Abstract
Proteomics and targeted gene disruption were used to investigate the catabolism of benzene, styrene, biphenyl, and ethylbenzene in Rhodococcus jostii RHA1, a well-studied soil bacterium whose potent polychlorinated biphenyl (PCB)-transforming properties are partly due to the presence of the related Bph and Etb pathways. Of 151 identified proteins, 22 Bph/Etb proteins were among the most abundant in biphenyl-, ethylbenzene-, benzene-, and styrene-grown cells. Cells grown on biphenyl, ethylbenzene, or benzene contained both Bph and Etb enzymes and at least two sets of lower Bph pathway enzymes. By contrast, styrene-grown cells contained no Etb enzymes and only one set of lower Bph pathway enzymes. Gene disruption established that biphenyl dioxygenase (BPDO) was essential for growth of RHA1 on benzene or styrene but that ethylbenzene dioxygenase (EBDO) was not required for growth on any of the tested substrates. Moreover, whole-cell assays of the ΔbphAa and etbAa1::cmrA etbAa2::aphII mutants demonstrated that while both dioxygenases preferentially transformed biphenyl, only BPDO transformed styrene. Deletion of pcaL of the β-ketoadipate pathway disrupted growth on benzene but not other substrates. Thus, styrene and benzene are degraded via meta- and ortho-cleavage, respectively. Finally, catalases were more abundant during growth on nonpolar aromatic compounds than on aromatic acids. This suggests that the relaxed specificities of BPDO and EBDO that enable RHA1 to grow on a range of compounds come at the cost of increased uncoupling during the latter's initial transformation. The stress response may augment RHA1's ability to degrade PCBs and other pollutants that induce similar uncoupling.
Rhodococci are mycolic acid-producing actinomycetes that degrade a wide variety of organic compounds (13). These catabolic capabilities are of interest for a range of bioremediation and biocatalytic applications (53). To better understand the physiology and metabolism of this important genus, we have conducted genomic studies of Rhodococcus jostii RHA1 (formerly Rhodococcus sp. strain RHA1, the species was recently identified by A. L. Jones and M. Goodfellow [personal communication]), a strain that possesses an exceptional ability to aerobically degrade polychlorinated biphenyls (PCBs) (47). Annotation of the 9.7-Mb RHA1 genome (31; http://www.rhodococcus.ca) predicted over 200 oxygenases and 30 pathways involved in the catabolism of aromatic compounds. Transcriptomic and proteomic studies have revealed a number of unique features of RHA1 metabolism, including a novel nitrile hydratase (37) and multiple steroid-degrading pathways (54).
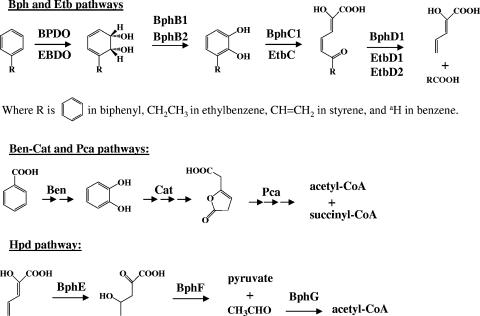
RHA1 transforms PCBs using the homologous Bph and Etb pathways (19), so named for their presumed respective specificities for biphenyl (47) and ethylbenzene (16, 57), respectively. The pathways utilize a meta-cleavage strategy in which vicinal dihydroxylation of an aromatic ring enables oxygenolytic extradiol (or meta) ring opening (Fig. 1). The upper Bph pathway aerobically transforms biphenyl to benzoate and 2-hydroxypenta-2,4-dienoate (HPD), while the Etb pathway transforms ethylbenzene to propionate and HPD. In RHA1, benzoate is further catabolized via the Ben-Cat and Pca pathways (40). In the former, enzymes encoded by benABCD transform benzoate to catechol, which is then transformed by enzymes encoded by catABC to β-ketoadipate enol-lactone via intradiol (or ortho) ring opening. The Pca pathway, encoded by pcaHGBLIJF, transforms the resulting β-ketoadipate enol-lactone to tricarboxylic acid cycle metabolites. For its part, HPD is transformed by Hpd or the “lower Bph” pathway encoded by bphEFG. The RHA1 genome contains eight clusters of bphEFG homologues (31), of which three sets were up-regulated during growth on biphenyl and ethylbenzene (12). Despite their names, the physiological substrates of the Bph and Etb pathways remain unclear. The involvement of the genes in the degradation of various compounds is further complicated by the occurrence of some of the etb genes in two nearly identical copies. Recent transcriptomic studies demonstrated that both the bph and etb genes (Fig. 2) are highly expressed in cells growing on biphenyl and ethylbenzene (12). It was suggested that this simultaneous induction of multiple isozymes may reflect either an adaptation to mixtures of compounds in natural environments or the lack of tight genetic regulation. In either case, the presence of both sets of genes appears to contribute to the potent PCB-degrading capability of RHA1, as the two dioxygenases have different preferences for PCB congeners (19).
FIG. 1.
The proposed pathways for the degradation of biphenyl (47), ethylbenzene (57), benzene, and styrene in R. jostii RHA1. The compounds are initially transformed by the Bph and Etb pathways comprising homologous enzymes as indicated: BPDO and EBDO, ring-hydroxylating dioxygenase; BphB1 and BphB2, cis-dihydrodiol dehydrogenase; BphC1 and EtbC, extradiol dioxygenase; BphD1, EtbD1, and EtbD2, meta-cleavage product hydrolase. The resulting HPD is transformed by the Hpd or “lower Bph” pathway comprising the following: BphE, HPD hydratase; BphF, 4-hydroxy-2-oxovalerate aldolase; and BphG, acetaldehyde dehydrogenase. Benzoate produced by biphenyl catabolism is catabolized via the Ben-Cat and Pca pathways (40). Gene names correspond to the indicated enzyme names, except for BPDO and EBDO, which are multicomponent enzymes encoded by each of the four bphA and etbA genes, respectively. a, Benzene is transformed to catechol by the first two steps of the Bph/Etb pathways, and catechol is further transformed by the Cat-Pca pathway.
FIG. 2.
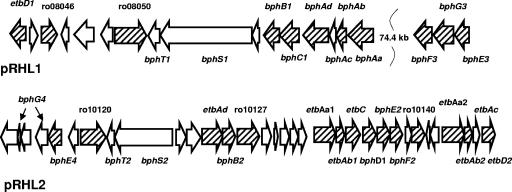
The genetic organization of key bph and etb genes in R. jostii RHA1. The numbers above the arrows indicate CDS (ro) numbers from the RHA1 genome assembly (http://www.rhodococcus.ca). These gene clusters are located on pRHL1 and pRHL2. Striped arrows indicate genes whose products were identified in this study, as listed in Table 4. Gene names correspond to enzyme names provided in Fig. 1, except for BPDO and EBDO, which are multicomponent enzymes encoded by bphAaAbAcAd and etbAaAbAcAd, respectively.
The best-characterized enzymes of the Bph and Etb pathways are the first ones: biphenyl and ethylbenzene dioxygenases (BPDO and EBDO). The catalytic subunits of these enzymes share 37% amino acid sequence identity. The two EBDO copies (large and small subunits) share 100% amino acid sequence identity and are thus expected to have identical catalytic properties. Phylogenetic analyses indicate that BPDO of RHA1 is most similar to dioxygenases that preferentially transform toluene and benzene (41). By contrast, EBDO is most similar to those that transform naphthalene, and it transforms this compound more efficiently than biphenyl (20). Moreover, bphD1, whose encoded enzyme preferentially transforms 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (HOPDA), the meta-cleavage product of biphenyl (57), is clustered with the etbA1 genes on pRHL2 (31). Similarly, one of the etbD genes is within 10 kb of the bphACB operon. The phylogenetic analyses, gene organization, and available biochemical data suggest that the optimal substrates of the Bph and Etb pathways in RHA1 may not have been identified. Other aromatic compounds that might be degraded by these enzymes include benzene and styrene.
Rhodococcal pathways responsible for the catabolism of benzene and styrene have been proposed based on metabolite studies, but the enzymes have not yet been identified. In Rhodococcus sp. strain 33, benzene is dihydroxylated and the resulting catechol is degraded via intradiol cleavage and the β-ketoadipate pathway, similar to what has been reported in other bacteria (38). By contrast, the single styrene catabolic pathway described in Rhodococcus differs from the pathway described in all other bacteria investigated to date. In the latter, styrene catabolism is initiated via oxidation of the vinyl side chain to either styrene oxide (2, 14) or phenylethanediol, which are further degraded via the phenylacetic acid and mandelic acid pathways (44), respectively. This side chain transformation is catalyzed by enzymes encoded by the styABCD operon. In Rhodococcus rhodochrous NCIMB 13259, styrene catabolism is analogous to biphenyl catabolism, in which a vinylcatechol is produced and subjected to meta-ring cleavage (56). Annotation of the RHA1 genome revealed neither a styABCD cluster nor genes dedicated to benzene catabolism (31).
Here, we investigated the roles of the Bph and Etb enzymes of RHA1 in the catabolism of four nonpolar aromatic compounds: biphenyl, ethylbenzene, benzene, and styrene. A quantitative proteomic approach was employed in combination with targeted gene disruption and whole-cell biotransformation studies to focus particularly on the two ring-hydroxylating dioxygenases and the role of the β-ketoadipate pathway in benzene and styrene catabolism. The results provide new insights into the physiological roles of the Bph and Etb pathways and the role of oxidative stress response in this catabolism.
MATERIALS AND METHODS
Chemicals.
Pharmalyte 3-10 and Immobiline Dry-Strips were purchased from GE Healthcare (Baie d'Urfé, Canada). Iodacetamide and 3-[(3-cholamidopropyl)dimethylamonio]-1-propanesulfonate (CHAPS) were from Acros Organics (New Jersey) and MP Biomedicals (Aurora, OH), respectively. Mini Complete protease inhibitor cocktail was purchased from Roche (Laval, Canada). Sypro Ruby was from Bio-Rad Laboratories Ltd. (Mississauga, Canada). Nucleotides were purchased from Qiagen (Mississauga, Canada). All chemicals were of analytical grade and were used without further purification.
Strains and media.
The Rhodococcus strains used in this study are listed in Table 1. All strains were grown at 30°C on W medium (30) supplemented with one of the following growth substrates: 20 mM pyruvate, 20 mM benzoate, 10 mM biphenyl, ethylbenzene vapors, benzene vapors, or styrene vapors. For substrates provided in the vapor phase, 2 ml of liquid was placed in a 6-ml glass tube suspended above the liquid medium in the incubation flask. Wild-type (WT) RHA1 was precultured in 250-ml flasks to mid-log phase (determined by the optical density at 600 nm [OD600]), used to inoculate fresh medium in 250- or 500-ml flasks, cultured to mid-log phase, and harvested. Mutant strains were first grown on solid W medium containing pyruvate as a carbon source. Colonies were used to inoculate W medium supplemented with an appropriate carbon source contained in 250-ml flasks. Growth rates were determined using exponentially growing cells and cultures inoculated with normalized quantities of cells based on OD, and were based on three replicate cultures. The genotypes of mutants were verified after each growth experiment using PCR and the primers listed in Table 2.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype/comments | Source |
|---|---|---|
| RHA1 | R. jostii RHA1; wild-type | 30 |
| HDT1 | Derivative of RHA1; aphII gene insertion mutant of etbAa1; etbAa1::aphII Kmr | 19 |
| HDB1 | Derivative of RHA1; aphII gene insertion mutant of etbAa2; etbAa2::aphII Kmr | 19 |
| HDBT1 | Derivative of HDB1; cmrA gene insertion mutant of etbAa1; aphII gene insertion mutant of etbAa2; etbAa1::cmrA etbAa2::aphII Cmr Kmr | 19 |
| RHA1_007 | Derivative of RHA1; ΔbphAa Aprr | This study |
| RHA1_008 | Derivative of HDBT1; ΔbphAa etbAa1::cmrA etbAa2::aphII Cmr Kmr Aprr | This study |
| RHA1_005 | Derivative of RHA1; ΔpcaL Aprr | 40 |
TABLE 2.
PCR primers
| Primer | Sequence (5′-3′) | Comments |
|---|---|---|
| BphAafor | CAACTTGGTGAGTGTGCC | Internal bphAa primer, forward |
| BphAarev | TGTAGAAGCCAGAACCATGC | Internal bphAa primer, reverse |
| EtbAa1for | CGATAGCGCATTGGCCATGACATT | Internal etbAa1 primer, forward |
| EtbAa1rev | ATCAGATCCCAACGCTGGTAA | Internal etbAa1 primer, reverse |
| EtbAa2for | AGAGCGTGGGCCAGTTGTTTAGTA | Internal etbAa2 primer, forward |
| EtbAa2rev | ATCAGATCCCAACGCTGGTAA | Internal etbAa2 primer, reverse |
Preparation of cell extracts.
Cell extracts were prepared essentially as described previously (40). Briefly, cell pellets were washed once with 0.14 M NaCl and once with Tris-EDTA buffer (10 mM Tris/HCl, 1 mM EDTA, pH 8.0) and then stored as aliquots at −80°C. The pelleted cells were suspended in lysis buffer (4% CHAPS, 30 mM Tris, pH 7.5, Mini Complete protease inhibitor cocktail, 1:100 [vol/vol]) and disrupted using a bead beater. Cell debris was removed by centrifugation, and the protein extract was either stored at −80°C or used immediately. For enzyme assays, cell-free protein extracts were prepared in the same manner except that the cell pellets were washed using 50 mM Tris-HCl, pH 7.0, and the extracts were used immediately. The protein concentration was determined using the 2D Quant kit (GE Healthcare).
Proteomic analyses.
The cytosolic proteome was resolved using two-dimensional (2D) gel electrophoresis essentially as described previously (40). Briefly, aliquots of cell extracts containing 90 μg protein were separated in the first dimension by isoelectric focusing using 24-cm nonlinear immobilized pH gradient (IPG) strips and a pH gradient of 3 to 7. Proteins were separated in the second dimension using 12% sodium dodecyl sulfate-polyacrylamide gels and the ETTAN DALTtwelve System (GE Healthcare). The gels were stained using Sypro Ruby and digitally imaged using a Typhoon 9400 (GE Healthcare). Spot detection and pattern matching were performed using Progenesis Workstation software (Nonlinear Dynamics, Durham, NC). The pattern of each growth condition was based on gels from three independent cultures. The relative abundance of each protein was determined from the integrated signal intensity of spots after subtraction of background values. The signal intensity of each spot was normalized against the total signal intensity detected on a gel. Only spots with a minimum normalized volume of 0.002 or greater were further analyzed. For the proteins that appeared on a gel as a horizontal series of spots, likely due to carbamylation, the pI and molecular weight (MW) of the major spot in the series were recorded, and the summed signal intensities of all the spots in the series were further compared. Protein spots of interest were identified using a Voyager Destr matrix-assisted laser desorption ionization-time of flight mass spectrometer (Applied Biosystems, Foster City, CA) based on peptide mass fingerprint analysis combined with a MASCOT search engine and the RHA1 protein database at the Proteomics Centre, University of Victoria. Identified proteins fulfilled four criteria: the MASCOT search score was above 55, a minimum of four peptides were matched, the protein sequence coverage was at least 20%, and the predicted MW and pI values were consistent with the experimentally observed values.
Enzyme assays.
Enzyme assays were performed using a Varian Cary IE UV-Visible spectrophotometer equipped with a cuvette holder with a thermostat. Catechol 1,2-dioxygenase (C12O) activity was measured by monitoring the formation of cis,cis-muconate at 260 nm (ɛ = 16.0 mM−1 cm−1), and catechol 2,3-dioxygenase (C23O) activity was determined by monitoring the formation of 2-hydroxymuconic semialdehyde at 375 nm (ɛ = 12.0 mM−1 cm−1) in assay mixtures containing 0.1 M phosphate, pH 7.0, and 100 μM catechol (28). The activity of dihydroxybiphenyl dioxygenase (DHBD) was measured by monitoring the formation of HOPDA at 434 nm (ɛ = 11.3 mM−1 cm−1) using an assay mixture containing 0.1 M phosphate, pH 7.0, and 100 μM DHB (8). One unit of enzyme activity was defined as the amount of enzyme required to produce 1 μmol of product per minute at 25°C.
Resting cell assays.
Cells grown on ethylbenzene at 30°C to mid-log phase (OD600 = 2.0) were harvested, washed twice with 50 mM sodium phosphate, pH 7.5, supplemented with 1 g/liter glucose, and suspended in the same buffer at an OD600 of 2.0. One-milliliter portions of this suspension were distributed in 12-ml glass vials sealed with Teflon caps. Each vial received one of the following compounds to a final concentration of 50 μM: biphenyl, ethylbenzene, benzene, styrene, chlorobenzene, toluene, or o-xylene. Assay mixtures were incubated for 0 (control) or 5 h at 30°C and 225 rpm, and then the substrate concentration was determined. Since biphenyl was depleted faster than the other compounds, it was assayed using only 1/10 of the biomass (OD600 = 0.2) and a 1-hour incubation. The reactions were stopped by adding a drop of 1 N HCl, and samples were immediately frozen at −80°C. Each sample was extracted twice with 1 ml of hexane. The pooled organic phases were dried over sodium sulfate and transferred to vials for further high-performance liquid chromatography (benzene) or gas chromatography-mass spectrometry (GC-MS) (all other substrates) analyses. Controls were performed using dead cells. Assays were performed at least in duplicate.
High-performance liquid chromatography analyses were performed using a Waters 2695 Separations Module equipped with a Waters 2996 Photodiode Array Detector and a C18 Waters Nova-Pak column (3.9 × 150 mm) (Waters Ltd., Mississauga, Ontario, Canada) operated at a flow rate of 1 ml/min. Benzene was eluted using 100% methanol. Samples (20 μl) were injected, and benzene was detected at 210 nm and quantified using a standard curve constructed using known amounts of benzene (r > 0.99). GC-MS was performed using an HP-5MS equipped with an Agilent column 19091S-433 (0.25 mm by 30 m by 0.25 μm) (Agilent, Mississauga, Ontario, Canada) operated at a flow rate of 53.5 ml/min and a pressure of 10.7 lb/in2. During the run, the oven temperature was ramped from 40 to 280°C. Samples (2 μl) were injected in splitless mode, and the amount of substrate was determined from the area of the corresponding peak at the appropriate m/z (standard curves had r values of >0.99). All standard samples were treated in the same manner as the reaction mixtures to correct for losses of substrates due to sample manipulation.
Gene replacement.
All mutants used in this study are listed in Table 1. The bphAa gene was deleted and replaced by an apramycin cassette using the λ-RED-based methodology as described previously (40). The etbAa1 and etbAa2 genes were disrupted in single mutants using a single-crossover insertion of aphII, encoding kanamycin resistance (19). In the double mutant, two genes were disrupted using aphII and cmrA, encoding chloramphenicol resistance (19). The triple mutant was constructed by deleting bphAa as summarized above in the double-insertion mutant HDBT1 lacking functional etbAa1 and etbAa2 (19). Genotypes were verified using PCR with the primers listed in Table 2. Disruption mutants were further verified by confirming the loss of the encoded protein in the proteomes of ethylbenzene-grown cells.
RESULTS
R. jostii RHA1 grew on biphenyl, ethylbenzene, benzene, styrene, and pyruvate as sole growth substrates in liquid medium at comparable rates (Table 3). RHA1 also used the following uncharged aromatic compounds as growth substrates: toluene, ο-xylene, and chlorobenzene. By contrast, RHA1 did not utilize naphthalene as a growth substrate.
TABLE 3.
Characteristics of growth of WT and mutant RHA1 on different substrates
| Strain (genotype) | Growth rate μ (h−1)a
|
|||||
|---|---|---|---|---|---|---|
| Biphenyl | Ethylbenzene | Benzene | Styrene | Benzoate | Pyruvate | |
| RHA1 WT | 0.044 (0.004) | 0.054 (0.003) | 0.0264 (0.0004) | 0.044 (0.006) | 0.14 (0.06) | 0.11 (0.01) |
| RHA1_005 (ΔpcaL) | 0.042 (0.005) | NAd | 0.04 (0.01)b | 0.03 (0.009) | 0.12 (0.04)b,g | 0.10 (0.02)g |
| RHA1_007 (ΔbphAa) | 0.032 (0.008) | 0.049 (0.008) | NDe | ND | 0.11 (0.02) | +h |
| HDT1 (etbAa1::aphII) | ND | 0.04 (0.01) | 0.016 (0.002) | 0.018 (0.001) | 0.14 (0.01) | + |
| HDB1 (etbAa2::aphII) | 0.04 (0.01) | 0.03 (0.01) | 0.047 (0.004) | 0.019 (0.006) | 0.13 (0.05) | + |
| HDBT1 (etbAa1::cmrA etbAa2::aphII) | ND | 0.03 (0.01)c | 0.033 (0.001)c | 0.03 (0.01) | 0.12 (0.01) | + |
| RHA1_008 (etbAa1::cmrA etbAa2::aphII ΔbphAa) | NDf | NDf | ND | ND | 0.10 (0.02) | + |
Growth rates represent those calculated during exponential growth phase and are based on three biologically independent replicates. Standard errors of the mean are indicated in parentheses.
The lag phase was 70 h longer than that detected in comparable cultures of WT cells.
The lag phase was 140 h longer than that detected in comparable cultures of WT cells.
NA, growth was not tested.
ND, growth was not detected.
Growth was first detected 3 to 4 weeks after inoculation. PCR analyses confirmed the restoration of etbAa1 and/or etbAa2.
data taken from reference 40.
+, growth was detected, but the rate was not measured.
Identification of catabolic enzymes.
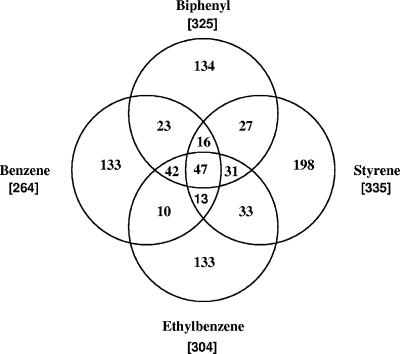
2D gel electrophoresis resolved approximately 1,400 protein spots in the cytosolic proteomes of RHA1 cells grown on each of the tested growth substrates. Representative sections of average gels are shown in Fig. 3. Pairwise comparison of the five averaged gels, including quantitative comparison of spot intensities, revealed that the proteomes of cells grown on the four aromatic compounds were more similar to each other than to the proteome of pyruvate-grown cells: any two of the former shared 68 to 73% of their protein spots, whereas any one of the former shared 57 to 62% of its protein spots with the pyruvate proteome. For each aromatic proteome, 40 to 47% of the detected proteins were at least twofold more abundant than during growth on pyruvate, in approximate agreement with the number of genes up-regulated in biphenyl- and ethylbenzene-grown cells by microarray analysis (12). Among the protein spots that were at least twofold more abundant in a given proteome, 41 to 59% were unique to that proteome and 15% were common to the four aromatic growth substrates (Fig. 4). Of the 15%, or 47 shared spots, 28 spots were not detected in pyruvate-grown cells. Among the subsets of more abundant proteins, those present in biphenyl- and benzene-grown cells showed the greatest overlap (44%), while those present in styrene- and benzene-grown cells showed the least (25%).
FIG. 3.
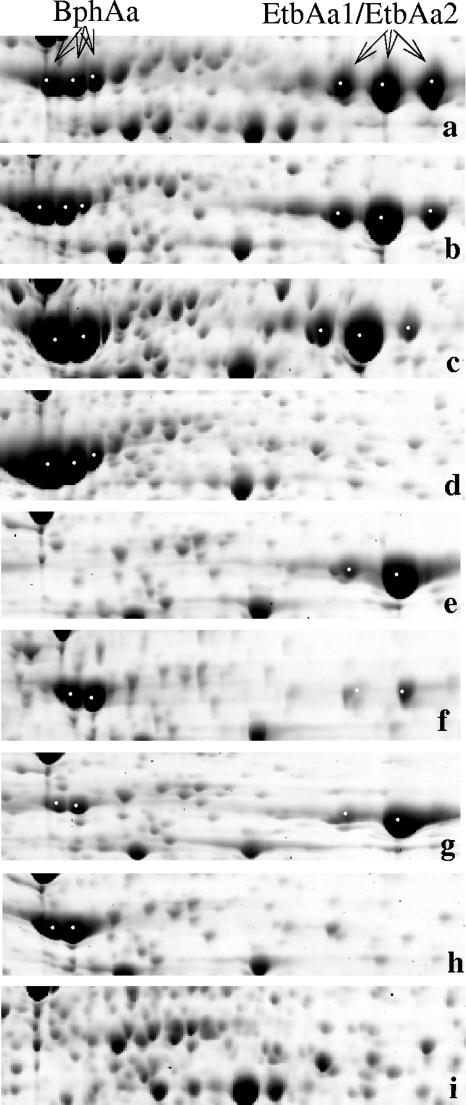
Sections of 2D gels showing parts of the cytosolic proteomes of R. jostii RHA1 grown on biphenyl (a), ethylbenzene (b), benzene (c), styrene (d), or pyruvate (i). The corresponding gel sections are shown for ethylbenzene-grown mutants: (e) RHA1_007 (ΔbphAa), (f) HDT1 (etbAa1::aphII), (g) HDB1 (etbAa2::aphII), and (h) HDBT1 (etbAa1::cmrA etbAa2::aphII). BphAa and EtbAa1/ EtbAa2 protein spots are indicated with arrows and are located in the left and right sides of the gel sections.
FIG. 4.
Venn diagram representation of the protein spots whose normalized intensities were at least twofold higher in at least one of the aromatic proteomes versus pyruvate proteome. The numbers in brackets indicate the total numbers of more abundant protein spots detected in the corresponding average gel.
Overall, we identified 151 proteins that occurred in at least one of the analyzed proteomes. The averaged normalized signal intensity of each identified protein for each condition, as well as protein experimental and theoretical pI and MW, and MS identification parameters are presented in Table S1 in the supplemental material. The identified proteins include 22 of the Bph and Etb pathways and 21 of the 47 proteins that were more abundant in the four aromatic proteomes (Tables 4 and 5). The identified proteins included three of the four polypeptides of the two ring-hydroxylating dioxygenases (BPDO and EBDO), two ring cleavage dioxygenases (BphC1 and EtbC), and three hydrolases (BphD1, EtbD1, and EtbD2).
TABLE 4.
Identification of the proteins involved in the catabolism of biphenyl, ethylbenzene, benzene, and styrene
| Protein name | Basis of identification | No. of peptides matched | Sequence coverage (%) | Signal intensitya
|
||||
|---|---|---|---|---|---|---|---|---|
| Pyruvate | Biphenyl | Ethylbenzene | Benzene | Styrene | ||||
| BphAab | MS | 11 | 26 | NDf | 1.9 | 3.4 | 6.5 | 9.1 |
| BphAbb | MS | 12 | 78 | ND | 1.5 | 2.4 | 3.3 | 5.9 |
| BphAdb | MS | 9 | 25 | ND | 0.47 | 0.50 | 0.90 | 1.1 |
| BphB1 | MS-MS | 24 | 47 | ND | 2.02 | 3.3 | 3.2 | 5.8 |
| BphB2 | MS | 8 | 46 | ND | 2.1 | 2.9 | 2.2 | ND |
| BphC1 | MS | 10 | 45 | ND | 1.3 | 1.3 | 2.7 | 4.7 |
| BphD1 | MS | 13 | 56 | ND | 3.6 | 4.3 | 3.2 | ND |
| BphE2 | MS | 4 | 30 | ND | 0.26 | 0.28 | 0.18 | ND |
| BphE3 | MS | 8 | 36 | ND | 0.062 | 0.10 | ND | 0.14 |
| BphE4 | MS | 6 | 34 | ND | 0.25 | 0.17 | 0.078 | ND |
| BphF2 | MS | 5 | 28 | ND | 0.15 | 0.23 | 0.14 | ND |
| BphF3 | MSd | 6 | 29 | ND | 0.28 | 0.35 | ND | 0.68 |
| BphG3 | MS | 14 | 70 | ND | 0.27 | 0.34 | ND | 0.42 |
| EtbAa1/EtbAa2b,c | MS | 12 | 35 | ND | 3.9 | 4.8 | 5.9 | ND |
| EtbAb2/EtbAb2b,c | MS | 4 | 31 | ND | 3.2 | 3.6 | 3.6 | ND |
| EtbAdb | MS | 10 | 24 | ND | 1.5 | 1.6 | 1.7 | ND |
| EtbC | MS | 12 | 44 | ND | 3.5 | 3.4 | 3.2 | ND |
| EtbD1 | MSd | 13 | 70 | ND | 1.5 | 2.5 | 2.2 | 2.9 |
| EtbD2 | MS | 11 | 48 | ND | 0.99 | 1.6 | 1.2 | ND |
| BenA | MSd | 14 | 44 | ND | NAg | ND | NA | ND |
| BenB | MSd | 6 | 31 | ND | 0.23 | ND | ND | ND |
| BenD | MSd | 8 | 39 | ND | 0.14 | ND | ND | ND |
| CatA | MS | 9 | 37 | ND | 1.5 | ND | 1.9 | 0.97 |
| CatB | MSd | 5 | 21 | ND | 0.15 | ND | 0.19 | 0.062 |
| PcaH | MS-MS | 7 | 33 | ND | 0.035 | ND | 0.094 | ND |
| PcaG | MS | 9 | 35 | ND | 0.062 | ND | 0.084 | ND |
| PcaB | MS-MS | 5 | 12 | 0.025 | 0.044 | ND | 0.017 | ND |
| PcaL | MS | 6 | 19 | 0.019 | 0.098 | 0.01 | 0.098 | 0.017 |
| PcaI | MS-MS | 8 | 47 | ND | 0.065 | ND | 0.11 | 0.015 |
| PcaJ | MSd | 4 | 23 | ND | 0.039 | 0.012 | 0.075 | 0.080 |
| PcaF | MS | 13 | 41 | 0.10 | 0.13 | 0.045 | 0.14 | 0.057 |
| PcaR | MSd | 7 | 43 | 0.012 | 0.035 | ND | 0.034 | 0.008 |
Spot signal intensities were normalized and averaged over three replica gels (each from an independent experiment).
BphAa, BphAb, BphAc, and BphAd comprise BPDO. EtbAa, EtbAb, EtbAc, and EtbAd comprise EBDO.
The EtbAa1/EtbAa2 and EtbAb1/EtbAb2 proteins are identical and therefore occupy the same spot on a 2D gel. The reported signal intensity value therefore represents the total of those of the two corresponding proteins.
Protein identity was confirmed by tandem MS (MS-MS) analysis.
eBenA protein spot overlaps with EtbAa, and therefore, its signal intensity value cannot be estimated in the biphenyl, ethylbenzene, and benzene proteome.
ND, not detected.
NA, not available.
TABLE 5.
Selected proteins induced at least twofold during RHA1 growth on biphenyl, ethylbenzene, benzene, and styrene
| Functional class | Protein namea | Gene ro no. | Best hit in NCBIb | Signal intensityc
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pyruvate | Biphenyl | Ethylbenzene | Benzene | Styrene | Benzoated | Phenylacetatee | ||||
| Stress response | Catalasef | 04309 | gi:2645461, 59, Micrococcus luteus (33) | 0.008 | 0.053 | 0.092 | 0.38 | 0.045 | 0.020 | 0.014 |
| Stress response | Alkyl hydroxyperoxide reductase, AhpC | 03275 | gi:75765541, 73, Salmonella typhimurium 602 (39) | ND | 0.062 | 0.19 | 0.89 | 0.009 | 0.009 | 0.099 |
| Stress response | ATP-dependent Clp protease, proteolytic subunit, Hsp100 family | 01371 | gi:17373662, 65, Streptomyces coelicolor A3(2) (5) | ND | 0.10 | 0.097 | 0.13 | 0.16 | 0.20 | 0.19 |
| Stress response | Fatty acid desaturase | 02258 | gi:8919145, 23, Mycobacterium avium subsp. paratuberculosis (3) | 0.045 | 0.016 | 0.14 | 0.021 | 0.022 | 0.018 | 0.083 |
| Stress response RNA metabolism | rRNA small subunit methyltransferase | 07164 | gi:4210751, 27, Lactococcus lactis subsp. lactis (23) | ND | 0.52 | 0.77 | 0.51 | ND | ND | ND |
| Catabolic | Quinone oxidoreductase | 08046 | gi:84617341, 35, Streptomyces achromogenes subsp. Rubradiris (49) | ND | 0.24 | 0.36 | 0.52 | 0.57 | ND | ND |
| Catabolic | 6-Oxohexanoate dehydrogenase | 10127 | gi:40787207, 99.7, Rhodococcus sp. strainDK17 (21) | ND | 0.037 | 0.12 | 0.12 | 0.043 | ND | ND |
| Catabolic | Medium-chain acyl-coenzyme A ligase | 08050 | gi:40787201, 99.6, Rhodococcus sp. strain DK17 (21) | ND | 0.60 | 0.81 | 0.66 | 0.48 | ND | ND |
| Catabolic | Oxidoreductase | 10140 | gi:40787219, 99.6, Rhodococcus sp. strain DK17 (21) | 0.016 | 0.25 | 0.33 | 0.26 | 0.008 | 0.019 | 0.040 |
Protein names are probable, as they are based on amino acid sequence identities, not demonstrated activities.
Accession number, percent identity, organism.
Spot signal intensities were normalized and averaged over three replica gels (each from independent experiment).
Data taken from reference 40.
Data taken from reference 36.
Boldface denotes products whose genes were up-regulated in biphenyl- and/or ethylbenzene-grown cells (12) (microarray data, NCBI GSE5280).
The 22 identified Bph and Etb proteins of biphenyl- and ethylbenzene-grown cells were among the most highly abundant in these proteomes. Biphenyl-grown cells also contained BenABD, CatAB, and PcaHGBLIJFR proteins responsible for benzoate, catechol, and β-ketoadipate catabolism, respectively, as previously established in benzoate-grown cells (40). The Ben, Cat, and Pca proteins were absent in ethylbenzene-grown cells, with the exception of PcaLJF, which were detected in very small quantities. These results are consistent with the well-established catabolism of biphenyl and ethylbenzene in RHA1. None of the Bph, Etb, or Ben catabolic proteins were detected in pyruvate-grown cells (Table 4 and Fig. 2).
The proteomes of benzene-grown cells contained 19 of the 22 identified Bph and Etb proteins. Proteins that were present included the ring-hydroxylating enzymes (BphA and EtbA), a dihydrodiol dehydrogenase (BphB1), two meta-cleavage dioxygenases (BphC1 and EtbC), three hydrolases (BphD1 and EtbD1D2), and two sets of the lower-pathway enzymes (BphE2F2 and BphE4) (Table 4 and Fig. 2). The third set of the last (BphF3G3E3) was not detected. These cells also contained the Cat and Pca enzymes, but none of the benzoate catabolic enzymes (BenABD). The Cat and Pca proteins were present in quantities similar to those in biphenyl-grown cells but at least twofold lower than in benzoate-grown cells (40). These data suggest that in RHA1, benzene may be dihydroxylated by BPDO and/or EBDO and that the resulting catechol may be degraded via ortho-cleavage and the β-ketoadipate pathway.
The proteome of styrene-grown cells was unique among the studied aromatic proteomes in that no Etb proteins were detected other than EtbD1. Significantly, etbD1 is clustered with bphT1S1BCA, not with other etb genes. Moreover, styrene-grown cells contained large amounts of the upper and lower Bph pathway enzymes (Table 4). Other proteins that were absent in these cells include the phenylacetate catabolic enzymes (36) and homologues of the StyABD enzymes. By contrast, styrene-grown cells contained significant quantities of the ortho-cleavage enzymes CatAB and the β-ketoadipate pathway enzymes PcaLIJF. Overall, these observations strongly suggest that RHA1 utilizes the Bph pathway to degrade styrene via 3-vinylcatechol (Fig. 1). However, based on these data, it is unclear whether the vinylcatechol is subject to meta- or ortho-cleavage.
The most abundant enzymes in the aromatic proteomes were the large subunits of either the BPDO or EBDO, BphAa and EtbAa. The relative abundance of BphAa was about fivefold higher in styrene-grown cells than in biphenyl-grown cells, with ethylbenzene- and benzene-grown cells carrying intermediate amounts (Table 4). The relative abundance of EtbAa was only slightly higher in ethylbenzene- and benzene-grown cells than in biphenyl-grown cells.
In addition to the Bph and Etb catabolic enzymes, we identified 31 other proteins that were at least twofold more abundant in one or more of the aromatic proteomes. Of particular note, proteins that were more abundant in all four aromatic proteomes are predicted to belong to oxidative-stress response, catabolism, and coenzyme A metabolism (Table 5). Interestingly, all of the catabolic proteins are encoded by genes located within the bph and etb gene clusters, and their abundance profiles were similar to the abundance profiles of the Bph and Etb proteins. Other important functional groups to which more abundant proteins belong include regulation and central metabolism (including gluconeogenesis and tricarboxylic acid cycle nucleoside metabolism, as well as the biosynthesis of branched amino acids, coenzymes, and polysaccharides) (see Table S1 in the supplemental material).
Analysis of gene disruption mutants.
To further investigate the physiological roles of the Bph and Etb pathways, we investigated the growth phenotypes and substrate-transforming capabilities of gene disruption mutants lacking one or more of bphAa, etbAa1, and etbAa2, which encode the large subunits of the initial dioxygenases (Table 1). Among the single gene disruption mutants, the clearest phenotype was observed in the ΔbphAa strain RHA1_007: this strain was unable to grow on either benzene or styrene (Table 3). By contrast, the strain grew at rates close to WT on either biphenyl or ethylbenzene. Disruption of etbAa1 and/or etbAa2 (strains with the single mutation HDT1 and HDB1 and a double mutant, HDBT1) did not affect growth on ethylbenzene, styrene, or benzene. Although disruption of etbAa1 resulted in loss of the ability to grow on biphenyl, this was likely due to a polar effect, as described below. Finally, disruption of all three genes encoding the large subunits of biphenyl or EBDO in RHA1_008 abolished the ability to grow on any of the four aromatic substrates. By contrast, growth on either benzoate or pyruvate was not affected. Overall, these results indicate that only biphenyl dioxygenase supports the growth of RHA1 on benzene and styrene, while both this enzyme and EBDO can support growth on ethylbenzene.
Quantitative proteomic comparison of ethylbenzene-grown mutants and WT cells (Fig. 3) revealed three additional properties of the mutants. First, the results corroborated PCR analyses verifying that the genotypes of the mutants were stable throughout the experiments. Second, these analyses revealed that both etbAa1 and etbAa2 were expressed in ethylbenzene-grown WT RHA1 cells. More specifically, the large subunit of the EBDO was detected in each of the etbAa1::aphII and etbAa2::aphII mutants, presumably arising from etbAa2 and etbAa1, respectively. Assuming that the abundance of the large subunit in the mutants reflects the relative expression of the genes in WT RHA1, the ratio of expression of etbAa1 and etbAa2 is approximately 4:1. Finally, these analyses revealed that the single-crossover insertion used to delete etbAa1 and etbAa2 exerted a polar effect on the downstream genes, etbAb1C, bphD1E2F2, and etbAb2D2, respectively, resulting in a 6- to 20-fold-decreased abundance of the encoded proteins (data not shown). This confirms the polar effect suggested previously (19). The decreased abundance of BphD1 in the etbAa1::aphII and etbAa1::cmrA etbAa2::aphII mutants likely explains the failure of these mutants to grow on biphenyl, as BphD1 is essential for this growth in RHA1 (48).
To investigate the catabolism of benzene and styrene by an ortho-cleavage pathway, we studied the growth of a ΔpcaL strain on these compounds. The pcaL gene encodes a bifunctional enzyme that is responsible for the transformation of carboxymuconolactone to β-ketoadipate, impairing growth on benzoate (40). The deletion of pcaL impaired the ability of RHA1 to grow on benzene in a manner similar to that observed on benzoate (Table 3). More specifically, no growth was observed for 70 h. Growth on benzoate after this time was due to induction of a homologue that compensated for loss of PcaL. Deletion of pcaL did not affect growth on either styrene or biphenyl. Overall, these results indicate that benzene is catabolized via the β-ketoadipate pathway, whereas styrene is transformed via meta-cleavage of 3-vinylcatechol. The latter is corroborated by our failure to detect the accumulation of vinyl-cis,cis-muconate, β-ketoadipate, or other metabolites arising from the ortho-cleavage of vinylcatechol in styrene-grown cells of WT RHA1 (data not shown).
Substrate preference of the ring-hydroxylating dioxygenases BPDO and EBDO.
To further investigate the catabolic activities of BPDO and EBDO, we performed resting-cell assays using two mutants: HDBT1 lacking the identical large subunits of EBDO and RHA1_007 lacking the large subunit of BPDO. Based on substrate depletion assays (Table 6), BPDO transformed substrates in the following order of preference: biphenyl ≫ toluene > benzene > chlorobenzene ∼ ethylbenzene > o-xylene > styrene. By contrast, EBDO transformed substrates in the following order of preference: biphenyl ≫ toluene > benzene > chlorobenzene ∼ o-xylene > ethylbenzene. The most significant differences between the mutants concerned toluene, ethylbenzene, and styrene, all of which were depleted significantly faster by the BPDO-containing mutant. Moreover, only this mutant detectably depleted styrene. Nevertheless, both mutants depleted biphenyl at significantly higher rates than any of the other tested compounds: the biphenyl depletion assays were performed using 1/10 of the cells used for the other assays, and 85 to 95% substrate depletion was detected after 1 h. However, the EBDO-containing mutant (RHA1_007) depleted biphenyl to a slightly greater extent.
TABLE 6.
Substrate preferences of the biphenyl and ethylbenzene dioxygenases
| Substrate | Substrate depletion (%)a
|
|
|---|---|---|
| RHA1_007 (ΔbphAa) | HDBT1 (etbAa1::cmrA etbAa2::aphII) | |
| Biphenylb | 95 (2) | 85 (2) |
| Ethylbenzene | <10 | 20 (5) |
| Benzene | 37 (1) | 40 (1) |
| Styrene | ND | <10 |
| Chlorobenzene | 32 (1) | 22 (6) |
| Toluene | 51 (8) | 86 (14) |
| o-Xylene | 25 (7) | 12 (5) |
RHA1_007 contains EBDO, and HDBT1 contains BPDO. The assays were performed at least in duplicate. Standard deviations are shown in parentheses. ND, not detected.
The biphenyl depletion assays were performed using 1/10 of the biomass used for all other substrates and an incubation time of only 1 hour (see Materials and Methods for details).
Enzyme activity.
To investigate the roles of ortho- and meta-cleavage in the catabolism of the nonpolar aromatic compounds, we measured the activities of C12O, C23O, and DHBD in extracts of cells grown on styrene, benzene, biphenyl, and pyruvate. As summarized in Table 7, all three ring cleavage activities were more abundant in cells grown on the aromatic substrates than on pyruvate. Moreover, the meta-cleavage activities (C23O and DHBD) were higher than the ortho-cleavage activity (C12O). All three activities were highest in styrene-grown cells: these activities were almost an order of magnitude higher than in biphenyl-grown cells. The meta- and ortho-cleavage activities were closest to parity in benzene-grown cells and agreed with the abundances of BphC1, EtbC, and CatA, respectively (Table 4).
TABLE 7.
Enzyme activities in cell lysates of RHA1 cells grown on different substrates
| Enzyme | Sp act (U mg−1) on growth substratea:
|
|||
|---|---|---|---|---|
| Biphenyl | Benzene | Styrene | Pyruvate | |
| C12O | 100 (9) | 800 (50) | 1000 (70) | 10 (1) |
| C23O | 800 (6) | 3000 (200) | 8600 (200) | 40 (7) |
| DHBD | 700 (10) | 1000 (200) | 3600 (300) | 30 (5) |
Data are based on three replicates (standard errors of the mean are in parentheses). C12O and C23O activities were measured by monitoring the formation of cis,cis-muconate and 2-hydroxymuconic semialdehyde, respectively, from catechol. The activity of DHBD was measured by monitoring the formation of HOPDA from DHB.
DISCUSSION
This study establishes that RHA1, a potent degrader of aromatic compounds, utilizes the Bph and Etb pathways to degrade a surprisingly broad range of compounds. Proteomic analyses revealed that one or both of these pathways were highly abundant during growth on biphenyl, ethylbenzene, benzene, and styrene. Targeted gene disruption established that BPDO catalyzes the initial transformation of all four compounds in vivo and that EBDO does not transform styrene. Whole-cell assays further substantiated that both enzymes are able to efficiently transform biphenyl and that BPDO is better able to transform certain smaller substrates, including toluene, ethylbenzene, and styrene, than EBDO.
The current proteomic analysis is highly congruent with a previous transcriptomic analysis (12). The latter established gene expression ratios for RHA1 grown on biphenyl and ethylbenzene versus pyruvate. The protein signal intensity values in Table 4 are highly correlated with the corresponding gene expression ratios, with correlation coefficients of 0.74 and 0.77 for biphenyl and ethylbenzene, respectively. All of the proteins that were detected in cells grown on one of these two aromatic substrates, but not pyruvate, had a large corresponding gene expression ratio—in nearly every case a ratio greater than 10. The proteomic data further demonstrate that etbD1 and etbD2 are simultaneously expressed during growth on biphenyl and ethylbenzene, which was not clear from the transcriptomic analysis. The only Bph and Etb proteins that were not detected as differentially expressed proteins were the bphAc- and etbAc-encoded ferredoxins of BPDO and EBDO, respectively. These genes were up-regulated in both biphenyl- and ethylbenzene-grown cells. Presumably, the proteins were not identified due to their small size.
The respective substrate preferences of BPDO and EBDO, while somewhat inconsistent with their names, are consistent with their phylogeny and other known properties. In phylogenetic analyses, BPDO clustered with enzymes that degrade compounds containing a single aromatic nucleus, such as halogenated and alkylated benzenes (41). Among characterized enzymes, BPDO shares the greatest amino acid sequence identity (98 to 99%) with toluene, isopropylbenzene, and benzene dioxygenases (34, 43, 50). Moreover, the substrate-binding pocket of the RHA1 enzyme (9) is not as large as that of other BPDOs, such as that of Pandoraea pnomenusa B-356 (11). By contrast, EBDO clusters with enzymes transforming larger substrates, such as phenanthrene dioxygenase from Burkholderia sp. strain RP007 (25) and tetralin dioxygenase from Sphingopyxis macrogoltabida strain TFA (32). The current results are also consistent with two recent studies of substrate preference in the RHA1 dioxygenases. First, EBDO preferentially transforms more highly chlorinated PCB congeners than BPDO (19). Second, EBDO transforms naphthalene, phenanthrene, dibenzofuran, and dibenzo-p-dioxin more efficiently than BPDO (20). Overall, these results complement the current results, demonstrating that while both BPDO and EBDO efficiently transform biphenyl, BPDO transforms monocyclic aromatic hydrocarbons, such as toluene, ο-xylene, and chlorobenzene, more effectively than EBDO.
Although the best substrates of BPDO and EBDO may not yet be identified, it is nonetheless remarkable that these two particular homologues have such a broad range of physiological substrates. Such dioxygenases are known to be subject to uncoupling in the presence of suboptimal substrates, resulting in the futile consumption of NADH and the production of H2O2. Benzene and PCBs cause uncoupling in naphthalene dioxygenase and BPDO, respectively (17, 26). The presence of a predicted catalase and alkyl hydroperoxide reductase (small subunit; AhpC) in the aromatic proteomes suggests that reactive oxygen species are produced under these growth conditions, consistent with the occurrence of uncoupling during the initial ring-hydroxylating reaction. More specifically, the predicted catalase was 2- to 25-fold more abundant under these growth conditions than during growth on either benzoate or phenylacetate (Table 5) (36, 40). The increased levels of catalases are substantiated by transcriptomic data, which revealed that the genes encoding each of RHA1's three predicted catalases (ro04309, ro05275, and ro05938) were two to six times more highly up-regulated during growth on biphenyl or ethylbenzene than on benzoate (12; microarray data, NCBI GSE5280). Further studies using purified BPDO and EBDO are required to determine the substrate specificities of these enzymes, as well as the coupling of the different substrate transformations to O2 and NADH consumption. More generally, our data contribute to the growing evidence that oxidative stress is associated with the aerobic catabolism of aromatic compounds (1, 4, 6, 27, 58).
The current study provides the first evidence of the independent regulation of bph and etb genes. In all studies conducted to date, the bph and etb genes of RHA1 have been coexpressed. These include transcriptomic studies of cells grown on ethylbenzene and biphenyl (12) and reports on coinduction of multiple isozymes of BphABCDEFG in the presence of a variety of aromatics, including biphenyl, ethylbenzene, benzene, toluene, and o-xylene (15, 19, 29, 45, 57). The current study indicates that during growth on styrene, only the bphABC genes are induced. The expression of the bph and etb genes is regulated by two-component regulatory systems in which BphS phosphorylates BphT in response to the presence of a range of aromatic compounds (51, 52). Phosphorylated BphT promotes transcription from at least five bph and etb promoters. RHA1 contains two copies of the BphST system, in which BphS and BphT share 92% and 97% amino acid identity, respectively. A third BphS paralog shares 65% sequence identity with the other two. All of the bphST genes are clustered with either the bph or etb catabolic genes. Although the BphS homologues have different effector specificities, their effects on the bph and etb promoters are similar (M. Fukuda, unpublished data). Thus, the mechanism responsible for the differential regulation of the bph and etb genes by styrene remains unclear and may involve another regulatory system.
The current study substantiates the previously proposed pathways for benzene and styrene, respectively, in rhodococci (38, 56). More specifically, our data indicate that benzene is transformed to catechol and is then subjected to ortho-cleavage and that styrene is transformed via direct oxidation of the aromatic ring to 3-vinylcatechol, which is transformed via meta-cleavage. The meta-cleavage activity present during growth on benzene presumably reflects the expression of the bph- and etb-encoded extradiol dioxygenases coproduced with BPDO and EBDO. The relative level of ortho-cleavage activity may be higher at physiologically relevant catechol concentrations, which are expected to be lower than those used in the assays. Regardless, our data distinguish benzene and styrene catabolism in rhodococci from that in pseudomonads and corynebacteria in two respects. First, the latter strains contain specific Ben and Sty pathways to catabolize these compounds (7, 18, 46, 55). Second, these strains initiate the degradation of styrene via the side chain. It is nevertheless unclear whether this mode of styrene catabolism is specific to rhodococci or actinomycetes.
Overall, this study expands our knowledge of the catabolic capabilities of Bph and Etb pathways in a model Rhodococcus strain. The simultaneous presence of multiple homologues of the Bph and Etb enzymes with overlapping, broad substrate preferences may facilitate the growth of the bacterium on mixtures of nonpolar aromatic compounds that are normally present in the environment. While this may lead to oxidative stress on the organism, it is not clear whether such stress is significantly higher than that which would occur were the cells to utilize a higher number of isozymes of narrower substrate specificity to grow on a mixture of compounds, each present at low levels. RHA1's response to the oxidative stress apparently associated with the broad-specificity ring-hydroxylating dioxygenases may contribute to the bacterium's potent PCB-degrading properties, as these pollutants cause uncoupling in BPDOs (17).
Supplementary Material
Acknowledgments
This work was supported by grants from Genome Canada and Genome BC. Robert Olafson, Derek Smith, and other members of the Proteomics Centre, University of Victoria, are thanked for their assistance with the MS. Gordon R. Stewart is thanked for his help with the GC-MS metabolite analysis. Matthew Myhre, Clinton Fernandes, and Michael McLeod assisted with bioinformatic analyses.
Footnotes
Published ahead of print on 26 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agullo, L., B. Camara, P. Martinez, V. Latorre, and M. Seeger. 2007. Response to (chloro)biphenyls of the polychlorobiphenyl-degrader Burkholderia xenovorans LB400 involves stress proteins also induced by heat shock and oxidative stress. FEMS Microbiol. Lett. 267167-175. [DOI] [PubMed] [Google Scholar]
- 2.Beltrametti, F., A. M. Marconi, G. Bestetti, C. Colombo, E. Galli, M. Ruzzi, and E. Zennaro. 1997. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl. Environ. Microbiol. 632232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull, T. J., J. Hermon-Taylor, I. Pavlik, F. El-Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 1462185-2197. [DOI] [PubMed] [Google Scholar]
- 4.Chavez, F. P., H. Lunsdorf, and C. A. Jerez. 2004. Growth of polychlorinated-biphenyl-degrading bacteria in the presence of biphenyl and chlorobiphenyls generates oxidative stress and massive accumulation of inorganic polyphosphate. Appl. Environ. Microbiol. 703064-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Crecy-Lagard, V., P. Servant-Moisson, J. Viala, C. Grandvalet, and P. Mazodier. 1999. Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol. Microbiol. 32505-517. [DOI] [PubMed] [Google Scholar]
- 6.Denef, V. J., M. A. Patrauchan, C. Florizone, J. Park, T. V. Tsoi, W. Verstraete, J. M. Tiedje, and L. D. Eltis. 2005. Growth substrate- and phase-specific expression of biphenyl, benzoate, and C1 metabolic pathways in Burkholderia xenovorans LB400. J. Bacteriol. 1877996-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fong, K. P., C. B. Goh, and H. M. Tan. 2000. The genes for benzene catabolism in Pseudomonas putida ML2 are flanked by two copies of the insertion element IS1489, forming a class-I-type catabolic transposon, Tn5542. Plasmid 43103-110. [DOI] [PubMed] [Google Scholar]
- 8.Fortin, P. D., A. T. Lo, M. A. Haro, S. R. Kaschabek, W. Reineke, and L. D. Eltis. 2005. Evolutionarily divergent extradiol dioxygenases possess higher specificities for polychlorinated biphenyl metabolites. J. Bacteriol. 187415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furusawa, Y., V. Nagarajan, M. Tanokura, E. Masai, M. Fukuda, and T. Senda. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 3421041-1052. [DOI] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Gomez-Gil, L., P. Kumar, D. Barriault, J. T. Bolin, M. Sylvestre, and L. D. Eltis. 2007. Characterization of biphenyl dioxygenase of Pandoraea pnomenusa B-356 as a potent polychlorinated biphenyl-degrading enzyme. J. Bacteriol. 1895705-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonçalves, E. R., H. Hara, D. Miyazawa, J. E. Davies, L. D. Eltis, and W. W. Mohn. 2006. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 726183-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurtler, V., B. C. Mayall, and R. Seviour. 2004. Can whole genome analysis refine the taxonomy of the genus Rhodococcus? FEMS Microbiol. Rev. 28377-403. [DOI] [PubMed] [Google Scholar]
- 14.Hartmans, S., M. J. van der Werf, and J. A. de Bont. 1990. Bacterial degradation of styrene involving a novel flavin adenine dinucleotide-dependent styrene monooxygenase. Appl. Environ. Microbiol. 561347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatta, T., T. Shimada, T. Yoshihara, A. Yamada, E. Masai, M. Fukuda, and H. Kiyohara. 1998. Meta-fission product hydrolases from a strong PCB degrader Rhodococcus sp. RHA1. J. Ferment. Bioeng. 85174-179. [Google Scholar]
- 16.Hauschild, J. E., E. Masai, K. Sugiyama, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1996. Identification of an alternative 2,3-dihydroxybiphenyl 1,2-dioxygenase in Rhodococcus sp. strain RHA1 and cloning of the gene. Appl. Environ. Microbiol. 622940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imbeault, N. Y. R., J. B. Powlowski, C. L. Colbert, J. T. Bolin, and L. D. Eltis. 2000. Steady-state kinetic characterization and crystallization of a polychlorinated biphenyl-transforming dioxygenase. J. Biol. Chem. 27512430-12437. [DOI] [PubMed] [Google Scholar]
- 18.Itoh, N., R. Morihama, J. Wang, K. Okada, and N. Mizuguchi. 1997. Purification and characterization of phenylacetaldehyde reductase from a styrene-assimilating Corynebacterium strain, ST-10. Appl. Environ. Microbiol. 633783-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki, T., K. Miyauchi, E. Masai, and M. Fukuda. 2006. Multiple-subunit genes of the aromatic-ring-hydroxylating dioxygenase play an active role in biphenyl and polychlorinated biphenyl degradation in Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 725396-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwasaki, T., H. Takeda, K. Miyauchi, T. Yamada, E. Masai, and M. Fukuda. 2007. Characterization of two biphenyl dioxygenases for biphenyl/PCB degradation in a PCB degrader, Rhodococcus sp. strain RHA1. Biosci. Biotechnol. Biochem. 71993-1002. [DOI] [PubMed] [Google Scholar]
- 21.Kim, D., J. C. Chae, G. J. Zylstra, Y. S. Kim, S. K. Kim, M. H. Nam, Y. M. Kim, and E. Kim. 2004. Identification of a novel dioxygenase involved in metabolism of o-xylene, toluene, and ethylbenzene by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 707086-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Kraus, J., and B. L. Geller. 2001. Cloning of genomic DNA of Lactococcus lactis that restores phage sensitivity to an unusual bacteriophage sk1-resistant mutant. Appl. Environ. Microbiol. 67791-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, K. 1999. Benzene-induced uncoupling of naphthalene dioxygenase activity and enzyme inactivation by production of hydrogen peroxide. J. Bacteriol. 1812719-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. E., J. S. Seo, Y. S. Keum, K. J. Lee, and Q. X. Li. 2007. Fluoranthene metabolism and associated proteins in Mycobacterium sp. JS14. Proteomics 72059-2069. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Z., H. Yang, Z. Huang, P. Zhou, and S. J. Liu. 2002. Degradation of aniline by newly isolated, extremely aniline-tolerant Delftia sp. AN3. Appl. Microbiol. Biotechnol. 58679-682. [DOI] [PubMed] [Google Scholar]
- 29.Masai, E., K. Sugiyama, N. Iwashita, S. Shimizu, J. E. Hauschild, T. Hatta, K. Kimbara, K. Yano, and M. Fukuda. 1997. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene 187141-149. [DOI] [PubMed] [Google Scholar]
- 30.Masai, E., A. Yamada, J. M. Healy, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 612079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLeod, M. P., R. L. Warren, W. W. Hsiao, N. Araki, M. Myhre, C. Fernandes, D. Miyazawa, W. Wong, A. L. Lillquist, D. Wang, M. Dosanjh, H. Hara, A. Petrescu, R. D. Morin, G. Yang, J. M. Stott, J. E. Schein, H. Shin, D. Smailus, A. S. Siddiqui, M. A. Marra, S. J. Jones, R. Holt, F. S. Brinkman, K. Miyauchi, M. Fukuda, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 10315582-15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno-Ruiz, E., M. J. Hernaez, O. Martinez-Perez, and E. Santero. 2003. Identification and functional characterization of Sphingomonas macrogolitabida strain TFA genes involved in the first two steps of the tetralin catabolic pathway. J. Bacteriol. 1852026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murshudov, G. N., A. I. Grebenko, J. A. Brannigan, A. A. Antson, V. V. Barynin, G. G. Dodson, Z. Dauter, K. S. Wilson, and W. R. Melik-Adamyan. 2002. The structures of Micrococcus lysodeikticus catalase, its ferryl intermediate (compound II) and NADPH complex. Acta Crystallogr. D 581972-1982. [DOI] [PubMed] [Google Scholar]
- 34.Na, K. S., A. Kuroda, N. Takiguchi, T. Ikeda, H. Ohtake, and J. Kato. 2005. Isolation and characterization of benzene-tolerant Rhodococcus opacus strains. J. Biosci. Bioeng. 99378-382. [DOI] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Navarro-Llorens, J. M., M. A. Patrauchan, G. R. Stewart, J. E. Davies, L. D. Eltis, and W. W. Mohn. 2005. Phenylacetate catabolism in Rhodococcus sp strain RHA1: a central pathway for degradation of aromatic compounds. J. Bacteriol. 1874497-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okamoto, S., and L. D. Eltis. 2007. Purification and characterization of a novel nitrile hydratase from Rhodococcus sp. RHA1. Mol. Microbiol. 65828-838. [DOI] [PubMed] [Google Scholar]
- 38.Paje, M. L., and I. Couperwhite. 1996. Benzene metabolism via the intradiol cleavage in a Rhodococcus sp. World J. Microbiol. Biotechnol. 12653-654. [DOI] [PubMed] [Google Scholar]
- 39.Parsonage, D., D. S. Youngblood, G. N. Sarma, Z. A. Wood, P. A. Karplus, and L. B. Poole. 2005. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry 4410583-10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patrauchan, M. A., C. Florizone, M. Dosanjh, W. W. Molm, J. Davies, and L. D. Eltis. 2005. Catabolism of benzoate and phthalate in Rhodococcus sp strain RHA1: redundancies and convergence. J. Bacteriol. 1874050-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pieper, D. H. 2005. Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol. 67170-191. [DOI] [PubMed] [Google Scholar]
- 42.Reference deleted.
- 43.Priefert, H., X. M. O'Brien, P. A. Lessard, A. F. Dexter, E. E. Choi, S. Tomic, G. Nagpal, J. J. Cho, M. Agosto, L. Yang, S. L. Treadway, L. Tamashiro, M. Wallace, and A. J. Sinskey. 2004. Indene bioconversion by a toluene inducible dioxygenase of Rhodococcus sp. I24. Appl. Microbiol. Biotechnol. 65168-176. [DOI] [PubMed] [Google Scholar]
- 44.Rustemov, S. A., L. A. Golovleva, R. M. Alieva, and B. P. Baskunov. 1992. New pathway of styrene oxidation by a Pseudomonas putida culture. Microbiology 611-5. [Google Scholar]
- 45.Sakai, M., K. Miyauchi, N. Kato, E. Masai, and M. Fukuda. 2003. 2-Hydroxypenta-2,4-dienoate metabolic pathway genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 69427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos, P. M., J. M. Blatny, I. Di Bartolo, S. Valla, and E. Zennaro. 2000. Physiological analysis of the expression of the styrene degradation gene cluster in Pseudomonas fluorescens ST. Appl. Environ. Microbiol. 661305-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seto, M., K. Kimbara, M. Shimura, T. Hatta, M. Fukuda, and K. Yano. 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 613353-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seto, M., N. Okita, K. Sugiyama, E. Masai, and M. Fukuda. 1996. Growth inhibition of Rhodococcus sp. strain RHA1 in the course of PCB transformation. Biotechnol. Lett. 181193-1198. [Google Scholar]
- 49.Sohng, J. K., T. J. Oh, J. J. Lee, and C. G. Kim. 1997. Identification of a gene cluster of biosynthetic genes of rubradirin substructures in S. achromogenes var. rubradiris NRRL3061. Mol. Cells 7674-681. [PubMed] [Google Scholar]
- 50.Stecker, C., A. Johann, C. Herzberg, B. Averhoff, and G. Gottschalk. 2003. Complete nucleotide sequence and genetic organization of the 210-kilobase linear plasmid of Rhodococcus erythropolis BD2. J. Bacteriol. 1855269-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda, H., N. Hara, M. Sakai, A. Yamada, K. Miyauchi, E. Masai, and M. Fukuda. 2004. Biphenyl-inducible promoters in a polychlorinated biphenyl-degrading bacterium, Rhodococcus sp. RHA1. Biosci. Biotechnol. Biochem. 681249-1258. [DOI] [PubMed] [Google Scholar]
- 52.Takeda, H., A. Yamada, K. Miyauchi, E. Masai, and M. Fukuda. 2004. Characterization of transcriptional regulatory genes for biphenyl degradation in Rhodococcus sp. strain RHA1. J. Bacteriol. 1862134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Geize, R., and L. Dijkhuizen. 2004. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7255-261. [DOI] [PubMed] [Google Scholar]
- 54.Van der Geize, R., K. Yam, T. Heuser, M. H. Wilbrink, H. Hara, M. C. Anderton, E. Sim, L. Dijkhuizen, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. USA 1041947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velasco, A., S. Alonso, J. L. Garcia, J. Perera, and E. Diaz. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J. Bacteriol. 1801063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warhurst, A. M., K. F. Clarke, R. A. Hill, R. A. Holt, and C. A. Fewson. 1994. Metabolism of styrene by Rhodococcus rhodochrous NCIMB 13259. Appl. Environ. Microbiol. 601137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 642006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamada, T., Y. Shimomura, Y. Hiraoka, and K. Kimbara. 2006. Oxidative stress by biphenyl metabolites induces inhibition of bacterial cell separation. Appl. Microbiol. Biotechnol. 73452-457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.