Abstract
The Bacillus subtilis SpoVE integral membrane protein is essential for the heat resistance of spores, probably because of its involvement in spore peptidoglycan synthesis. We found that an SpoVE-yellow fluorescent protein (YFP) fusion protein becomes localized to the forespore during the earliest stages of engulfment, and this pattern is maintained throughout sporulation. SpoVE belongs to a well-conserved family of proteins that includes the FtsW and RodA proteins of B. subtilis. These proteins are involved in bacterial shape determination, although their function is not known. FtsW is necessary for the formation of the asymmetric septum in sporulation, and we found that an FtsW-YFP fusion localized to this structure prior to the initiation of engulfment in a nonoverlapping pattern with SpoVE-cyan fluorescent protein. Since FtsW and RodA are essential for normal growth, it has not been possible to identify loss-of-function mutations that would greatly facilitate analysis of their function. We took advantage of the fact that SpoVE is not required for growth to obtain point mutations in SpoVE that block the development of spore heat resistance but that allow normal protein expression and targeting to the forespore. These mutant proteins will be invaluable tools for future experiments aimed at elucidating the function of members of the SEDS (“shape, elongation, division, and sporulation”) family of proteins.
Bacterial shape is determined by an extracellular structure composed of peptidoglycan (PG), a rigid polymer of repeated subunits of a disaccharide peptide monomer. This giant macromolecule is found on the outside of the cytoplasmic membrane of nearly all eubacteria. A series of essential and highly conserved enzymes convert UDP-GlcNAc to lipid II, the UDP-muramyl pentapeptide (47). To form mature PG, lipid II is translocated across the cytoplasmic membrane via an uncharacterized mechanism and is added to the preexisting cell wall through transpeptidation and transglycosylation reactions mediated by members of the PBP (penicillin binding protein) family of proteins. Little is known about the mechanism of lipid II transport across the cytoplasmic membrane other than that it is probably dependent on a protein(s) since it does not occur spontaneously across lipid bilayers (46).
The integral membrane proteins RodA and FtsW are members of what has been termed the SEDS (“shape, elongation, division, and sporulation”) family of integral membrane proteins (16, 18) that are present in all cell wall-containing bacteria. Several lines of evidence are consistent with the participation of RodA and FtsW in the translocation of lipid II during cell elongation and cell division, respectively (17, 19), although this hypothesis has not been subjected to a direct test. For instance, depletion of RodA leads to a block in lateral cell growth (16), although RodA is not strictly essential for cell viability. Null mutants exhibit slow growth and small cell diameters and are viable in minimal medium (7). Also, temperature-sensitive Escherichia coli ftsW mutations lead to blocks at both early and late stages of cell division (23, 24), suggesting that FtsW acts during both initiation and septum maturation (3).
RodA and FtsW are likely to function as part of essential elongation and division complexes for PG synthesis (17). Each complex is thought to include one protein from the SEDS family and one PBP (although several PBPs may associate with a single SEDS protein) (19). Mutations in E. coli ftsI (encoding PBP3) that lead to a reduced ability to divide can be suppressed by rodA mutations that, by themselves, interfere with normal cell growth (1). FtsW from Mycobacterium tuberculosis interacts with PBP3 (6), and E. coli FtsW interacts with PBP2 in two different bacterial two-hybrid assays (21). The formation of PG on isolated E. coli membranes requires the presence of high levels of both RodA and PBP2 proteins (19). These observations are consistent with the proposed role of the SEDS proteins in lipid II translocation. However, the essential nature of FtsW and RodA makes testing this hypothesized function difficult. Thus, either temperature-sensitive (24) or depletion (3, 16) strains must be used to evaluate the properties of FtsW or RodA. One issue complicating the interpretation of experiments based on such strains is that a reduced rate of lipid II translocation could be due to the absence of the proteins or to a general loss in viability.
Fortunately, a third SEDS family member functions during the nonessential process of sporulation in Bacillus subtilis (18, 20). Bacterial endospores can survive extremes of heat and desiccation, largely because of the presence of the spore cortex. This structure is composed of a form of PG that is similar, although not identical, to vegetative PG, with fewer peptide side chains, a concomitant reduction in the cross-linking of the glycan strands, and the presence of muramic δ-lactam residues (52-54). Sporulation in B. subtilis begins with an asymmetric division that creates a smaller forespore and a larger mother cell. During the process of engulfment, the forespore compartment becomes completely enclosed in the mother cell, and it is surrounded by two membranes, possibly separated by a thin layer of PG. The assembly of the spore PG then occurs in the space between the two membranes, resulting from the action of genes expressed in the mother cell compartment (36). spoVE, which during sporulation is expressed under the control of the mother cell-specific transcription factor σE (45), is not essential for vegetative growth, yet it is absolutely required for the production of cortex PG. Thus, during sporulation, ΔspoVE mutants fail to form a cortex (15, 34, 50) and they accumulate cytoplasmic PG precursors (50), suggesting a defect at an early step in PG polymerization and consistent with the view that the SEDS proteins may function in lipid-linked precursor translocation. SpoVE may be part of a third putative PG biosynthetic complex of B. subtilis and other endospore-forming bacteria which includes a sporulation-specific, σE-controlled PBP, called SpoVD, required for synthesis of the cortex (4). It is interesting to note that both spoVE (which occupies the position of ftsW) and spoVD are located in the dcw cluster of spore-forming bacteria, perhaps because the coordinated expression of dcw genes is important for the synthesis of the spore cortex (38).
The large number of mother cell-expressed membrane proteins that localize to the outer forespore membrane (48, 49) suggests that SpoVE might localize in a similar fashion. Some mother cell-expressed membrane proteins can be found distributed in all accessible membranes, particularly in the absence of appropriate “anchoring” proteins (39). Since spore cortex synthesis occurs underneath the outer forespore membrane, it is thus reasonable to expect that SpoVE would localize to this membrane.
Finally, since the spore cortex layer is essential to providing the heat resistance of spores, the loss of heat resistance is a convenient assay for SpoVE function (28). Since heat resistance can be accurately measured over several orders of magnitude, SpoVE mutants can be quantitatively differentiated. Therefore, the ability to examine the phenotypes of loss-of-function spoVE mutants will greatly facilitate studies of the role of SpoVE in cortex biogenesis and is also likely to provide insight into the function of the SEDS family of proteins.
MATERIALS AND METHODS
Standard procedures were used to prepare and handle recombinant DNA and to transform E. coli cells. The B. subtilis strains were derivatives of PY79 unless noted otherwise (55), and the E. coli strains used were DH5, TG1, or CC118(DE3)pLysS. B. subtilis was transformed using competent cells made by the two-step method (14). Sporulation for microscopy used either Difco sporulation medium (DSM) (14) or CH medium for growth and A+B medium for resuspension (44). The heat resistance of spores was assayed following sporulation by exhaustion in DSM medium in 2-ml cultures for 24 h. Serial dilutions were plated before and after the cells were heated to 80°C or 55°C for 20 min. The numbers of CFU were compared from before and after heat treatment.
Strain construction.
Strains are listed in Table 1. For details of construction, see Table S1 in the supplemental material.
TABLE 1.
Bacterial strains
| Strain | Genotype | Origin/reference |
|---|---|---|
| MB24 | trpC2 metC3 | Laboratory stock |
| SL666 | spoVE85 | 33 |
| AH3561 | spoVE85 amyE::spoVE-yfp | This work |
| AH3700 | spoVE85 amyE::spoVEI58N-yfp | This work |
| AH3701 | spoVE85 amyE::spoVEC82R-yfp | This work |
| AH3702 | spoVE85 amyE::spoVES103N-yfp | This work |
| AH3703 | spoVE85 amyE::spoVEE126G-yfp | This work |
| AH3704 | spoVE85 amyE::spoVEC160R-yfp | This work |
| AH3705 | spoVE85 amyE::spoVEG292R-yfp | This work |
| AH3706 | spoVE85 amyE::spoVEG355D-yfp | This work |
| PY79 | wild type | Laboratory stock |
| JDB223 | ftsW::ftsW-gfpΩspc | This work |
| JDB646 | spoVE::neo | P. Eichenberger |
| JDB1376 | spoVE::neo amyE::PspoVE-spoVE-cfpΩcat | This work |
| JDB1407 | amyE::PspoVE-ftsW-gfpΩcat | This work |
| JDB1483 | amyE::PspoVE-PftsW(102)-spoVE-gfpΩcat | This work |
| JDB1678 | spoVE::neo ftsW-yfpΩspc amyE::PspoVE-spoVE-cfpΩcat | This work |
| JDB1752 | ΔspoVE::tet | This work |
| JDB1755 | spoVE::neo amyE::PspoVE-ftsW-gfpΩcm | This work |
| JDB1813 | ΔspoVE::tet amyE::PspoVE-spoVE-gfpΩcm | This work |
| JDB1835 | ΔspoVE::tet amyE::PspoVE-spoVE-gfpΩspcΩcat | This work |
| JDB1845 | ΔspoVE::tet amyE::PspoVE-spoVEG335A-gfpΩspcΩcat | This work |
| JDB1846 | ΔspoVE::tet amyE::PspoVE-spoVES341A-gfpΩspcΩcat | This work |
| JDB1847 | ΔspoVE::tet amyE::PspoVE-spoVEG343A-gfpΩspcΩcat | This work |
| JDB1848 | ΔspoVE::tet amyE::PspoVE-spoVEN322A-gfpΩspcΩcat | This work |
| JDB1849 | ΔspoVE::tet amyE::PspoVE-spoVEE271A-gfpΩspcΩcat | This work |
| JDB1850 | ΔspoVE::tet amyE::PspoVE-spoVEW69A-gfpΩcat | This work |
| JDB1851 | ΔspoVE::tet amyE::PspoVE-spoVEK76A-gfpΩcat | This work |
| JDB1852 | ΔspoVE::tet amyE::PspoVE-spoVET173A-gfpΩcat | This work |
| JDB1853 | ΔspoVE::tet amyE::PspoVE-spoVE-gfpΩcat | This work |
| JDB1854 | ΔspoVE::tet amyE::PspoVE-spoVEI58N-yfpΩcat | This work |
| JDB1855 | ΔspoVE::tet amyE::PspoVE-spoVEC82R-yfpΩcat | This work |
| JDB1856 | ΔspoVE::tet amyE::PspoVE-spoVES103N-yfpΩcat | This work |
| JDB1857 | ΔspoVE::tet amyE::PspoVE-spoVEE116G-yfpΩcat | This work |
| JDB1858 | ΔspoVE::tet amyE::PspoVE-spoVEC160R-yfpΩcat | This work |
| JDB1859 | ΔspoVE::tet amyE::PspoVE-spoVEG292R-yfpΩcat | This work |
| JDB1860 | ΔspoVE::tet amyE::PspoVE-spoVEG355D-yfpΩcat | This work |
| JDB1926 | ΔspoVE::tet amyE::PspoVE-spoVEG335A-gfpΩcat | This work |
| JDB1927 | ΔspoVE::tet amyE::PspoVE-spoVES341A-gfpΩcat | This work |
| JDB1928 | ΔspoVE::tet amyE::PspoVE-spoVEG343A-gfpΩcat | This work |
| JDB1929 | ΔspoVE::tet amyE::PspoVE-spoVEN322A-gfpΩcat | This work |
| JDB1930 | ΔspoVE::tet amyE::PspoVE-spoVEE271A-gfpΩcat | This work |
| JDB1933 | ΔspoVE::tet amyE::PspoVE-spoVE-gfpΩcat | This work |
Plasmid construction.
Table S2 in the supplemental material lists plasmids and details of their construction. The oligonucleotides used are listed in Table S3 in the supplemental material.
Fluorescence microscopy.
Samples of DSM cultures for visualization of fluorescence were prepared as previously described (37). For visualization of DNA and membranes, 4,6-diamidino-2-phenylindole dihydrochloride (DAPI; 1 mg/ml stock) and membrane dye FM4-64 (100 mg/ml stock; Molecular Probes) were added to cells at a final concentration of 10 μg/ml prior to mounting the cells on agarose-coated slides. All samples were observed with a 63× objective lens. Phase contrast or fluorescence images were acquired with a Leica DMRA2 microscope-coupled CoolSNAP HQ photometrics camera (Roper Scientific, Tucson, AZ), recorded, and processed for publication using Adobe Photoshop, version 6.0.
For cultures in resuspension medium, 100 μl of sporulating cells were taken at designated times after resuspension. To each sample, 0.5 μl of FM4-64 (100 mg/ml) was added just before the cells were collected by centrifugation. The pellet was resuspended in 10 μl phosphate-buffered saline and added to a poly-l-lysine-pretreated coverslip. All microscopy was performed on a Nikon Eclipse 90i with a 100× objective using phase contrast and captured by using a Hamamatsu Orca-ER camera using Nikon Elements BR software. The exposure was 800 ms for all pictures taken. For the SpoVE-cyan fluorescent protein (CFP), FtsW-yellow fluorescent protein (YFP) time course, the strain (JDB1678) was sporulated by resuspension and, at the designated times, samples were taken and prepared as mentioned above. Line scans of fluorescence on images were performed using Nikon BR Elements software.
Time-lapse movies.
Strain JDB1835 was grown in CH medium to an optical density at 600 nm (OD600) of 0.6, resuspended in an equal volume of A+B resuspension medium (44) for 1.5 h, and then put on a 1.5% (wt/vol) low-melting-point agarose (Sigma) pad made from fresh A+B and cut into an ∼1-cm2 square. The pads were sealed in a glass-bottomed dish (Wilco Wells, Netherland) with bacteria facing down and were imaged with an epifluorescence inverted Olympus BX-61 microscope every 7 min using a xenon lamp and a green fluorescent protein (GFP) filter cube. Under these conditions, the bacteria grew an average of three cell divisions and then sporulated.
Generation of spoVE mutants with the mutations I58N, C82R, S103N, E116G, C160R, G292R, and G355D.
Plasmid pTC30 (lab stock) was constructed by inserting a fragment containing PspoVE-His6-spoVE between the HindIII and EcoRI sites of the pMLK83 shuttle vector (22). Plasmid pTC30 was used to transform the mutator strain E. coli XL1-Red (Stratagene), and transformants were selected on LB plates containing ampicillin (100 μg/ml). The transformants were then pooled and grown overnight at 37°C in LB broth containing ampicillin (100 μg/ml). Randomly mutated plasmid DNA extracted from this culture was then used to transform B. subtilis strain SL666 (33) with selection for neomycin resistance. Mutations leading to an Spo− phenotype were identified following the plating of transformants on DSM medium. Sporulation-deficient transformants were then streaked onto tryptose blood agar base plates containing 1% starch to confirm that the plasmid had integrated at the amyE locus. From a total of 34 mutants obtained, only 7 showed missense mutations within the spoVE coding sequence and 1 in the ribosome binding site. The remaining mutations were insertions and deletions and were discarded.
Generation of spoVE mutants with the mutations E271A, N322A, G335A, S341A, G343A, W69A, K76A, and T173A.
We introduced alanine substitutions at specific positions in the SpoVE coding sequence by using a QuikChange site-directed mutagenesis kit (Stratagene). We used plasmid pCB41 carrying amyE-flanking sequences; PspoVE-spoVE-gfp as the template; and oligonucleotides oJD104 (G335A), oJD106 (S341A), ojd108 (G343A), and ojd142 (N322A) to generate plasmids carrying the following mutations: pAF150, spoVE(E271A); pAF155, spoVE(N322A); pAF147, spoVE(G335A); pAF148, spoVE(S341A); and pAF149, spoVE(G343A). We used pAF032 as the template and oligonucleotides oAF302, oAF304, and oAF306, respectively, to generate plasmids carrying the following mutations: pAFM302, spoVE(W69A); pAFM304, spoVE(K76A); and pAFM308, spoVE(T173A).
Construction of spoVE′-phoA fusions and assay for AP activity.
The phoA gene was excised from pPHO7 (13) with XhoI and BamHI and inserted between the SalI and BamHI sites of pLITMUS29, generating pAH310. phoA was then released from pAH310 with SpeI and StuI and inserted between the SpeI and HincII sites of pBluescriptIISK(+) to yield pAH312. A series of 10 3′ truncations of spoVE were then constructed by PCR using chromosomal DNA from MB24 as the template, spoVE-10D as the forward primer, and various reverse primers chosen so that PhoA is fused after each of the transmembrane segments, predicted with the TMpred, TopPred, and TM hidden Markov model (HMM) programs. All PCR products were digested with PstI and BamHI, BglII, or BclI and introduced into PstI- and BamHI-cut pAH312. The resulting plasmids, which were sequenced to ensure that no undesired mutation was introduced, were used to transform E. coli CC118(DE3)pLysS with selection for ampicillin. PhoA activity was detected as blue colonies on LB agar plates containing 5-bromo-4-chloro-3-indolyl-phosphate (XP), ampicillin, and chloramphenicol at concentrations of 40 μg/ml, 100 μg/ml, and 30 μg/ml, respectively. The activity of alkaline phosphatase (AP) was also assayed in liquid cultures by measuring the rate of p-nitrophenyl phosphate hydrolysis. The cells were grown at 37°C in LB to an OD600 of 0.4 to 0.6, at which time isopropyl-β-d-thiogalactopyranoside (to 0.5 mM) was added to the cultures. Growth continued for 2.5 h, after which the cells were harvested and assayed for activity (29).
Construction of spoVE′-lacZ fusions and assay for β-galactosidase activity.
Fusions of the SpoVE amino-terminal region to LacZ were constructed by amplification of the spoVE′ fragments present in the pAH312 derivatives described above by using primers spoVE1D and spoVE-lacZR. The resulting fragments were digested with EcoRI and BamHI and inserted between the EcoRI and BamHI sites of pNM480 (31a). The resulting plasmids were used to transform E. coli CC118(DE3)pLysS with selection for ampicillin. β-Galactosidase activity was detected as blue colonies on LB agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), ampicillin, and chloramphenicol at concentrations of 80 μg/ml, 100 μg/ml, and 30 μg/ml, respectively. β-Galactosidase activity was assayed on liquid cultures by measuring the rate of hydrolysis of o-nitrophenyl-β-galactopyranoside as described previously (31).
Immunoblot analysis.
For DSM cultures, samples (10 ml) were collected and lysed as described before (41). Samples (30 μg) of total protein were resolved on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and subjected to immunoblot analysis (41). Anti-GFP antibody (laboratory stock) was used at a dilution of 1:1,000. A rabbit secondary antibody conjugated to horseradish peroxidase (Sigma) was used at a dilution of 1:10,000. The immunoblots were developed with ECL plus (GE Health).
For cultures in resuspension medium, at each time point, the OD600 was taken. The volumes of the samples were normalized to yield an OD600 of 0.50. After being pelleted, cells were resuspended and protoplasted in 100 μl of Spizizen's minimal medium (SMM) with 1 mg/ml lysozyme for 5 min at room temperature. Protoplasts were then lysed after collection by being resuspended in 100 μl of 1× sample buffer and solubilized for 30 min at 37°C. An amount of 20 μl of each sample was loaded on a 13% polyacrylamide gel for SDS-PAGE. The gels were transferred to Biotrace-NT (Pall Life Sciences) and probed using anti-GFP rabbit serum (1:25,000) (generous gift of Howard Shuman), anti-rabbit-horseradish peroxidase (1:25,000; Pierce), and then ECL plus (GE Health).
RESULTS
Localization of SpoVE.
Like all other genes known to be required for spore cortex formation (50), spoVE is under the control of σE, a mother cell-specific transcription factor (45). Therefore, SpoVE is likely to be found at the outer forespore membrane, surrounding the developing spore, and facing the mother cell cytoplasm. We examined this possibility by constructing a C-terminal fusion of SpoVE with YFP and observing its pattern of localization in sporulating cells. This fusion, expressed from the spoVE promoter (PspoVE-spoVE-yfp) and designed for recombination at the ectopic amyE chromosomal locus, was fully functional because, when introduced into a strain carrying an spoVE85 mutation (33), it restored wild-type levels of spore heat resistance. Consistent with its dependence on σE, SpoVE-YFP is not observed in cells upon entry to sporulation (Fig. 1A, panels b and c). Two hours after the onset of the process, however, some cells have entered sporulation, as indicated by the formation of asymmetric septa (Fig. 1A, panel e), and at 3 h, most cells contain either polar septa or engulfing forespores (Fig. 1A, panel h). SpoVE-YFP fluorescence was first detected at 2 h, closely associated with septa or engulfing forespores (Fig. 1A, panel f). Strikingly, at 2 h or 3 h, YFP fluorescence was rarely found associated with straight septa but, rather, was typically found associated with curved septa (Fig. 1A, panels f and i). Finally, at 6 h, nearly all of the cells appear to have completed the engulfment process (Fig. 1A, panel k) and, again, a YFP signal can be observed around those forespores (Fig. 1A, panel l). Thus, SpoVE localizes in a pattern that closely matches the membrane dynamics of the engulfing forespores. Further, the pattern at later time points (post-3 h) is consistent with the role of SpoVE in spore cortex synthesis, a late (postengulfment) event in sporulation (50). The examination of SpoVE-YFP expression by immunoblotting analysis of samples taken at equivalent time points demonstrated that the protein appeared at 3 h, consistent with the results of microscopy (data not shown).
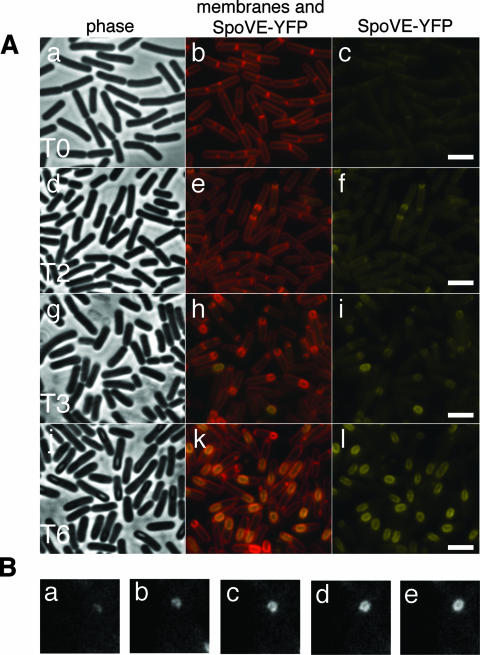
FIG. 1.
SpoVE-YFP localization throughout sporulation. (A) Strain AH3561 (spoVE85 amyE::spoVE-yfp) was induced to sporulate in DSM medium, and samples were collected at the onset (0 h [T0]) and throughout sporulation and labeled with vital stain FM4-64 for visualization of membranes by fluorescence microscopy. The first column shows phase contrast images and the second column depicts the colocalization of the membranes (red) and the YFP signal (yellow). The third column shows the localization of SpoVE-YFP alone (yellow). Scale bars (white) represent 1 μm. (B) Strain JDB1813 carrying an SpoVE-GFP fusion was visualized by time-lapse microscopy every 7 min as it proceeded through sporulation. Shown is a single cell with images taken at five consecutive time points (a, t = 0; b, t = 7 min; c, t = 14 min; d, t = 21 min; e, t = 28 min).
Since these experiments were performed under time-course conditions, it was not possible to follow SpoVE localization in a single cell. We therefore employed time-lapse microscopy to visualize the distribution of a functional SpoVE-GFP fusion every 7 min in a single cell as it proceeded through sporulation (Fig. 1B). Initially, SpoVE-GFP was observed as a single, slightly curved band (Fig. 1B, panel a), consistent with our time course observations (Fig. 1A, panel f). As the cell continued to undergo sporulation, the SpoVE-GFP signal followed the expected pattern of a protein associated with an engulfing forespore (Fig. 1B, panels b, c, and d), and at the final time point (Fig. 1B, panel e), the signal surrounded the (presumably) engulfed forespore. This experiment confirms the time-course data in that SpoVE appears absent from polar septa prior to the initiation of engulfment (when the septa become curved) and in that its distribution thereafter closely mimics the engulfing outer forespore membrane.
Localization of SpoVE and FtsW in vegetative cells.
The apparent absence of SpoVE from straight polar septa was intriguing because another SEDS protein, E. coli FtsW, associates with division septa (3, 23) and because SpoVE and its paralog B. subtilis FtsW (also expected to localize at division sites) are highly similar along their entire primary structure. This prompted us to examine both the localization of B. subtilis FtsW and whether the production of SpoVE under vegetative conditions would result in its localization to division sites at midcell. We first examined the localization of a functional FtsW-GFP fusion integrated at the ftsW locus of B. subtilis during growth and sporulation. We observed this strain during vegetative growth and found that FtsW-GFP localized, as expected, to midcell (Fig. 2A, panels a and b). In contrast, no GFP signal was observed at a time during sporulation (3 h) (Fig. 2A, panels c and d) when most cells exhibit an SpoVE-GFP signal at the forespore.
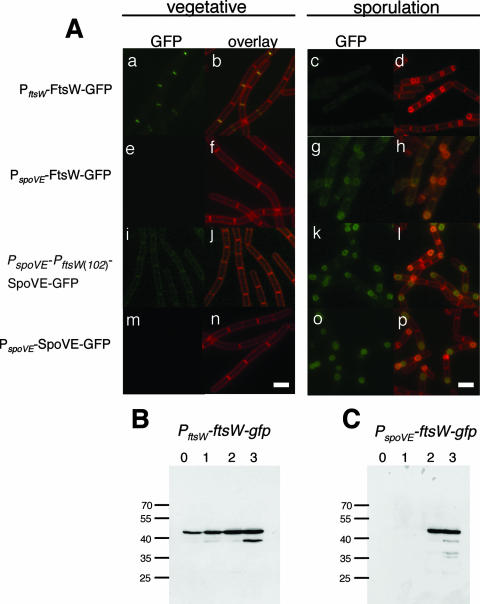
FIG. 2.
Localization of SpoVE-GFP and FtsW-GFP. (A) Strains JDB223 (ftsW::ftsW-gfp; a to d), JDB1755 (spoVE::neo amyE::PspoVE-ftsW-gfp; e to h), JDB1483 (amyE::PspoVE-PftsW(102)-spoVE-gfp; i to l), and JDB1835 (ΔspoVE::tet amyE::PspoVE-spoVE-gfp; m to p) were grown in CH medium and samples were taken before the cells were resuspended at an OD600 of 0.5 (“vegetative”; a, b, e, f, i, j, m, n) or at 3 h of sporulation in A+B medium (“sporulation” [T3]; c, d, g, h, k, l, o, p). The samples were observed by fluorescence microscopy. Cells were labeled with FM4-64 to visualize membranes. The first and third columns show GFP signal; the second and fourth columns show overlay of GFP and membrane signals. Scale bars (white) represent 1 μm. (B, C) Strains JDB223 (ftsW::ftsW-gfp) and JDB1755 (spoVE::neo amyE::PspoVE-ftsW-gfp) were induced to sporulate by resuspension, and samples were collected at the onset of sporulation (0 h) and subsequently at hourly intervals as indicated above the panels. Samples were resolved using a 12% SDS-PAGE gel and subjected to immunoblot analysis with an anti-GFP antibody. Molecular mass markers, in kDa, are on the left of the panels.
The absence of an FtsW-GFP signal in sporulating cells did not reflect an absence of FtsW-GFP protein, because immunoblotting using anti-GFP antibodies revealed that FtsW-GFP expression under the control of PftsW was relatively constant during sporulation (Fig. 2B). When FtsW-GFP was placed under the control of the heterologous promoter PspoVE, no fluorescent signal was seen during vegetative growth (Fig. 2A, panels e and f), consistent with the sporulation-specific activity of the promoter. However, during sporulation, a GFP signal was observed throughout all mother cell membranes (Fig. 2A, panels g and h), in contrast with the enrichment of SpoVE that we observed at the outer forespore membrane (Fig. 2A, panels o and p). The FtsW-GFP signal and the comparatively high level of FtsW-GFP protein revealed by immunoblotting (Fig. 2C; compare with Fig. 2B) indicate that the protein was present in sporulating cells but was not targeted to the forespore.
When we placed SpoVE-GFP under the control of PftsW, the protein lacked discrete septal localization; instead, we observed a relatively faint signal in all cell membranes (data not shown). To confirm that this pattern was not the result of a nonfunctional fusion protein, we inserted PspoVE upstream of PftsW such that SpoVE-GFP was under the control of both promoters. During vegetative growth, this construct again resulted in no septal localization (Fig. 2A, panels i and j), although a membrane-associated fluorescent signal that was absent in cells expressing SpoVE-GFP under the control of its endogenous PspoVE promoter was observed (Fig. 2A, panels m and n). When the PspoVE-PftsW-spoVE-gfp strain was sporulated, a GFP signal was observed at the forespore (Fig. 2A, panels k and l), similar to that seen with PspoVE-spoVE-GFP strain (Fig. 2A, panels o and p), indicating that the SpoVE construct was functional. Thus, SpoVE-GFP appears to be excluded from both vegetative and polar septa, while FtsW-GFP fails to localize to the engulfing membranes during sporulation. These differences in the subcellular distribution of SpoVE-GFP and FtsW-GFP likely reflect the activity of different targeting signals contained in their primary sequence.
FtsW and SpoVE have nonoverlapping patterns of localization.
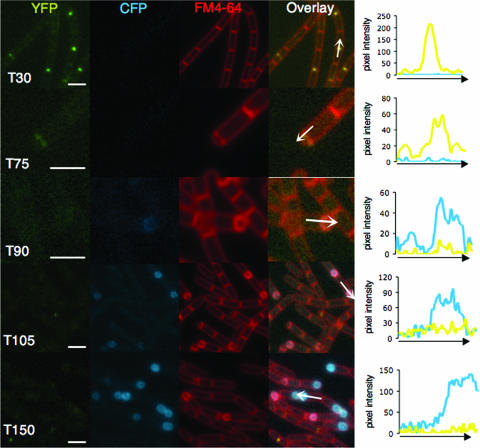
The likely involvement of FtsW in polar division suggests that the protein localizes to sites of asymmetric division. However, this localization must be transient since FtsW-GFP localization to the forespore membranes was not detected by h 3 of sporulation when asymmetric division is complete in most cells and the process of engulfment commences (see above). This suggests that FtsW is no longer detected in those cells initiating the engulfment process and in which the localization of SpoVE is first detected. To investigate this possibility, we examined the cellular distribution of FtsW and SpoVE simultaneously in sporulating cells. We constructed a strain expressing both a complementing FtsW-YFP fusion under the control of PftsW and a complementing SpoVE-CFP fusion under the control of PspoVE. We quantified the fluorescence in >20 cells at each time point by measuring the pixel intensity for both the CFP and YFP channels along a line drawn lengthwise along the cell. The traces chosen represent the results for a typical cell. At 30 min following resuspension, some cells contained a discrete focus of the YFP signal at midcell, consistent with the completion of the last round of division, but lacked a CFP signal, consistent with the expected lack of σE expression at this time (Fig. 3). At 75 min, cells began to contain the asymmetric septa associated with a YFP signal, but there was still no CFP signal. Starting at 90 min following resuspension, some of these septa started to curve and were now associated with a CFP signal, but there was little YFP signal. This difference between the CFP and YFP signals became even more pronounced at later time points (Fig. 3, 105 and 150 min). Thus, FtsW and SpoVE have nonoverlapping patterns of localization at the sporulation septum, with FtsW-YFP present at the time of asymmetric septation but disappearing at or before the time of the initiation of engulfment, followed by the observation of SpoVE-CFP only at curving asymmetric septa. This morphological event is coupled to the activation of σE (35), consistent with the known dependence of spoVE expression on σE.
FIG. 3.
Fluorescence microscopy of FtsW and SpoVE. Strain JDB1678 (ftsW-yfp spoVE::neo amyE::spoVE-cfp) was sporulated by resuspension. Time (T) is in minutes. Cells were labeled with FM4-64 to visualize membranes. Consecutive images were acquired using YFP, CFP, and FM4-64 filter sets. The overlay was generated from the YFP, CFP, and FM4-64 images by using the Nikon NIS program. Measurement of pixel intensities was performed by using the Line Scan function of the Nikon NIS program. The values for the CFP and YFP channels are plotted as a function of distance along the arrows shown in the overlays. Note that different scales were used for different samples; scale bars (white) represent 1 μm.
The loss of FtsW-GFP signal upon the initiation of membrane curvature could result from protein degradation or, alternatively, from the dilution of the GFP signal as the protein becomes delocalized from the septal membrane and enters the much larger mother cell membrane. The FtsW-GFP levels, as determined by immunoblotting cell lysates from sporulating strains carrying FtsW-GFP alone, were not different at times early in sporulation than at later times (Fig. 2B). We confirmed that the absence of a GFP signal did not reflect a substantial decrease in overall cellular FtsW levels because immunoblotting using anti-GFP antibodies revealed that FtsW-GFP expression under the control of PftsW was relatively constant (albeit lower) during sporulation compared with the expression of FtsW under PspoVE control (Fig. 2B and C). Thus, FtsW is likely to become delocalized rather than degraded.
Generation of SpoVE point mutants.
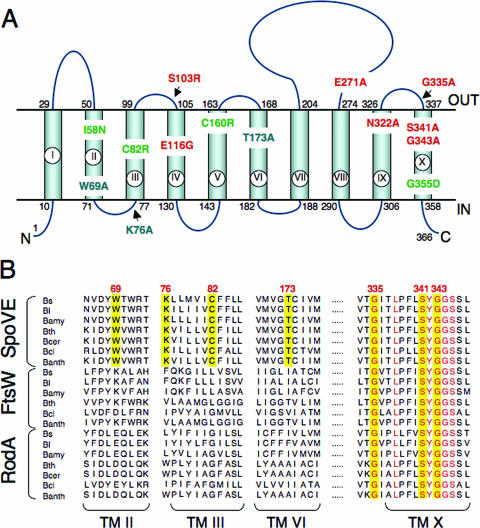
Unlike FtsW and RodA, SpoVE is not essential for normal growth; a strain carrying an spoVE mutation appears completely normal for vegetative growth but results in the production of heat-sensitive spores. We took advantage of this characteristic to identify mutations in spoVE that block function using two strategies. First, we randomly mutagenized plasmids containing an spoVE gene by passage through an E. coli mutator strain. Pooled plasmids from the mutagenesis were then transformed into a strain that carried an spoVE::kan insertion mutation. The plasmid contained amyE sequences flanking the spoVE gene, favoring integration to (and usually disrupting) the amyE locus. Amylase− transformants were screened for the Spo− phenotype on DSM medium, and in this manner we identified seven spoVE missense mutations, spoVE(I58N), spoVE(C82R), spoVE(S103N), spoVE(E116G), spoVE(C160R), spoVE(G292R), and spoVE(G355D) (Fig. 4A; see Fig. S1 in the supplemental material for conservation of these positions among SEDS orthologs). Note that spoVE(G355D) is the same mutation found in strain SL666 (Table 1), bearing the spoVE85 classical allele (33).
FIG. 4.
Topology model for B. subtilis SpoVE and localization of point mutations. (A) B. subtilis SpoVE membrane topology and the location of SpoVE mutations. The beginning and end of each TM segment (TM I to X) is indicated by the residue number. The model is derived from TopPredII predictions (51) and LacZ and PhoA fusion protein analysis. SpoVE mutations are color coded according to how SpoVE-GFP or SpoVE-YFP mutant proteins carrying the respective mutations accumulate and localize during sporulation: green, have reduced accumulation; red, accumulate and localize; and blue, accumulate but are mislocalized. (B) Partial alignment of SEDS proteins (SpoVE, FtsW, and RodA) from selected members of the genus Bacillus. The alignment shows parts of TM II, III, VI, and X (as represented in panel A for the B. subtilis SpoVE protein) and was obtained using CLUSTAL-W (www.ebi.ac.uk). SpoVE mutations W69A, K76, C82R, and T173A are in residues conserved only among predicted SpoVE proteins, whereas mutations G335A, S341A, and G343A are in conserved residues among the Bacillus SEDS proteins. Residues in the positions corresponding to all six mutations are shown against a yellow background. Other conserved residues in TM X are shown in red. Bs, B. subtilis; Bl, B. licheniformis; Bamy, B. amyloliquefaciens; Bth, B. thuringiensis; Bcer, B. cereus; Bcl, B. clausii; Banth, B. anthracis.
As a second, complementary strategy of spoVE mutagenesis, we used Pfam (10) to generate an HMM (40) of SEDS proteins from diverse bacterial species (∼500) that identified well-conserved residues (Fig. 4B). We then introduced alanine substitutions into several of these positions in B. subtilis SpoVE by site-directed mutagenesis of a plasmid carrying an SpoVE-GFP fusion that was subsequently integrated into the chromosome at the ectopic amyE locus [spoVE(E271A), spoVE(G335A), spoVE(S341A), and spoVE(G343A) (Fig. 4A)]. We also generated a second HMM, this time using only SpoVE proteins, and again identified residues (Fig. 4B) that are conserved in SpoVE proteins from diverse spore-formers but not in other SEDS proteins. We then introduced alanine substitutions in several of these positions in a B. subtilis SpoVE-GFP plasmid-borne fusion that was subsequently integrated at amyE [mutations spoVE(W69A), spoVE(K76A), and spoVE(T173A) (Fig. 4A)].
Characterization of SpoVE point mutations: heat resistance.
SpoVE is absolutely required for the development of heat resistance since no CFU are obtained following exposure of ΔspoVE::tet spores to 80°C for 20 min (Table 2). We took advantage of this requirement to assess the functionality of each of the spoVE point mutants we generated and determined the heat resistance of spores carrying a mutant spoVE allele in the absence of the wild-type copy. All mutations (I58N, C82R, S103N, E116G, C160R, G292R, and G355D) generated by random mutagenesis resulted in a complete loss of spore heat resistance (Table 2). Similarly, most of the spores carrying alanine substitutions introduced by site-directed mutagenesis in the C terminus of SpoVE (E271A, G335A, S341A, and G343A) were no longer heat resistant (Table 2). The only exception to this pattern was spores with the N322A mutation that were somewhat heat resistant, and this resistance was dependent on temperature, as a less-stringent heat treatment (55°C for 20 min) increased spore survival to about 10% compared to 0.19% at 80°C (data not shown). In contrast to the phenotype of most of the C-terminal mutations, spores carrying mutations in the N-terminal end of SpoVE (W69A, K76A, and T173A) were somewhat heat resistant, albeit at <1% of the survival rate of the wild type (Table 2), and none of these mutants exhibited a temperature dependence of survival. The inability of each of these SpoVE mutants to complement fully the loss of heat resistance of an spoVE::tet mutation demonstrates that both mutagenesis strategies successfully identified spoVE mutations that led to a loss of function. However, the heat-kill data do not permit discrimination between potential causes of this loss of function, including increased protein degradation or aberrant protein localization. To address these possibilities, we examined the localization and expression of each of these mutant SpoVE proteins.
TABLE 2.
Phenotypes of SpoVE mutantsa
| Genotype | CFU/ml preheat | CFU/ml postheat | % Sporulation | Protein accumulationb | Localization ratioc | Localizationd |
|---|---|---|---|---|---|---|
| Wild type (PY79) | 1.9 × 108 | 1.4 × 108 | 74 | N/A | N/A | N/A |
| ΔspoVE::tet | 8.8 × 107 | 0 | 0 | N/A | N/A | N/A |
| spoVE-gfp | 2.1 × 108 | 1.4 × 108 | 67 | ++++ | 3.51 ± 0.20 | OFM |
| spoVE(G335A)-gfp | 1.3 × 108 | 0 | 0 | ++++ | 3.34 ± 0.21 | OFM |
| spoVE(S341A)-gfp | 1.6 × 108 | 0 | 0 | +++++ | 3.32 ± 0.33 | OFM |
| spoVE(G343A)-gfp | 1.2 × 108 | 0 | 0 | +++++ | 3.28 ± 0.19 | OFM |
| spoVE(N322A)-gfp | 9.9 × 107 | 1.6 × 105 | 0.16 | +++++ | 3.20 ± 0.12 | OFM |
| spoVE(E271A)-gfp | 1.2 × 108 | 0 | 0 | +++++ | 3.11 ± 0.22 | OFM |
| spoVE(I58N)-yfp | 2.6 × 108 | 0 | 0 | ++ | ND | OFM |
| spoVE(C82R)-yfp | 3.0 × 108 | 0 | 0 | ND | ND | ND |
| spoVE(S103N)-yfp | 2.5 × 108 | 0 | 0 | +++++ | ND | OFM |
| spoVE(E116G)-yfp | 1.4 × 108 | 0 | 0 | +++++ | ND | OFM |
| spoVE(C160R)-yfp | 9.2 × 107 | 0 | 0 | + | ND | ND |
| spoVE(G292R)-yfp | 1.3 × 108 | 0 | 0 | ++ | ND | MCM |
| spoVE(G355D)-yfp | 1.6 × 108 | 0 | 0 | + | ND | MCM |
| spoVE(W69A)-gfp | 1.0 × 108 | 6.0 × 101 | 0.00006 | ++ | 1.97 ± 0.15 | MCM |
| spoVE(K76A)-gfp | 7.3 × 107 | 3.8 × 102 | 0.0005 | ++ | 1.94 ± 0.13 | MCM |
| spoVE(T173A)-gfp | 9.7 × 107 | 1.8 × 102 | 0.0002 | ++ | 1.89 ± 0.28 | MCM |
Data represent the results of one typical heat kill experiment. Each strain was grown in DSM for 24 h at 37°C and then exposed to 80°C for 20 min.
SpoVE-GFP or SpoVE-YFP accumulation at h 6 of sporulation. ND, not detectable.
Ratio between all four mother cell membranes (including forespore) and the two sporangial cell membranes. See text and Fig. 5 for details.
SpoVE localization as assessed by analysis with GFP and YFP fusions. See Fig. 5. N/A, not assessed; OFM, outer forespore membrane; MCM, mother cell membranes.
Characterization of SpoVE point mutations: protein localization.
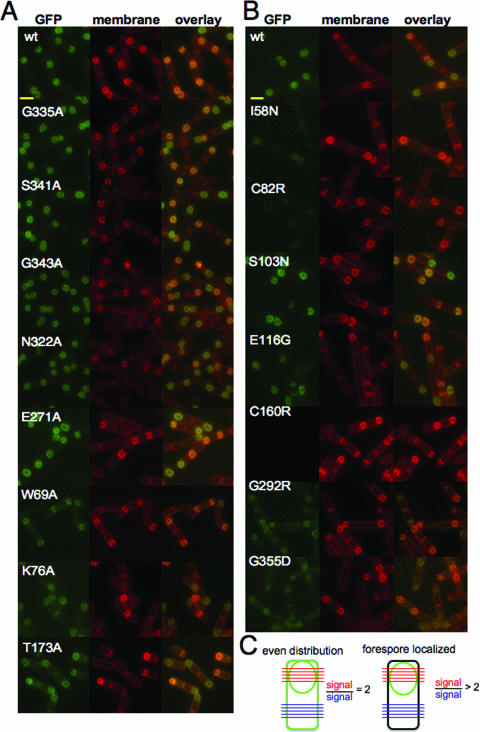
We took advantage of the ability of the SpoVE-YFP and SpoVE-GFP fusion proteins to fully complement either spoVE85 or ΔspoVE::tet mutations, respectively, to observe the cellular distribution of the SpoVE point mutants. Since the fluorescent signal derived from an SpoVE-YFP fusion is enriched at the outer forespore membrane (Fig. 1), we examined cells expressing mutant SpoVE-GFP fusion proteins at h 2.5 of sporulation using a resuspension protocol. The fluorescent signals of SpoVE-GFP mutants containing alanine substitutions in C-terminal residues (G335A, S341A, G343A, N322A, and E271A) were enriched at the forespore membrane, similar to the localization of wild-type SpoVE-GFP (Fig. 5A). By contrast, SpoVE-GFP mutants with alanine substitutions (W69A, K76A, and T173A) in their N-terminal domain were no longer concentrated at the forespore (Fig. 5A). In the case of the SpoVE-YFP mutant proteins, those carrying the S103N and E116G mutations were still enriched at the forespore, but other mutant proteins appeared to be expressed at lower levels (I58N, G292R, and G355D) or not at all (C82R and C160N) and were not appreciably enriched at the forespore (Fig. 5B).
FIG. 5.
Fluorescence microscopy of spoVE point mutants. (A) Strains expressed either wild-type SpoVE-GFP (JDB1835) or SpoVE-GFP mutants generated using site-directed mutagenesis: G335A (JDB1845), S341A (JDB1846), G343A (JDB1847), N322A (JDB1848), E271A (JDB1849), W69A (JDB1850), K76A (JDB1851), and T173A (JDB1852). Scale bars (yellow) represent 1 μm. (B) Strains expressed wild-type SpoVE-YFP (JDB1853) or SpoVE-YFP mutants generated by random mutagenesis: I58N (JDB1854), C82R (JDB1855), S103N (JDB1856), E116G (JDB1857), C160R (JDB1858), G292R (JDB1859), and G355D (JDB1860). (C) We quantified SpoVE localization using ImageJ (NIH [National Institutes of Health]) to measure the average pixel intensity for five lines spanning the forespore (red) and for five lines across the cell at the other end (blue). When the red/blue ratio is 2, then GFP signal is evenly distributed in all membranes (left). When the red/blue ratio is >2, the GFP signal is concentrated at the forespore (right). All strains were sporulated by resuspension, cells were labeled by using FM4-64, and samples for microscopy were obtained at 2.5 h.
Since the double membrane of the forespore can obscure qualitative impressions of forespore localization, we measured the ratio of the mean GFP fluorescence along a line across the middle of the forespore to the mean GFP fluorescence along a line across the middle of the mother cell (Fig. 5C). The first line crosses four mother cell-accessible membranes, and the second line crosses two mother cell-accessible membranes. Thus, if there is an equal distribution across all mother cell-accessible membranes, then the ratio equals 2. Alternatively, a ratio of >2 indicates that the fluorescent signal is enhanced at the forespore and, therefore, that the protein preferentially localizes to the outer forespore membrane. For wild-type SpoVE-GFP, the ratio was 3.51 ± 0.20 (mean ± standard error of the mean). For the SpoVE mutants that appeared to be enriched at the forespore, the ratios were as follows: G335A, 3.34 ± 0.21; S341A, 3.32 ± 0.33; G343A, 3.28 ± 0.19; N322A, 3.20 ± 0.12; and E271A, 3.21 ± 0.22. For the SpoVE mutants that appeared evenly distributed, the ratios were as follows: W69A, 1.97 ± 0.15; K76A, 1.94 ± 0.13; and T173A, 1.89 ± 0.28. Thus, quantification of the fluorescent signals yielded results consistent with the qualitative observations (Table 2).
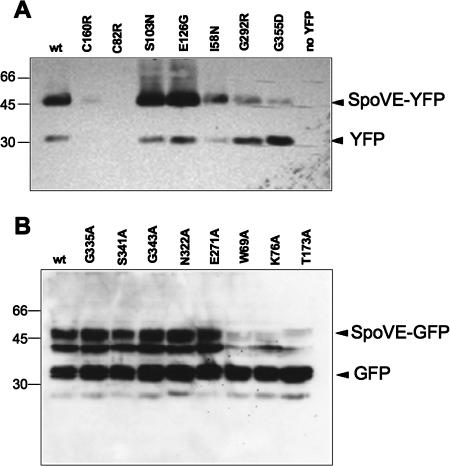
Although inspection of the fluorescence microscopy images indicated that most SpoVE-GFP mutant proteins and some SpoVE-YFP mutant proteins were expressed at wild-type levels, a number of the SpoVE-YFP mutant proteins did not appear to be expressed at all or were expressed at lower levels (Fig. 5A and B). Since proper interpretation of the loss-of-function phenotype observed in our measurements of spore heat resistance (Table 2) depends on knowledge of protein expression levels, we examined lysates from sporulating cells expressing different SpoVE-GFP mutants by Western blot analysis using an anti-GFP antibody. The SpoVE-YFP mutants with mutations C82R and C160R were not detected in lysates, similar to the results for a strain lacking an SpoVE-YFP fusion (Fig. 6A). The fluorescent signal for one of the SpoVE mutants, with mutation I58N, was only slightly lower than that of the wild-type SpoVE-YFP; however, two SpoVE mutants with similarly weak signals, with mutations G292R and G355D, had both a reduced level of full-length protein and an increased level of free GFP compared to the results for the wild-type strain. SpoVE mutants with mutations S103N and E126N that had wild-type fluorescent signals were expressed at levels similar to, if not higher than, that of the wild-type protein. Thus, the expression levels were consistent with the observed fluorescent signal, suggesting that the loss of heat resistance of spores carrying most spoVE-yfp alleles was a result of reduced accumulation of the various mutant forms of SpoVE-YFP.
FIG. 6.
Accumulation of mutant SpoVE proteins during sporulation. (A) Strains were induced to sporulate in DSM medium and collected 6 h after the onset of sporulation for Western blot analysis using an anti-gfp antibody. wt (wild type), spoVE85 amyE::spoVE-yfp (AH3561); C160R, spoVE85 amyE::spoVE(C160R)-yfp (AH3704); C82R, spoVE85 amyE::spoVE(C82R)-yfp (AH3701); S103N, spoVE85 amyE::spoVE(S103N)-yfp (AH3702); E126G, spoVE85 amyE::spoVE(E126G)-yfp (AH3703); I58N, spoVE85 amyE::spoVE(I59N)-yfp (AH3700); G292R, spoVE85 amyE::spoVE(G292R)-yfp (AH3705); G355D, spoVE85 amyE::spoVE(G355D)-yfp (AH3706); no YFP, wild-type spoVE (MB24). (B) Strains were sporulated by resuspension and collected at h 3 of sporulation for Western blot analysis using an anti-gfp antibody. wt (wild type), ΔspoVE::tet amyE::PspoVE-spoVE-gfp (JDB1835); G335A, ΔspoVE::tet amyE::PspoVE-spoVE(G335A)-gfp (JDB1845); S341A, ΔspoVE::tet amyE::PspoVE-spoVE(S341A)-gfp (JDB1846); G343A, ΔspoVE::tet amyE::PspoVE-spoVE(G343A)-gfp (JDB1847); N322A, ΔspoVE::tet amyE::PspoVE-spoVE(N322A)-gfp (JDB1848); E271A, ΔspoVE::tet amyE::PspoVE-spoVE(E271A)-gfp (JDB1849); W69A, ΔspoVE::tet amyE::PspoVE-spoVE(W69A)-gfp (JDB1850); K76A, ΔspoVE::tet amyE::PspoVE-spoVE(K76A)-gfp (JDB1851); T173A, ΔspoVE::tet amyE::PspoVE-spoVE(T173A)-gfp (JDB1852). Molecular mass markers, in kDa, are on the left of the panels.
When we examined the expression of the SpoVE-GFP mutant proteins, all of which resulted in a substantial fluorescence signal (Fig. 5A), we observed full-length protein in all cases following immunoblotting with anti-GFP antibodies, although the levels were greatly reduced for the mutants with mutations W69A, K76A, and T173A compared to the level of wild-type SpoVE-GFP. We observed an additional band at ∼33 kDa that presumably is free GFP and one at ∼38 kDa that we assume is a degradation product of SpoVE-GFP. The reduced heat resistance of spores carrying W69A, K76A, and T173A mutations (Table 2) could therefore be attributed to the lower protein levels, although the inability of the proteins to be correctly targeted to the forespore (Fig. 5A) could also be a factor in this phenotype. By contrast, the fact that spores carrying SpoVE G335A, S341A, G343A, and N322A mutations were completely sensitive to heat (Table 2) cannot be explained by defects in protein expression. Thus, these mutations must block the correct activity of SpoVE, either by interfering with necessary protein-protein interactions or by preventing the correct activity of the protein.
Analysis of the membrane topology of SpoVE.
Although the high sequence conservation along the entire lengths of SpoVE and FtsW suggests that they perform similar functions, they act at distinct times of the cell cycle (Fig. 2 and 3) and therefore likely interact with different protein(s). Consistent with this expectation, FtsW interacts with components of the cell division apparatus (e.g., FtsZ [5]) that are not present at later stages (i.e., post-asymmetric division) where SpoVE acts (27). The precise membrane topology of an SEDS protein may reflect these interactions, and we wanted to locate the functionally important residues identified in our mutagenesis of spoVE. However, because the topological models available have been determined for FtsW orthologs (5), we derived a topological model for SpoVE by constructing a series of fusions of spoVE to phoA and to lacZ. Since AP is only active when present on the outside of the cell and, conversely, β-galactosidase is only active in the cytoplasm, these two kinds of fusions provide a complementary readout of membrane protein topology (29). The AP or β-galactosidase activity of E. coli cells harboring spoVE′-phoA and -lacZ plasmids was determined by blue and white screening on plates containing the chromogenic substrates XP and X-Gal, respectively, or in cells grown in liquid medium and harvested during the exponential growth phase (see Materials and Methods). The end points of the various PhoA or LacZ fusions and the corresponding activities are indicated in Table 3. Fusions D40, G106, P162, S205, F228, and T262 showed high AP activity (>88.5 relative units [RU]; note that the background level of AP activity was 14.8 RU), suggesting the periplasmic localization of the PhoA moiety, whereas fusions K76, P146, P300, and Y366 showed reduced AP activity (<62.9 RU) (Table 3), revealing the cytoplasmic localization of PhoA. Conversely, β-galactosidase activity was only detected with fusions K76, P146, P300, and Y366 (Table 3), indicating the cytoplasmic localization of the LacZ moiety (>228.5 RU; background levels of β-galactosidase activity were 8.5 RU). The accumulation of all fusion proteins was confirmed by immunoblot analysis using antibodies against LacZ, PhoA, and GFP (data not shown). Together with the fluorescence of strains expressing either an N- or C-terminal fusion of SpoVE to GFP, which also serves as a reporter for cytoplasmic localization (9), the results suggest a topology with both the N and C termini of SpoVE facing the cytoplasm and separated by 10 transmembrane (TM) segments, with an extracellular loop of 71 residues located between TM VII and TM VIII (Fig. 4A). The model is in agreement with that proposed for the FtsW proteins of E. coli (25), Streptococcus pneumoniae (11), and Mycobacterium tuberculosis (5), suggesting structural conservation at least among the sporulation and cell division members of the SEDS family. The positions of the various point mutations characterized in this study are shown in Fig. 4A. Most of the mutations map to predicted TM domains, but a few are in hydrophilic loops located in the cytoplasm (K76A) or in the extracellular space between the mother cell and the forespore (S103N, E271A, and G335A) (Fig. 4A). All of these affect highly conserved or invariant residues among SEDS proteins. However, three mutations (W69A, C82R, and T173A) affect residues absolutely conserved only among SpoVE orthologs (Fig. 4B).
TABLE 3.
Description and enzyme activities of SpoVE′-PhoA and SpoVE′-LacZ fusion proteins
DISCUSSION
Many bacterial proteins display distinct patterns of subcellular distribution, and where the function of a protein is known, its pattern is often functionally relevant (42). Here we have examined the sporulation-specific SpoVE integral membrane protein of B. subtilis. Consistent with its essential role in spore heat resistance (34) and its involvement in spore cortex synthesis (50), we find that SpoVE, as assayed by a fully complementing SpoVE-YFP fluorescent protein fusion, is enriched at the forespore (Fig. 1). Since spoVE is under the control of the mother cell-specific transcription factor, σE, it is likely that this enrichment is restricted to the outer forespore membrane and not to the inner forespore membrane as well.
SpoVE localizes to the asymmetric septum very early in the process of membrane engulfment (Fig. 1), as is seen with several other proteins. This early localization is consistent with the targeting of SpoVE to this septum early in its synthesis. However, our experiments do not rule out SpoVE being initially inserted into all accessible membranes, where it undergoes free diffusion before becoming “captured” by an anchoring protein in the septum like SpoIIQ (2, 8). While we do not know if this mechanism is responsible for SpoVE forespore targeting, this possibility is certainly appealing.
Regardless of the mechanism that targets SpoVE to the forespore, our experiments demonstrate that this mechanism is specific to SpoVE and not to other members of the SEDS family. First, the forced production of FtsW during sporulation does not lead to a pattern of localization similar to that of SpoVE (Fig. 2A). Second, FtsW and SpoVE have nonoverlapping patterns of localization at the asymmetric septum (Fig. 3) and this difference occurs despite the presence of FtsW protein expressed from its own promoter (Fig. 2A). Thus, the observed differences in the subcellular distributions of SpoVE and FtsW likely reflect the activities of the different targeting signals intrinsic to their primary sequences. The fact that FtsW seems to disappear from the spore membranes as sporulation proceeds suggests that the role of the protein during this process is restricted to septation. In any event, the similarity between FtsW and SpoVE along their entire primary sequences should facilitate the identification of domains and/or residues responsible for these differences.
Here, we have taken advantage of the involvement of SpoVE in the nonessential process of spore formation to identify point mutants in an SEDS protein that appear to be true loss-of-function mutations. Although several spoVE alleles have been isolated before (33, 34), they were not characterized, and with the exception of spoVE85 that we sequenced and found to be identical to the G355D mutation generated in E. coli, they were probably lost. Therefore, this is the first isolation and characterization of loss-of-function mutations in an SEDS protein. We used both random and directed mutagenesis methods to identify residues that are important for heat resistance, an easily scored phenotype that is dependent on SpoVE. We characterized the localization and expression of these mutant SpoVE proteins and succeeded in determining which of our original mutations did not interfere with either proper expression or localization. Some of the mutations we obtained (W69A, K76A, C82R, and T173A) were in residues that are highly conserved within each group of proteins, SpoVE-, FtsW-, or RodA-like (Fig. 4B). For instance, the W at position 69 of SpoVE is replaced by a K in FtsW orthologs and by an L in RodA proteins, and C82 of SpoVE tends to be an L in FtsW proteins and a G in RodA proteins (Fig. 4B). Conceivably, these residues could be involved in protein-protein interactions that are specific to PG synthesis complexes operating during elongation, division, or cortex synthesis. If so, these interactions could be involved in proper protein localization, as the W69A and C82R mutant forms of SpoVE show deficient localization (Fig. 5 and Table 2). In contrast, some of the mutations found in TM X of SpoVE affect residues that are absolutely conserved among the entire SEDS family. These residues, e.g., G335, S341, and G343, may affect a functional aspect, such as a specific protein-protein interaction that is essential for the activity of all SEDS proteins. Consistent with this view, mutations G335A, S341A, and G343A, which affect invariant residues, do not affect the accumulation or localization of SpoVE (Fig. 5 and Table 3). Thus, we may have identified mutations that block the correct functioning of SpoVE, either by interfering with necessary protein-protein interactions or by preventing the correct enzymatic activity of the protein. Presently, and in the absence of a functional assay, we are unable to differentiate between those possible loss-of-function mechanisms.
Although the phylogenetically conserved SEDS family of essential proteins involved in cell division and growth was first characterized nearly 20 years ago, their function remains largely unclear. The phenotypes of ftsW mutants, in particular, led to the hypothesis that FtsW could be the missing “flippase” responsible for the translocation of lipid II across the cytoplasmic membrane (17). Although PG synthesis is a very well characterized metabolic pathway, with crystal structures for most of the enzymes determined (43) and the small-molecule inhibitors specific to many of the enzymatic steps identified (12), the mechanism of lipid II translocation and, in fact, the proteins that mediate this process remain unknown. Despite the evident appeal of the model that posits SEDS proteins in this role, and additional observations consistent with predicted protein interactions (e.g., FtsW and PBP3 in mycobacteria [6]), no direct evidence for this proposed role has emerged. One reason for the absence likely lies in the requirement for FtsW (and to a lesser extent, RodA) for growth under normal conditions. Temperature-sensitive mutants (24) or depletion strains (3, 16) of FtsW or RodA could be useful in this regard, although the likelihood that FtsW and RodA use the same substrate (30) would complicate such analysis. Thus, the generation of a spectrum of point mutants and of a null mutant of the SpoVE SEDS protein as described herein will facilitate the development of strategies aimed at demonstrating its function using either in vivo (26) or in vitro (32) approaches.
Supplementary Material
Acknowledgments
We thank Colin Manoil for the gift of strains, Teresa Costa for the construction of pTC30, and Anabela Isidro for the anti-GFP antibody.
This work was funded by grants POCTI/BCI/48647/2002 and POCI/BIA-BCM/60855/2004 from the Fundação para a Ciência e a Tecnologia (F.C.T.) to A.O.H. and by an Irma T. Hirschl scholar award and startup funds from the Department of Microbiology, Columbia University, to J.D. G.R. was the recipient of a postdoctoral fellowship (SFRH/BPD/20668/2004) from the F.C.T. A.F. was supported by NIH training grant AI007161-29.
Footnotes
Published ahead of print on 2 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Begg, K. J., B. G. Spratt, and W. D. Donachie. 1986. Interaction between membrane proteins PBP3 and RodA is required for normal cell shape and division in Escherichia coli. J. Bacteriol. 1671004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaylock, B., X. Jiang, A. Rubio, C. P. Moran, Jr., and K. Pogliano. 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 182916-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle, D. S., M. M. Khattar, S. G. Addinall, J. Lutkenhaus, and W. D. Donachie. 1997. ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 241263-1273. [DOI] [PubMed] [Google Scholar]
- 4.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235209-220. [DOI] [PubMed] [Google Scholar]
- 5.Datta, P., A. Dasgupta, S. Bhakta, and J. Basu. 2002. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J. Biol. Chem. 27724983-24987. [DOI] [PubMed] [Google Scholar]
- 6.Datta, P., A. Dasgupta, A. K. Singh, P. Mukherjee, M. Kundu, and J. Basu. 2006. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol. Microbiol. 621655-1673. [DOI] [PubMed] [Google Scholar]
- 7.de Pedro, M. A., W. D. Donachie, J. V. Holtje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol. 1834115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doan, T., K. A. Marquis, and D. Z. Rudner. 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 551767-1781. [DOI] [PubMed] [Google Scholar]
- 9.Feilmeier, B. J., G. Iseminger, D. Schroeder, H. Webber, and G. J. Phillips. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 1824068-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerard, P., T. Vernet, and A. Zapun. 2002. Membrane topology of the Streptococcus pneumoniae FtsW division protein. J. Bacteriol. 1841925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, D. W. 2002. The bacterial cell wall as a source of antibacterial targets. Expert Opin. Ther. Targets 61-19. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez, C., and J. C. Devedjian. 1989. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 173999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular biological methods for Bacillus. Wiley, New York, NY.
- 15.Henriques, A. O., H. de Lencastre, and P. J. Piggot. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74735-748. [DOI] [PubMed] [Google Scholar]
- 16.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28235-247. [DOI] [PubMed] [Google Scholar]
- 17.Holtje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda, M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 1716375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishino, F., W. Park, S. Tomioka, S. Tamaki, I. Takase, K. Kunugita, H. Matsuzawa, S. Asoh, T. Ohta, B. G. Spratt, et al. 1986. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J. Biol. Chem. 2617024-7031. [PubMed] [Google Scholar]
- 20.Joris, B., G. Dive, A. Henriques, P. J. Piggot, and J. M. Ghuysen. 1990. The life-cycle proteins RodA of Escherichia coli and SpoVE of Bacillus subtilis have very similar primary structures. Mol. Microbiol. 4513-517. [DOI] [PubMed] [Google Scholar]
- 21.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 1872233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karow, M. L., and P. J. Piggot. 1995. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene 16369-74. [DOI] [PubMed] [Google Scholar]
- 23.Khattar, M. M., S. G. Addinall, K. H. Stedul, D. S. Boyle, J. Lutkenhaus, and W. D. Donachie. 1997. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J. Bacteriol. 179784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khattar, M. M., K. J. Begg, and W. D. Donachie. 1994. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J. Bacteriol. 1767140-7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lara, B., and J. A. Ayala. 2002. Topological characterization of the essential Escherichia coli cell division protein FtsW. FEMS Microbiol. Lett. 21623-32. [DOI] [PubMed] [Google Scholar]
- 26.Lara, B., D. Mengin-Lecreulx, J. A. Ayala, and J. van Heijenoort. 2005. Peptidoglycan precursor pools associated with MraY and FtsW deficiencies or antibiotic treatments. FEMS Microbiol. Lett. 250195-200. [DOI] [PubMed] [Google Scholar]
- 27.Levin, P. A., and R. Losick. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10478-488. [DOI] [PubMed] [Google Scholar]
- 28.Mallidis, C. G., and J. Scholefield. 1987. Relation of the heat resistance of bacterial spores to chemical composition and structure. II. Relation to cortex and structure. J. Appl. Bacteriol. 63207-215. [DOI] [PubMed] [Google Scholar]
- 29.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 3461-75. [DOI] [PubMed] [Google Scholar]
- 30.Matsuhashi, M. 1994. Utilization of lipid-linked precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and mechanism of their regulation. In J.-M. Ghuysen and R. Hackenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 31.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 31a.Minton, N. P. 1984. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene 31269-73. [DOI] [PubMed] [Google Scholar]
- 32.Pastoret, S., C. Fraipont, T. den Blaauwen, B. Wolf, M. E. Aarsman, A. Piette, A. Thomas, R. Brasseur, and M. Nguyen-Disteche. 2004. Functional analysis of the cell division protein FtsW of Escherichia coli. J. Bacteriol. 1868370-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piggot, P. J., K. F. Chak, and U. D. Bugaichuk. 1986. Isolation and characterization of a clone of the spoVE locus of Bacillus subtilis. J. Gen. Microbiol. 1321875-1881. [DOI] [PubMed] [Google Scholar]
- 34.Piggot, P. J., and J. G. Coote. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40908-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogliano, J., N. Osborne, M. D. Sharp, A. Abanes-De Mello, A. Perez, Y. L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 311149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popham, D. L. 2002. Specialized peptidoglycan of the bacterial endospore: the inner wall of the lockbox. Cell. Mol. Life Sci. 59426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Real, G., S. Autret, E. J. Harry, J. Errington, and A. O. Henriques. 2005. Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis. Mol. Microbiol. 55349-367. [DOI] [PubMed] [Google Scholar]
- 38.Real, G., and A. O. Henriques. 2006. Localization of the Bacillus subtilis murB gene within the dcw cluster is important for growth and sporulation. J. Bacteriol. 1881721-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudner, D. Z., Q. Pan, and R. M. Losick. 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc. Natl. Acad. Sci. USA 998701-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuster-Bockler, B., J. Schultz, and S. Rahmann. 2004. HMM Logos for visualization of protein families. BMC Bioinform. 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seyler, R. W., Jr., A. O. Henriques, A. J. Ozin, and C. P. Moran, Jr. 1997. Assembly and interactions of cotJ-encoded proteins, constituents of the inner layers of the Bacillus subtilis spore coat. Mol. Microbiol. 25955-966. [DOI] [PubMed] [Google Scholar]
- 42.Shapiro, L., H. H. McAdams, and R. Losick. 2002. Generating and exploiting polarity in bacteria. Science 2981942-1946. [DOI] [PubMed] [Google Scholar]
- 43.Smith, C. A. 2006. Structure, function and dynamics in the mur family of bacterial cell wall ligases. J. Mol. Biol. 362640-655. [DOI] [PubMed] [Google Scholar]
- 44.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 11329-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theeragool, G., A. Miyao, K. Yamada, T. Sato, and Y. Kobayashi. 1993. In vivo expression of the Bacillus subtilis spoVE gene. J. Bacteriol. 1754071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Dam, V., R. Sijbrandi, M. Kol, E. Swiezewska, B. de Kruijff, and E. Breukink. 2007. Transmembrane transport of peptidoglycan precursors across model and bacterial membranes. Mol. Microbiol. 641105-1114. [DOI] [PubMed] [Google Scholar]
- 47.van Heijenoort, J. 2001. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 18503-519. [DOI] [PubMed] [Google Scholar]
- 48.van Ooij, C., P. Eichenberger, and R. Losick. 2004. Dynamic patterns of subcellular protein localization during spore coat morphogenesis in Bacillus subtilis. J. Bacteriol. 1864441-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Ooij, C., and R. Losick. 2003. Subcellular localization of a small sporulation protein in Bacillus subtilis. J. Bacteriol. 1851391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasudevan, P., A. Weaver, E. D. Reichert, S. D. Linnstaedt, and D. L. Popham. 2007. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol. Microbiol. 651582-1594. [DOI] [PubMed] [Google Scholar]
- 51.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225487-494. [DOI] [PubMed] [Google Scholar]
- 52.Warth, A. D., and J. L. Strominger. 1972. Structure of the peptidoglycan from spores of Bacillus subtilis. Biochemistry 111389-1396. [DOI] [PubMed] [Google Scholar]
- 53.Warth, A. D., and J. L. Strominger. 1969. Structure of the peptidoglycan of bacterial spores: occurrence of the lactam of muramic acid. Proc. Natl. Acad. Sci. USA 64528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickus, G. G., A. D. Warth, and J. L. Strominger. 1972. Appearance of muramic lactam during cortex synthesis in sporulating cultures of Bacillus cereus and Bacillus megaterium. J. Bacteriol. 111625-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Youngman, P., J. B. Perkins, and R. Losick. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol. Gen. Genet. 195424-433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.