upstream of a loxP site-flanked Kan resistance marker, to yield pAG3. Sequencing of a fragment of 1,450 bp downstream of mamC revealed 204 bp downstream of the 5′ end of an ApaI restriction site, which are missing in the partial 35-kb sequence of the magnetosome island deposited in GenBank (accession no. BX571797) that was used for primer construction. Consequently, the digestion of the PCR product of the 1,450-bp fragment downstream of mamC with ApaI and SacI yielded a 1,246-bp fragment that was inserted downstream of the loxP-flanked Kan resistance marker of ApaI- and SacI-digested pAG3, producing pAG4. Allelic exchange vector pAG4 was introduced into M. gryphiswaldense strain R3/S1 by conjugation from E. coli S17-1, and transconjugants were selected on solid ACAM containing Kan. Kan-resistant transconjugants were found at a frequency of 2.2 × 10−6 per recipient cell. Several randomly selected clones were propagated for one passage in liquid medium and streaked onto solid medium without antibiotics. Colonies from those plates were screened by PCR for the loss of mamC, which occurred at a frequency of 1.0 × 10−1. For one clone, designated ΔC::Kan, the replacement of mamC by a loxP-flanked Kan resistance marker was confirmed by Southern blot analysis. For the excision of the Kan marker gene from clone ΔC::Kan, plasmid pCM157 was introduced by conjugation from E. coli S17-1 and transconjugants were selected on tetracycline. After one passage on solid medium with tetracycline, 96% of the tetracycline-resistant ΔC::Kan-derived clones were Kan sensitive. For one clone, designated ΔC, the loss of the Kan resistance gene was confirmed by Southern blot analysis. Plasmid pCM157 was cured from ΔC by transfer to medium lacking tetracycline. The excision of the marker by Cre left behind a loxP scar at the position of the mamC gene.
Complementation studies.
For the genetic complementation of the ΔC and ΔGFDC mutant strains, a series of pBBR1MCS-2-based plasmids harboring the full-length mamGFDC cluster (pAS31) or deletion-containing variants (pAS32 to pAS44) were generated. Sequence deletions within the recombinant mamGFDC cluster in plasmid pAS100 were generated by restriction digestion. The generated mamGFDC cluster variants were then cloned between the XhoI and SacI sites of pBBR1MCS-2 for expression in M. gryphiswaldense. The construction of plasmid pAS100, which harbors a 2,941-bp XhoI-SacI fragment consisting of a 705-bp sequence upstream of mamGFDC containing the native promoter of the mamGFDC operon, the 2,077-bp mamGFDC cluster containing silent mutations, and a 159-bp sequence downstream of mamGFDC, is illustrated in Fig. S1 in the supplemental material. For the construction of the 2,941-bp fragment, primers annealing to the 5′ and 3′ regions of mamC (5′ region, primers b and b*; 3′ region, primers c and c*), mamF (5′ region, primers d and d*; 3′ region, primers e and e*), and mamD (5′ region, primers f and f*; 3′ region, primers h and h*) and to regions within mamD (primers g and g*) and mamC (primers i and i* and j and j*), upstream of the mamG start codon (primer a), and downstream of the mamC stop codon (primer k) were deduced from the magnetosome island sequence deposited under accession number BX571797. Primers annealing within the mamGFDC cluster contained mismatches to generate silent point mutations which either created or removed a restriction site: primers b/b* and c/c* created a NaeI and an Eco47III site within mamG, primers d/d* and e/e* created a PvuII and a PsiI site within mamF, primers f/f* and h created a NruI and a BfrBI site within mamD, primers g/g* removed a PvuII site contained in mamD, primer h* created an EcoRI site 18 bp upstream of mamC, and primers i/i* and j/j* removed NaeI sites contained in mamC. The assembly of the 2,941-bp XhoI-SacI sequence fragment was accomplished by four rounds of PCR. The first round produced 10 sequence fragments: AB* (corresponding to primer pair a/b*), BC* (primer pair b/c*), CD* (primer pair c/d*), DE* (primer pair d/e*), EF* (primer pair e/f*), FG* (primer pair f/g*), GH* (primer pair g/h*), HI* (primer pair h/i*), IJ* (primer pair i/j*), and JK (primer pair j/k). Next, sequence fragments from the first PCR round were fused in three successive rounds of fusion PCR (13) until two sequence fragments (AE* and EK) remained, and these fragments were ligated between the XhoI and SacI sites of pSP72 to produce pAS100. Sequence deletions in modified variants of the mamGFDC cluster were created in pAS100 by parallel digestion with two restriction enzymes and subsequent religation of the vector backbone. For instance, for the excision of mamC, pAS100 was digested with EcoRI and recirculated, producing pAS104, while for the creation of a large deletion in mamG, pAS100 was digested with NaeI and Eco47III, producing pAS101. pBBR1MCS-2-based expression vectors containing single gene constructs were pAS32 (mamG), pAS33 (mamF), pAS34 (mamD), and pAS35 (mamC), vectors containing double gene constructs were pAS36 (mamGD), pAS37 (mamGC), pAS38 (mamFD), pAS39 (mamFC), and pAS40 (mamDC), and vectors containing triple gene constructs were pAS41 (mamGFD), pAS42 (mamGFC), pAS43 (mamGDC), and pAS44 (mamFDC). For a negative control, we used vector pBBR1MCS-2 without an insert. Complementation constructs were introduced into the recipient mutant strains of M. gryphiswaldense by biparental conjugation with E. coli S17-1 as a donor. The expression of single, double, and triple complementation constructs was verified by reverse transcription-PCR, showing that the created deletions within the mamGFDC operon did not inhibit the transcription of genes located downstream in the operon.
Electron microscopy and size analysis of membrane vesicles and magnetite crystals.
Transmission electron microscopy (TEM) was performed either on unstained cells adsorbed onto carbon-coated copper grids by using a Zeiss EM 10 microscope or on thin sections by using a Zeiss EM 912 microscope equipped with an integrated OMEGA energy filter operated in the zero-loss mode.
For the analysis of thin sections, cells were fixed with 2.5% glutardialdehyde in 75 mM sodium cacodylate-2 mM MgCl2 (pH 7.0) for 1 h at room temperature. Postfixation was performed for 1 h with 1% osmium tetroxide in fixative buffer. Then cells were stained en bloc with 1% uranyl acetate in 20% acetone for 30 min. Dehydration was performed with a graded acetone series. Samples were then infiltrated and embedded in Spurr's low-viscosity resin. Ultrathin sections were cut with a diamond knife and mounted onto uncoated copper grids. The thin sections were poststained with aqueous lead citrate (100 mM, pH 13.0).
For crystal analysis, M. gryphiswaldense cultures were grown under microoxic conditions for 24 h at 28°C in FSM with 50 μM iron. Crystal parameters (crystal sizes and shapes) were measured from digitized TEM micrographs using ImageJ 1.36b and the plug-in Watersheds_514 developed by M. Pinchon and N. Bonnet, which allows the semiautomatic segmentation of particles from the images (http://helios.univ-reims.fr/Labos/INSERM514/ImageJ/). Twin crystals, which were occasionally observed (at a frequency of approximately 7%), were omitted from the analysis because the segmentation algorithm often failed to detect the correct crystal edges. The Mann-Whitney significance test (http://elegans.swmed.edu/∼leon/stats/utest.html) was used to determine the significance of differences in crystal size distributions (CSDs) and shape distributions.
RESULTS
Loss of MamC has only minor effects on magnetite crystal formation.
Cells of the ΔmamC mutant exhibited a magnetic reaction under the microscope and in the light-scattering assay and had a dark brown appearance virtually identical to that of wild-type cells (Fig. 1A). In electron micrographs, magnetosomes were found arranged in chains, and the sizes and shapes of magnetite crystals were very similar to those of wild-type crystals (Fig. 1B). However, size measurements of 225 magnetosome particles from ΔmamC mutant cells revealed that mature magnetite crystals were on average slightly smaller than those from the wild type (Fig. 1C; Table 2). Complementation of the mutant strain by pAS35 restored the formation of magnetosomes at sizes close to the wild-type range. An analysis of solubilized MMPs from the mutant by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting revealed the absence of the highly abundant 12.4-kDa MamC band from the resolved polypeptide pattern, which was otherwise virtually unchanged from that of the wild type (data not shown). In electron micrographs, isolated magnetosome particles from the mutant appeared identical to wild-type magnetosomes with respect to the presence of an organic membrane layer, the interparticle spacing, and their tendency to rearrange in chains (Fig. 1B), suggesting that the absence of MamC did not markedly affect the formation of a functional MM.
FIG. 1.
Phenotypic analysis of ΔmamC and ΔmamGFDC mutant strains. (A) Appearance of pellets of mutant and wild-type cells. For comparison, a pellet of the magnetosome-free M. gryphiswaldense MSR-1B mutant is shown. (B) Transmission electron micrographs of wild-type, ΔmamC, and ΔmamGFDC cells. Insets show higher-magnification images of prevalent magnetosome chains (I), prevalent crystal shapes (II), and purified magnetosomes which were negatively stained with uranylacetate (III). Arrowheads indicate MM junctions between isolated crystals. (C) CSDs and shape factor distributions (SFD) for the wild type, the mutant strains ΔmamC and ΔmamGFDC, and the complemented mutants.
TABLE 2.
Statistical parameters and results of the Mann-Whitney significance test for CSDs of magnetite crystals from wild-type and mutant strains of M. gryphiswaldense
| Strain | No. of crystals | CSDa (nm) | Mean crystal diam (nm) | Median crystal diam (nm) | Maximum crystal diam (nm) | Significance of differenceb from CSD of:
|
|
|---|---|---|---|---|---|---|---|
| Wild type | Strain ΔGFDC | ||||||
| Wild type | 236 | 35-40 | 34.8 | 36.2 | 50.4 | 2.77E−38* | |
| ΔGFDC | 235 | 25-30 | 24.1 | 25.3 | 41.5 | 2.77E−38* | |
| ΔGFDC_GFDC | 245 | 35-40 | 33.7 | 34.8 | 57.8 | 2.11E−01 | 2.55E−27* |
| ΔGFDC_MCS2 | 139 | 25-30 | 28.1 | 28.4 | 42.0 | 1.19E−17* | 4.80E−06* |
| ΔGFDC_G | 169 | 30-35 | 30.6 | 31.7 | 42.7 | 3.87E−10* | 7.60E−17* |
| ΔGFDC_F | 160 | 30-35 | 31.3 | 31.4 | 46.4 | 2.52E−07* | 9.71E−18* |
| ΔGFDC_D | 179 | 30-35 | 32.6 | 33.1 | 42.7 | 4.32E−06* | 2.26E−28* |
| ΔGFDC_C | 230 | 30-35 | 33.3 | 33.1 | 47.7 | 1.21E−03 | 5.48E−34* |
| ΔGFDC_GD | 110 | 30-35 | 30.3 | 30.9 | 45.1 | 3.81E−08* | 4.81E−11* |
| ΔGFDC_GC | 187 | 30-35 | 31.8 | 32.3 | 45.2 | 1.64E−08* | 1.61E−24* |
| ΔGFDC_FD | 199 | 35-40 | 35.5 | 36.9 | 54.9 | 4.56E−01 | 1.42E−37* |
| ΔGFDC_FC | 177 | 30-35 | 31.3 | 32.6 | 43.4 | 4.29E−08* | 3.90E−20* |
| ΔGFDC_DC | 184 | 35-40 | 32.9 | 33.5 | 44.3 | 1.36E−04* | 2.75E−29* |
| ΔGFDC_GFD | 141 | 30-35 | 31.0 | 32.8 | 55.1 | 3.82E−06* | 1.57E−12* |
| ΔGFDC_GFC | 204 | 35-40 | 34.7 | 35.5 | 51.4 | 4.20E−01 | 1.44E−37* |
| ΔGFDC_GDC | 182 | 35-40 | 33.0 | 34.5 | 46.3 | 1.87E−03 | 2.62E−27* |
| ΔGFDC_FDC | 143 | 35-40 | 34.9 | 35.5 | 49.6 | 7.00E−01 | 5.00E−30* |
| ΔC::Kan | 225 | 30-35 | 31.9 | 33.4 | 49.1 | 6.12E−07* | 5.24E−25* |
| ΔC_C | 230 | 35-40 | 37.5 | 37.8 | 56.0 | 5.76E−04* | 2.35E−06* |
Range of sizes most frequently represented among the crystal population.
*, Mann-Whitney probability test result is statistically highly significant (P < 1E−03).
A ΔmamGFDC mutant produces magnetosome crystals only 75% of the wild-type size.
The unexpected finding that the loss of the most abundant magnetosome protein, MamC, had only a minor effect on magnetosome formation prompted us to generate a deletion mutant lacking the entire mamGFDC operon, which was designated strain ΔGFDC. Cells of ΔGFDC exhibited a magnetic response if checked by microscopic observation. However, in contrast to the dark brown wild-type and ΔmamC mutant colonies, colonies of strain ΔGFDC had only a slightly brownish color (Fig. 1A). TEM micrographs of mutant cells revealed the presence of small magnetosome crystals that frequently had a cuboidal shape and were aligned in irregular, widely spaced chains (Fig. 1B; Table 2). An analysis of more than 220 crystals confirmed that mutant CSDs were shifted towards smaller sizes (the Mann-Whitney P value determined for CSDs of wild-type and ΔGFDC crystals was <1E−03, indicating that the difference was statistically significant). For the mutant, crystals between 25 and 30 nm occurred at the highest frequency, whereas crystals larger than 30 nm were in low abundance, accounting for only 24.3% of the analyzed crystal population. In contrast, crystals between 35 and 40 nm in diameter were the most abundant among the wild-type crystals, and thus, crystals larger than 30 nm occurred at a significantly higher frequency, 77.5%, among the wild-type population. Maximum sizes of crystals without obvious defects, such as twinning, were measured at 41.5 nm in mutant cells and 50.1 nm in the wild type. In addition, mutant crystals showed anisotropic shapes more often than wild-type crystals, as only 37.4% of the analyzed mutant crystals were equidimensional (shape factor [SF], >0.94), whereas 50.8% of wild-type crystals had an SF of >0.94 (Fig. 1C).
Complementation of strain ΔGFDC with plasmid pAS31 harboring the entire mamGFDC cluster restored the sizes of mature magnetite crystals to the wild-type range. CSDs and SF distributions for wild-type and complemented mutant strains were similar (P > 1E−01), which substantiates the idea that the effects on the ΔGFDC magnetosome crystals resulted from the loss of the MamGFDC proteins (Fig. 1C; Table 2).
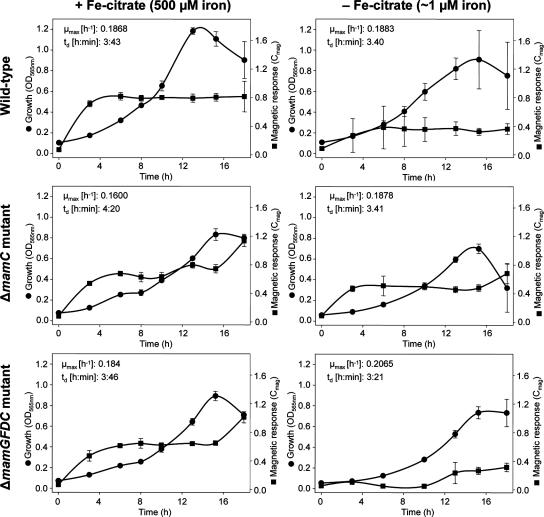
The formation of small magnetosomes could not be compensated for by increased iron, as indicated by TEM and Cmag measurements. Almost identical doubling times (approximately 3 h 40 min) were determined for the wild type at low (∼1 μM) and high (500 μM) iron concentrations (Fig. 2), for strain ΔGFDC at a high iron concentration, and for the ΔmamC mutant under low iron conditions. Even though strain ΔGFDC grew slightly faster at a low iron concentration than at a high iron concentration and the growth of the ΔmamC mutant was slightly slower at 500 μM Fe than at lower concentrations, we found no substantial effect on growth caused by the deletion of the mamC or mamGFDC genes. The development of magnetic responses after transfer to iron-sufficient conditions occurred similarly in iron-starved wild-type and ΔmamC cultures (Fig. 2). Freshly inoculated cultures were nearly nonmagnetic (Cmag < 0.1), and magnetic responses increased within the first 3 h of cultivation to a level which remained almost unchanged during further growth, indicating that the dynamics of magnetite formation were unaffected in the ΔmamC mutant. Likewise, the lack of the MamGFDC proteins did not affect the development of the magnetic response at a high iron concentration. At low iron concentrations, however, the magnetic response of the ΔmamGFDC mutant cultures was close to the detection limit during the first 10 h of cultivation, which may have resulted from the slower growth of crystals to sizes of permanent magnetic remanence. At iron concentrations nonlimiting for magnetite formation, Cmag values of ΔmamGFDC cultures were slightly lower than those of the wild type due to the less regular chain arrangement and smaller crystal size in the mutant cultures.
FIG. 2.
Growth and magnetic responses of wild-type and ΔmamC and ΔmamGFDC mutant strains under high- and low-iron conditions. + Fe citrate, with Fe citrate; − Fe citrate, without Fe citrate; μmax, growth rate; td, doubling time; OD565nm, optical density at 565 nm.
The ΔmamGFDC mutant forms wild type-like MM vesicles.
A possible reason for the observed growth inhibition of magnetite crystals in the ΔmamGFDC mutant may be the formation of aberrantly shaped or sized membrane vesicles, which may constrain the growth of crystals by size limitation. Isolated ΔmamGFDC magnetosomes like those from the wild type were associated with an organic envelope (Fig. 1B, inset III), suggesting that the formation of the MM was not prevented by the deletion. To examine the structure of the MM prior to magnetite synthesis, we analyzed thin-sectioned iron-starved cells by TEM. Empty and partially filled magnetosome vesicles were visible in micrographs of the mutant. These vesicles had the same shape and bilaminar structure as those in the wild type (Fig. 3). Slightly elongated vesicles were occasionally observed, but these were present in both the mutant and the wild type. In both wild-type and mutant cells, the membrane layer had a thickness of approximately 6 nm. Measurements of about 50 vesicles in mutant and wild-type cells revealed that the sizes of the vesicles varied and that the average diameter of mutant vesicles was slightly decreased compared to that of wild-type vesicles (wild-type average diameter, 44.9 nm; ΔGFDC mutant average diameter, 40.3 nm) (Fig. 3). A statistical comparison of vesicle size distributions, however, revealed that the difference was below the level of significance (P > 1E−2). In addition, in both strains the mean diameter of empty vesicles significantly exceeded the mean diameter of mature magnetite particles, suggesting that the growth of crystals was not limited by spatial constraints.
FIG. 3.
Transmission electron micrographs of magnetosome vesicles and distributions of the diameters of empty magnetosome vesicles in iron-starved cells of the ΔmamGFDC mutant (empty bars) and the wild type (filled bars) (P > 1E−02). Ultrathin sections were prepared from wild-type and ΔmamGFDC mutant strains grown under iron-sufficient (50 μM ferric citrate) and iron-limited (<1 μM iron) conditions. Arrows indicate empty and partially filled MM vesicles.
Transcomplementation with the mamG, mamF, mamD, and mamC genes gradually restores wild-type size.
After we had confirmed that the phenotype of the ΔmamGFDC mutant could be restored to that of the wild type by in-trans complementation (Fig. 1C; Table 2), we utilized the complementation assay to assess the contributions of the individual mamGFDC genes with respect to the observed effects on crystal size and shape development. Instead of generating numerous different knockout mutants, 13 variants of the mamGFDC operon were constructed, which permitted the in-trans expression of all individual genes of the operon as well as any combination of them in the ΔmamGFDC mutant. A comparison of crystal sizes from different complemented mutants with those produced by the wild type and the ΔmamGFDC mutant showed in most cases that differences between CSDs were statistically significant, indicating that the complementation constructs had a measurable effect on crystal size. Mutant strains complemented with only one of the four mamGFDC genes (strains ΔGFDC_G, ΔGFDC_F, and ΔGFDC_D) or with any two genes (strains ΔGFDC_GD, ΔGFDC_GC, ΔGFDC_FC, and ΔGFDC_DC) produced mature crystals larger than those produced by the ΔmamGFDC mutant but smaller than those produced by the wild type, suggesting that crystal size is not controlled by a single gene of the mamGFDC cluster (Fig. 4; Table 2). In contrast, strains complemented with any three of the four mamGFDC genes (strains ΔGFDC_GFC, ΔGFDC_GDC, and ΔGFDC_FDC) produced mature crystals of essentially wild-type size (P > 1E−03) (Fig. 4; Table 2). As a general trend, these data strongly suggest that the restoration of wild type-like crystal sizes requires at least three of the four MamGFDC proteins, almost independently of their combination, which points towards a cumulative action of the MamGFDC proteins in the regulation of crystal size.
FIG. 4.
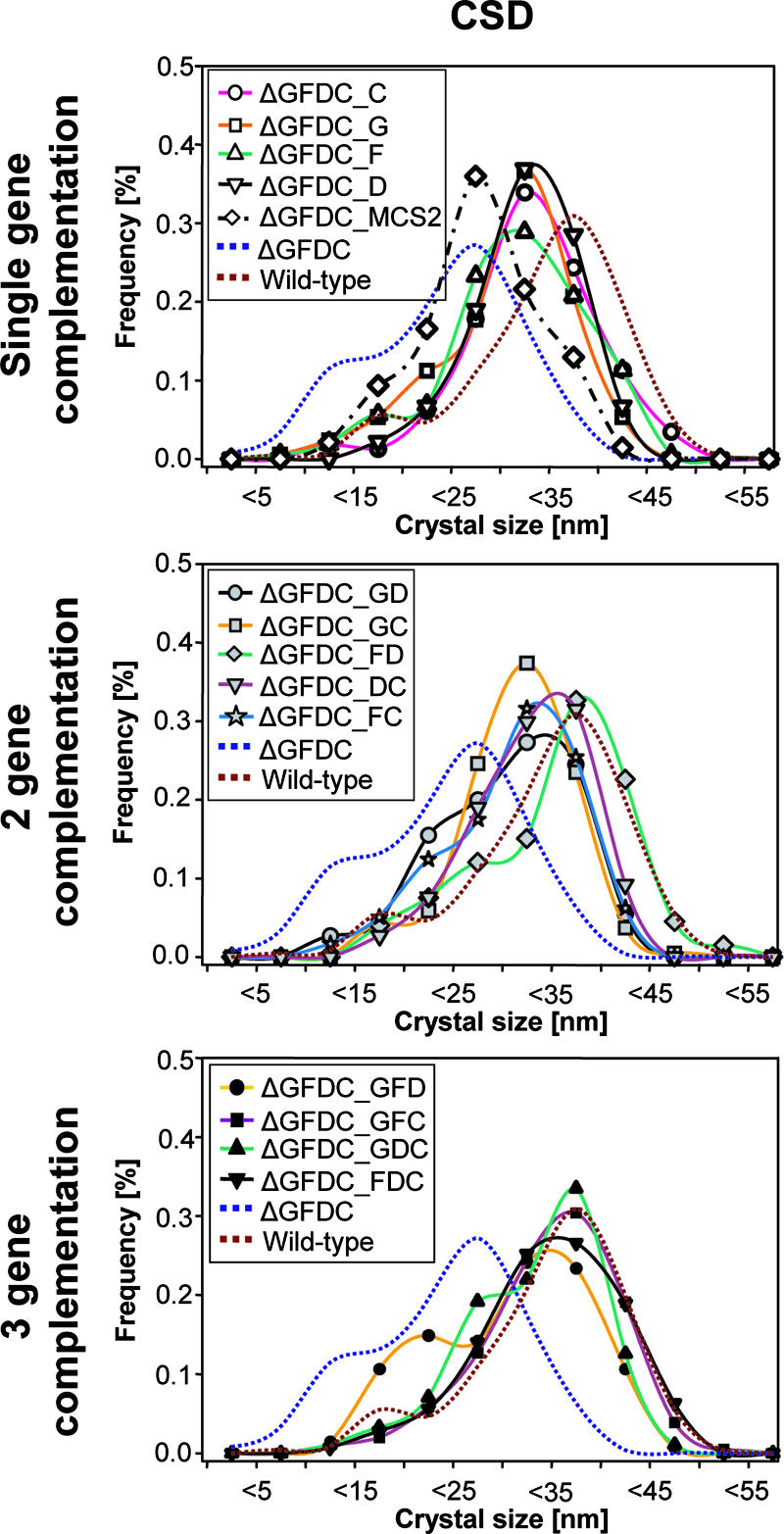
Complementation analysis of the ΔmamGFDC mutant showing size distributions of magnetite crystals produced by ΔmamGFDC strains complemented in trans with engineered variants of the mamGFDC cluster.
DISCUSSION
In this study, we analyzed the function of the abundant MMPs encoded in the mamGFDC operon by deletion mutagenesis and complementation analysis. Surprisingly, our analysis revealed that neither MamC nor MamG, MamF, and MamD, which together account for approximately 35% of all MMPs, are essential for magnetite biomineralization. We found that the loss of the most abundant MMP, MamC, had only a minor effect on the sizes of mature crystals. Even the absence of all four proteins did not entirely abolish magnetosome formation. However, the loss of MamGFDC had a significant effect on crystal size and chain organization, indicating that these proteins may have regulatory or accessory functions. It was impossible to assign these effects to one of the individual MamGFDC proteins, and our complementation study of the ΔmamGFDC mutant suggested that they have overlapping and partially redundant functions and may act collectively on the crystal size.
Our data suggest that the mode of action of MamGFDC is correlated to the expression of the heterogeneous mamGFDC genes, and surprisingly, the in-trans expression of additional copies of the entire mamGFDC operon in the wild type yielded magnetite particles even larger than those produced by the wild type without the additional copies. It remains unclear, however, by which mechanism the MamGFDC proteins regulate the size of the magnetosome crystals. In principle, there are several different factors which may affect the growth of magnetite crystals, such as the sizes and the shapes of the vesicles which spatially constrain crystal growth. It could have been envisioned, for instance, that the absence of four abundant integral membrane proteins would have a marked effect on the surface and curvature of MM vesicles. However, magnetosome vesicles of wild-type and ΔmamGFDC mutant cells had very similar sizes, shapes, and structures, and vesicles in both strains were larger on average than mature magnetite crystals. This finding argues against the idea that the smaller crystals in the mamGFDC mutant were caused simply by a reduced vesicle size. However, our method of size determination from thin sections potentially bears the risk of underestimation, as vesicles may not always have been sliced exactly along their maximum widths, and to some extent the determined size distribution may represent vesicles that have been cut more peripherally or tangentially. Therefore, methods such as cryoelectron tomography should be used in future studies to determine the spatial dimensions more precisely from a statistical number of three-dimensional vesicles.
Another possible explanation would be a reduced flux of iron from the cell exterior into the magnetosome vesicles. However, crystal growth inhibition was independent from the availability of iron in the medium, and the heterogeneous MamGFDC proteins lack any similarity to known transporters, which seems to argue against their direct involvement in iron transport into the MM vesicles. It has been suggested by Komeili and coworkers that magnetosome vesicles need to be activated for magnetosome formation, for example, by the action of the magnetosome protein MamA (16). Our observation that any combination of several different, unrelated proteins was capable of gradually restoring the wild-type phenotype seems to argue against a similar role for MamGFDC proteins. Alternatively, the MamGFDC proteins may act on the growth of magnetite crystals by regulating the physicochemical conditions within the interior of vesicles, such as, for instance, the charge distribution at the inner surfaces of vesicles or the intravesicular pH and redox conditions. For example, previous studies showed that the sizes and shapes of crystals of M. gryphiswaldense are strongly affected by redox conditions during magnetite biomineralization, and the inhibition of crystal growth was observed under highly oxidizing conditions, resulting in small and imperfect particles resembling those in the ΔmamGFDC mutant strain (12). It may also be possible that MamGFDC have an anti-inhibitory effect on crystallization by direct binding to the nascent crystallites, thereby preventing the premature termination of crystal growth. A further possible reason for the observed phenotype may be that MamGFDC act indirectly by interaction with or the recruitment of other proteins supporting the growth of crystals.
Although the mode by which the MamGFDC proteins act on crystal growth could not be fully unveiled in this first functional-analysis approach, our study sheds light on the in vivo functions of four abundant constituents of the MM, which have remained largely unknown. Remarkably, several major magnetosome proteins have only regulatory or modulating functions rather than being essential for magnetite biomineralization. For example, the loss of MamA protein also had only a minor effect, resulting in slightly reduced numbers of particles per cell, and MamA has been speculated to function in the activation of magnetosome vesicles for magnetite biomineralization by an unknown mechanism (15). One possible explanation for these weak phenotypes may be that some steps of the magnetosome-forming pathway are genetically redundant and further as-yet-unidentified genes may partially compensate for the loss of the major magnetosome proteins.
Intriguingly, the selective expression of different magnetosome proteins resulted in distinct mean particle sizes that consistently differed by only a few nanometers, while the number of magnetosomes per cell was not affected. Thus, fine tuning of MamGFDC expression may provide a powerful strategy for the precise control of the particle size. The synthesis of size-controlled magnetic nanoparticles by genetic engineering would be of considerable interest in a number of technological applications, since various physical characteristics of magnetic nanoparticles, such as sedimentation stability and magnetic remanence, are functions of particle size (11, 38).
Supplementary Material
Acknowledgments
We are grateful to Wolfgang Heyser and Anke Toltz (University of Bremen, Bremen, Germany) for support and access to the electron microscope. We thank Michael Winkelhofer (Ludwig-Maximilians-University of Munich, Munich, Germany) for helpful hints on statistical analysis. The continued support of Friedrich Widdel (Max-Planck-Institute for Marine Microbiology, Bremen, Germany) is greatly acknowledged. We thank René Uebe, Rita Dunker, and Frank Schreiber (Max-Planck-Institute, Bremen, Germany) for help with complementation experiments.
This work was supported by the BMBF BioFuture program and the Max Planck Society.
Footnotes
Published ahead of print on 26 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arakaki, A., J. Webbs, and T. Matsunaga. 2003. A novel protein tightly bound to bacterial magnetite particles in Magnetospirillum magnetotacticum strain AMB-1. J. Biol. Chem. 2788745-8750. [DOI] [PubMed] [Google Scholar]
- 2.Balkwill, D., D. Maratea, and R. P. Blakemore. 1980. Ultrastructure of a magnetotactic spirillum. J. Bacteriol. 1411399-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2217-230. [DOI] [PubMed] [Google Scholar]
- 4.Bazylinski, D. A., A. Garratt-Reed, and R. B. Frankel. 1994. Electron-microscopic studies of magnetosomes in magnetotactic bacteria. Microsc. Res. Tech. 27389-401. [DOI] [PubMed] [Google Scholar]
- 5.Blakemore, R. P. 1975. Magnetotactic bacteria. Science 190377-379. [DOI] [PubMed] [Google Scholar]
- 6.Bochicchio, B., A. Pepe, and A. M. Tamburro. 2001. On (GGLGY) synthetic repeating sequences of lamprin and analogous sequences. Matrix Biol. 20243-250. [DOI] [PubMed] [Google Scholar]
- 7.Frankel, R. B., T. J. Williams, and D. A. Bazylinski. 2006. Magneto-aerotaxis, p. 1-24. In D. Schüler (ed.), Magnetoreception and magnetosomes in bacteria, vol. 3. Springer, Heidelberg, Germany. [Google Scholar]
- 8.Gorby, Y. A., T. J. Beveridge, and R. Blakemore. 1988. Characterization of the bacterial magnetosome membrane. J. Bacteriol. 170834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grünberg, K., E. C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 701040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 674573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hergt, R., R. Hiergeist, M. Zeisberger, D. Schüler, U. Heyen, I. Hilger, and W. A. Kaiser. 2005. Magnetic properties of bacterial magnetosomes as potential diagnostic and therapeutic tools. J. Magn. Magn. Mater. 29380-86. [Google Scholar]
- 12.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61536-544. [DOI] [PubMed] [Google Scholar]
- 13.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 14.Hoell, A., A. Wiedenmann, U. Heyen, and D. Schüler. 2004. Nanostructure and field-induced arrangement of magnetosomes studied by SANSPOL. Physica B 350e309-e313. [Google Scholar]
- 15.Komeili, A., Z. Li, D. Newman, and G. Jensen. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311242-245. [DOI] [PubMed] [Google Scholar]
- 16.Komeili, A., H. Vali, T. J. Beveridge, and D. Newman. 2004. Magnetosome vesicles are present prior to magnetite formation and MamA is required for their activation. Proc. Natl. Acad. Sci. USA 1013839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 18.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 331062-1067. [DOI] [PubMed] [Google Scholar]
- 19.Palmeros, B., J. Wild, W. Szybalski, S. Le Borgne, G. Hermandez-Chavez, G. Gosset, F. Valle, and F. Bolivar. 2000. A family of removable cassettes designed to obtain antibiotic-resistance-free genomic modifications of Escherichia coli and other bacteria. Gene 247255-264. [DOI] [PubMed] [Google Scholar]
- 20.Pradel, N., C. Santini, A. Bernadac, Y. Fukumori, and L. Wu. 2006. Biogenesis of actin-like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc. Natl. Acad. Sci. USA 10317485-17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prozorov, T., S. K. Mallapragada, B. Narasimhan, L. Wang, P. Palo, M. Nilsen-Hamilton, T. J. Williams, D. A. Bazylinksi, R. Prozozov, and P. C. Canfield. 2007. Protein-mediated synthesis of uniform superparamagnetic magnetite nanocrystals. Adv. Funct. Mater. 17951-957. [Google Scholar]
- 22.Richter, M., M. Kube, D. A. Bazylinski, T. Lombardot, F. O. Glöckner, R. Reinhardt, and D. Schüler. 2007. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes with putative functions in magnetosome biomineralization and magnetotaxis. J. Bacteriol. 1894899-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 25.Scheffel, A., M. Gruska, D. Faivre, A. Linaroudis, P. L. Graumann, J. M. Plitzko, and D. Schüler. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440110-115. [DOI] [PubMed] [Google Scholar]
- 26.Scheffel, A., and D. Schüler. 2007. The acidic repetitive domain of the Magnetospirillum gryphiswaldense MamJ protein displays hypervariability but is not required for magnetosome chain assembly. J. Bacteriol. 1896437-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheffel, A., and D. Schüler. 2006. Magnetosomes in magnetotactic bacteria, p. 167-191. In J. M. Shively (ed.), Complex intracellular structures in prokaryotes, vol. 1. Springer, Berlin, Germany. [Google Scholar]
- 28.Schübbe, S., C. Würdemann, J. Peplies, U. Heyen, C. Wawer, F. O. Glöckner, and D. Schüler. 2006. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 725757-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schüler, D. 2004. Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense. Arch. Microbiol. 1811-7. [DOI] [PubMed] [Google Scholar]
- 30.Schüler, D., R. Uhl, and E. Baeuerlein. 1995. A simple light scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol. Lett. 132139-145. [Google Scholar]
- 31.Schultheiss, D., M. Kube, and D. Schüler. 2004. Inactivation of the flagellin gene flaA in Magnetospirillum gryphiswaldense results in nonmagnetotactic mutants lacking flagellar filaments. Appl. Environ. Microbiol. 703624-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 17989-94. [DOI] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1784-791. [Google Scholar]
- 34.Sudo, S., T. Fujikawa, T. Nagakura, T. Ohkubo, K. Sakaguchi, M. Tanaka, K. Nakashima, and T. Takahashi. 1997. Structures of mollusc shell framework proteins. Nature 387563-564. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka, M., Y. Okamura, A. Arakaki, T. Tanaka, H. Takeyama, and T. Matsunaga. 2006. Origin of magnetosome membrane: proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics 65234-5247. [DOI] [PubMed] [Google Scholar]
- 36.Taoka, A., R. Asada, H. Sasaki, K. Anzawa, L.-F. Wu, and Y. Fukumori. 2006. Spatial localizations of Mam22 and Mam12 in the magnetosomes of Magnetospirillum magnetotacticum. J. Bacteriol. 1883805-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ullrich, S., M. Kube, S. Schübbe, R. Reinhardt, and D. Schüler. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 1877176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winklhofer, M., and N. Petersen. 2006. Paleomagnetism and magnetic bacteria, p. 255-273. In D. Schüler (ed.), Magnetoreception and magnetosomes in bacteria, vol. 3. Springer, Heidelberg, Germany. [Google Scholar]
- 39.Würdemann, C., J. Peplies, S. Schübbe, A. Ellrott, D. Schüler, and F. O. Glöckner. 2006. Evaluation of gene expression analysis using RNA-targeted partial genome arrays. Syst. Appl. Microbiol. 29349-357. [DOI] [PubMed] [Google Scholar]
- 40.Yoshino, T., and T. Matsunaga. 2006. Efficient and stable display of functional proteins on bacterial magnetic particles using Mms13 as a novel anchor molecule. Appl. Environ. Microbiol. 72465-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zurovec, M., and F. Sehnal. 2002. Unique molecular architecture of silk fibroin in the waxmoth, Galleria mellonella. J. Biol. Chem. 27722639-22647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.