Abstract
φRSA1 is a wide-host-range bacteriophage isolated from Ralstonia solanacearum. In this study, the complete nucleotide sequence of the φRSA1 genomic DNA was determined. The genome was 38,760 bp of double-stranded DNA (65.3% G+C) with 19-bp 5′-extruding cohesive ends (cos) and contained 51 open reading frames (ORFs). Two-thirds of the φRSA1 genomic region encodes the phage structural modules, and they are very similar to those reported for coliphage P2 and P2-like phages. A φRSA1 minireplicon with an 8.2-kbp early-expressing region was constructed. A late-expression promoter sequence motif was predicted for these φRSA1 genes as 5′ TGTTGT-(X)13-ACAACA. The genomic sequence similarity between φRSA1 and related phages φ52237 and φCTX was interrupted by three AT islands, one of which contained an insertion sequence element, suggesting that they were recombinational hot spots. φRSA1 was found to be integrated into at least three different strains of R. solanacearum, and the chromosomal integration site (attB) was identified as the 3′ portion of the arginine tRNA(CCG) gene. In the light of the φRSA1 gene arrangement, one possible prophage sequence previously detected on the chromosome of R. solanacearum strain GMI1000 was characterized as a φRSA1-related prophage (designated φRSX). φRSX was found to be integrated at the serine tRNA (GGA) gene as an att site, and its size was determined to be 40,713 bp. φRSX ORFs shared very high amino acid identity with their φRSA1 counterparts. The relationships and evolution of these P2-like phages are discussed.
Ralstonia solanacearum is a soil-borne gram-negative bacterium known to be the causative agent of bacterial wilt in many important crops (20, 51). This bacterium has an unusually wide host range, with more than 200 species belonging to more than 50 botanical families (21). R. solanacearum strains represent a heterogeneous group subdivided into five races on the basis of their host range or six biovars based on their physiological and biochemical characteristics (21). Recently, the complete genome sequence of R. solanacearum GMI1000 was reported (44). The 5.8-Mbp genome consists of two replicons, a 3.7-Mbp chromosome and a 2.1-Mbp megaplasmid. Such a bipartite genome structure seems to be a characteristic of R. solanacearum, as a megaplasmid has been detected in most of the strains of this species (42). Both replicons of strain GMI1000 contain a number of regions known as alternative codon usage regions (ACURs), most of which differ significantly in G+C content from the average of 67% found in the entire genome, with G+C contents varying from 50% to 70%. These regions are often associated with mobile genetic elements such as prophages, insertion sequences (ISs), and IS-related sequences, suggesting that ACURs may have been acquired through horizontal gene transfer and have an important role in genomic evolution (44). At least four possible prophage sequences were detected on the chromosome of strain GMI1000, but the nature of these is largely unknown.
Recently, Yamada et al. (52) detected and isolated various kinds of bacteriophage that specifically infect R. solanacearum strains belonging to different races and/or biovars. Two of the phages, φRSS1 and φRSM1, are filamentous Ff-like phages (inoviruses) and contained single-stranded DNA genomes of 6,662 and 9,004 bases, respectively (30). Both phages have an integrative nature, and some strains of R. solanacearum contained prophages of these at a specific att sequence. φRSL1, another phage, with a head-tail structure resembling that of phages belonging to the myoviruses, contained an approximately 240-kb double-stranded DNA genome. This phage has a wider host range and only replicates via a lytic cycle. A template phage, φRSA1, spontaneously appeared from a strain of R. solanacearum (MAFF211272) and showed the widest host range; all of the strains tested, including those of races 1, 3, and 4 and biovars 3, 4, and N2, produced plaques on assay plates. φRSA1 particles have a unique morphology with a head and a tail, to the bottom of which a tail sheath is connected. A similar structure was also reported for Burkholderia cepacia phage KS5 (46). The genome of φRSA1 is a 39-kb linear DNA. A lysogenic state of this kind of phage was detected by genomic Southern blot analysis in 3 of 15 strains of the different races and different biovars (52).
In this study, the complete nucleotide sequence of the φRSA1 genomic DNA was determined. In the light of the φRSA1 gene arrangement, one possible prophage sequence previously detected on the chromosome of R. solanacearum strain GMI1000 was characterized as a φRSA1-related prophage (designated φRSX).
MATERIALS AND METHODS
Bacterial strains and phages.
Wild-type R. solanacearum strains M4S and MAFF106611 and strain MAFF211272 were from the Leaf Tobacco Research Center, Japan Tobacco Inc., and the National Institute of Agrobiological Sciences, Japan, respectively. Bacterial cells were cultured in CPG medium (26) at 28°C with shaking at 200 to 300 rpm. Phages were propagated and purified from single-plaque isolates. Routinely, the φRSA1 phage was propagated by using strain M4S as the host. A 16- to 24-h culture of bacterial cells grown in CPG medium was diluted 100-fold with 100 ml fresh CPG medium in a 500-ml flask. To collect sufficient amounts of phage particles, a total of 2 liters of bacterial culture was grown. When the cultures reached 0.2 U of optical density at 600 nm, the phage was added at a multiplicity of infection of 0.001 to 1.0. After further growth for 9 to 18 h, the cells were removed by centrifugation with an R12A2 rotor in a Hitachi himac CR21E centrifuge at 8,000 × g for 15 min at 4°C. To increase phage recovery, EGTA (to a final concentration of 1 mM) was added to the φRSA1-infected culture at 6 to 9 h postinfection. The supernatant was passed through a 0.2-μm-pore-size membrane filter, and phage particles were precipitated by centrifugation with a P28S rotor in a Hitachi XΠ100β centrifuge at 40,000 × g for 1 h at 4°C and dissolved in SM buffer (50 mM Tris-HCl at pH 7.5, 100 mM NaCl, 10 mM MgSO4, 0.01% gelatin). Purified phages were stained with Na-phosphotungstate before observation in a Hitachi H600A electron microscope as described by Yamada et al. (52). λ phage particles were used as an internal standard marker for size determination. Escherichia coli XL10 Gold and pBluescript II SK+ were obtained from Stratagene (La Jolla, CA).
DNA manipulations and sequencing.
Standard molecular biological techniques for DNA isolation, digestion with restriction enzymes and other nucleases, and construction of recombinant DNAs were followed as described by Sambrook and Russell (45). Phage DNA was isolated from purified phage particles by phenol extraction. In some cases, purified phage particles were embedded in 0.7% low-melting-point agarose (InCert agarose; FMC Corp.). After treatment with proteinase K (1 mg/ml; Merck) and 1% Sarkosyl, it was subjected to pulsed-field gel electrophoresis with a CHEF MAPPER electrophoresis apparatus (Bio-Rad) as described by Higashiyama and Yamada (24). Shotgun cloning and sequencing were performed at Hitachi High-Tech Fields Corp. as follows. φRSA1 whole genomic DNA was fragmented by sonication. DNA fragments in the ∼2-kb range were blunt ended and cloned with the pTS1/HincII vector (NipponGene) in E. coli cells. Shotgun sequencing of the clones (with an averaged insert of 2.0 kb) was performed with a BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems) in an Applied Biosystems 3700 DNA analyzer. A total of 920 sequences larger than 150 bases were assembled by the use of a phred/phrap/consed program (http://www.phrap.org). The analyzed sequences corresponded to 6.0 times the final genome size of 38,760 bp. Potential open reading frames (ORFs) larger than 300 bp were identified by using the online program Orfinder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and the DNASIS program (version 3.6; Hitachi Software Engineering Co. Ltd.). To assign possible functions to ORFs, searches through the databases were performed with the BLAST, BLASTX, and BLASTP programs (1).
To identify the attP and attB sequences of φRSA1, genomic DNA of φRSA1 lysogenic strain MAFF211272 of R. solanacearum was digested with HincII and hybridized with a φRSA1 DNA probe as described below. Two hybridizing bands of 2.6 and 5.7 kbp which possibly contain each of the integration junctions were cut out from the gel, ligated to the EcoRV site of pBluescript II SK+, and cloned in E. coli XL10 Gold. The nucleotide sequences determined for the clones were compared with the φRSA1 genomic sequence.
An autonomously replicating plasmid (minireplicon) was constructed from φRSA1 DNA as follows. Regions on the right side of the φRSA1 genome which contain possible early genes, including those for DNA replication, were amplified by PCR with a combination of forward primers (ORF33-P1, ORF36-P2, ORF39-P3, and ORF35-P5; Table 1) and reverse primer ORF49-P4 (Table 1). Twenty-five rounds of PCR were performed with 1 ng of φRSA1 DNA as the template under standard conditions in an MY Cycler (Bio-Rad). After digestion with HindIII at the reverse primer sites, the amplified fragment was connected to a Kmr cassette cut out with HindIII and SmaI from plasmid pUC4-KIXX (Amersham Biosciences). It was then introduced into cells of strains M4S and MAFF106611 by electroporation with a Gene Pulser Xcell (Bio-Rad) with a 2-mm cell at 2.5 kV in accordance with the manufacturer's instructions. Transformants were selected on CPG plates containing 15 μg/ml kanamycin (Meiji Seika, Tokyo, Japan).
TABLE 1.
φRSA1-specific oligonucleotide primers used for cloning and plasmid construction
| Oligonucleotide primer | Sequence (5′ to 3′)a | Location (nt)b |
|---|---|---|
| attP-Lc | GCAGTATGTGTCCTGGGTGTTTGTCTACCG | 36488-36517 |
| attP-Rc | CCTCTTATCAGAACGCCCCACCTCCC | 37203-37178 |
| ORF33-P1 | GATCCAGCCGGAGAAGTTGGAAGAATCGGG | 25247-25276 |
| ORF36-P2 | GCGAGCCGGTCTCCGTAGTGCATTTTCAAT | 28000-28029 |
| ORF39-P3 | CTCTTCCCCTCACGTTTTTTTCGCGCCTTG | 28997-29026 |
| ORF49-P4 | AGAGCGACAAAGCTTGATTTCCTTTGCTTG | 35718-35689 |
| ORF35-P5 | GTTCAGGCTCACGGAATTCTTCGAAGAACC | 27485-27514 |
Sequence underlined is changed to a HindIII site.
Numbering corresponds to the sequence with DDBJ accession no. AB276040.
A 720-bp region containing attP of φRSA1 was amplified with these primers and used as a probe to confirm the exact integration of φRSA1 in strain MAFF211272 by genomic Southern blot analysis.
Southern blot hybridization.
Genomic DNA of R. solanacearum cells was prepared by the minipreparation method as described by Ausubel et al. (2). After digestion with various restriction enzymes, DNA fragments were separated by agarose gel electrophoresis, blotted onto a nylon membrane (Biodyne, Pall Gelman Laboratory, Closter, NJ), hybridized with a probe (φRSA1 genomic DNA) labeled with fluorescein (Gene Images random prime labeling kit; Amersham Biosciences, Uppsala, Sweden), and detected with a Gene Images CDP-Star detection module (Amersham Biosciences). Hybridization was performed in a buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 5% liquid block, and 5% dextran sulfate for 16 h at 65°C. The filter was washed at 60°C in 1× SSC-0.1% SDS for 15 min and then in 0.5× SSC-0.1% SDS for 15 min with agitation, in accordance with the manufacturer's protocol. The hybridization signals were detected by exposing the filter to X-ray film (RX-U; Fuji Film, Tokyo, Japan).
In planta virulence assay of R. solanacearum strains.
Cells of R. solanacearum were grown in CPG medium for 1 to 2 days at 28°C. After centrifugation, cells were resuspended in distilled water at a density of 108/ml. The cell suspension was injected with a needle into the major stem of tobacco plants (Nicotiana tabacam SR1, 4 weeks old with four to six leaves) at a site 1 cm above the soil level (just above the cotyledons). As a control, distilled water was injected in the same manner. Each bacterial strain was injected into five plants. Plants were cultivated in a Sanyo Growth Cabinet (Sanyo, Osaka, Japan) at 25°C (16 h light, 8 h dark) for 3 to 4 weeks before detailed examination. Symptoms of wilting were graded from 1 to 5 as described by Winstead and Kelman (50). Extracellular polysaccharide production by R. solanacearum cells was assayed by the method of Gatt and Berman (15).
Nucleotide sequence accession number.
The φRSA1 genomic sequence was deposited in DDBJ under accession no. AB276040.
RESULTS AND DISCUSSION
Host range and lysogenic nature of φRSA1.
φRSA1 has a wide host range; all 15 strains of the different races or biovars of R. solanacearum produced plaques (with variable frequency) on assay plates (52). With strain M4S as the host, φRSA1 titers were usually not so high; a value of 2 × 108 to 1 × 109 PFU/ml was obtained, but phage recovery was dramatically increased (100-fold) by addition of 1 mM EGTA to the φRSA1-infected host culture. This EGTA effect was also reported for φCTX, which uses the lipopolysaccharide core as a receptor site on the cell surface and requires Ca2+ ions for binding to the receptor (38). In that case, EGTA treatment reduced phage adsorption, and when the phage was added at the final stage of infection, its recovery from the cell debris greatly increased. On the basis of these similar observations, we suspect that the lipopolysaccharide receptor and Ca2+ ions are involved in the adsorption of φRSA1 to R. solanacearum.
φRSA1 spontaneously appeared from strain MAFF211272 (52), but UV light irradiation of this strain did not induce phage production; the lysates prepared by UV irradiation contained a rather reduced number of infectious phage particles compared with the culture supernatant of untreated cells (3.7 × 103 PFU/ml without UV irradiation and 3.4 × 103 PFU/ml with UV irradiation for 16 s). This result indicated that φRSA1 is not UV inducible.
Determination of the genomic DNA sequence.
In our previous study, the φRSA1 genomic DNA gave a ladder pattern of a 39-kbp unit by pulsed-field gel electrophoresis (52), indicating a linear molecule of 39 kbp. To determine the nucleotide sequence of the entire φRSA1 genomic DNA, shotgun cloning and sequencing were performed (as described in Materials and Methods). A total of 920 sequences larger than 150 bases were assembled by the use of the Phred/Phrap/Consed program. The analyzed sequences corresponded to 6.0 times the final genome size of 38,760 bp. The φRSA1 genome has a G+C content of 65.3%. Potential ORFs that consist of more than 100 codons and start with ATG or GTG were identified with the online program Orfinder and the DNASIS program (version 3.6; Hitachi Software Engineering Co. Ltd.). To assign possible functions to ORFs, searches through the databases were done with the BLAST, BLASTX, and BLASTP programs (1). When two ORFs in different reading frames overlapped, the ORFs that had homologies to known sequences or a codon usage typical of the host R. solanacearum were selected. Accordingly, a total of 51 potential ORFs were assigned on the genome (Table 2 and Fig. 1). Most ORFs started with ATG and ended with TGA, but five ORFs (based on the database sequence) started with GTG (ORF13, -20, -22, -24, and -32). Five ORFs ended with TAA (ORF2, -22, -28, -43, and -46), and four ORFs ended with TAG (ORF17, -21, -26, and -36). In two cases, extended overlapping of two ORFs was observed, i.e., ORF23 and -24 and ORF47 and -48. As indicated in Table 2, in each case, significant homology was detected for each ORF in the database and so they are included in the final assignment.
TABLE 2.
Predicted ORFs found in the RSA1 genome
| Coding sequence | Position (5′ to 3′) | G+C content (%) | Protein length (aa) | Mol. mass (Da) | Amino acid sequence identity/similarity to best homologs | BLAST score (E value) | Accession no. |
|---|---|---|---|---|---|---|---|
| ORF1 | 353-742 | 65.9 | 130 | 13,173 | Janthinobacterium lividum hypothetical protein | 113 (3e-24) | A1X1P8 |
| PAAR motif protein (Burkholderia multivorans) | 82 (1e-14) | A0UPR0 | |||||
| ORF2 | 747-1247 | 68.9 | 167 | 18,701 | Janthinobacterium lividum hypothetical protein | 168 (8e-41) | A1X1P7 |
| ORF3 | 1260-2189 | 68.1 | 310 | 33,466 | Burkholderia thailandensis hypothetical protein | 79 (3e-13) | Q2SWS5 |
| ORF4 | 2102-2914 | 65.5 | 271 | 30,022 | Janthinobacterium lividum hypothetical protein | 176 (1e-42) | A1X1P4 |
| ORF5 | 4008-2905 | 65.4 | 368 | 41,541 | Bacteriophage protein RSc1941 | 642 (0.0) | Q8XY22 |
| Bacteriophage P2 gpQ probable capsid portal protein | 396 (e-109) | P25480 | |||||
| ORF6 | 5786-4008 | 65.3 | 593 | 67,089 | Terminase RSc1939 (ATPase subunit-related protein) | 707 (0.0) | Q8XY24 |
| Putative ATPase subunit of terminase (gpP) | 504 (e-141) | Q8Z358 | |||||
| Bacteriophage P2 gpP | 431 (e-119) | P25479 | |||||
| ORF7 | 5922-6764 | 65.2 | 281 | 30,415 | Bacteriophage protein RSc1938 | 461 (e-128) | Q8XY25 |
| Phage capsid scaffolding protein (gpO) | 201 (4e-50) | Q2T5K2 | |||||
| Bacteriophage P2 gpO | 192 (2e-47) | P25478 | |||||
| ORF8 | 6821-7837 | 63.5 | 339 | 37,784 | Bacteriophage protein RSc1937 | 537 (e-151) | Q8XY26 |
| P2 family phage major capsid protein (gpN) | 382 (e-104) | Q45YE6 | |||||
| Bacteriophage P2 gpN | 346 (8e-94) | P25477 | |||||
| ORF9 | 7837-8556 | 68.0 | 240 | 26,476 | Bacteriophage protein RSc1936 | 439 (e-122) | Q8XY27 |
| Terminase, endonuclease subunit (gpM) | 180 (5e-44) | P25476 | |||||
| ORF10 | 8656-9132 | 70.0 | 159 | 17,076 | Bacteriophage protein RSc1935 | 322 (5e-87) | Q8XY28 |
| Head completion/stabilization protein L (gpL) | 120 (3e-26) | P25475 | |||||
| ORF11 | 9135-9338 | 72.1 | 68 | 7,307 | Bacteriophage protein RSc1934 | 138 (1e-31) | Q8XY29 |
| Phage tail X (gpX) | 91 (2e-17) | Q41S37 | |||||
| ORF12 | 9357-9758 | 70.6 | 134 | 13,018 | Phage-related transmembrane protein RSc1933 | 179 (5e-44) | Q8XY30 |
| ORF13 | 9758-10069 | 70.7 | 104 | 11,253 | Phage-related transmembrane protein RSc1932 | 173 (3e-42) | Q8XY31 |
| Putative holin | 50 (3e-05) | A1AE02 | |||||
| ORF14 | 10069-10872 | 70.1 | 268 | 28,697 | Phage-related protein (hydrolase) RSc1931 | 500 (e-140) | Q8XY32 |
| Phage-encoded peptidoglycan-binding protein | 290 (5e-77) | A0UZD9 | |||||
| Bacteriophage φCTX ORF11 | 249 (8e-65) | Q92XL6 | |||||
| ORF15 | 10872-11366 | 67.5 | 165 | 17,049 | Signal peptide protein RSc1930 | 169 (6e-41) | Q8XY33 |
| ORF16 | 11366-11797 | 67.4 | 144 | 16,042 | Tail completion-like protein RSc1929 | 286 (2e-76) | Q8XY34 |
| Bacteriophage P2 gpR | 126 (3e-28) | P36933 | |||||
| ORF17 | 11797-12240 | 68.0 | 148 | 16,679 | Tail completion-like protein RSc1928 | 170 (2e-41) | Q8XY35 |
| Bacteriophage P2 tail completion protein gpS | 67 (4e-10) | P36934 | |||||
| ORF18 | 13153-12215 | 45.6 | 313 | 36,444 | Burkholderia pseudomallei hypothetical protein | 189 (1e-46) | Q6S459 |
| ORF19 | 13397-14011 | 69.2 | 205 | 21,753 | Phage-related protein RSc1925 (baseplate assembly-like protein) | 384 (e-105) | Q8XY38 |
| Baseplate assembly protein V (gpV) | 160 (3e-38) | A0UFJ5 | |||||
| Bacteriophage P2 gpV | 108 (2e-22) | P31340 | |||||
| ORF20 | 14011-14355 | 69.4 | 115 | 12,612 | Phage-related protein RSc1924 (baseplate assembly-like protein) | 216 (3e-55) | Q8XY39 |
| gpW/gp25 family protein | 124 (1e-27) | A0U2D3 | |||||
| Bacteriophage P2 gpW | 89 (9e-17) | P51768 | |||||
| ORF21 | 14361-15266 | 69.8 | 302 | 32,456 | Baseplate assembly-like protein RSc1923 | 517 (e-145) | Q8XY40 |
| Baseplate J-like protein (gpJ) | 320 (8e-86) | Q1QXR7 | |||||
| Bacteriophage P2 gpJ | 317 (5e-85) | P51767 | |||||
| ORF22 | 15262-15876 | 70.4 | 205 | 22,168 | Tail-related protein RSc1922 | 370 (e-101) | Q8XY41 |
| Phage tail protein gpI (tail formation-like protein) | 227 (2e-58) | Q1QXR8 | |||||
| Bacteriophage P2 gpI | 216 (4e-55) | P26701 | |||||
| ORF23 | 15884-17545 | 70.8 | 554 | 57,813 | Tail fiber-related protein RSc1921 | 951 (0.0) | Q8XY42 |
| Tail fiber protein gpH | 269 (3e-70) | Q87ZM6 | |||||
| Bacteriophage P2 gpH | 148 (9e-34) | P26700 | |||||
| ORF24 | 16871-17545 | 67.8 | 225 | 23,947 | Phage tail fiber protein RSc1692 | 108 (2e-22) | Q8XYR6 |
| ORF25 | 17561-18310 | 72.8 | 250 | 25,888 | Tail fiber assembly-like protein RSc1920 | 362 (7e-99) | Q8XY43 |
| ORF26 | 18310-18771 | 68.8 | 154 | 17,140 | Hypothetical protein RSc1919 | 259 (2e-68) | Q8XY44 |
| ORF27 | 18868-20040 | 67.7 | 391 | 42,397 | Phage-related tail sheath protein RSc1918 | 758 (0.0) | Q8XY45 |
| P2 major tail sheath protein gpFI | 585 (e-165) | Q66BD0 | |||||
| Bacteriophage P2 gpFI | 510 (e-143) | P22501 | |||||
| ORF28 | 20075-20581 | 62.5 | 169 | 18,802 | Phage-related protein (major tail protein) RSc1917 | 341 (7e-93) | Q8XY46 |
| Putative P2 tail tube protein gpFII | 174 (1e-42) | Q66BD1 | |||||
| Bacteriophage P2 gpFII | 147 (2e-34) | P22502 | |||||
| ORF29 | 20630-20983 | 63.2 | 118 | 12,796 | Phage-related protein RSc1916 | 225 (5e-58) | Q8XY47 |
| Bacteriophage P2 gpE | 83 (5e-15) | Q66BM8 | |||||
| ORF30 | 21081-23657 | 69.6 | 859 | 90,965 | Phage-related tail transmembrane protein RSc1914 | 1,280 (0.0) | Q8XY49 |
| Bacteriophage P2 tail protein gpT | 109 (6e-22) | Q66BN0 | |||||
| ORF31 | 23749-24168 | 65.2 | 140 | 15,695 | Phage-related tail protein RSc1913 | 281 (7e-75) | Q8XY50 |
| Bacteriophage P2 gpU | 155 (5e-37) | Q41SZ1 | |||||
| ORF32 | 24168-25370 | 66.2 | 401 | 43,433 | Phage related protein RSc1912 | 669 (0.0) | Q8XY51 |
| Bacteriophage P2 gpD | 322 (3e-86) | Q66BD6 | |||||
| ORF33 | 26135-25392 | 61.8 | 248 | 26,462 | Hypothetical transmembrane protein | 134 (4e-30) | Q8XSE3 |
| ORF34 | 27434-26217 | 62.6 | 406 | 47,730 | Transposase ISRSO15 | 803 (0.0) | Q8XFK1 |
| ORF35 | 28022-27609 | 59.9 | 138 | 15,091 | XRE family, transcriptional regulator | 103 (2e-21) | A0UFH8 |
| Putative phage DNA-binding protein | 102 (6e-21) | Q63YP6 | |||||
| DNA-binding repressor RSc1907 | 99 (7e-20) | Q8XY56 | |||||
| ORF36 | 28062-28490 | 59.7 | 143 | 15,823 | Unknown | ||
| ORF37 | 28539-28730 | 58.3 | 64 | 7,380 | Unknown | ||
| ORF38 | 28730-28975 | 66.5 | 82 | 9,126 | Putative transcription activator RSc1904 | 124 (2e-27) | Q8XY59 |
| Phage transcription activator Ogr/Delta | 77 (4e-13) | A0UFH5 | |||||
| ORF39 | 29093-29644 | 65.9 | 184 | 20,512 | Hypothetical phage protein | 235 (9e-61) | Q63YQ0 |
| Prophage antirepressor | 95 (1e-18) | Q3R0Y7 | |||||
| ORF40 | 29657-29887 | 61.9 | 77 | 8,082 | Hypothetical protein RSc1903 | 75 (1e-12) | Q8XY60 |
| ORF41 | 29887-30048 | 65.8 | 54 | 5,540 | Hypothetical protein RSc1902 | 81 (1e-14) | Q8XY61 |
| ORF42 | 30048-30251 | 63.4 | 68 | 7,171 | Transmembrane protein RSc1901 | 58 (2e-07) | Q8XY62 |
| ORF43 | 30254-30487 | 64.5 | 78 | 8,612 | Transmembrane protein Rsc1900 | 147 (2e-34) | Q8XY63 |
| ORF44 | 30483-30692 | 70.0 | 70 | 7,810 | Hypothetical protein RSc1899 | 103 (3e-21) | Q8XY64 |
| ORF45 | 30770-31243 | 65.5 | 158 | 17,212 | Unknown | ||
| ORF46 | 31512-31799 | 65.3 | 96 | 11,432 | Pseudomonas resinovorans/pCAR1 ORF80 | 128 (1e-28) | Q8GHW9 |
| ORF47 | 31802-34603 | 65.6 | 934 | 103,175 | Zinc finger, CHC family protein | 1,454 (0.0) | A0UFH0 |
| φ52237 hypothetical protein (AAZ72605.1) | 1437 (0.0) | Q45Y14 | |||||
| Conserved hypothetical phage protein | 1,433 (0.0) | Q63YQ8 | |||||
| ORF48 | 33589-31811 | 66.4 | 593 | 63,364 | Hypothetical protein φ52237 (AAZ72604.1) | 332 (1e-80) | Q45Y15 |
| ORF49 | 34778-35494 | 61.7 | 239 | 25,241 | Unknown | ||
| ORF50 | 35709-36788 | 64.3 | 360 | 41,524 | Site-specific recombinase, phage integrase | 514 (e-144) | Q62FM0 |
| tRNA | 36831-36875 | 71.1 | attP [Arg tRNA(CCG)] R. solanacearum GMI1000 chromosomal sequence | 90 (7e-17) | AL646073 | ||
| ORF51 | 37124-38182 | 56.4 | 353 | 38,895 | Unknown |
FIG. 1.
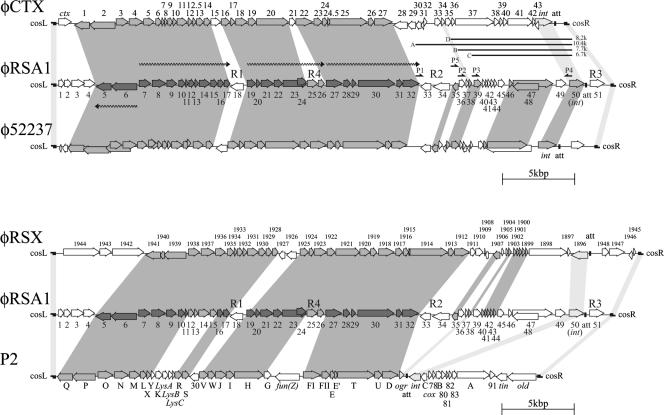
(Top) Alignment of φRSA1 ORFs with those of φCTX (38) and φ52237 (accession no. DQ087285). (Bottom) Alignment of φRSA1 ORFs with those of P2 (GenBank accession no. NC001895) and a prophage (designated φRSX in this work) previously found in the genome of R. solanacearum GMI1000 (9, 44). ORFs are indicated by arrows. Predicted transcription units for φRSA1 late genes are shown by wavy lines. Gray shading indicates significant amino acid or nucleotide sequence similarity among the phages. Light gray shading indicates marginally similar regions. R1, R2, R3, and R4 are regions with relatively low G+C contents (AT-rich regions), corresponding to ACURs (44). P1, P2, P3, P5, and P4 are PCR primers for minireplicon formation. A, B, C, and D are fragments of 10.4, 7.7, 6.7, and 8.2 kbp, respectively, used for minireplicon formation.
φRSA1 gene organization and homology to other phage genomes.
The databases were searched with the BLAST and BLASTX programs for sequences homologous to the nucleotide sequence of φRSA1 DNA. Extensive homologies were detected in the genomic sequences of Burkholderia pseudomallei phage φ52237 (accession no. DQ087285), Pseudomonas aeruginosa phage φCTX (accession no. AB008550), and coliphage P2 (accession no. AF063097) and the chromosomal DNA of R. solanacearum GMI1000 (accession no. AL646052).
To avoid confusion, the P2 definition of the genes was followed in the identification of φRSA1 genes in this study. Interestingly, R. solanacearum was formerly classified as Pseudomonas solanacearum and had another synonym, Bacillus solanacearum (47, 51), based on classical taxonomic characteristics. Currently, the three bacterial genera Ralstonia (Betaproteobacteria), Burkholderia (Betaproteobacteria), and Pseudomonas (Gammaproteobacteria) are clearly distinguished from each other on the basis of their 16S rRNA sequences (51). In this context, we are interested in the interrelationships among the three phages φRSA1, φCTX, and φ52237.
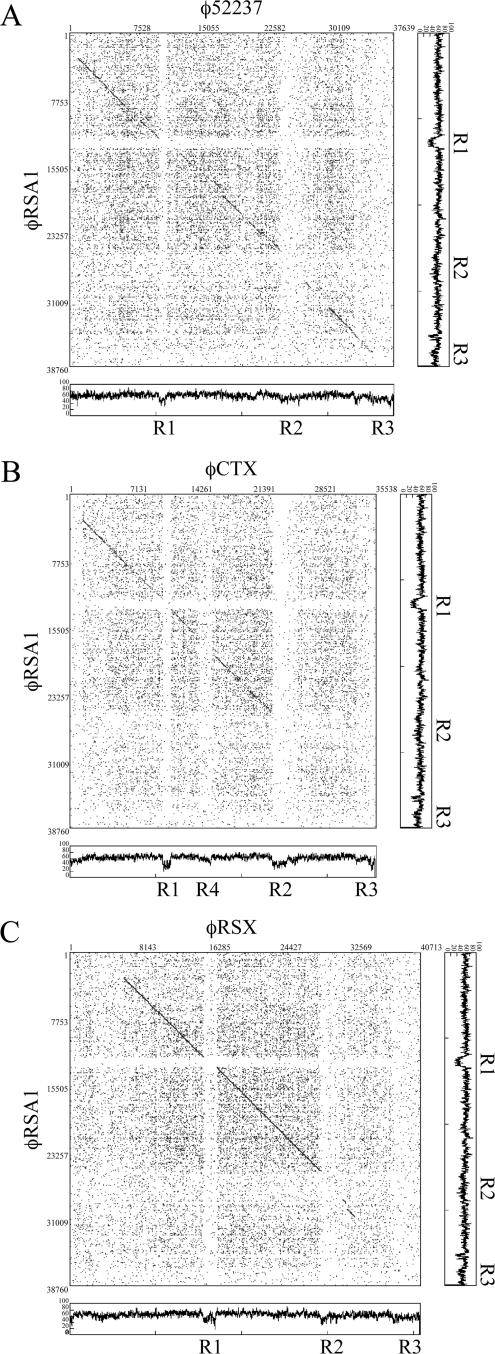
Extended comparison of the φRSA1 sequence with these sequences by the matrix plot method revealed characteristic features of the phage gene organization, as shown in Fig. 2. Between the φRSA1 and φ52237 (37,639 bp) sequences, an extended colinearity was obvious throughout almost the entire genomic region (Fig. 2A). The sequence homology was broken by three small regions around φRSA1 positions 12,500 to 13,500 (region R1), 26,000 to 29,000 (region R2), and 37,000 to 38,760 (region R3), whose G+C contents were 50.5% (region R1), 60.6% (region R2), and 56.1% (region R3) and relatively low compared with the overall average value of the genome (65.3%). These regions with low G+C contents (AT islands) correspond to φ52237 positions 10,000 to 12,000 (58.6% G+C), 24,000 to 27,000 (57.3% G+C), and 35,600 to 37,630 (53.1% G+C), whose G+C contents are also lower than the overall average value of the φ52237 genome (64.8%). In the comparison between φRSA1 and φCTX, colinearity was limited to the initial two-thirds of the genomes (Fig. 2B). Interestingly, the sequence homology was broken by the same AT islands of the φRSA1 genome. On the φCTX genome (35,652 bp), regions R1 to R3 approximately corresponded to positions 10,500 to 12,000 (51% G+C), 23,500 to 25,500 (46.2% G+C), and 35,000 to 35,652 (47.1% G+C). In addition, another break was seen around positions 15,000 to 16,500 (58.0% G+C; region R4). The G+C contents of these φCTX regions are again lower than the average genomic G+C content of 62.6%. In each comparison, these AT islands can be recognized as blank lattices against a background of dotted areas, suggesting that they are regions containing unrelated sequences probably acquired recently.
FIG. 2.
Matrix comparison of the genomic nucleotide sequences of φRSA1 and φ52237 (A), φRSA1 and φCTX (B), and φRSA1 and φRSX (C). Nucleotide positions are shown along the genomic sequence. Fifteen matches between 20 nucleotide sequences are marked by dots (DNASIS). The base distribution along the phage genomes (percent G+C, 60-bp windows) are also shown. Regions R1 to R4 are the same as in Fig. 1.
φRSA1 region R1 corresponds to a region containing ORF18 (function unknown) between structural genes gpS (orf17) and gpV (orf19). In the same location, φCTX contains ORF15 (function unknown). Region R2 of φRSA1 corresponds to the junction where the homology with φCTX ends. This AT island of approximately 3.0 kb separates the gpD gene (orf32) and the ogr gene (orf38), which are immediately linked to each other on the coliphage P2 genome. A similar AT island of 3.4 kb was found to be inserted in the same region in φCTX, where six ORFs with unknown functions are located (38). These ORFs showed no homology with any of five ORFs (ORF33 to -37) in φRSA1 region R2. It is interesting that ORF34, located close to ORF32 (the gpD homologue), showed 100% amino acid sequence identity with transposase ISRSO15, which was found in the chromosomal DNA (positions 2,780,153 to 2,781,370; accession no. AL646070), as well as the megaplasmid DNA (positions, 111,895 to 113,185; accession no. AL646085) of R. solanacearum GMI1000. ORF34 is on an IS of 1,319 bp with a terminal repeat of seven A residues. A cluster of ORFs, ORF33 to -35, is located in reverse orientation compared to the conserved genes for structural components (described below). These facts strongly suggest that this region was transferred horizontally. The third AT island (region R3) around the right cos of φRSA1 contains only ORF51 with an unknown function.
The entire genomic comparison for these phages suggests that φRSA1 is closely related to B. pseudomallei phage φ52237 and P. aeruginosa phage φCTX, which belong to the P2-like phages. So far, genomic sequences of a number of P2-related phages from various bacteria have been reported in the literature or in the databases, including P2 (GenBank accession no. NC001895), 186 (GenBank accession no. U32222), φ108 (7), Fels-2 (35), HP1 (14), HP2 (41), K139 (29), and φMja-PHL101 (25). Many of these phages infect bacteria belonging to the family Enterobacteriaceae and have the same genomic organization (with minor variations) that has been best characterized for coliphage P2 (39). The well-established P2 gene organization is compared with that of φRSA1 in Fig. 1 (bottom). The gray shading is based on a matrix comparison of the nucleotide sequences (more than 12 matches between 15 nucleotide sequences are marked) (data not shown). As shown here, the left two-thirds of the φRSA1 genome that showed a high degree of homology with the φCTX and φ52237 genomes (Fig. 2B) also shared homology with phage P2, even with a difference in G+C content (50.2% in P2 DNA). This conserved region corresponds to the P2 late region, including genes for phage structural components, assembly, and regulation (6). The homology-disrupting AT islands corresponding to R1, R4, and R3, which contain four nonessential genes, orf30, fun(Z), tin, and old, were also found in P2 (Fig. 1, bottom) (6). A region containing lys genes is also distinct (see below). Contrasting to the highly conserved structural modules, the right one-third of P2 DNA, containing genes for early functions and int/att, is remarkably different from that of φRSA1. Especially the locations of int and att are quite different between the two phages; they are immediately to the right of ogr in P2 but to the left of cosR in φRSA1 (see below). In this respect, two types of P2-like phages are obvious, P2-type and φRSA1-type phages, including φCTX and φ52237. φ108 and φMja-PHL101, which infect Pasteurella maltocida and Mannheimia haemolytica, respectively (both belong to the family Pasteurellaceae), are also of the latter type.
It is noteworthy that the G+C content of the segment corresponding to the P2 early genes and int/att is slightly lower than the rest of the genome in all of these phages, even in P2 (data not shown). It might be possible that this region was derived from foreign genomes. The patchy genetic relationship between φRSA1 and these phages indicates frequent recombinations within the φRSA1 ancestral genome during its evolution, as suggested for tailed phages and their prophages (8, 9, 22).
Genes for capsid synthesis and DNA packaging.
φRSA1 possesses homologues of almost all of the P2 genes required for capsid synthesis and DNA packaging in the left end of the genome in the same order as in P2 (Table 2 and Fig. 1). ORF5 and ORF6 correspond to homologues of the portal protein, gpQ, and the large subunit of terminase, gpP, respectively. orf6 and orf5 are adjacent in the same direction as in the PQ operon of P2, suggesting a single transcription unit. orf7, -8, -9, and -10 encode homologues of the scaffold protein, gpO; the major capsid protein, gpN; the small subunit of terminase, gpM; and the head completion protein, gpL, respectively, in the same order as in P2. An icosahedral morphology and dimension of the φRSA1 head resembling the P2 head reflect the high homology seen in the amino acid sequences and sizes of these capsid proteins, for example, 50% amino acid identity between the ORF7 and P2 gpO proteins and 56% amino acid identity between the ORF8 and P2 gpN proteins. As described below, the DNA sequence of the φRSA1 cos site is similar to those of the P2-like phages, according to the high homology seen in the amino acid sequence and size between ORF6 and ORF9 and P2 gpP and gpM, respectively (59% amino acid identity between ORF6 and P2 gpP and 45% identity between ORF9 and P2 gpM).
Genes for tail synthesis.
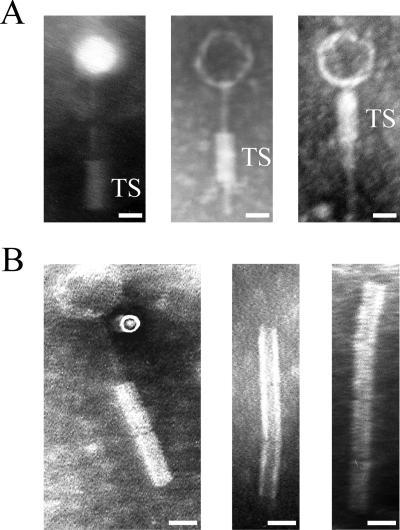
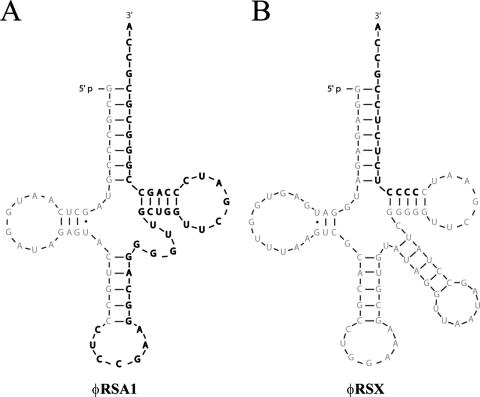
One characteristic feature of φRSA1 particles is their unusual tail and tail sheath structure. As shown in Fig. 3, a tail 110 ± 8 nm in length and 3 ± 0.2 nm in diameter is associated with a tail sheath (40 ± 6 nm in length and 17 ± 1.5 nm in diameter) movable along the tail (Fig. 3A). A tail sheath was often observed attached at the bottom of the tail but sometimes at intermediate positions along the tail, giving unique structures which resemble the morphology reported for B. cepacia phage KS5 (46). One possibility is that after attachment to the host cells the sheath contracts or moves up along the tail to bring the tail tube into the surface of the cell. Tail fibers and collar structures were not discerned from the electron micrographs (52). Sometimes, structures resembling the tail sheath were observed connected in a chain (Fig. 3B).
FIG. 3.
Electron micrographs showing morphology of φRSA1 particles. A tail sheath (TS) with a constant size (40 nm in length) moves along the thin tail structure (A). Sometimes structures resembling the tail sheath are connected in a chain (B). Bars, 20 nm.
φRSA1 also possesses homologues of most P2 tail genes in a gene arrangement similar to that of P2. In P2, a total of 16 essential genes were identified arranged within three operons for the tail assembly (16, 17). φRSA1 ORF11, corresponding to P2 gpX (with 60% amino acid identity), is located next to ORF10 (gpL homologue) at the junction of the head gene cluster and the tail and lysis gene cluster. ORF16 and ORF17 (corresponding to P2 tail completion proteins gpR and gpS, respectively) are found after the lysis genes. Moderate amino acid identity was observed between these genes (46% amino acid identity between ORF16 and P2 gpR and 40% identity between ORF17 and P2 gpS). The second gene cluster for tail assembly comprised seven ORFs (ORF19 to ORF25). ORF19 to -21 are homologues of the P2 baseplate assembly proteins gpV (41% amino acid identity), gpW (39% amino acid identity), and gpJ (62% amino acid identity), respectively. ORF22 showed homology to P2 tail formation protein gpI (52% amino acid identity). ORF23 is a homologue of P2 tail fiber protein gpH, and ORF24 is a C-terminal portion of ORF23, which is described as a putative tail fiber protein (RSc1692; accession no. Q8XYR6). Although the 150-amino-acid N-terminal portion of ORF23 showed high amino acid identity (54%) with P2 gpH, the remaining part showed only marginal homology. The C-terminal part of P2 gpH is made up of several modules that are found in the tail fiber proteins of a variety of double-stranded DNA phages in various combinations (16). ORF25 is located at a position corresponding to that of P2 tail fiber assembly protein gpG, but its size, 250 amino acids (aa), is larger than that of gpG (175 aa) and the amino acid identity is low, only 25%. This is also the case in φCTX; there was no φCTX ORF with significant homology to P2 gpG; φCTX ORF21 seemed to have replaced it (38). In contrast to the highly conservative nature of other structural proteins in P2-like phages, this tail fiber assembly protein may be highly specific to each phage.
Another gene cluster for tail formation includes six ORFs (ORF27 to -32) corresponding to the P2 gene cluster gpFI-gpFII-gpE-gpT-gpU-gpD. High amino acid identity was observed between ORF27 and P2 major tail sheath protein gpFI (62% identity) and between ORF28 and P2 tail tube protein gpFII (47% identity), but all other homologues shared moderate homologies ranging from 35% to 44% amino acid identity. As described above, the φRSA1 particles showed characteristic tail structures somewhat different from those of P2 and typical P2-related phages, which reflects the differences observed in ORFs included in these tail assembly structural modules.
Lysis genes.
φRSA1 encodes four ORFs (orf12 to orf15) in the region corresponding to the P2 region for lysis function, where five genes (gpY, gpK, lysA, lysB, and lysC) are located. orf12 and orf13 encode highly hydrophobic small (possibly transmembrane) proteins related to holins which form the channels in the cytoplasmic membranes for the translocation of lytic enzymes (53), although no significant amino acid sequence identity was detected between these ORFs and P2 holin gpY. φRSA1 ORF14 showed high amino acid sequence homology with various phage lytic enzymes (unpublished data). ORF14 exhibited an amino acid sequence almost identical to that of an ORF (RSc1931) detected on the chromosome of R. solanacearum GMI1000 (98% identity), and an amino acid identity as high as 57% with lytic enzymes of φ52237 and φCTX was observed. A composite structure of the φCTX lytic enzyme consisting of the N-terminal region conserved in the gram-positive bacteria and their phages and the C-terminal region shared by lytic enzymes of lipid-containing phages was reported (38). The high observed homology with these enzymes suggests that the φRSA1 ORF14 protein may function in cells of P. aeruginosa and B. pseudomallei as well. In contrast to this, P2 endolysin gpK (166 aa) did not show significant amino acid sequence homology to these enzymes. Homologues of P2 lysA (which affects the timing of lysis), lysB (for the regulation of lysis), and lysC (a transcription attenuator) are not clear in φRSA1, although ORF14 showed marginal homology with both P2 lysA (30% amino acid identity) and lysB (30% identity).
Regulatory genes for late gene expression and possible late promoters.
The expression of the P2-related phage late genes is absolutely dependent on the Ogr proteins (4, 10). The Ogr protein family represents a unique group of prokaryotic zinc finger DNA-binding transcriptional activators which are highly conserved in P2-related phages. A set of four cysteine residues is conserved in all Ogr proteins in the arrangement C-(X)2-C-(X)22-C-(X)4-C, where a zinc ion is coordinated by four cysteines (32). The Ogr protein interacts with the α subunit of RNA polymerase and activates the late-gene promoters (3, 31). On the P2 genome, the ogr gene is located immediately downstream of the gene cluster gpF-gpE-gpT-gpU-gpD close to attP and the int gene (11). A possible ogr homologue, φRSA orf38, that has 34% amino acid identity with P2 Ogr and contains precisely the conserved zinc finger motif, was found approximately 3.4 kb downstream from the gpD homologue (orf32) on the φRSA1 genome (Table 2 and Fig. 1). As described above, this is caused by an insertion of a 3.4-kb region (region R2) with five ORFs, including ISRSO15.
The consensus binding sequence for Ogr proteins has been identified in each late promoter region in the P2 family as TGT-(N)12-ACA, which is central approximately at the −55 position from the first ATG codon (28). In φRSA1, four possible late promoter regions (5′ noncoding regions of orf6, orf7, orf19, and orf27) were examined for such sequence motifs or common sequences. All of these regions contained a sequence motif [5′ TGTTGT-(X)13-ACAACA] that is centered around positions −50 to −89 from the initiation codon (unpublished data). This sequence is related to the P2 consensus sequence, indicating that φRSA1 Ogr may have the same promoter recognition mechanism as the Ogr proteins of the P2 family. No promoter sequence of the σ70 type was present in these possible late promoter regions of φRSA1.
Early genes and genes for lysogeny.
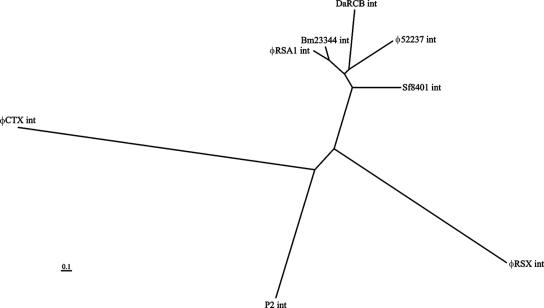
On the right side of ORF38 (Ogr homologue), there are 12 ORFs (ORF39 to -51) arranged in the same direction and 1 ORF (ORF48) that overlaps ORF47 in the reverse orientation. Because both ORF47 and -48 showed homologous sequences in the databases, they are included in Table 2 and Fig. 1. In P2, this genomic region contains early-expressed genes, including genes for DNA replication (34) such as gpA and gpB, genes for lysogeny such as the integrase gene and cox (43), and genes for lysogenic conversion such as old (37) and tin (36). These φRSA1 ORFs did not show significant similarity to the P2 genes in the corresponding region, whereas their homologous sequences were found in the databases for various phages or prophages, especially of Ralstonia species and Burkholderia species. For example, ORF39 showed 65% amino acid identity with B. pseudomallei phage φ52237 ORF15 (accession no. AAZ72616.1), 65% identity with the prophage sequence in B. pseudomallei strain K96243 (accession no. Q63YQ0), and 41% identity with the prophage antirepressor sequence of Xylella fastidiosa strain Ann-1 (accession no. Q3R0Y7). φRSA1 ORF47 (the largest ORF of φRSA1) showed 74% amino acid identity with φ52237 ORF4 (accession no. AAZ72605.1), 74% identity with the prophage sequence of B. pseudomallei strain K96243 (accession no. Q63YQ8), 51% identity with the R. solanacearum NCBI305 prophage sequence RSc0967 (accession no. Q8Y0S6), and 51% identity with the prophage sequence of R. eutropha JMP134 (accession no. Q46Z72). Although the function of this well-conserved ORF is not known, a 150-amino-acid portion at the N terminus showed 42% amino acid identity with some bacterial DNA primases (accession no. Q43KY5), suggesting its involvement in DNA replication. φRSA1 ORF50 is a homologue of int of the P2-related phages (unpublished data). Its amino acid sequence shows various degrees of homology to int sequences of P2-related phages and prophages. Based on the sequence alignment data, a phylogenetic tree was constructed as shown in Fig. 4, which suggests a close relatedness of integrases of φRSA1 and the prophage integrated in the chromosome of B. mallei ATCC 23344 (accession no. Q62FM0).
FIG. 4.
Phylogenetic tree based on the alignment phage int ORFs. The amino acid sequence of φRSA1 ORF50 is compared with int sequences of Bm23344 (accession no. Q62FM0), φ52237 (accession no. Q45Y16), DaRCB (accession no. Q47H88), Sf8401 (accession no. Q0T7U1), P2 (accession no. P36932), φRSX (accession no. Q8Y3C8), and φCTX (accession no. Q38644). The analysis was performed by the multiple-alignment program CLUSTAL W.
To verify the possible functions for DNA replication of ORFs in this region, φRSA1 minireplicons were constructed. A P2 minireplicon was constructed with a small region containing gene A and ori (34). However, as suggested by the genome comparison in Fig. 1 (bottom), the gene organization required for DNA replication may be different in φRSA1. In fact, such a minireplicon could not be obtained for φCTX (38), and to our knowledge, no successful example has been reported from “φRSA1-type” phages so far. φRSA1 DNA fragments containing ORF49 and its upstream ORFs were amplified by PCR with a combination of primers as shown in Table 1 and Fig. 1. Three fragments of 10.4 kbp (amplified with primers ORF33-P1 and ORF49-P4), 7.7 kbp (amplified with primers ORF36-P2 and ORF49-P4), and 6.7 kbp (amplified with primers ORF39-P3 and ORF49-P4), as indicated in Fig. 1 (fragments A, B, and C, respectively), were connected to a Kmr cassette of 1.5 kbp and introduced into cells of R. solanacearum strains M4S and MAFF106611 by electroporation. The resulting minireplicon of 11.5 kbp was stably maintained in the cells and conferred Kmr on the cells, indicating that ori and functions necessary for φRSA1 DNA replication are encoded within this region. However, neither the 7.7-kbp nor the 6.7-kbp φRSA1 fragment could support stable replication of the DNA constructs. Some ORFs (ORF33, ORF34, and ORF35) in the 10.4-kbp fragments may be involved in plasmid maintenance. In this context, it is interesting that ORF35 resembles the DNA-binding repressor proteins of bacteriophages (Table 2). The actual involvement of ORF35 in the maintenance of the φRSA1 minireplicon was confirmed by removing a 2.0-kbp fragment containing ORF33 and ORF34 from the 10.4-kbp construct by PCR with primers ORF35-P5 and ORF49-P4 (Table 1). The resulting construct of 8.2 kbp (D in Fig. 1, top) was stably maintained in cells of both strains M4S and MAFF106611 on selection plates containing kanamycin. φRSA1 ORF35 showed amino acid sequence similarity to φ RSX RSc1907 (E value of 7e-20) and φ52237 gp23 (Q63YP6; E value of 6e-21). This is the first successful example of a minireplicon of φRSA1-type phages.
φRSA1 cos and att sequences.
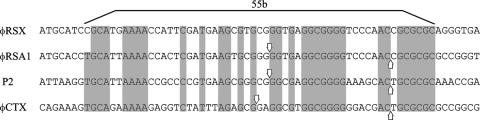
On pulsed-field gel electrophoresis, the φRSA1 DNA showed a ladder pattern with a monomer size of approximately 39 kb, like coliphage λ (52), suggesting the presence of cohesive ends (cos) on the linear molecule. When φRSA1 DNA was digested with HincII, more than 20 fragments were generated, among which was a 5.7-kb fragment that dissociated into two fragments (2.4 and 3.3 kb) after heating at 70°C for 15 min. The 5.7-kb HincII fragment extracted from agarose gel after electrophoresis was heated to generate the two bands, which were treated with T4 DNA polymerase to form blunt ends. After connecting to the EcoRV site of pBluescript II SK+, the fragments were cloned and sequenced. By comparing the nucleotide sequences to each other and with the φRSA1 genomic sequence, it was revealed that a 19-base sequence of the 3′ end of the 2.4-kb fragment is the same as a 19-base sequence of the 5′ end of the 3.3-kb fragment, indicating that the φRSA1 DNA contains a 19-base single-stranded extrusions on the left (5′-GGTGAGGCGGGGTCCCAAC-3′) and on the right (3′-CCACTCCGCCCCAGGGTTG-5′). In Fig. 5, the φRSA1 cos sequence is compared with those of a few phages of the P2 family, including P2 and φCTX. A 55-bp core sequence (54) was also found to be well conserved in φRSA1. Both φRSA1 and P2 have 19-base 5′-extruding cohesive ends, but φCTX has a 21-base 5′ extrusion (19).
FIG. 5.
Comparison of the cos region of φRSA1 with those of P2-related phages. Bases within the 55-bp core sequence that are shared by phages (54) are shaded. Downward and upward vertical arrows indicate cos cleavage sites. The exact cohesive end of φRSX is unknown.
φRSA1 was spontaneously released from R. solanacearum strain MAFF211272, and a φRSA1-related sequence was detected on the chromosomal DNA of this strain, indicating that φRSA1 contains an attachment site (attP) where it recombines with a homologous sequence on the bacterial genome (attB) via site-specific recombination (12). As described above, φRSA1 encodes a site-specific integrase (ORF50, int), and an attP sequence was predicted in the vicinity of this gene, as seen in most P2-related phages. Chromosomal DNA fragments of strain MAFF211272 that contain flanking regions of the φRSA1 prophage were cloned as follows. Two HincII fragments (2.6 and 6.0 kb), which showed different patterns compared with HincII fragments in Southern blot hybridization, were extracted from the agarose gel, connected to pBluescript II SK+, and cloned into E. coli XL10 Gold. The nucleotide sequence of the 2.6-kb fragment contained a φRSA1 sequence region containing ORF50 (int), followed by 88 bases overlapping the gene for arginine tRNA(CCG), after which a chromosomal sequence appeared. On the 6.0-kb fragment, a chromosomal sequence was connected to a φRSA1 sequence corresponding to a 45-base 3′ portion of the arginine tRNA(CCG) gene, followed by ORF51. These results demonstrated that φRSA1 used a 45-base 3′ portion of the arginine tRNA(CCG) gene as attP and was integrated into the arginine tRNA(CCG) gene on the chromosome of strain MAFF211272 as indicated in Fig. 6. It is also interesting that φRSA1 att sequence was present in the chromosomal DNA of B. pseudomallei strain 1026b (accession no. AY471582), indicating that a φRSA1-related phage may also infect this bacterial species and integrate at the same att site. For comparison, we examined the nucleotide sequence of B. pseudomallei phage φ52237 for a possible att sequence and found in the corresponding region (positions 2845 to 2889, accession no. DQ087285) a 45-base 3′ portion of the phenylalanine tRNA(GAA) gene. Therefore, φRSA1 and φ52237 use different tRNA genes as an att site. P2-like phages and their satellite phages 186, P4, Hp1, R73, and φCTX use tRNA genes as attB as well (18, 19, 33, 40, 48).
FIG. 6.
attP sequences of φRSA1 and φRSX. The φRSA1 and φRSX attachment sites correspond to a 45-base 3′ portion of the arginine tRNA(CCG) (A) and a 15-base 3′ portion of the serine tRNA(GGA) (B), respectively. The attP core nucleotide sequence is in bold letters.
Other genes.
In addition to unknown genes located in the AT islands, such as orf18, orf36, orf37, and orf51 as described above, φRSA1 contained four additional ORFs (ORF1 to ORF4) between cosL and ORF5 (P2 gpQ homologue), as shown in Table 2 and Fig. 1. Interestingly this position is also suggested as another hot spot on the P2-like phage genomes where foreign genes can be picked up without disturbing the functions of other genes (13, 38). φRSA1 ORF1, -2, and -4 show some amino acid sequence similarity to proteins of Janthinobacterium lividum, and ORF3 is similar to a hypothetical protein (accession no. Q2SWS5) of B. thailandensis. These genes might have been acquired from previous hosts, suggesting a wide host range of φRSA1 extending to Burkholderia-Janthinobacterium species.
Homology to the prophage sequence detected on the R. solanacearum GMI1000 chromosome.
On the 3.7-Mb chromosome of R. solanacearum GMI1000, at least four possible prophage sequences were detected (44). One of them, a 40.7-kb region located at positions 2,084,442 to 2,125,154 (designated φRSX), showed significant nucleotide similarity to φRSA1. This prophage sequence was briefly described as RS6 previously (9). Figure 2C shows a matrix comparison of the nucleotide sequences of the φRSA1 genome and the φRSX chromosomal region. As shown in the comparison of φRSA1 and φCTX (Fig. 2B), the two-thirds portion of the left side of the φRSA1 genome that encodes the structural modules showed a high level of homology to the φRSX sequence and the homology is interrupted by the AT island R2 as described above. Therefore, the possible prophage φRSX can also be a member of the family of P2-like phages and retains conserved structural modules, even with a somewhat larger entire-genome size. This is supported by additional information as follows. In the φRSX region, there is a gene for serine tRNA(GGA) at positions 2,084,367 to 2,084,457 immediately upstream of the possible integrase ORF (RSc1896). A 15-bp sequence at the 3′ end of this gene was found to be repeated at positions 2,125,140 to 2,125,154, which separates the phage-related sequences from the chromosomal sequences, suggesting that the sequence of serine tRNA(GGA) is attL and the 15-bp sequence at the 3′ end of this gene is attR on the chromosome of strain GMI1000, as depicted in Fig. 6B. In the vicinity of this attR region, there is a sequence closely related to cos at positions 2,121,802 to 2,121,858. This possible cos sequence of φRSX is compared with those of other phages in Fig. 5. As shown here, only five nucleotides of the conserved 55 bp are different between φRSA1 and φRSX.
All of the φRSA1 ORFs for the P2-like phage structural proteins identified (Table 2) are also highly conserved in φRSX (compared with φRSA1 in Fig. 1, bottom). In the database search for homology to these φRSA1 proteins, the highest BLAST scores were always obtained with φRSX proteins (RSc1899 to RSc1941), as indicated in Table 2; in each comparison of the φRSA1 and φRSX counterparts, amino acid identity ranged from 85% (gpX; φRSA1 ORF11 versus φRSX RSc1934) to 100% (gpS; φRSA1 ORF17 versus φRSX RSc1928), indicating a very close relatedness of φRSA1 and φRSX. In contrast to this high degree of similarity in the organization of structural genes, RSA1 regions R1, R2, and R3 also interrupted the sequence homology between φRSA1 and φRSX (Fig. 2C). In the φRSX sequence (67.0% average G+C content), region R1 corresponds to an ACUR of 1,512 bp (51.5% G+C) containing RSc1926 and RSc1927 (without known functions), and φRSX region R2 coincided with another ACUR of 2,657 bp (56.7% G+C) containing six ORFs (RSc1905, RSc1906, RSc1907, RSc1908, RSc1909, and RSc1910). Among them, only RSc1907 showed a relatively low amino acid sequence similarity to φRSA1 ORF35 (39% identity). RSc1907 is noted as a putative phage DNA-binding repressor (accession no. Q8XY56). Within the φRSX-R2 region, there is no IS (ISRSO15) that was observed in the corresponding region of the φRSA1 sequence. The φRSX sequence corresponding to region R3 overlaps another ACUR of 3,427 bp (56.5% G+C) containing two unknown ORFs (RSc1947 and RSc1948). The φRSX ORFs around this region, including RSc1896 (int), RSc1897, RSc1898, RSc1945, and RSc1946, did not show significant homology to φRSA1 ORFs in the corresponding positions. For example, the amino acid identity between φRSA1 ORF50 (int) and RSc1896 (int) is only 29% (unpublished data).
In addition to these nonconserved regions, a region between cosL and gpQ is also divergent between φRSA1 and φRSX. There is a vgr-like gene (RSc1944) located close to cos of φRSX. Wang et al. (49) considered the vgr gene a structural component of at least some Rhs (recombination hot spot) elements. The functions of two other ORFs (RSc1942 and RSc1943) located in this φRSX region are not known. There is a long repeat sequence within the coding region of RSc1942.
These nonconserved regions may have been rearranged very recently, possibly by horizontal gene transfers in the divergence of φRSA1 and φRSX. In the light of the gene arrangement found in P2-like phage φRSA1, the prophage φRSX sequence was characterized in this work.
Prophage effects of φRSA1 on lysogenic cells.
Some temperate phages and prophages are known to carry additional cargo genes (termed morons or lysogenic conversion genes). Many morons from prophages in pathogenic bacteria encode proven or suspected virulence factors (5, 8, 23, 27). The cytotoxin gene (ctx) was found to be inserted at an AT island in φCTX of P. aeruginosa (38). The ctx gene appeared to have jumped in between cos and orf1 (gpQ equivalent) (Fig. 1, top), suggesting that this region is a hot spot for such genes. Within this corresponding region of φRSA1, there are four ORFs (ORF1 to ORF4) in the same orientation as described above. In the φRSX sequence, integrated in R. solanacearum GMI1000, three ORFs, RSc1944, RSc1943, and RSc1942, are also found in the same orientation as described above and comapped in Fig. 1, bottom. Compared to cells without φRSA1 sequences, the lysogenic cells showed no obvious changes in growth rate, cell morphology, colony morphology, pigmentation, or extracellular polysaccharide production in culture. In our preliminary infection experiments, no obvious enhanced pathogenicity was observed with φRSA1 lysogenic cells by in planta virulence assay with tobacco plants. However, these extra genes are still interesting subjects for further analyses concerning pathogenesis-related physiological functions.
Conclusion.
(i) A bacteriophage, φRSA1, with an especially wide host range of R. solanacearum has been characterized as a P2-related phage by genomic analysis. With the established gene organization, this phage might be useful as a biocontrol agent or a biodiagnosis tool for bacterial wilt disease. (ii) One putative prophage sequence (previously named RS6 [9]) detected on the chromosome of R. solanacearum strain GMI1000 has been characterized in detail as a φRSA1-related prophage (designated φRSX in this work). Extra genes found in the hot-spot regions of these phage genomes may be candidates for further studies related to the pathogenesis of this wilt disease bacterium. (iii) A stable minireplicon has been constructed from an 8.2-kbp early region of φRSA1 DNA. This is the first example of a minireplicon from φRSA1-type phages and will serve as a good system to study the unknown replication mechanism of related phages.
Acknowledgments
We are grateful to W. C. Nierman for allowing us to use his φ52237 sequence data in comparative analyses prior to their publication.
This study was supported by the Industrial Technology Research Grant Program in 04A09505 from the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
Footnotes
Published ahead of print on 26 October 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., Hoboken, NJ.
- 3.Ayers, D. J., M. G. Sunshine, E. W. Six, and G. E. Christie. 1994. Mutations affecting two adjacent amino acid residues in the alpha subunit of RNA polymerase block transcriptional activation by the bacteriophage P2 Ogr protein. J. Bacteriol. 1767430-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birkeland, N. K., G. E. Christie, and B. H. Lindqvist. 1988. Directed mutagenesis of the bacteriophage P2 ogr gene defines an essential function. Gene 73327-335. [DOI] [PubMed] [Google Scholar]
- 5.Brüssow, H., C. Canchaya, and W.-D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calendar, R., S. Yu, H. Myung, V. Barreiro, R. Odegrip, K. Carlson, L. Davenport, G. Mosig, G. E. Christie, and E. Haggård-Ljungquist. 1998. The lysogenic conversion genes of coliphage P2 have unusually high AT content, p. 241-252. In M. Syvanen and C. Kado (ed.), Horizontal gene transfer. Chapman & Hall, Ltd., London, United Kingdom.
- 7.Campoy, S., J. Aranda, G. Alvareg, J. Barbe, and M. Llagostera. 2006. Isolation and sequencing of a temperate transducing phage for Pasteurella multocida. Appl. Environ. Microbiol. 723154-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brussow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49277-300. [DOI] [PubMed] [Google Scholar]
- 10.Christie, G. E., E. Haggård-Ljungquist, R. Feiwell, and R. Calendar. 1986. Regulation of bacteriophage P2 late gene expression: the ogr gene. Proc. Natl. Acad. Sci. USA 833238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie, G. E., L. M. Temple, B. A. Bartlett, and T. S. Goodwin. 2002. Programmed translational frameshift in the bacteriophage P2 FETUD tail gene operon. J. Bacteriol. 1846522-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig, N. L. 1988. The mechanism of conservative site-specific recombination. Annu. Rev. Genet. 2277-105. [DOI] [PubMed] [Google Scholar]
- 13.Dodd, I. B., and J. B. Egan. 1996. The Escherichia coli retrons Ec67 and Ec86 replace DNA between the cos site and a transcription terminator of a 186-related prophage. Virology 219115-124. [DOI] [PubMed] [Google Scholar]
- 14.Esposito, D., W. P. Fitzmaurice, R. C. Benjamin, S. D. Goodman, A. S. Waldman, and J. J. Scocca. 1996. The complete nucleotide sequence of bacteriophage HP1 DNA. Nucleic Acids Res. 242360-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatt, R., and E. R. Berman. 1966. A rapid procedure for the estimation of amino sugars on a microscale. Anal. Biochem. 15167-171. [DOI] [PubMed] [Google Scholar]
- 16.Haggård-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 1741462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haggård-Ljungquist, E., K. Jacobsen, S. Rishovd, W. Six, O. Nilssen, M. G. Sunshine, B. H. Lindqvist, K.-J. Kim, V. Barreiro, E. V. Koonin, and R. Calendar. 1995. Bacteriophage P2: genes involved in baseplate assembly. Virology 213109-121. [DOI] [PubMed] [Google Scholar]
- 18.Hauser, M. A., and J. J. Scocca. 1990. Location of the host attachment site for phage HP1 within a cluster of Haemophilus influenzae tRNA genes. Nucleic Acids Res. 185305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi, T., H. Matsumoto, M. Ohnishi, and Y. Terawaki. 1993. Molecular analysis of a cytotoxin-converting phage, φCTX, of Pseudomonas aeruginosa: structure of the attP-cos-ctx region and integration into the serine tRNA gene. Mol. Microbiol. 7657-667. [DOI] [PubMed] [Google Scholar]
- 20.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 2965-87. [DOI] [PubMed] [Google Scholar]
- 21.Hayward, A. C. 2000. Ralstonia solanacearum, p. 32-42. In J. Lederberg (ed.) Encyclopedia of microbiology, vol. 4. Academic Press, San Diego, CA. [Google Scholar]
- 22.Hendrix, R. W., G. F. Hatfull, and M. C. M. Smith. 2003. Bacteriophages with tails: chasing their origins and evolution. Res. Microbiol. 154253-257. [DOI] [PubMed] [Google Scholar]
- 23.Hendrix, R. W., J. G. Lawrence, G. F. Hartfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8504-508. [DOI] [PubMed] [Google Scholar]
- 24.Higashiyama, T., and T. Yamada. 1991. Electrophoretic karyotyping and chromosomal gene mapping of Chlorella. Nucleic Acids Res. 196191-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Highlander, S. K., S. Weissenberger, L. E. Alvarez, G. M. Weinstock, and P. B. Berget. 2006. Complete nucleotide sequence of a P2 family lysogenic bacteriophage, φMhaA1-PHL101, from Mannheimia haemolytica serotype A1. Virology 35079-89. [DOI] [PubMed] [Google Scholar]
- 26.Horita, M., and K. Tsuchiya. 2002. MAFF microorganism genetic resources manual no. 12, p. 5-8. National Institute of Agricultural Sciences, Tsukuba, Japan.
- 27.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hartful, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 29927-51. [DOI] [PubMed] [Google Scholar]
- 28.Julien, B., and R. Calendar. 1995. Purification and characterization of the bacteriophage P4 δ protein. J. Bacteriol. 1773743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapfhammer, D., J. Blass, S. Evers, and J. Reidl. 2002. Vibrio cholerae phage K139: complete sequence and comparative genomics of related phages. J. Bacteriol. 1846592-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawasaki, T., S. Nagata, A. Fujiwara, H. Satsuma, M. Fujie, S. Usami, and T. Yamada. 2007. Genomic characterization of the filamentous integrative bacteriophages φRSS1 and φRSM1, which infect Ralstonia solanacearum. J. Bacteriol. 1895792-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King, R. A., D. L. Anders, and G. E. Christie. 1992. Site-directed mutagenesis of an amino acid residue in bacteriophage P2 Ogr protein implicated in interaction with Escherichia coli RNA polymerase. Mol. Microbiol. 63313-3320. [DOI] [PubMed] [Google Scholar]
- 32.Lee, T.-C., and G. E. Christie. 1990. Purification and properties of the bacteriophage P2 ogr gene product. A prokaryotic zinc-binding transcriptional activator. J. Biol. Chem. 2657472-7477. [PubMed] [Google Scholar]
- 33.Lindqvist, B. H., G. Deho, and R. Calendar. 1993. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol. Rev. 57683-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, Y., S. Saha, and E. Haggård-Ljungquist. 1993. Studies of bacteriophage P2 DNA replication. The DNA sequence of the cis-acting gene A and ori region and construction of a P2 mini-chromosome. J. Mol. Biol. 231361-374. [DOI] [PubMed] [Google Scholar]
- 35.Mirold, S., and W.-D. Hardt. 2003. The SopEΦ phage integrates into the ssrA gene of Salmonella enterica serovar Typhimurium A36 and is closely related to the Fels-2 prophage. J. Bacteriol. 1855182-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosig, G., S. Yu, H. Myung, E. Haggård-Ljungquist, L. Davenport, K. Carlson, and R. A. Calendar. 1997. A novel mechanism of virus-virus interactions: bacteriophage P2 Tin protein inhibits phage T4 DNA synthesis by poisoning the T4 single-stranded DNA binding protein gp32. Virology 23072-81. [DOI] [PubMed] [Google Scholar]
- 37.Myung, H., and R. Calendar. 1995. The old exonuclease of bacteriophage P2. J. Bacteriol. 177497-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama, K., S. Kanaya, M. Ohnishi, Y. Terawaki, and T. Hayashi. 1999. The complete nucleotide sequence of CTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol. Microbiol. 31399-419. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson, A. S., and Haggård-Ljungquist. 2006. The P2-like bacteriophages, p. 365-390. In R. Calendar (ed.), The bacteriophages. ASM Press, Washington, DC.
- 40.Pierson, L. S., III, and K. L. Kahn. 1987. Integration of satellite bacteriophage P4 in Escherichia coli. DNA sequences of the phage and host regions involved in site-specific recombination. J. Mol. Biol. 196487-496. [DOI] [PubMed] [Google Scholar]
- 41.Pontarollo, R. A., C. R. Rioux, and A. A. Potter. 1997. Cloning and characterization of bacteriophage-like DNA from Haemophilus sommus homologous to phage P2 and HP1. J. Bacteriol. 1791872-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenberg, C., F. Casse-Delbart, I. Dusha, M. David, and C. Boucher. 1982. Megaplasmids in the plant associated bacteria Rhizobium meliloti and Pseudomonas solanacearum. J. Bacteriol. 150402-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saha, S., E. Haggård-Ljungquist, and K. Nordstrom. 1989. Activation of prophage P4 by the P2 Cox protein and the site of action of the Cox protein on the two phage genomes. Proc. Natl. Acad. Sci. USA 863973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Ariat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisene, S. Claudel-Renard, N. Cunnac, C. Gaspin, M. Lavie, A. Molsan, C. Robert, W. Saurin, T. Schlex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415497-502. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Seed, K. D., and J. J. Dennis. 2005. Isolation and characterization of bacteriophages of Burkholderia cepacia complex. FEMS Microbiol. Lett. 251273-280. [DOI] [PubMed] [Google Scholar]
- 47.Smith, E. F. 1986. A bacterial disease of tomato, pepper, eggplant, and Irish potato (Bacillus solanacearum nov. sp.). U.S. Dept. Agric. Div. Vegetable Physiol. Pathol. Bull. 121-28. [Google Scholar]
- 48.Sun, J., M. Inouye, and S. Inouye. 1991. Association of a retroelement with a P4-like cryptic prophage (retronphage φR73) integrated into the selenocystyl tRNA gene of Escherichia coli. J. Bacteriol. 1734171-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Y.-D., S. Zhao, and C. W. Hill. 1998. Rhs elements comprise three subfamilies which diverged prior to acquisition by Escherichia coli. J. Bacteriol. 1804102-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winstead, N. N., and A. Kelman. 1952. Inoculation techniques for evaluating resistance to Pseudomonas solanacearum. Phytopathology 42628-634. [Google Scholar]
- 51.Yabuuchi, E., V. Kosako, I. Yano, H. Hotta, and Y. Nishiuchi. 1995. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia picketii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol. Immunol. 39897-904. [DOI] [PubMed] [Google Scholar]
- 52.Yamada, T., T. Kawasaki, S. Nagata, A. Fujiwara, S. Usami, and M. Fujie. 2007. Isolation and characterization of bacteriophages that infect the phytopathogen Ralstonia solanacearum. Microbiology 1532630-2639. [DOI] [PubMed] [Google Scholar]
- 53.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziermann, R., and R. Calendar. 1990. Characterization of the cos sites of bacteriophages P2 and P4. Gene 969-15. [DOI] [PubMed] [Google Scholar]